Abstract
Peroxisomes are single membrane-delimited subcellular organelles that carry out numerous vital metabolic reactions in nearly all eukaryotes. Peroxisomes alter their morphology, abundance, and enzymatic constituents in response to environmental cues, yet little is known about the underlying mechanisms. In this work, we investigated the regulatory role of light in peroxisome proliferation in Arabidopsis (Arabidopsis thaliana). We provide evidence that light induces proliferation of peroxisomes in Arabidopsis seedlings and that the peroxisomal protein PEX11b plays an important role in mediating this process. The far-red light receptor phytochrome A (phyA) and the bZIP transcription factor HY5 HOMOLOG (HYH) are both required for the up-regulation of PEX11b in the light. We further demonstrate that the phyA and hyh mutants exhibit reduced peroxisome abundance, a phenotype that can be rescued by overexpressing PEX11b in these plants. The HYH protein is able to bind to the promoter of PEX11b, suggesting that the PEX11b gene is a direct target of HYH. We conclude that HYH and PEX11b constitute a novel branch of the phyA-mediated light signaling cascade, which promotes peroxisome proliferation during seedling photomorphogenesis.
Peroxisomes are ubiquitous eukaryotic organelles enclosed by a single membrane and devoid of DNA. They perform a variety of metabolic functions that differ depending on the species, cell type, developmental stage, and prevailing environmental conditions (Purdue and Lazarow, 2001). Plant peroxisomes can be categorized into several variants that are structurally similar but metabolically distinct, such as glyoxysomes in seeds and germinating seedlings, leaf peroxisomes, glyoxysome-related gerontosomes in senescent tissue, nodule-specific peroxisomes, and unspecialized peroxisomes. Functions attributed to plant peroxisomes include metabolism of lipids, reactive oxygen species, and nitrogen, photorespiration, plant hormone biosynthesis and metabolism, and plant pathogen interaction (Beevers, 1979; Olsen and Harada, 1995; Zolman et al., 2000; Hayashi and Nishimura, 2003; Lipka et al., 2005; McCartney et al., 2005; Nyathi and Baker, 2006; Reumann and Weber, 2006).
Although direct evidence has not been obtained from plant systems, cell biological and phylogenetic evidence suggested that peroxisomes can be formed de novo in the endoplasmic reticulum (ER) and have an ER origin in evolutionary history (Hoepfner et al., 2005; Gabaldon et al., 2006; Schluter et al., 2006; Titorenko and Mullen, 2006). Peroxisome biogenesis involves the formation of the peroxisome membrane, import of cytoplasmically synthesized proteins into the peroxisomal matrix, and multiplication and maintenance of peroxisomes, processes mediated by a group of peroxisome proteins mostly known as PEROXINS or PEX proteins (Heiland and Erdmann, 2005). Earlier studies in yeast (Saccharomyces cerevisiae) identified 32 PEX proteins, which have approximately 20 and 23 homologs in mammals and plants, respectively, and are referred to as PexNp in yeasts and PEXN in mammals and plants (Purdue and Lazarow, 2001; Charlton and Lopez-Huertas, 2002).
In addition to de novo formation from the ER, peroxisomes can multiply by division, constitutively or under induced conditions (the latter is also called proliferation). Yeast peroxisomes are believed to divide through a specific order of partially overlapping events, including organelle elongation, membrane constriction, and fission (Fagarasanu et al., 2007). The division process involves eight PEX proteins, which, based on sequence similarity, can be categorized into several subgroups: Pex11p, Pex25p/Pex27p, Pex28p/Pex29p, and Pex30p/Pex31p/Pex32p. The biochemical functions of these PEX proteins, all of which are associated with the peroxisomal membrane, have yet to be clearly defined (Thoms and Erdmann, 2005). The Arabidopsis (Arabidopsis thaliana) genome does not contain obvious sequence homologs to most of the aforementioned yeast PEX proteins known to specifically control peroxisome multiplication. One exception is Pex11p, which is primarily involved in peroxisome elongation/tubulation, the early stages of peroxisome division (Erdmann and Blobel, 1995; Marshall et al., 1995; Thoms and Erdmann, 2005; Fagarasanu et al., 2007). The five Arabidopsis PEX11 isoforms, PEX11a to PEX11e, were shown to promote peroxisome elongation and population increase with some level of specificity and redundancy (Lingard and Trelease, 2006; Orth et al., 2007). In addition to PEX proteins, the large GTPase dynamin-related protein (DRP) and FISSION1 (FIS1) proteins in yeast and mammals function in the late stages of peroxisome division, whereby the integral membrane protein FIS1 recruits the mechanochemical enzyme DRP to execute the tubulation and separation of peroxisomal membranes (Koch et al., 2003, 2005; Kobayashi et al., 2007). Interestingly, at least in yeast and mammals, the division machinery of peroxisomes and mitochondria seems to share the same DRP and FIS1 orthologs (Koch et al., 2005; Thoms and Erdmann, 2005; Schrader and Yoon, 2007). The Arabidopsis DRP3A protein was shown to be required for the division of both peroxisomes and mitochondria (Mano et al., 2004). Additional AtDRP proteins were also suggested to be involved in peroxisome division (Hu, 2007).
Peroxisome abundance is strongly affected by external signals. Yeast peroxisomes proliferate in response to nutritional stimuli, such as oleate and methanol and nitrogen sources, to metabolize these substrates for carbon or nitrogen utilization (Gurvitz and Rottensteiner, 2006). Likewise in mammals, when exposed to peroxisome proliferators—a pool of more than 70 chemicals ranging from hypolipidemic agents, phthalate ester plasticizers, solvents, herbicides, and dietary factors to hormones—mammalian peroxisome activity and abundance also increase (Citron, 1995; Desvergne and Wahli, 1999). Signal transduction of some of these external stimuli is mediated by transcription factors, such as the yeast Adr1p, Oaf1p, and Pip2p proteins, and the mammalian peroxisome proliferator-activated receptor-α and the retinoid X receptor proteins (Desvergne and Wahli, 1999; Gurvitz and Rottensteiner, 2006), all of which lack apparent orthologs in plants. Through electron microscopic analysis, ozone, herbicide, clofibrate, and high light were found to increase peroxisome abundance in plant cells with undetermined mechanisms (de Felipe et al., 1988; Ferreira et al., 1989; Palma et al., 1991; Oksanen et al., 2003).
We are particularly interested in the role of light, one of the major environmental cues, in peroxisome proliferation. Light affects many aspects of plant development, from seed germination, seedling photomorphogenesis, shade avoidance, photoperiodism, circadian clock entrainment, phototropism, chloroplast movement, stomata opening, to flowering (Jiao et al., 2007). When exposed to light, Arabidopsis seedlings undergo a process called photomorphogenesis, in which cotyledons open, hypocotyl growth is inhibited, chloroplasts develop, and genes involved in photosynthesis and related functions are activated (Wang and Deng, 2002a). During this process, glyoxysomes are converted into leaf peroxisomes by replacing enzymes specific for lipid metabolism during germination with proteins involved in the glycolate recycling during photorespiration (Olsen and Harada, 1995). Previously, we identified a gain-of-function mutation in a peroxisomal gene (PEX2) in Arabidopsis that partially suppressed the photomorphogenic (de-etiolated) phenotype of the mutant defective in the nuclear protein DET1 (Hu et al., 2002). However, the mechanism for this suppression appears to be complex and still remains to be determined.
Here, we investigated the impact of light on peroxisome proliferation in living plant cells and identified the underlying signal transduction pathway mediating this process. We present evidence that light promotes peroxisome proliferation, at least in part, by activating the expression of the peroxisomal gene PEX11b through the photoreceptor phytochrome A (phyA) and the bZIP transcription factor HYH.
RESULTS
Light Induces Peroxisome Proliferation
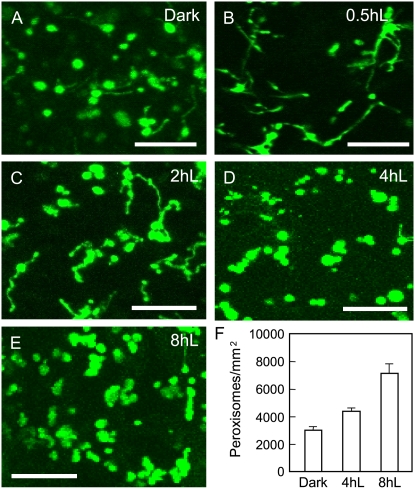
To address the question of whether light plays a role in peroxisome proliferation, we analyzed transgenic Arabidopsis seedlings constitutively expressing a fusion of the yellow fluorescent protein (YFP) with C-terminal PEROXISOME-TARGETING SIGNAL TYPE1 (PTS1; consisting of Ser-Lys-Leu). These YFP-PTS1 plants displayed fluorescently labeled peroxisomes as numerous punctate structures in virtually all cell types (Fan et al., 2005; Orth et al., 2007). Three-day-old YFP-PTS1 seedlings grown in the dark were illuminated with 70 μmol m−2 s−1 continuous white light for different time intervals and scored for obvious alterations in size, shape, and abundance of peroxisomes in cotyledon cells with a confocal laser-scanning microscope. Dark-grown seedlings exhibited a morphologically heterogeneous population of peroxisomes: Most were spherical and a few were elongated (Fig. 1A). After 0.5 h of illumination, a strong increase in the population of elongated peroxisomes was evident (Fig. 1B). By 2 h of light treatment, membrane constriction and organelle fission seemed to have occurred in many elongated peroxisomes, which led to segmentation of the organelles (Fig. 1C). At 4 and 8 h, the number of elongated/segmented peroxisomes decreased, whereas the number of spherical peroxisomes, some of which were still clustered together, was markedly elevated (Fig. 1, D and E). The abundance of peroxisomes did not change significantly beyond 8 h of illumination (data not shown). During the course of this experiment, peroxisomes should be going through the transition from glyoxysomes to leaf-type peroxisomes. For simplicity, we use the general term peroxisomes to refer to all the organelles in this article.
Figure 1.
Light induces peroxisome proliferation in Arabidopsis seedlings. A to E, Confocal microscopic images of cotyledon cells from 3-d dark-grown seedlings expressing the YFP-PTS1 peroxisomal marker. Seedlings were exposed to continuous white light at 70 μmol m−2 s−1 for the indicated lengths of time. Note the elongated and segmented peroxisomes appearing at 0.5 and 2 h, respectively, after light exposure. Bars = 10 μm. F, Peroxisome quantification. Error bars indicate sd, n = 4, P < 0.0002. [See online article for color version of this figure.]
The number of peroxisomes was quantified using ImageJ software, which has been used in many studies to quantitatively analyze the number and morphology of cells and subcellular structures (Soyombo et al., 2006; Shi et al., 2007). Because counting peroxisomes at 0.5 and 2 h was not feasible due to the significant degree of elongation and reticulation of peroxisomes, we only compared the number of peroxisomes at the following time points: dark, 4-, and 8-h light exposure. A significant increase in peroxisome abundance was observed after 8-h light treatment: from 2,963.9 ± 277/mm2 in etiolated seedlings to 4,349 ± 182.8/mm2 at 4 h and 7,063.7 ± 736.8/mm2 at 8 h of illumination (Fig. 1F; see “Materials and Methods” for details of counting). These data demonstrate a role of light in up-regulating the change in peroxisome abundance via a multistep process: organelle elongation, constriction, and fission.
PEX11b Plays a Critical Role in Light-Induced Peroxisome Proliferation
As a first step in understanding light induction of peroxisome proliferation at the mechanistic level, we focused our attention on the PEX11 proteins, the only Arabidopsis proteins shown to date to be specifically involved in peroxisome proliferation during dark-to-light transition. Overexpression of each of the five PEX11 isoforms led to peroxisome elongation and population increase to various degrees (Lingard and Trelease, 2006; Orth et al., 2007), whereas silencing of individual PEX11 genes reduced peroxisomal abundance (Orth et al., 2007). PEX11b was the prime candidate for a mediator in this process given that it was the only Arabidopsis PEX11 gene strongly up-regulated by light (Orth et al., 2007).
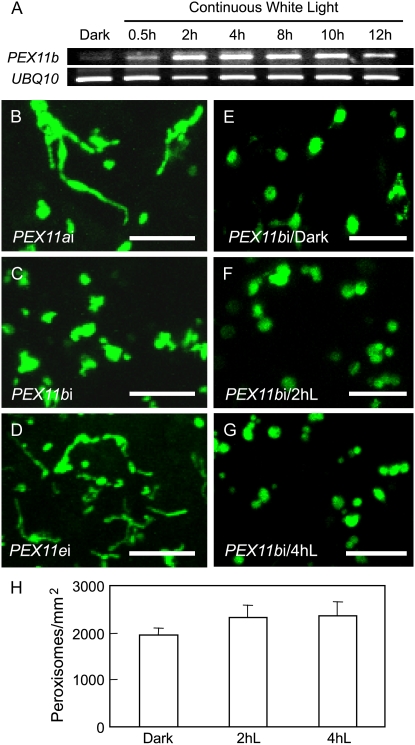
We examined the kinetics of light induction of PEX11b by reverse transcription (RT)-PCR with total RNA isolated from 3-d dark-grown seedlings exposed to white light for different time periods. The expression level of PEX11b was very low in dark-grown seedlings, but increased steadily after exposure to light, reached its maximum at 4 h, and started to drop by 12 h (Fig. 2A). The kinetics of PEX11b expression in light largely correlated with those of the light-dependent alterations in peroxisome morphology and abundance observed in the YFP-PTS1 plants (Fig. 1), implicating a specific role for PEX11b in light-mediated peroxisome elongation and population increase.
Figure 2.
PEX11b is required for light-induced peroxisome proliferation. A, RT-PCR analysis of the expression kinetics of PEX11b along with UBQ10 in the light. B to G, Confocal micrographs of cotyledon cells from 3-d dark-grown seedlings expressing YFP-PTS1. Bars = 10 μm. Seedlings analyzed in A to G were exposed to continuous white light at 70 μmol m−2 s−1 for 2 h (B–D) or as indicated (A and E–G). H, Quantification of peroxisomes in the cotyledons of the PEX11b RNAi plant. Error bars indicate sd, n = 4, P < 0.05. [See online article for color version of this figure.]
This correlation prompted us to examine loss-of-function PEX11b mutants for defects in peroxisome proliferation. To this end, we analyzed YFP-PTS1-expressing RNA interference (RNAi) plants generated in our previous study, each having a strongly reduced transcript level of PEX11a, PEX11b, or PEX11e (lines 1, 4, and 7 in Orth et al., 2007). These lines showed a decrease in overall peroxisome abundance, but exhibited no obvious deficiency in plant growth, lipid mobilization in germinating seedlings (evaluated by a sugar dependence assay), or photorespiration, most likely due to functional redundancy of the PEX11 genes (Orth et al., 2007). After 2-h light treatment, significantly elongated peroxisomes were present in the PEX11a and PEX11e RNAi lines, whereas peroxisome morphology hardly changed in the PEX11b RNAi plant (Fig. 2, B–D). The phenotype of abnormally shaped and aggregated peroxisomes is typical for the PEX11b RNAi plant (Orth et al., 2007), which may reflect partial peroxisome division deficiency in this mutant. Moreover, the number of peroxisomes/mm2 in the PEX11b RNAi plant increased only approximately 20% from dark (1,966.7 ± 141.2) to 2-h (2,326.8 ± 249.3) or 4-h (2,354.5 ± 315.7) light treatment (Fig. 2, E–H), in contrast to the approximately 47% increase in the wild type (dark to 4 h light; Fig. 1F). Taken together, our data have revealed PEX11b as an important peroxisomal mediator in light-induced peroxisome proliferation.
phyA and HYH Are Required for PEX11b-Mediated Regulation of Peroxisome Proliferation
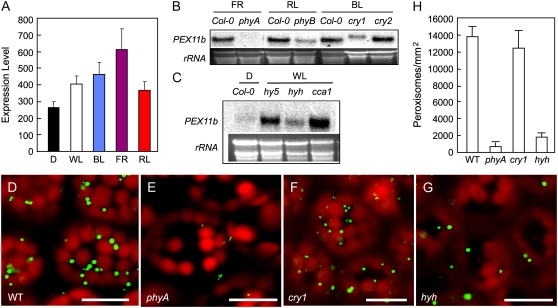
For optimal growth and development, higher plants are able to monitor the wavelength, quantity, direction, and duration of light via several families of photoreceptors, transduce the signals through complex protein networks, including transcription cascades, and mount a wide array of responses (Chen et al., 2004; Jiao et al., 2007). To determine which wavelengths of the light spectrum specifically affect the expression of PEX11b, we searched GENEVESTIGATOR (http://www.genevestigator.ethz.ch), a reference expression database and analysis tool (Zimmermann et al., 2004), and obtained quantitative data of PEX11b expression in different light conditions based on experiments performed with the Arabidopsis full genome chip (ATH1) arrays. In reference to transcript level in the dark, far-red light conferred the strongest up-regulation of PEX11b (2.3-fold), followed by blue (1.7-fold), white (1.5-fold), and red (1.3-fold) light (Fig. 3A), suggesting that, whereas all three wavelengths of light may activate the expression of PEX11b, far-red light appears to play the strongest role.
Figure 3.
phyA and HYH play important roles in mediating light-induced expression of PEX11b and in controlling peroxisome abundance. A, GENEVESTIGATOR analysis of PEX11b expression level under various light conditions. Error bars indicate sd, n = 6, P < 0.04. B to C, RNA-blot analyses of PEX11b expression in wild-type and mutant seedlings exposed to different wavelengths of light. Plants were germinated in the dark for 3 d before exposure to white (70 μmol m−2 s−1), far-red (2 μmol m−2 s−1), red (90 μmol m−2 s−1), or blue (30 μmol m−2 s−1) light for 2 h. Ethidium bromide-stained rRNA was used as a loading control. The transcript level of PEX11b did not vary between wild-type Col-0, Landsberg erecta, and Wassilewskija plants under different light wavelengths (data not shown); thus RNA from Col-0 was used here as a control for all mutants. D to G, Confocal images of cotyledon cells from 14-d light-grown wild type, phyA, cry1, and hyh mutants each expressing YFP-PTS1. Bars = 20 μm. H, Quantification of peroxisomes in the corresponding cells shown in D to G. Error bars indicate sd, n = 4.
To identify components in the light signaling pathways responsible for activating PEX11b, we obtained null mutants of various light signaling genes and analyzed light-induced PEX11b expression in these lines using RNA-blot and RT-PCR analyses. First, we checked several photoreceptors known to perceive different wavelengths of light during seedling development, namely, phyA for far-red, phyB for red, and cryptochrome 1 (cry1) and cry2 for blue light (lowercase letters are used in the light research field to refer to the holoproteins; Chen et al., 2004). The light induction of PEX11b expression was strongly decreased in phyA and moderately reduced in cry1 and phyB, whereas no significant changes were detected in cry2 (Fig. 3B). PEX11b was identified as one of the late up-regulated genes by phyA in previous microarray studies (Tepperman et al., 2001; Hudson and Quail, 2003). Analysis of the microarray data from Hudson and Quail (2003) revealed that the expression of PEX11b in wild-type seedlings was up-regulated with increasing time of far-red light irradiation, but was diminished in the phyA mutant under the same conditions (Supplemental Fig S1). Thus, the impact of far-red light and phyA on PEX11b expression is substantiated.
To identify phyA signaling intermediates involved in the regulation of PEX11b, we further examined null mutants of several well-characterized players in the phyA signaling pathway, including the novel nuclear protein FHY1, the homologous nuclear proteins FHY3 and FAR1, the GRAS family member PAT1, and the basic helix-loop-helix (bHLH) transcription factor HFR1 (Hudson et al., 1999; Bolle et al., 2000; Fairchild et al., 2000; Desnos et al., 2001; Wang and Deng, 2002b; Duek and Fankhauser, 2003). After far-red light treatment, the expression of PEX11b in these mutants did not differ significantly from that of the wild type, in contrast to the strongly decreased induction in phyA (Supplemental Fig S2). In parallel, we also analyzed mutants of several light signaling components that are downstream from multiple photoreceptors, including the key photomorphogenic transcription regulators LONG HYPOCOTYL5 (HY5; Oyama et al., 1997) and HY5 HOMOLOG (HYH; Holm et al., 2002), and CCA1—a central component of the light-entrained circadian clock oscillator (Wang and Tobin, 1998). A partial, yet significant, reduction in PEX11b expression was detected in the hyh mutant when compared with the other mutants (Fig. 3C).
Given that the light up-regulation of PEX11b was strongly decreased in phyA compared with other photoreceptor mutants, and that the light induction of PEX11b was relatively weak in hyh among all the downstream light signaling mutants tested, we further examined these two mutants for possible peroxisome phenotypes. To this end, the YFP-PTS1 marker gene was individually crossed into the phyA, cry1, and hyh mutant background; cry1 served as a control. Confocal microscopic analysis of the YFP-PTS1-expressing mutants revealed a drastic decrease in peroxisome abundance in phyA and hyh, but not in cry1, in both germinating seedlings (data not shown) and adult plants (Fig. 3, D–G). When quantitatively compared with the wild-type plants (13,850.4 ± 1,046.4/mm2), there was a 22.5-fold decrease in peroxisome abundance in phyA (615.5 ± 523.2/mm2; P < 0.001), a 7.5-fold decrease in hyh (1,846.7 ± 369.3/mm2; P < 0.001), but no significant reduction in cry1 (12,311.4 ± 2,154.5/mm2; P = 0.1047; Fig. 3H). These results led us to the conclusion that the far-red light receptor phyA and the bZIP transcription factor HYH may play direct roles in peroxisome proliferation through activating the expression of PEX11b.
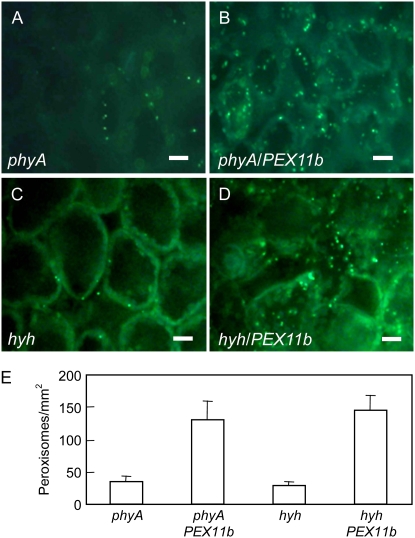
We further tested the hypothesis that phyA and HYH promote peroxisome proliferation in light through PEX11b by overexpressing the PEX11b gene in the phyA and hyh mutants to see whether the peroxisomal phenotype in these mutants could be rescued. We conducted an in vivo transient assay in the leaves of the YFP-PTS1-expressing phyA and hyh plants by introducing a PEX11b overexpression construct via leaf Agrobacterium infiltration. Two days postinoculation, we observed many elongated peroxisomes, as well as an increase in the total number of peroxisomes in the phyA and hyh mutants overexpressing PEX11b; this phenotype is similar to what we observed previously in wild-type plants overexpressing PEX11b (Orth et al., 2007). For quantification purposes, epifluorescence microscopic images exhibiting obvious increases in peroxisome abundance, but not elongation, are shown here (Fig. 4, A–D). The number of peroxisomes increased to approximately 4-fold in phyA (33.5 ± 8.7–130.7 ± 29.3/mm2) and approximately 5-fold in hyh (29.3 ± 5.2–145.4 ± 23.7/mm2) after PEX11b was expressed (Fig. 4E). The lower peroxisome abundance in these plants compared with those shown in Figure 3 may have been mainly caused by stress from agrobacterial infection. These data provide additional evidence for the notion that PEX11b is downstream in the phyA- and HYH-mediated pathway in light-mediated peroxisome proliferation.
Figure 4.
Overexpressing PEX11b rescues the peroxisome abundance phenotype in phyA and hyh mutants. A to D, Epifluorescence microscopic images of 3-week-old phyA and hyh plants infiltrated with agrobacteria containing the empty vector (A and C) or the PEX11b gene (B and D). Bars = 10 μm. E, Peroxisome quantification in the corresponding cells shown in A to D. Error bars indicate sd, n = 6, P < 0.0005. [See online article for color version of this figure.]
HYH Interacts with the Promoter of PEX11b
HYH and its homolog, HY5, are transcription factors that contain a bZIP DNA-binding domain and play redundant, as well as specific, roles in gene regulation during photomorphogenesis (Oyama et al., 1997; Holm et al., 2002; Vandenbussche et al., 2007). The fact that the up-regulation of PEX11b expression by light was reduced in hyh, but not in hy5, suggests that PEX11b may be one of the specific targets for HYH.
The promoters of light-inducible genes in plants harbor various types of cis-elements, collectively named light-response elements (LREs), which are bound by transcription factors and mediate light-dependent gene activation (Ueda et al., 1989; Park et al., 1996; Lescot et al., 2002). To assess whether putative LREs are present in the promoter of PEX11b, we performed detailed in silico searches of the PlantCARE and the PLACE plant cis-element databases, using a 219-bp promoter region immediately upstream of the transcription start site (TSS) of this gene as query. We found a total number of 19 putative LREs (Supplemental Fig. S3), including seven GATA boxes (Lam and Chua, 1989), six GT1 boxes (Villain et al., 1996), three I boxes (Giuliano et al., 1988), one AE box (Park et al., 1996), one ATCT motif (Conley et al., 1994), and one GGATAA element (Tepperman et al., 2001). GATA, GT1, and I boxes are cis-elements commonly existing in light-regulated promoters and shown to be essential for light-controlled transcriptional activity (Tobin and Kehoe, 1994; Terzaghi and Cashmore, 1995; Millar and Kay, 1996). The GGATAA element frequently occurs in the promoters of phyA-regulated genes (Tepperman et al., 2001).
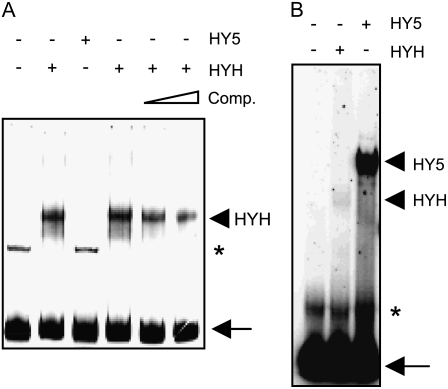
Considering that the PEX11b promoter is laden with LREs commonly found in light-responsive promoters, it is possible that HY5 and HYH may differ in their binding affinity to the PEX11b promoter. To test this hypothesis, we performed an electrophoretic mobility shift assay. A DNA fragment containing 200 bp of the PEX11b promoter immediately upstream of the TSS was used as a probe to determine whether it could be bound by purified HYH-His fusion protein. As shown in Figure 5A, we detected binding of HYH-His with the DNA probe, which was competed away efficiently with 50 and 100 molar excess unradiolabeled probes. Despite the fact that HYH and HY5 share 88.5% amino acid identity in the DNA-binding domain (Holm et al., 2002), no interaction was observed between HY5-His and the 200-bp promoter fragment (Fig. 5A).
Figure 5.
HYH interacts with the PEX11b promoter in gel-shift assays. A, Interaction between HYH and the 200-bp PEX11b promoter. The triangle above the last two lanes of the gel indicates increasing concentrations of the cold competitor (Comp.). Content in lane 2 is the same as in lane 4. B, Interaction between HYH/HY5 and the G box from the RBCS1A promoter. In both A and B, protein-DNA complexes were resolved on a 5% native 0.5× Tris-borate/EDTA polyacrylamide gel. The pluses and minuses above the gels symbolize the presence or absence of the proteins indicated. Arrowheads point to positions of the putative protein-DNA complexes. Asterisks indicate nonspecific binding; arrows indicate unbound probes.
To ensure that the recombinant protein HY5-His used in our study was functional in DNA binding, we subsequently tested whether HY5-His was able to bind G box, an LRE absent from the 200-bp promoter of the PEX11b gene, but was shown previously to be bound by both HY5 and HYH (Holm et al., 2002). Another gel-shift assay was performed with a 23-bp oligonucleotide containing the G-box motif from the RBCS1A promoter (Holm et al., 2002) as the probe. Both recombinant proteins were able to bind to this fragment; much stronger binding was observed for HY5-His (Fig. 5B). Although binding of HYH to the G box was previously shown not to be as strong as that of HY5 (Holm et al., 2002), this binding was even weaker in our assay. This is possibly due in part to the fact that the full-length HYH, instead of a truncated version (Holm et al., 2002), was used in our study.
Based on these data, we conclude that HYH is able to bind specifically to the PEX11b promoter and therefore is most likely a direct transcription activator of PEX11b in the light.
DISCUSSION
Dark-germinated Arabidopsis seedlings contain maximal activity of the glyoxylate cycle enzyme isocitrate lyase at 3 d, which gradually declines afterward (P. Yang and J. Hu, unpublished data). Consistent with this finding, we found more (basal) elongated peroxisomes in the dark-grown cotyledon cells than in adult tissue (observed in our previous studies, such as Fan et al. [2005] and Orth et al. [2007]). This observation suggests that peroxisomes divide at a higher rate in 3-d seedlings due to the active growth of seedlings at this stage. During photomorphogenesis, as seedlings undergo the transition from heterotrophic to autotrophic growth, organelles involved in photosynthesis and related functions (such as photorespiration) need to develop and genes required in these processes need to be activated. It was suggested that, during such a transitional stage, glyoxysomes are converted into leaf peroxisomes by replacement of glyoxysome-specific enzymes with those characteristic of leaf-type peroxisomes (Trelease et al., 1971; Olsen and Harada, 1995). In this study, we observed in Arabidopsis seedlings peroxisome elongation after 0.5-h light exposure, constriction and fission of these organelles at 2 h, and a sheer increase in peroxisome abundance at 4 and 8 h (Fig. 1). Therefore, we believe that, during seedling photomorphogenesis, a strong increase in peroxisome abundance occurs simultaneously with the import of leaf peroxisomal enzymes into the organelles to meet the high demand of leaf photosynthesis and photorespiration.
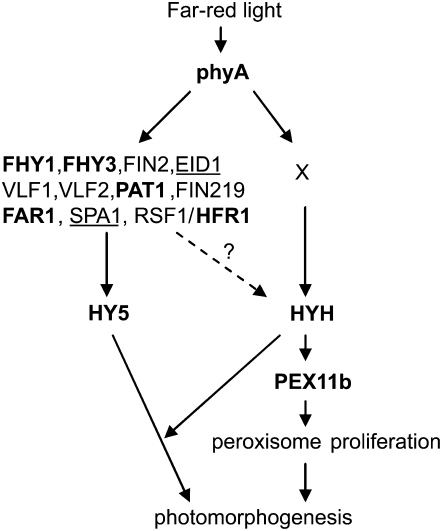
Given their sessile nature, plants have evolved a complex web of signaling events to perceive and respond to environmental cues, such as light (Chen et al., 2004; Jiao et al., 2007). Light-regulated plant development is one of the most extensively studied areas in plant biology. As a result, many players in light signaling have been identified through genetic and biochemical means and numerous mutants are available. To this end, we were able to take advantage of the mutant collections and employed molecular, genetic, and cell biological approaches to reveal the signal transduction pathway responsible for light-dependent peroxisome proliferation. First, PEX11b was strongly up-regulated by light, especially far-red light. Second, silencing of the PEX11b gene caused deficiency in light-induced peroxisome proliferation. Notably, the number of peroxisomes in 3-d cotyledons from the PEX11b RNAi line showed approximately 33% (1,966.7 versus 2,963.9) decrease from the wild type (Figs. 1F and 2H). This reduction in basal peroxisome abundance was not as dramatic as the decrease shown in 4-week-old mesophyll cells from our previous study (approximately 75% in Orth et al., 2007), indicating possible tissue-specific functions of the PEX11b gene. Third, the light induction of PEX11b expression was significantly reduced in the phyA and hyh mutants. Fourth, phyA and hyh displayed decreased numbers of peroxisomes, a phenotype that could be rescued by overexpressing the PEX11b gene in these mutants. Last, HYH was able to interact with the 200-bp promoter of PEX11b. Based on these results, a new branch of the phyA pathway in seedling development has been identified in which HYH induces peroxisome proliferation by directly activating the PEX11b gene (Fig. 6). Although null mutants of the photoreceptors cry1 and phyB also exhibited some level of reduction in the light-dependent up-regulation of PEX11b, they showed no obvious peroxisome phenotypes (Fig. 3; data not shown). Thus, the influence of cry1 and phyB on peroxisome proliferation through PEX11b might be minor compared with that of phyA.
Figure 6.
A branch of the phyA-dependent light signaling network is controlled by HYH and PEX11b in Arabidopsis seedling photomorphogenesis. In far-red light, phyA promotes photomorphogenesis through multiple pathways, which contain a number of signaling intermediates. HY5 and HYH are downstream components in the phyA network and act as transcription factors that regulate overlapping and specific sets of phyA-responsive genes. The PEX11b gene, which encodes a peroxisomal protein directly involved in the proliferation of peroxisomes, is one of the specific target genes for HYH. Protein X could be an unidentified player acting in between phyA and HYH. Alternatively (dotted arrow with a question mark), some of the known signaling intermediates in the phyA network may also be involved in regulating PEX11b through HYH. Proteins analyzed in this study are in bold; negative regulators of phyA signaling are underlined.
In addition to the identification of PEX11b as one of the late up-regulated genes by phyA in previous microarray studies (Tepperman et al., 2001; Hudson and Quail, 2003), several early studies in mustard (Sinapis alba) plants also pointed to the role of phyA in the transformation of glyoxysomes to leaf peroxisomes. For example, transient formation of glyoxyperoxisomes (peroxisomes associated with both lipid bodies and plastids) was observed in cotyledons when mustard seedlings were exposed to continuous far-red light; formation of such functional intermediates of peroxisomes was attributed to a transitional stage from glyoxysomal to leaf peroxisomal metabolism (Schopfer et al., 1976). The activity of leaf peroxisomal enzymes, such as glycolate oxidase, urate oxidase, and allantoinase, increased in mustard cotyledons after continuous far-red light irradiation (Poucke et al., 1970; Hong and Schopfer, 1980, 1981). These reports agree well with our finding that phyA plays a key role in the light-dependent proliferation of peroxisomes during the transition from glyoxysomes to leaf peroxisomes in Arabidopsis seedlings.
A previous kinetic study of the action of phyA and phyB showed that phyA was responsible for initiating hypocotyl growth inhibition at the onset of continuous red light. This role of phyA started to decrease after 3 h of irradiation, at which point the phyA protein began to be degraded and the contribution of phyB to hypocotyl inhibition became predominant (Parks and Spalding, 1999). This finding also correlates with our results, which showed that the expression of PEX11b reached its maximum at 4 h of white-light exposure and declined afterward (Fig. 2), and supports the view that phyA plays a primary role in the up-regulation of the PEX11b gene during early hours of light exposure (dark-to-light transition) in Arabidopsis seedlings.
As a photoreceptor primarily perceiving far-red light, the phyA protein is mainly cytoplasmic in the dark and translocates into the nucleus after being activated by light, where it interacts with various transcriptional factors to regulate the transcription of far-red light-responsive genes (Wang and Deng, 2003). However, despite our data showing the significance of far-red light and phyA in regulating peroxisome abundance, we failed to detect the obvious reduction in PEX11b expression and any peroxisomal phenotype in several mutants of the phyA pathway, namely, pat1, hfr1, fhy1, fhy3, and far1 (Supplemental Fig. S2; data not shown). It is possible that additional constituents of the phyA pathway not tested in this study are involved in regulating peroxisome proliferation through PEX11b. The proteins examined here may also be redundant in function with each other or with other untested proteins. In partial support of these predictions, FHY3 and FAR1 are closely related in sequence (Hudson et al., 1999; Wang and Deng, 2002b), HFR1 is a member of the bHLH family known to contain many members (such as the PIFs) with roles in phytochrome signaling (Duek and Fankhauser, 2005), and PAT1 shares high sequence similarity with several members of the GRAS protein family (Bolle et al., 2000). Finally, despite being a less likely scenario, the possibility that phyA regulates PEX11b partially through its cytoplasmically localized protein pool cannot be completely excluded.
HY5 and HYH are master regulators in the light signaling transcription cascades in photomorphogenesis, affecting the expression of numerous downstream genes (Oyama et al., 1997; Holm et al., 2002). In a recent study using in vivo chromatin immunoprecipitation combined with microarray analysis (CHIP-chip), 3,894 putative in vivo binding sites for HY5 were mapped in the Arabidopsis genome. These HY5 binding sites included G box, Z box and its variants, CG hybrid, and CA hybrid (Lee et al., 2007). Although PEX11b was among the putative target sites for HY5, we found only two G boxes and two CA hybrid motifs present in the PEX11b promoter, all of which are approximately 1.7 to 2.0 kb upstream from the TSS and thus absent from the promoter region analyzed in our study. Despite only minor differences in the DNA-binding domain (three exchanges over 24 amino acids; Holm et al., 2002), PEX11b expression was not reduced in the hy5 null mutant (Fig. 3C) and HY5 was unable to interact with the 200-bp promoter of PEX11b in our gel-shift assay (Fig. 5A). We postulate that the three unique amino acids in the DNA-binding domain, and possibly residues immediately adjacent to this domain, may facilitate the specific DNA-binding activity of HYH to the 200-bp promoter of PEX11b.
PEX11 proteins from various organisms play mostly conserved roles in promoting peroxisome elongation and population increase (Thoms and Erdmann, 2005) and are targets for peroxisome proliferation agents. For example, PEX11 in yeast is up-regulated by oleic acid (Erdmann and Blobel, 1995; Marshall et al., 1995). One of the three mouse PEX11 isoforms, PEX11α, is induced by peroxisome proliferation factors, such as clofibrate and 4-phenylbutyrate (Abe et al., 1998; Li et al., 2002). Our work has revealed light to be a significant peroxisome proliferator in plants, which strongly up-regulates the expression of the Arabidopsis PEX11b gene. It will be important to identify the specific cis-elements in the promoter of PEX11b required for such activation in the light. Yeast one-hybrid screens using the promoter of PEX11b as bait may also be necessary to isolate additional nuclear proteins interacting with this promoter. We expect that a more complete model for light-mediated peroxisome proliferation in plants will be established in the near future.
MATERIALS AND METHODS
Plant Growth, Light Conditions, and Genetic Crosses
The wild-type Arabidopsis (Arabidopsis thaliana) plants used in this study were from the Columbia-0 (Col-0) ecotype. The phyA (SALK_014575), hfr1 (SALK_037727), pat1 (SALK_064220), far1 (SALK_031652), fhy1 (SALK_130614), fhy3 (SALK_002711), and cry2-1 (Mockler et al., 2003) mutants were in the Col-0 background. The cry1-1 (hy4-1) mutant was in the Landsberg erecta background. The hy5-Ks50 (Oyama et al., 1997), hyh (Holm et al., 2002), and cca1-1 (Alabadi et al., 2002) mutants were in the Wassilewskija background. These mutants were confirmed by their respective light phenotypes and genotyped by PCR analysis to ensure their homozygosity. RT-PCR analysis was also performed to confirm that they were true null mutants.
Seeds were surface sterilized with 20% commercial bleach and 0.025% Triton X-100, washed five times with sterile water, plated on 0.5× Murashige and Skoog medium solidified with 0.6% phytagar, stratified at 4°C for 2 d, and subject to 1-h white light treatment to induce synchronous germination. Plates were then kept for 3 d in the dark at 21°C and subsequently exposed to white (70 μmol m−2 s−1), far-red (2 μmol m−2 s−1), red (90 μmol m−2 s−1), or blue (30 μmol m−2 s−1) light for the designated length of time. After the seedling stage, plants were transferred to soil and grown in growth chambers at 21°C with 100 μmol m−2 s−1 white-light conditions and 16/8-h photoperiod.
Genetic crosses were made between YFP-PTS1 plants and phyA, cry1, or hyh. F3 plants homozygous for YFP-PTS1 and the mutant alleles were selected after confirmation by fluorescence microscopy and RT-PCR analysis with gene-specific primers.
Databases Used in This Study
Databases used in this study include PLACE (http://www.dna.affrc.go.jp/PLACE) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
Epifluorescence and Confocal Laser-Scanning Microscopy
Epifluorescence microscopy was performed with an Axio Imager M1 microscope (Carl Zeiss) for visualization of the YFP-PTS1 protein (excitation 500 ± 12 nm; emission 542 ± 13.5 nm). A confocal laser-scanning microscope (Zeiss LSM 510 META) was also used to obtain images of YFP-PTS1 proteins. A 488-nm argon ion laser was used for excitation of YFP and chlorophyll. Emission filters of 505- to 530-nm bandpass and 650-nm longpass were used for YFP and chlorophyll, respectively. Images were acquired at 63×.
Peroxisome Quantification
Peroxisome quantification data were obtained from microscopic images analyzed by ImageJ software using the procedure described by Shi et al. (2007) with slight modifications. Specifically, YFP-tagged peroxisomes were imaged with a confocal (Zeiss LSM 510 META) or an epifluorescence (Zeiss Axio Imager M1) microscope. The confocal images shown in Figures 1 to 3 were each taken from four optical sections, each 0.6 μm in depth. The epifluorescence images shown in Figure 4 were obtained from six optical sections; each section was 1 μm in depth. Using ImageJ software, the following procedures were performed. First, RGB color images were converted to 8-bit black-and-white images. Second, the threshold function was used to designate the black pixels (peroxisomes) to be counted. Third, the “analyze particles” function was used to count the number of pixel groups in each image. Peroxisome quantification was performed on four to six individual images taken from different cotyledon/leaf areas of a plant. For quantification, a peroxisome having a clearly defined boundary, which means that fission has occurred, was counted as an individual organelle no matter how closely it was attached to another organelle. In contrast, a segmented peroxisome (constricted but no fission has taken place) was considered as a single organelle.
RNA Gel-Blot and RT-PCR Analyses
Total RNA was isolated from 3-d dark-grown seedlings using the Qiagen RNeasy plant mini prep kit. Total RNA (15 μg) was loaded onto the gel and blotted to nylon membranes. The PEX11b probe was derived from a 0.7-kb fragment of the cDNA clone (Orth et al., 2007) and was 32P-dCTP labeled by the random priming kit (Invitrogen). Hybridizations and washings were conducted according to standard protocols (Sambrook and Russell, 2001).
For RT-PCR analysis, total RNA was reverse transcribed with the Omniscript RT kit (Qiagen). PEX11b-specific primers At3g47430F (5′-CAGTGATCCGTTTCTTGGCG-3′) and At3g47430R (5′-GGCCAGTTCCTATACCAACC-3′) were used to amplify a 0.43-kb product from PEX11b. UBQ10-1 (5′-TCAATTCTCTCTACCGTGATCAAGATGCA-3′) and UBQ10-2 (5′-GGTGTCAGAACTCTCCACCTCAAGAGTA-3′) from the UBQ10 gene (At4g05320) were used to amplify a cDNA product (approximately 320 bp) as a loading control.
For PEX11b and UBQ10 amplification, PCR was performed with the following conditions: 94°C for 3 min, 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 4 min.
Constructs for Protein Expression and Purification
P35S:PEX11b, a PEX11b overexpression construct in the pCAMBIA vector (Orth et al., 2007), was used for the transient expression experiments.
Specific oligonucleotides were synthesized for amplification of the following genes: HYH N terminus, 5′-CACGCCATGGGCATGTCTCTCCAACGACC-3′ and HYH C terminus, 5″-ATAAGAATGCGGCCGCGTGATTGTCATCAGTTTTAGG-3′ for HYH (At3g17609); HY5 N terminus, 5′-CATGCCATGGGCATGCAGGAACAAGCGAC-3′ and HY5 C terminus, 5′-CCGCTCGAGAAGGCTTGCATCAGCATTAG-3′ for HY5 (At5g11260). Using these primer pairs, complete coding regions of HYH and HY5 were PCR amplified with the pfu turbo enzyme (Stratagene) using Arabidopsis total cDNA from light-grown seedlings as template. The products were separately cloned into NcoI and NotI sites of the bacterial pET-28a expression vector (Novagen). Recombinant full-length HYH and HY5 proteins fused to 6×-His in the pET28a+ vector (Novagen) were expressed in bacteria and purified with nickel nitilotriacetic acid agarose (Qiagen) according to the manufacturer's protocol.
Transient Expression Assay
The Agrobacterium-mediated transient expression assay was performed as described in Wieland et al. (2006). Overnight bacterial cultures of the Agrobacterium tumefaciens strain C58C1 containing the plasmid of interest were harvested by centrifugation. Cells were resuspended in induction buffer (10 mm MgCl2 and 150 μm acetosyringone) to an OD600 of 0.5, and incubated for 2 h at room temperature. Agrobacteria were then hand-infiltrated into 3-week-old Arabidopsis leaves with a 1-mL needleless syringe. Infiltrated plants were grown for 2 to 3 d in growth chambers before microscopic study of the YFP fusion proteins was performed.
Electrophoretic Mobility Shift (Gel-Shift) Assay
The 200-bp promoter of PEX11b was PCR amplified from genomic DNA using the primer pair forward (5′-CCGCTCGAGGCACAAATTCTCGGATTTC-3′) and reverse (5′-GCTCTTGTTGTCTCATGTTTTGATATTCAAGCTTGGG-3′). The amplified product was digested with HindIII for 2 h, gel purified, and used for labeling with 32P α-dCTP.
For binding assay with the G box, oligonucleotides were synthesized for the G-box sense strand, 5′-CCGCTCGAGAATTATCTTCCACGTGGCATTATTCC-3′, and antisense strand, 5′- GGAATAATGCCACGTGGAAGATAATTCTCGAGCGG-3′, with an XhoI site at the 5′ end of each oligonucleotide. Equal molar quantities of the two strands were annealed by incubating sequentially at 95°C for 30 s, 72°C for 2 min, 37°C for 2 min, and, finally, 25°C for 2 min. Annealed oligonucleotides were then digested overnight with XhoI, extracted with phenol-chloroform, and ethanol precipitated. The precipitated pellet was then washed with 70% ethanol, dried, resuspended in Tris-HCl (pH 8.0), and used for labeling with 32P α-dCTP.
The DNA-binding assays were performed as described by Yadav et al. (2002) at room temperature in a final volume of 25 μL of binding buffer [15 mm HEPES, pH 7.5, 35 mm KCl, 1 mm EDTA, 6% glycerol (v/v), 1 mm dithiothreitol, 1 mm MgCl2, and 0.5 μg poly(dI-dC)]. The samples were incubated at room temperature for 15 min and later run on a 5% polyacrylamide gel at 12 to 15 mAmp. After electrophoresis, the gel was dried and autoradiographed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_190327 (PEX11b), NP_850605 (HYH), and NP_568246 (HY5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of PEX11b expression at different time periods after far-red light treatment in wild-type and phyA mutant plants.
Supplemental Figure S2. RT-PCR analysis of PEX11b and UBQ10 in 3-d dark-grown phyA signaling pathway mutants after exposure to far-red light.
Supplemental Figure S3. Analysis of the PEX11b promoter region.
Supplementary Material
Acknowledgments
We would like to express our thanks to Dr. Sigrun Reumann, Dr. Meng Chen, Navneet Kaur, and Dr. Vandana Yadav for comments on the manuscript; Dr. Chunjie Tian for help with statistical analysis; the Arabidopsis Biological Resource Center for phyA, fhy1, fhy3, far1, pat1, hfr1, cca1, and cry1 seeds; Dr. Magnus Holm for hyh, hy5, and cry2 seeds; and Karen Bird and Marlene Cameron for editorial and graphic assistance.
This work was supported by the U.S. Department of Energy (grant to J.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jianping Hu (huji@msu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe I, Okumoto K, Tamura S, Fujiki Y (1998) Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett 431 468–472 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Más P, Harmer SL, Key SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12 757–761 [DOI] [PubMed] [Google Scholar]
- Beevers H (1979) Microbodies in higher plants. Annu Rev Plant Physiol 30 159–193 [Google Scholar]
- Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Charlton W, Lopez-Huertas E (2002) PEX genes in plants and other organisms. In A Baker, IA Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 385–426
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Citron M (1995) Focus. Environ Health Perspect 103 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley TR, Park SC, Kwon HB, Peng HP, Shih MC (1994) Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Mol Cell Biol 14 2525–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe M, Lucas MM, Pozuelo JM (1988) Cytochemical study of catalase and peroxidase in the mesophyll of Lolium rigidum plants treated with isoproturon. J Plant Physiol 132 67–73 [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvergne B, Wahli W (1999) Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 20 649–688 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34 827–836 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10 51–54 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol 128 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA (2007) Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol 23 321–344 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M, Bird B, Davies DD (1989) The effect of light on the structure and organization of Leman peroxisomes. J Exp Bot 40 1029–1035 [Google Scholar]
- Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (2006) Origin and evolution of the peroxisomal proteome. Biol Direct 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H (2006) The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochim Biophys Acta 1763 1392–1402 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2003) Entering a new era of research on plant peroxisomes. Curr Opin Plant Biol 6 577–582 [DOI] [PubMed] [Google Scholar]
- Heiland I, Erdmann R (2005) Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J 272 2362–2372 [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (2005) Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122 85–95 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YN, Schopfer P (1980) Density of microbodies on sucrose gradients during phytochrome-mediated glyoxysome peroxisome transformation in cotyledons of mustard seedlings. Plant Physiol 66 194–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YN, Schopfer P (1981) Control by phytochrome of urate oxidase and allantoinase activities during peroxisome development in the cotyledons of mustard (Sinapis alba L.) seedlings. Planta 152 325–335 [DOI] [PubMed] [Google Scholar]
- Hu J (2007) Plant peroxisome multiplication: highly regulated and still enigmatic. J Int. Plant Biol 49 1112–1118 [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297 405–409 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8 217–230 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Tanaka A, Fujiki Y (2007) Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res 313 1675–1686 [DOI] [PubMed] [Google Scholar]
- Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M (2003) Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem 278 8597–8605 [DOI] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M (2005) A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 16 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Baumgart E, Dong GX, Morrell JC, Jimenez-Sanchez G, Valle D, Smith KD, Gould SJ (2002) PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol Cell Biol 22 8226–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Trelease RN (2006) Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci 119 1961–1972 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M (2004) An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J 38 487–498 [DOI] [PubMed] [Google Scholar]
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM (1995) Pmp27 promotes peroxisomal proliferation. J Cell Biol 129 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT (2005) Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17 3513–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu XH, Parikh D, Cheng Y, Dolan S, Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA 100 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y, Baker A (2006) Plant peroxisomes as a source of signaling molecules. Biochim Biophys Acta 1763 1478–1495 [DOI] [PubMed] [Google Scholar]
- Oksanen E, Haikio E, Sober J, Karnosky DF (2003) Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol 161 791–799 [DOI] [PubMed] [Google Scholar]
- Olsen L, Harada J (1995) Peroxisomes and their assembly in higher plants. Annu Rev Plant Biol 46 123–146 [Google Scholar]
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma JM, Garrido M, Rodriguez-Garcia MI, del Rio LA (1991) Peroxisome proliferation and oxidative stress mediated by activated oxygen species in plant peroxisomes. Arch Biochem Biophys 287 68–74 [DOI] [PubMed] [Google Scholar]
- Park SC, Kwon HB, Shih MC (1996) Cis-acting elements essential for light regulation of the nuclear gene encoding the A subunit of chloroplast glyceraldehyde 3-phosphate dehydrogenase in Arabidopsis thaliana. Plant Physiol 112 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Spalding EP (1999) Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc Natl Acad Sci USA 96 14142–14146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poucke VM, Cerff R, Barthe F, Mohr H (1970) Simultaneous induction of glycolate oxidase and glyoxylate reductase in white mustard seedlings by phytochrome. Naturwissenschaften 57 132–133 [DOI] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (2001) Peroxisome biogenesis. Annu Rev Cell Dev Biol 17 701–752 [DOI] [PubMed] [Google Scholar]
- Reumann S, Weber AP (2006) Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 1763 1496–1510 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A (2006) The origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol 23 838–845 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Bajracharya D, Bergfeld R, Falk H (1976) Phytochrome-mediated transformation of glyoxysomes into peroxisomes in the cotyledons of mustard (Sinapis alba L.) seedlings. Planta 133 73–80 [DOI] [PubMed] [Google Scholar]
- Schrader M, Yoon Y (2007) Mitochondria and peroxisomes: Are the ‘Big Brother’ and the ‘Little Sister’ closer than assumed? Bioessays 29 1105–1114 [DOI] [PubMed] [Google Scholar]
- Shi Q, Hjelmeland AB, Keir ST, Song L, Wickman S, Jackson D, Ohmori O, Bigner DD, Friedman HS, Rich JN (2007) A novel low-molecular weight inhibitor of focal adhesion kinase, TAE226, inhibits glioma growth. Mol Carcinog 46 488–496 [DOI] [PubMed] [Google Scholar]
- Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, Kiselyov K (2006) TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J Biol Chem 281 7294–7301 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Photomorphogenesis. Seeing the light in plant development. Curr Biol 5 466–468 [DOI] [PubMed] [Google Scholar]
- Thoms S, Erdmann R (2005) Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J 272 5169–5181 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Mullen RT (2006) Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J Cell Biol 174 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin EM, Kehoe DM (1994) Phytochrome regulated gene expression. Semin Cell Biol 5 335–346 [DOI] [PubMed] [Google Scholar]
- Trelease RN, Becker WM, Gruber PJ, Newcomb EH (1971) Microbodies (glyoxysomes and peroxisomes) in cucumber cotyledons: correlative biochemical and ultrastructural study in light- and dark-grown seedlings. Plant Physiol 48 461–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Pichersky E, Malik VS, Cashmore AR (1989) Level of expression of the tomato rbcS-3A gene is modulated by a far upstream promoter element in a developmentally regulated manner. Plant Cell 1 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49 428–441 [DOI] [PubMed] [Google Scholar]
- Villain P, Mache R, Zhou DX (1996) The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem 271 32593–32598 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002. a) Phytochrome signaling mechanism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0074.1, www.aspb.org/publications/arabidopsis/
- Wang H, Deng XW (2002. b) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2003) Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci 8 172–178 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wieland WH, Lammers A, Schots A, Orzaez DV (2006) Plant expression of chicken secretory antibodies derived from combinatorial libraries. J Biotechnol 122 382–391 [DOI] [PubMed] [Google Scholar]
- Yadav V, Kundu S, Chattopadhyay D, Negi P, Wei N, Deng XW, Chattopadhyay S (2002) Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J 31 741–753 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.