Abstract
Rpa34 and Rpa49 are nonessential subunits of RNA polymerase I, conserved in species from Saccharomyces cerevisiae and Schizosaccharomyces pombe to humans. Rpa34 bound an N-terminal region of Rpa49 in a two-hybrid assay and was lost from RNA polymerase in an rpa49 mutant lacking this Rpa34-binding domain, whereas rpa34Δ weakened the binding of Rpa49 to RNA polymerase. rpa34Δ mutants were caffeine sensitive, and the rpa34Δ mutation was lethal in a top1Δ mutant and in rpa14Δ, rpa135(L656P), and rpa135(D395N) RNA polymerase mutants. These defects were shared by rpa49Δ mutants, were suppressed by the overexpression of Rpa49, and thus, were presumably mediated by Rpa49 itself. rpa49 mutants lacking the Rpa34-binding domain behaved essentially like rpa34Δ mutants, but strains carrying rpa49Δ and rpa49-338::HIS3 (encoding a form of Rpa49 lacking the conserved C terminus) had reduced polymerase occupancy at 30°C, failed to grow at 25°C, and were sensitive to 6-azauracil and mycophenolate. Mycophenolate almost fully dissociated the mutant polymerase from its ribosomal DNA (rDNA) template. The rpa49Δ and rpa49-338::HIS3 mutations had a dual effect on the transcription initiation factor Rrn3 (TIF-IA). They partially impaired its recruitment to the rDNA promoter, an effect that was bypassed by an N-terminal deletion of the Rpa43 subunit encoded by rpa43-35,326, and they strongly reduced the release of the Rrn3 initiation factor during elongation. These data suggest a dual role of the Rpa49-Rpa34 dimer during the recruitment of Rrn3 and its subsequent dissociation from the elongating polymerase.
Fast-growing Saccharomyces cerevisiae and Schizosaccharomyces pombe cells mobilize some 70% of their transcriptional capacity to produce the 6.8-kb precursor of the 18S, 5.8S, and 25S rRNAs by transcribing the highly repeated ribosomal DNA (rDNA) locus (44, 50). Genetic studies (43) have established that this is the only role or, at least, the only essential role of yeast RNA polymerase I (Pol I). rDNA transcription starts with the binding of Pol I to its specificity factor Rrn3 (7, 39-41, 48, 57). The Rrn3-Pol I dimer is then directed to a preinitiation complex residing at the rDNA promoter region. This complex combines the TATA box-binding protein (also operating in the Pol II and Pol III systems) with the core and upstream activation factors that are specific to the Pol I system (31, 32). In mammals, Pol I first binds transcription initiation factor IA, akin to Rrn3 and functionally interchangeable with that protein in vivo (7, 40, 41). The Pol I-transcription initiation factor IA dimer is then targeted to the rDNA promoter by interacting with SL1, a factor made of the TATA box-binding protein associated with four protein subunits, TAFI95, TAFI68, TAFI48, and TAFI41 (11, 22, 23, 40, 59); TAFI68 is distantly related to the yeast core factor at the level of the core factor's Rrn7/TAFI68 subunit (9). Pol I is also associated with the upstream binding factor (UBF; mammals) and Hmo1 (yeast) HMG box proteins, which stimulate rDNA transcription by a still poorly understood mechanism (19, 46, 52).
Pol I has 14 polypeptide subunits in S. cerevisiae (20, 27) and S. pombe (2, 28). Five of these subunits, including the two largest ones, are akin to the bacterial core enzyme (58). Seven others are related or even identical to subunits of the Pol II and Pol III core structures (1, 16, 29) and are homologous to archaeal RNA polymerase subunits (35, 55). Rpa49 and Rpa34, on the other hand, are Pol I-specific subunits with unknown functions (20, 36) that are also homologous to PAF53 and PAF49 in the mammalian Pol I system (47, 56). We show here that these subunits bind each other in the Pol I structure and play an important role in transcription by improving the recruitment of the Rrn3-Pol I complex to the rDNA and by triggering the release of Rrn3 from the elongating Pol I.
MATERIALS AND METHODS
Tables listing the plasmids, yeast strains, and DNA primers used in this study are included in the supplemental material. Yeast strains were constructed by meiotic crossing, DNA transformation, and plasmid shuffling through selection on fluoroorotic acid (FOA) medium with 0.1% 5-fluoroorotate (8). Null alleles with KanMX4 insertions were obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html) and were checked by ad hoc PCR amplifications. The rpa135(D395N) and rpa135(L656P) alleles were isolated after UV mutagenesis of the rpa34::HIS3 strain OG1-1C transformed by plasmid pOG1-A34 (20) and were selected by a method based on the lack of white sectoring in the UV-mutagenized clones (33), which signals their inability to lose pOG1-A34, therefore indicating that the rpa34::HIS3 allele is now essential for growth.
Newly constructed plasmids were prepared from PCR-amplified DNA. In most cases, we used the Gateway cloning system with previously described destination vectors (54). All inserts were resequenced to detect spurious mutations generated by DNA amplification. Chimeras including the Rpa43 coding sequences of S. cerevisiae and S. pombe were constructed by overlap extension PCR using Pfu DNA polymerase. Primer sequences are available upon request.
Chromatin immunoprecipitation (ChIP) assays were systematically done on three independent cultures of 100 ml harvested at an A600 of 0.6, with slight modifications of a previously described experimental protocol (34) and with the rDNA primers listed in the tables in the supplemental material. Yeast cells were cultivated in yeast extract-peptone-dextrose (YPD) medium, except in experiments involving mycophenolate, in which we used synthetic dextrose medium supplemented with histidine, arginine, tryptophan, and adenine sulfate, each at 20 mg/liter, and leucine and lysine, each at 30 mg/liter (SD+aa medium). rDNA copy numbers were estimated by comparing the kinetics of PCR amplification on rDNA primers and on PYK1 primers used as a single-gene control, giving an average value (± the standard deviation) of 186 ± 54 copies.
RESULTS
The Pol I-specific subunits Rpa49 and Rpa34 bind each other in two-hybrid assays.
Viable rpa49Δ mutants fail to grow at 25°C and are sensitive to mycophenolate, and the rpa49Δ mutation is lethal in cells lacking the nonessential Rpa14 subunit or the HMG box protein Hmo1 (13, 19, 20, 36). Rpa49 has close homology to the human PAF53 protein (Fig. 1) and can be replaced in vivo by Rpa51, its S. pombe counterpart (42). A homology search revealed a gene for an Rpa49-like product in most eukaryotic genomes, with conserved motifs scattered over the entire subunit. In particular, all Rpa49-like subunits contain a highly conserved C-terminal domain, located between positions 326 and 371 in S. cerevisiae (detailed sequence alignments are provided in Fig. S1 in the supplemental material). Proteins distantly related to Rpa34 have also been identified in fission yeast, mice, and humans (2, 47). However, their sequence homology is restricted to two short conserved domains and to C-terminal KKE/D repeats often found in nucleolar proteins (21), and the mammalian subunit (PAF49) is much larger than its fungal counterpart (see Fig. S2 and S3 in the supplemental material).
FIG. 1.
Properties of the Rpa49 subunit. (A) Homology between Rpa49 (S. cerevisiae) and its S. pombe (Rpa51) and human (PAF53) counterparts, shown by Blosum 62 matrices drawn at a 5/35 stringency. H. sapiens, Homo sapiens. (B) Two-hybrid screening with Gal4BD-Rpa34. Six Gal4AD-Rpa49 plasmids were isolated by genome-wide screening of a previously described two-hybrid library (17) by using Gal4BD-Rpa34. Vertical lines denote the limits of the RPA49 inserts. β-Galactosidase (β-Gal) was tested as previously described (17). Data are shown for the Gal4AD::RPA49(36,119) plasmid and for a reciprocal two-hybrid interaction between Gal4BD-Rpa34 and Gal4AD-Rpa49. The RPA49 open reading frame is shown with gray boxes representing four conserved parts of Rpa49, corresponding to positions 60 to 89, 128 to 154, 201 to 216, and 326 to 371 in S. cerevisiae (see Fig. S1 in the supplemental material). (C) Deletion mapping of the Rpa34-interacting domain on Rpa49. N-terminal and C-terminal deletion forms of Rpa49 were cloned as Gal4BD fusions into the pVV213 vector (54) and tested against Gal4BD-Rpa34 in a two-hybrid assay.
Rpa49 and Rpa34 were fused to the binding domain of Gal4 (Gal4BD) and tested for two-hybrid interactions in a genome-wide screen based on a library of genomic fragments fused to the Gal4 activation domain (Gal4AD) (17, 18). The Gal4BD-Rpa49 screen yielded two Gal4AD clones encoding the entire Rpa34 subunit, whereas the Gal4BD-Rpa34 fusion yielded six clones with products overlapping between positions 63 and 119 of Rpa49 (Fig. 1A). Subunits with N-terminal deletions (encoded by rpa49-63,416, rpa49-89,416, and rpa49-119,416) and C-terminal deletions (encoded by rpa49-1,260 and rpa49-1,366) were also fused to pVV213, a Gal4BD fusion vector based on a Gateway cassette. This new round of two-hybrid assays, done with a slightly different vector, confirmed that the interaction between Gal4BD-Rpa34 and Gal4BD-Rpa49 was lost through the deletion of the N terminus of Rpa49 and was insensitive to C-terminal deletions (Fig. 1B).
The genome-wide screening performed with Gal4BD-Rpa34 also yielded two Top1 clones (suggesting that the synthetic lethality of rpa34Δ and top1Δ [20] may involve direct Top1-Rpa34 binding) and four clones encoding Gno1 (two isolates), Nop56, and Nop58. Like Rpa34, these three proteins are nucleolar and contain KKD/E repeats that are commonly encountered in nucleolar proteins. Gno1 contributes to the first cleavage steps of pre-rRNA processing (24). Nop56 and Nop58 are components of the U3 ribonucleoprotein complex required for 18S rRNA biogenesis (14).
The N-terminal part of Rpa49 enables Rpa34 to bind Pol I.
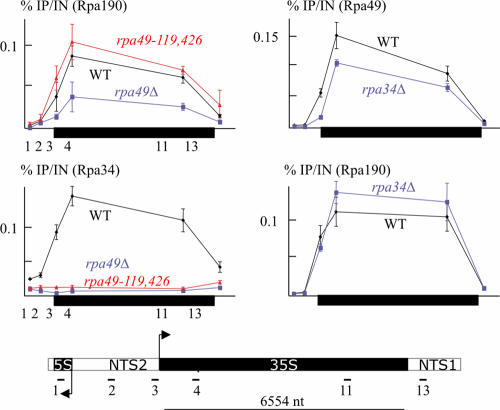
A Pol I subform lacking Rpa34 and Rpa49 is catalytically active on nonspecific DNA templates and can be chromatographically separated from the 14-subunit enzyme (27). Moreover, Rpa49 dissociates from a highly purified preparation of Pol I in rpa34Δ mutants (20), suggesting that Rpa49 and Rpa34 may stabilize each other within the Pol I structure. We further investigated this point by ChIP assays of rpa49 mutants. As shown in Fig. 2, Rpa34 was entirely lost from the rDNA-bound Pol I in an N-terminal deletion mutant (rpa49-119,416) lacking the putative Rpa34-binding domain. A fortiori, it was also lost in rpa49Δ mutants, which also displayed an overall reduction in the amount of Pol I residing on rDNA (as detected by anti-Rpa190 antibodies), consistent with the slow growth of these mutants at 30°C. rpa34Δ, on the other hand, slightly reduced the Rpa49 signal but had no effect on Pol I itself, in keeping with the wild-type growth rate of rpa34Δ mutants (Fig. 2).
FIG. 2.
Association of Rpa34 and Rpa49 with Pol I in rpa34Δ or rpa49 mutants. ChIP assays were done on three independent cultures (comprising 100 ml of YPD) harvested at an A600 of 0.6. Pol I was revealed by polyclonal anti-Rpa190 and anti-Rpa34 or anti-Rpa49 rabbit antibodies (49). DNA occupancy was defined by the ratio between the immunoprecipitation (IP) signal and the DNA input (IN) signal. The diagram at the bottom shows the approximate positions of the primers used for reverse transcription-PCR (RT-PCR) amplification. Arrows denote the start sites and transcription orientations of the 5S and 35S rRNA transcripts. NTS1 and NTS2, nontranscribed spacers 1 and 2; nt, nucleotides. (Left panels) Effect of rpa49 mutations on the binding of Rpa34 to Pol I. Strain D704-6C was transformed with pVV200 plasmids bearing the mutant (rpa49-119,416) and wild-type (WT) RPA49 alleles. (Right panels) Effect of rpa34Δ on the binding of Rpa49 in YPH499 (WT) and T4-1C (rpa34Δ).
The adverse phenotypes of rpa34Δ mutants are suppressed by Rpa49 overexpression.
Unlike rpa49Δ, rpa34Δ caused no growth defect in its own right, and rpa34Δ mutants were not particularly sensitive to mycophenolate or 6-azauracil (Fig. 3A). Like rpa49Δ mutants, however, rpa34Δ mutants were sensitive to caffeine (trimethylxanthine), and the mutation was synthetically lethal with rpa14Δ and top1Δ (Fig. 3B and C). It should be noted that we previously obtained a viable top1Δ rpa49Δ double mutant (20), but further analysis has now shown that this was an exceptional case, almost certainly due to some unrecognized suppression.
FIG. 3.
Growth phenotypes of RNA Pol I mutants. (A) Sensitivity of RNA Pol I mutants to low temperature (25°C), mycophenolate (MPA), and 6-azauracil (AZA). Strains YPH499 (wild type [WT]), D704-6C (rpa49Δ), D360-1A (rpa14Δ), T4-1C (rpa34Δ), OG36-1A [rpa135(L656P)], SL46-1D [rpa135(D395N)], SL9-2C (rpa12Δ), and D101-CHIM2 (rpa43-chim2) were serially diluted and spotted onto YPD and onto synthetic complete medium lacking uracil but containing mycophenolate (10 μg/ml) or 6-azauracil (20 μg/ml). Plates were incubated for 3 days at 30°C. The box on the right summarizes the synthetic growth defects seen in rpa14Δ and top1Δ strains as monitored by plasmid shuffle tests (8) based on the counterselection of URA3 centromeric plasmids (with RPA12, RPA14, RPA34, or RPA49) on FOA medium. Lethality is indicated by a minus sign. Mutant strains that grew on FOA were streaked onto YPD for 3 days at 30°C. The + and (+) symbols denote a growth pattern identical or partially impaired relative to that of the corresponding wild-type control tested before the FOA chase. (B) Suppression of the caffeine sensitivity of the rpa34Δ mutant by RPA49 overexpression. The YPH499 wild type (WT), the isogenic mutant strain T4-1C (rpa34Δ), and T4-1C transformed with pGEN-A49 (rpa34Δ 2μ RPA49) were spotted onto YPD and onto YPD with caffeine at 1 g/liter and grown for 3 days at 30°C. (C) Suppression of rpa34Δ synthetically lethal defects by RPA49 overexpression. Strains OG1-L656P [rpa135(L656P) rpa34Δ], OG1-D395N [rpa135(D395N) rpa34Δ], and OG17-1C (top1Δ rpa34Δ) bearing the pOG1-A34 plasmid (2μm RPA34 URA3) were transformed with pGEN-A49 (2μm RPA49 TRP1) by using pGEN as an empty vector control (−). Suppression was detected by a plasmid shuffle assay on FOA, as described above. (D) Suppression of the rpa14Δ rpa34Δ synthetic lethality by RPA49 overexpression. Strains Y11277 (rpa34Δ::KanMX4) and D360-1A (rpa14Δ::HIS3) were crossed and transformed with pSLA49 (2μm RPA49 URA3), and meiotic tetrads were isolated by microdissection. The results presented show a tetratype with two parental and two recombinant segregants after replica plating onto 5-FOA, demonstrating that the rpa14Δ rpa34Δ strain (white frame) is unable to lose pSLA49. (E) Synthetic lethality of rpa49Δ with the RRN3::RPA43 fusion. Strains D781-10B [(RPA49 rpa43Δ)/2μm TRP1 RRN3::RPA43/CEN URA3 RPA43] and D780-6D [(rpa49Δ rpa43Δ)/2μm TRP1 RRN3::RPA43/CEN URA3 RPA43] were tested for their ability to lose the YCpA43-12 (CEN URA3 RPA43) plasmid by using a plasmid shuffle assay on FOA. The white frame around the D780-6D replica indicates the inability to lose YcpA43-12 in the rpa49Δ context.
Using an ad hoc procedure to select mutations that were synthetically lethal with rpa34Δ (see Materials and Methods), we obtained two additional Pol I mutants, the rpa135(L656P) and rpa135(D395N) strains. These mutants grew like the wild type but were moderately [rpa135(D395N)] or strongly [rpa135(L656P)] sensitive to mycophenolate and 6-azauracil. The rpa135(L656P) and rpa135(D395N) mutations were also lethal with rpa49Δ, but this synthetic lethality was specific to the Rpa49 subunit since strains carrying the rpa135(L656P) and rpa135(D395N) mutations combined with rpa12Δ and rpa14Δ were viable (Fig. 3A and B). Intriguingly, the rpa135-(L656P) mutation corresponds to a highly conserved position shared by the archaeal and eukaryotic polymerases, and this conservation extends to bacteria in the case of position D395 (Fig. 4). The equivalent positions belong to the lobe and external domains of the yeast Pol II crystal structure (12) and are fairly close to the Rpb9 subunit paralogous to Rpa12.
FIG. 4.
Positions of the rpa135(D395N) and rpa135(L656P) mutations as deduced from the Pol II structure. Local sequence alignments are shown for the second-largest subunit of the three yeast RNA polymerases, for the corresponding archaeal subunits of Methanocaldococcus jannaschii (MjB′ and MjB″), and for the β subunit of Thermus aquaticus (Taq β). Two views of the Pol II spatial structure (Protein Data Bank coordinate 1WCM) are provided, showing the positions of Rpa12 (blue ribbons) and of the lobe and external folds. Asterisks indicate mutant positions.
rpa34Δ mutants, therefore, had a wild-type growth pattern but were sensitive to caffeine and presented a small number of synthetically lethal defects shared with rpa49Δ mutants. Importantly, all the currently identified phenotypes associated with the rpa34Δ mutation were suppressed when Rpa49 was overexpressed from a multicopy plasmid (Fig. 3C). Together with the above-cited data showing that rpa34Δ slightly reduced the Rpa49 ChIP signal, these observations argue that Rpa34 alters the spatial organization of the corresponding Rpa49, therefore stabilizing this subunit in the Pol I structure, and that this activity accounts for all the adverse effects currently associated with rpa34Δ. It should be noted that previous large-scale screening for synthetically lethal yeast mutations had suggested that Rpa34 might contribute to cell wall synthesis, due to synthetic lethality occurring with rpa34Δ in chitin synthase-defective mutants (53). However, we were unable to reproduce these observations.
The conserved C terminus of Rpa49 is critical in vivo.
Rpa49 mutants were constructed by progressive N- and C-terminal deletions (Fig. 5A). Deleting the first 89 amino acids of Rpa49 (rpa49-63,416 and rpa49-89,416) had no adverse effects under any conditions tested, despite the strongly conserved motif between positions 60 and 90 (see Fig. S1 in the supplemental material). The rpa49-119,416 mutation, which removed the next 20 amino acids, also produced a wild-type phenotype but was nearly lethal with rpa14Δ, consistent with the data discussed above (Fig. 2) showing that Pol I was unable to recruit the Rpa34 subunit in the rpa49-119,416 mutant. We then turned to the rpa49-338::HIS3 strain, a previously described mutant (36) with a C-terminal insertion of the sequence encoded by the HIS3 cassette. This mutant forms a truncated Rpa49 lacking the highly conserved C-terminal domain mentioned above (see Fig. S1 in the supplemental material). Since the truncated Rpa49 subunit is still stably incorporated into the multisubunit structure of RNA Pol I (36), the defects associated with rpa49-338::HIS3 directly result from the absence of the Rpa49 C terminus.
FIG. 5.
Mutagenesis of RPA49. (A) Growth defects of rpa49 mutants. Strain D704-7C (rpa49Δ) was transformed with pVV200 plasmids (54) bearing the relevant wild-type sequences and rpa49 mutations corresponding to progressive N- and C-terminal deletions. Plasmid pVV214 was used as an empty vector control. Strain 49::HIS3 (see the tables in the supplemental material) was used to test the rpa49-338::HIS3 allele, in which the HIS3 insertion interrupts the Rpa49 coding sequence at H338 with a sequence encoding a GSAARSCSLACT extension beyond H338 (36). Four gray boxes represent conserved parts of Rpa49 (Fig. 1). Yeast cultures were serially diluted and spotted onto YPD (at 25 and 30°C and at 30°C with 1 g of caffeine [CAF]/liter) and synthetic complete medium with or without mycophenolate (MPA) at 20 μg/ml. Plates were incubated at 30°C for 3 days. The synthetic lethality of rpa49 mutations was tested in strains D699-13C (rpa14Δ rpa49Δ) and D684-2B (hmo1Δ rpa49Δ) bearing the pFB74 (RPA49 URA3) plasmid and transformed with the same plasmids indicated above. The resulting transformants were replica plated onto FOA medium to chase pFB74. Synthetically lethal effects were revealed by the lack of growth on FOA (8). In the case of rpa49-338::HIS3, synthetic lethality was deduced from meiotic crosses based on 12 tetrads. Lethality is indicated by a minus sign. (−) indicates very slow growth. The + and (+) symbols denote a growth pattern identical or partially impaired relative to that of the corresponding wild-type control. (B) Effect of rpa49 mutations on rDNA occupancy by Pol I. ChIP assays were done on D704-6C (rpa49Δ) transformed with the appropriate pVV200-derived plasmids. Pol I was revealed by polyclonal anti-Rpa190 rabbit antibodies (49). DNA occupancy was defined by the ratio between the immunoprecipitation (IP) signal and the DNA input (IN) signal. The diagram below shows the approximate positions of the primers used for RT-PCR amplification (see the tables in the supplemental material). Arrows denote the start sites and transcription orientations of the 5S and 35S rRNA transcripts. WT, wild type; NTS1 and NTS2, nontranscribed spacers 1 and 2.
The rpa49Δ and rpa49-338::HIS3 mutations produced the synthetically lethal defects and caffeine sensitivity mentioned above for rpa34Δ mutants but also engendered many phenotypes of their own, causing an almost complete lack of growth at 25°C, strong sensitivity to mycophenolate and 6-azauracil, and synthetic lethality with hmo1Δ (Fig. 5A). A mutant with a somewhat shorter C-terminal deletion, corresponding to rpa49-1,366, also failed to grow at 25°C but had slightly milder drug sensitivity, and a strain carrying rpa49-1,366 and hmo1Δ was viable. The importance of the Rpa49 C-terminal domain was also underscored by results from ChIP assays showing strongly reduced Pol I occupancy of rDNA in rpa49Δ and rpa49-1,366 mutants (Fig. 5B).
rpa49Δ and rpa49-338::ITHIS3 have a dual effect on the recruitment and release of Rrn3.
Earlier ChIP assays have shown that Rrn3 is present on the rDNA promoter but not on the rDNA 3′ end, as befits a Pol I-associated initiation factor (5). Using Rrn3-hemagglutinin (HA)-tagged strains, we showed that Rrn3 was strictly restricted to the promoter region. The rpa43-chim2 strain, a slow-growing mutant expressing a form of Rpa43 constructed by domain swapping with the S. pombe subunit (F. Beckouet, S. Labarre-Mariotte, Y. Nogi, and P. Thuriaux, unpublished results), exhibited strongly reduced Rrn3-HA occupancy at the rDNA promoter (Fig. 6A) but little or no effect on the Rrn7 and Rrn9 subunits of the core and upstream activating factors. This finding further supports earlier data showing that Rpa43 is specialized in transcription initiation and that Rrn3 needs to bind Rpa43 in order to be recruited to the rDNA promoter region as a preformed Rrn3-Pol I complex (34, 48, 57). Rrn3 appears to be released from rDNA (and therefore from Pol I) at the onset of transcription, although we cannot rule out that it undergoes a dramatic change in conformation that masks the Rrn3-HA tag at elongation. These two possibilities cannot be distinguished in ChIP assays, but the in vitro data of Bier et al. (5) strongly argue that Rrn3 no longer binds the elongating Pol I.
FIG. 6.

Effect of RNA Pol I mutations on Rrn3-HA and Rrn7-HA occupancy. (A) Rrn3 occupancy in rpa43-chim2, rpa49Δ, and rpa49-338::HIS3 strains. ChIP assays were done on strain SL97-17C (rpa43Δ Rrn3-HA) bearing plasmid pGEN-A43 (wild type [WT]) or pGA43-chim2 (rpa43-chim2) and strains SL112-1C (rpa49Δ) and D612-5A (rpa49-338::HIS3) by using the same conditions described above. Rrn3-HA was revealed with anti-hemagglutinin A mouse antibody (12CA5). A schematic representation of the rpa43-chim2 strain is shown below. Numbers denote amino acid positions and correspond to the limits of the domains swapped between S. cerevisiae and S. pombe. IP, immunoprecipitation signal; IN, DNA input signal. (B) Rrn7 occupancy. ChIP assays were done on strains SL105-7A (rpa43Δ Rrn7-HA) and SL111-4A (rpa49Δ Rrn7-HA) bearing the same plasmids and under the same conditions described above.
Turning to the rpa49Δ and rpa49-338::HIS3 mutants, we also found the Rrn3-HA signal at the promoter to be decreased, but much less so than that in the rpa43-chim2 strain. However, in contrast to the Rrn3 in the wild-type or rpa43-chim2 strain, a substantial fraction of the Rrn3 in the rpa49Δ and rpa49-338::HIS3 mutants remained bound to the elongating Pol I, with a decreasing signal toward the 3′ end of the transcript. The core and upstream activating factors (Rrn7-HA and Rrn9-HA), on the other hand, were still restricted to the promoter domain (as shown in Fig. 6B for Rrn7-HA). Intriguingly, rpa49Δ and rpa49-338::HIS3 were lethal when Rpa43 was replaced by an Rrn3-Rpa43 fusion in which Rrn3 was, by construction, permanently associated with the elongating Pol I (34) and which was previously shown to yield a viable strain when introduced into a wild-type background (data for rpa49Δ are shown in Fig. 7C).
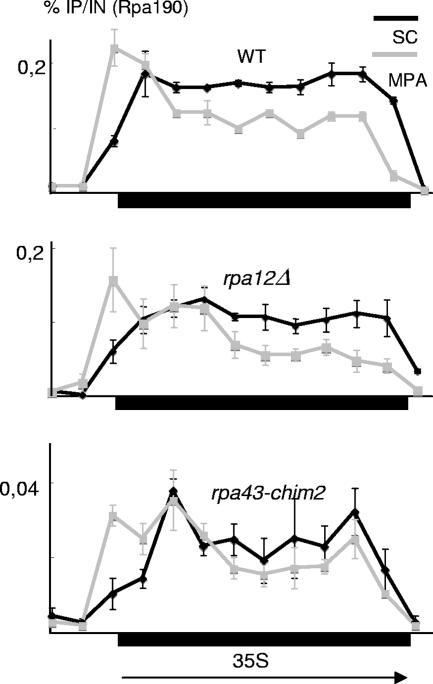
FIG. 7.
Effect of mycophenolate (MPA) on wild-type (WT), rpa12Δ, and rpa43-chim2 strains. Cells were cultivated in SD+aa medium until they reached an A600 of 0.2, treated (or left untreated) with 20 μg of mycophenolate/ml, and further grown for 30 min. In the case of Pol I, ChIP assays were done on strains SL97-17C [(rpa43::LEU2 Rrn3-HA)/YCPA43], used here as the wild-type control, SL9-2C (rpa12Δ), and D101-chim2 (rpa43-chim2). Pol I was revealed by polyclonal anti-Rpa190 rabbit antibodies (49). Five additional primers (5, 6, 7, 9, 10) were used for RT-PCR amplification and are listed in the tables in the supplemental material. The diagram at the bottom shows the approximate positions of the primers used for RT-PCR amplification (see the tables in the supplemental material). Arrows denote the start sites and transcription orientations of the 5S and 35S rRNA transcripts. SC, synthetic complete medium; IP, immunoprecipitation signal; IN, DNA input signal; NTS1 and NTS2, nontranscribed spacers 1 and 2.
The data cited above suggest that rpa49Δ and rpa49-338::HIS3 have a dual effect on Rrn3, partially reducing its association with the rDNA promoter but also impairing its release from the elongating Pol I. We therefore anticipated that conditions dissociating Pol I from its rDNA template should also dissociate the Pol I-bound Rrn3 of rpa49Δ and rpa49-338::HIS3 mutants, and we exploited the effect of mycophenolate on the elongating Pol I to test this possibility. As shown in Fig. 7, mycophenolate increased the Pol I signal seen at the rDNA promoter region, which may be due to a lower rate of elongation, and progressively decreased the amount of rDNA-bound Pol I as the Pol I moved toward the 3′ end of the rDNA, indicating a processivity defect similar to the one recently described for Pol II (38). In contrast, mycophenolate had no effect on Rrn3 occupancy, indicating that Rrn3 leaves the promoter before or shortly after the start of transcription (Fig. 8, right panels).
FIG. 8.
Effect of mycophenolate (MPA) on rpa49Δ, rpa49-338::HIS3, and rpa135(L656P) mutants. (Left panels) Effect on Pol I (Rpa190) in wild-type (WT), rpa49Δ, rpa49-338::HIS3, and rpa135(L656P) strains. Cells were cultivated in SD+aa medium until they reached an A600 of 0.2, treated (or left untreated) with 20 μg of mycophenolate/ml, and further grown for 30 min. ChIP assays were done on strains SL97-17C [(rpa43::LEU2 Rrn3-HA)/YCPA43], used here as the wild-type control, SL112-1C (Rrn3-HA rpa49Δ), SL107-3b (Rrn3-HA rpa49-338::HIS3), and OG36-1a [rpa135(L656P)]. Pol I was revealed by polyclonal anti-Rpa190 rabbit antibodies (49). (Right panels) Effect on Rrn3-HA. Cells were cultivated in SD+aa medium until they reached an A600 of 0.2, treated (or left untreated) with 20 μg of mycophenolate/ml, and further grown for 30 min. ChIP assays were done on strains SL97-17C [(rpa43::LEU2 Rrn3-HA)/YCPA43], used here as the wild-type control, SL112-1C (Rrn3-HA rpa49Δ), SL107-3b (Rrn3-HA rpa49-338::HIS3), and D769-2C [Rrn3-HA rpa135(L656P)]. Rrn3-HA was revealed with anti-hemagglutinin A mouse antibody (12CA5). SC, synthetic complete medium; IP, immunoprecipitation signal; IN, DNA input signal.
rpa43-chim2 and rpa12Δ strains responded essentially like the wild type when treated with mycophenolate (Fig. 7), consistent with their normal (rpa43-chim2) and mildly increased (rpa12Δ) drug sensitivities in vivo. However, a very different response by rpa49Δ and rpa49-338::HIS3 mutants was seen (Fig. 8, left panels). These mutants were strongly sensitive to mycophenolate, and their Pol I was almost entirely absent from rDNA after 30 min of exposure to this drug. In addition, and in sharp contrast to the wild-type situation, there was a massive loss of Rrn3, even at the promoter region, supporting the view that Rrn3 and Pol I were physically associated in these mutants and that Rrn3 was therefore lost together with Pol I itself (Fig. 8, right panels). As a control, these assays were also done with a strain carrying rpa135(L565P), shown as described above to cause sensitivity to mycophenolate and to be synthetically lethal with rpa49Δ and rpa49-338::HIS3. As shown in Fig. 8 (lower panels), there was an almost complete loss of Pol I in rpa135(L565P) cells exposed to mycophenolate, but Rrn3 remained bound to the rDNA promoter, as in the wild type.
An rpa43 mutation suppresses the cold-sensitive defect of rpa49 mutants.
In the course of this study, we observed that the rpa43-35,326 mutation, yielding a wild type-like mutant with a deletion of the nonconserved N terminus of Rpa43 (Beckouet et al., unpublished), acted as a gain-of-function mutation suppressing the cold sensitivity of rpa49 mutants (Fig. 9, upper panels). Consistent with this suppression, Pol I occupancy was restored to a nearly wild-type level, which coincided with better Rrn3 occupancy. However, rpa43-35,326 had little or no effect on the mycophenolate sensitivity of rpa49 mutants and did not change their defective Rrn3 release (Fig. 9, lower panels). These data further argue that rpa49Δ and rpa49-338::HIS3 have a dual effect on Rrn3, interfering with its recruitment during transcription initiation and preventing its release from the elongating Pol I. The former defect was suppressed by rpa43-35,326, thus possibly reflecting a direct interaction of Rpa49 with the N terminus of Rpa43. The defect in Rrn3 release, on the other hand, was independent of rpa43-35,326 and correlated with strong sensitivity to mycophenolate.
FIG. 9.
Suppression of rpa49 mutations by an N-terminal deletion of Rpa43 (rpa43-35,326). (Upper panels) Strains D710-8A (rpa43Δ rpa49Δ) and SL107-3B (rpa43Δ rpa49-338::HIS3) bearing the YCP43-12 (URA3 RPA43) plasmid were transformed with plasmids pGEN-RPA43 (TRP1 RPA43) and pFB63 (TRP1 rpa43-35,326). Transformants were replica plated onto FOA medium to chase YCP43-12, tested on YPD (25°C) and synthetic complete medium with mycophenolate (MPA) at 0, 5, and 20 g/liter (30°C), and incubated for 3 days at 25°C, as described in the legend to Fig. 3. Strain YPH500 was used as a wild-type (WT) control. (Lower panels) ChIP assays were done on strains SL97-17C (WT) and SL107-3b (rpa49-338::HIS3) bearing plasmid pFB63 (rpa43-35,326) or pGEN-RPA43 (RPA43+) by using the same conditions described above, except that cells were cultivated in SD+aa medium and were harvested at an A600 of 0.2. IP, immunoprecipitation signal; IN, DNA input signal.
DISCUSSION
Rpa49 and Rpa34 are noncatalytic components of Pol I (27), and their genetic inactivation in S. cerevisiae yields viable strains (20, 36). However, rpa49Δ mutants have severe growth defects (19, 20), and the rpa49Δ mutation was identified in genome-wide screening for mycophenolate-sensitive mutants (13), a first clue to the importance of Rpa49 for transcript elongation. We found here that Rpa34 and Rpa49 specifically interacted in a two-hybrid assay, which assigned the main Rpa34-binding domain to the N-terminal part of Rpa49. This binding may recruit Rpa34 onto Pol I, as this subunit was lost from Pol I in rpa49 mutants lacking the binding domain. rpa34Δ had only a minor effect on the association of Rpa49 with Pol I, suggesting that Rpa49 directly binds the Pol I core structure. Previous data suggested that Rpa34 and Rpa49 do not make contact with each other in the Pol I envelope (6), but results from recent structural studies support the idea that Rpa34 directly binds Rpa49, and modeling data also suggest that this binding operates at the level of the N-terminal domain of Rpa49 (33a). In S. pombe, the C terminus of the Rpa49-like subunit is critical in vivo (42), and we found here that a strain carrying rpa49-338::HIS3, a premature-termination mutation removing the last 28 amino acids of Rpa49, behaved like the rpa49Δ null mutant. The most important part of the Rpa34-Rpa49 dimer, therefore, appears to be the C terminus of Rpa49, which is strongly conserved in all eukaryotes.
Our data indicate a dual effect of rpa49Δ and rpa49-338::HIS3 on the recruitment of Rrn3 and on its binding to the elongating Pol I. These two phenotypes were genetically dissociated by rpa43-35,326, encoding an N-terminal deletion of the Rpa43 subunit which specifically suppressed the recruitment defect and also corrected the cold-sensitive growth defect of rpa49Δ and rpa49-338::HIS3 mutants. This suppression implies that Rpa43 and Rpa49 cooperate to load Pol I-Rrn3 onto the rDNA promoter but does not necessarily call for direct physical interaction between these two subunits. It may be, for example, that Rpa49 favors the recruitment of the Pol I-Rrn3 complex by interacting with the core factor. Importantly, rpa43-35,326 had no effect on the mycophenolate sensitivity of rpa49Δ and rpa49-338::HIS3 mutants and did not affect their defective release of Rrn3. However, the rpa49Δ and rpa49-338::HIS3 mutations were lethal in strains carrying an RRN3::RPA43 fusion product in which Rrn3 was, by construction, permanently associated with the elongating Pol I. Moreover, they were synthetically lethal with top1Δ (yielding a strain lacking a type I DNA topoisomerase) or hmo1Δ, a mutation inactivating Hmo1, an HMG box protein residing on rDNA (19, 25), and these results suggest that the progression of the mutant Pol I strongly depends on the topology (Top1) or spatial organization (Hmo1) of rDNA. The findings of a recent study (Kuhn et al., submitted) also indicate that a Pol I form lacking Rpa34 and Rpa49 has elongation defects in vitro.
rpa34Δ produced no growth defect of its own and led only to synthetically lethal effects that were shared by rpa49Δ and were suppressed by the overexpression of Rpa49. These findings raise the question of whether Rpa34 has any role other than to stabilize Rpa49 in the Pol I structure, thus optimizing the biological role of that subunit. Indeed, the two-hybrid interaction found here between Rpa34 and three other nucleolar proteins, Gno1, Nop56, and Nop58, tentatively suggests that Rpa34 may have a function of its own by favoring the cotranscriptional recruitment of the pre-rRNA maturation machine to the nascent transcript, since these putative partners intervene in the early maturation of the 35S pre-rRNA (14, 21, 24). Intriguingly, these proteins are also endowed with KKE/D repeats, like Rpa34 itself, although the biological meaning of these domains remains to be established.
Mammalian cells contain two Pol I-associated factors, PAF53 and PAF49, with strong homology to Rpa49 (PAF53) and weaker homology to Rpa34 (PAF49) (26, 47). In vitro, PAF53 and PAF49 stimulate the faithful transcription of rDNA by Pol I and may functionally and/or physically interact with UBF, an HMG box protein associated with animal Pol I (3). The name UBF, which stands for upstream binding factor, points to a role in the preinitiation steps of rDNA transcription (3), but recent evidence suggests that UBF contributes to promoter escape (46) or may even modulate the progression of Pol I along its rDNA template, since UBF actually binds the entire Pol I-transcribed region (45, 51). rpa49Δ is suppressed by the overexpression of Hmo1, which is probably the yeast UBF (4, 19), and is lethal in an hmo1Δ context. In other words, the existence of Hmo1 explains why the rpa49Δ null allele still supports transcription, albeit inefficiently. Hmo1 binds the whole Pol I-transcribed unit and is also a Pol II factor that binds many members of the ribosomal protein regulon (4, 25, 30). It is worth noting that, in vitro, Hmo1 unwinds DNA in the presence of DNA topoisomerase I (37) and that top1Δ and hmo1Δ are both synthetically lethal with rpa49Δ. The functional interplay between Rpa49-Rpa34 and Hmo1 needs to be investigated more thoroughly, but we would be surprised if it did not reflect a conserved mechanism operating in species from yeast to humans.
Supplementary Material
Acknowledgments
This work was supported by the Association pour la Recherche contre le Cancer (F.B.), by an ATIP grant from CNRS, and by the Agence Nationale de la Recherche (O.G.).
We thank M. Riva for polyclonal antibodies; M. Fromont-Racine for a two-hybrid yeast library; M. Goussot for technical help in two-hybrid screening; C. Carles, S. Chédin, M. Kwapisz, C. Mann, M. Riva, E. Shematorova, J. Soutourina, and M. Wéry for useful suggestions; and P. Cramer for kindly communicating unpublished data.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armache, K. J., H. Kettenberger, and P. Cramer. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 1006964-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslett, M., and V. Wood. 2006. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100%. Yeast 23913-919. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., R. M. Learned, H. M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates ribosomal RNA synthesis. Science 2411192-1197. [DOI] [PubMed] [Google Scholar]
- 4.Berger, A. B., L. Decourty, G. Badis, U. Nehrbass, A. Jacquier, and O. Gadal. 2007. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell. Biol. 278015-8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bier, M., S. Fath, and H. Tschochner. 2004. The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS Lett. 56441-46. [DOI] [PubMed] [Google Scholar]
- 6.Bischler, N., L. Brino, C. Carles, M. Riva, H. Tschochner, V. Mallouh, and P. Schultz. 2002. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 214136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf, H. Delius, M. Vingron, and I. Grummt. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., F. Lacroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197345-346. [DOI] [PubMed] [Google Scholar]
- 9.Boukhgalter, B., M. Liu, A. Guo, M. Tripp, K. Tran, C. Huynh, and L. Pape. 2002. Characterization of a fission yeast subunit of an RNA polymerase I essential transcription initiation factor, SpRrn7h/TAF(I)68, that bridges yeast and mammals: association with SpRrn11h and the core ribosomal RNA gene promoter. Gene 291187-201. [DOI] [PubMed] [Google Scholar]
- 10.Cadwell, C., H. Y. Yoon, Y. Zebarjadian, and J. Carbon. 1997. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 176175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comai, L., N. Tanese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor SL1. Cell 68965-976. [DOI] [PubMed] [Google Scholar]
- 12.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase at 2.8 angstrom resolution. Science 2921863-1876. [DOI] [PubMed] [Google Scholar]
- 13.Desmoucelles, C., B. Pinson, C. Saint-Marc, and B. Daignan-Fornier. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 27727036-27044. [DOI] [PubMed] [Google Scholar]
- 14.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 229-11. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Tornero, C., B. Bottcher, M. Riva, C. Carles, U. Steuerwald, R. W. Ruigrok, A. Sentenac, C. W. Muller, and G. Schoehn. 2007. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell 25813-823. [DOI] [PubMed] [Google Scholar]
- 17.Flores, A., J. F. Briand, C. Boschiero, O. Gadal, J. C. Andrau, L. Rubbi, V. Van Mullem, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 967815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16277-282. [DOI] [PubMed] [Google Scholar]
- 19.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 215498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadal, O., S. Mariotte-Labarre, S. Chédin, E. Quémeneur, C. Carles, A. Sentenac, and P. Thuriaux. 1997. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machinery. Mol. Cell. Biol. 171787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier, T., T. Berges, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 177088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorski, J. J., S. Pathak, K. Panov, T. Kasciukovic, T. Panova, J. Russell, and J. C. Zomerdijk. 2007. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 261560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grummt, I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 171691-1702. [DOI] [PubMed] [Google Scholar]
- 24.Guglielmi, B., and M. Werner. 2002. The yeast homolog of human PinX1 is involved in rRNA and snoRNA maturation, not in telomere elongation inhibition. J. Biol. Chem. 27735712-35719. [DOI] [PubMed] [Google Scholar]
- 25.Hall, D. B., J. T. Wade, and K. Struhl. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 263672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanada, K. I., C. Z. Song, K. Yamamoto, K. I. Yano, Y. Maeda, K. Yamagushi, and M. Muramatsu. 1996. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 152217-2226. [PMC free article] [PubMed] [Google Scholar]
- 27.Huet, J., J. M. Buhler, A. Sentenac, and P. Fromageot. 1975. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl. Acad. Sci. USA 723034-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imazawa, Y., K. Hisatake, H. Mitsuzawa, M. Matsumoto, T. Tsukui, K. Nakagawa, T. Nakadai, M. Shimada, A. Ishihama, and Y. Nogi. 2005. The fission yeast protein Ker1p is an ortholog of RNA polymerase I subunit A14 in Saccharomyces cerevisiae and is required for stable association of Rrn3p and RPA21 in RNA polymerase I. J. Biol. Chem. 28011467-11474. [DOI] [PubMed] [Google Scholar]
- 29.Jasiak, A. J., K. J. Armache, B. Martens, R. P. Jansen, and P. Cramer. 2006. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol. Cell 2371-81. [DOI] [PubMed] [Google Scholar]
- 30.Kasahara, K., K. Ohtsuki, S. Ki, K. Aoyama, H. Takahashi, T. Kobayashi, K. Shirahige, and T. Kokubo. 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 276686-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10887-903. [DOI] [PubMed] [Google Scholar]
- 32.Keys, D. A., L. Vu, J. S. Steffan, J. A. Dodd, R. Yamamoto, Y. Nogi, and M. Nomura. 1994. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 82349-2362. [DOI] [PubMed] [Google Scholar]
- 33.Kranz, J. E., and C. Holm. 1990. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 876629-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Kuhn, C.-D., S. R. Geiger, S. Baumli, M. Gartmann, J. Gerber, S. Jennebach, T. Mielke, H. Tschochner, R. Beckmann, and P. Cramer. 2007. Functional architecture of RNA polymerase I. Cell 1311260-1272. [DOI] [PubMed] [Google Scholar]
- 34.Laferté, A., E. Favry, A. Sentenac, M. Riva, C. Carles, and S. Chédin. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 202030-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer, D., J. Hain, P. Thuriaux, and W. Zillig. 1995. Transcription in Archaea: similarity to that in Eucarya. Proc. Natl. Acad. Sci. USA 925768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liljelund, P., M. Mariotte, J. M. Buhler, and A. Sentenac. 1992. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 899302-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, J., R. Kobayashi, and S. J. Brill. 1996. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 27133678-33685. [DOI] [PubMed] [Google Scholar]
- 38.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17831-840. [DOI] [PubMed] [Google Scholar]
- 39.Milkereit, P., and H. Tschochner. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 173692-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 201373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorefield, B., E. A. Greene, and R. H. Reeder. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. USA 974724-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa, K., K. Hisatake, Y. Imazawa, A. Ishiguro, M. Matsumoto, L. Pape, A. Ishihama, and Y. Nogi. 2003. The fission yeast RPA51 is a functional homolog of the budding yeast A49 subunit of RNA polymerase I and required for maximizing transcription of ribosomal DNA. Genes Genet. Syst. 78199-209. [DOI] [PubMed] [Google Scholar]
- 43.Nogi, Y., L. Vu, and M. Nomura. 1991. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 887026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nomura, M. 1999. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J. Bacteriol. 1816857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Sullivan, A. C., G. J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panov, K. I., J. K. Friedrich, J. Russell, and J. C. Zomerdijk. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 253310-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panov, K. I., T. B. Panova, O. Gadal, K. Nishiyama, T. Saito, J. Russell, and J. C. Zomerdijk. 2006. RNA polymerase I-specific subunit CAST/hPAF49 has a role in the activation of transcription by upstream binding factor. Mol. Cell. Biol. 265436-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyroche, G., P. Milkereit, N. Bischler, H. Tschochner, P. Schultz, A. Sentenac, C. Carles, and M. Riva. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 195473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riva, M., S. Mémet, J. Y. Micouin, J. Huet, I. Treich, J. Dassa, R. Young, J. M. Buhler, A. Sentenac, and P. Fromageot. 1986. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc. Natl. Acad. Sci. USA 831554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudra, D., and J. R. Warner. 2004. What better measure than ribosome synthesis? Genes Dev. 182431-2436. [DOI] [PubMed] [Google Scholar]
- 51.Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21629-639. [DOI] [PubMed] [Google Scholar]
- 52.Stefanovsky, V. Y., G. Pelletier, D. P. Bazett-Jones, C. Crane-Robinson, and T. Moss. 2001. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 293241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2942364-2368. [DOI] [PubMed] [Google Scholar]
- 54.Van Mullem, V., M. Wery, X. De Bolle, and J. Vandenhaute. 2003. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast 20739-746. [DOI] [PubMed] [Google Scholar]
- 55.Werner, F., and R. O. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10635-646. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto, K., M. Yamamoto, K. Hanada, Y. Nogi, T. Matsuyama, and M. Muramatsu. 2004. Multiple protein-protein interactions by RNA polymerase I-associated factor PAF49 and role of PAF49 in rRNA transcription. Mol. Cell. Biol. 246338-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 153964-3973. [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98811-824. [DOI] [PubMed] [Google Scholar]
- 59.Zomerdijk, J. C. B. M., H. Beckmann, L. Comai, and R. Tjian. 1994. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 2662015-2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.