Abstract
In Arabidopsis thaliana, transamination steps in the leucine biosynthetic and catabolic pathways and the methionine (Met) chain elongation cycle of aliphatic glucosinolate formation are catalyzed by branched-chain aminotransferases (BCATs) that are encoded by a small gene family of six members. One member of this family, the plastid-located BCAT3, was shown to participate in both amino acid and glucosinolate metabolism. In vitro activity tests with the recombinant protein identified highest activities with the 2-oxo acids of leucine, isoleucine, and valine, but also revealed substantial conversion of intermediates of the Met chain elongation pathway. Metabolite profiling of bcat3-1 single and bcat3-1/bcat4-2 double knockout mutants showed significant alterations in the profiles of both amino acids and glucosinolates. The changes in glucosinolate proportions suggest that BCAT3 most likely catalyzes the terminal steps in the chain elongation process leading to short-chain glucosinolates: the conversion of 5-methylthiopentyl-2-oxo and 6-methylthiohexyl-2-oxo acids to their respective Met derivatives, homomethionine and dihomo-methionine, respectively. The enzyme can also at least partially compensate for the loss of BCAT4, which catalyzes the initial step of Met chain elongation by converting Met to 4-methylthio-2-oxobutanoate. Our results show the interdependence of amino acid and glucosinolate metabolism and demonstrate that a single enzyme plays a role in both processes.
Plants de novo synthesize many compounds that cannot be made by humans or animals and that have to be taken up as essential compounds in the diet. For instance, several amino acids, products of primary metabolism, make an important contribution to a healthy nutrition. Many other plant compounds, mostly secondary metabolites, are not essential but have beneficial impacts on human health. For instance, glucosinolates, a group of about 120 different metabolites seem to play a role in the prevention of certain types of cancer (Smith et al., 2003; Keck and Finley, 2004). Glucosinolates are derived from various amino acids; thus their biosynthesis is initiated from products of primary metabolism (Wittstock and Halkier, 2002; Grubb and Abel, 2006; Halkier and Gershenzon, 2006). Although enormous progress has been made in understanding plant biochemical pathways in recent years, the interdependence of primary and secondary metabolism is not fully understood.
Val, Leu, and Ile, the small group of branched-chain amino acids (BCAAs), are essential for humans (Singh, 1999; Binder et al., 2007). In plants the first three steps in the two parallel pathways toward Val and Leu or Ile are each catalyzed by single enzymes, resulting in the generation of 3-methyl-2-oxopentanoate (3MOP) that is transaminated to Ile, or of 3-methyl-2-oxobutanoate (3MOB) that is either transaminated to Val or that serves as substrate for the biosynthesis of Leu. The latter requires four additional reactions including a final transamination reaction converting 4-methyl-2-oxopentanoate (4MOP) to Leu. All of the final transamination steps are catalyzed by the pyridoxal-5′-phosphate-dependent branched-chain aminotransferases (BCATs; Mehta and Christen, 2000; Binder et al., 2007). In Arabidopsis (Arabidopsis thaliana), these enzymes are encoded by a small gene family of six transcribed members (Diebold et al., 2002). BCAT1 is imported into mitochondria and is most likely involved in the degradation of Leu (Schuster and Binder, 2005). BCAT2, BCAT3, and BCAT5 are targeted to plastids (Diebold et al., 2002) and are accordingly assigned to the biosynthesis of BCAAs (Singh, 1999; Binder et al., 2007). Surprisingly, the cytosolic BCAT4 has a completely different function acting at the interface between primary and secondary metabolism (Schuster et al., 2006). This promiscuous enzyme exhibits highest activities with Met and its 2-oxo acid, 4-methylthio-2-oxobutanoate (MTOB), whereas of the standard substrates only Leu and 4MOP are converted at substantial rates. The protein catalyzes the committed step of the Met chain elongation pathway, the first part of the biosynthesis of Met-derived aliphatic glucosinolates. Thus the members of the BCAT protein family can function in different pathways.
In our analysis of BCATs in Arabidopsis, we have now analyzed the plastid-located BCAT3. In vitro activity tests and metabolite profiling of bcat3-1 single and bcat3-1/bcat4-2 double knockout mutants reveals that this protein is involved in the biosynthesis of BCAAs, but at the same time it is also active in Met chain elongation. In the latter it seems to have a function in the transamination of short-chain keto acids to the respective Met derivatives. However, BCAT3 can also partially compensate for the function of BCAT4 by initiating the Met chain elongation pathway in plastids.
RESULTS
Expression Pattern Analysis Suggests That BCAT3 Is Like BCAT4 Involved in Glucosinolate Biosynthesis
Recently we found that BCAT4 (At3g19710) catalyzes the transamination of Met to MTOB, the committed step in the Met chain elongation pathway, the first part of the biosynthesis of Met-derived glucosinolates (Schuster et al., 2006). This pathway includes several transamination steps and it is clear that at least one other aminotransferase is required for the generation of Met derivatives with different chain lengths (Halkier and Gershenzon, 2006).
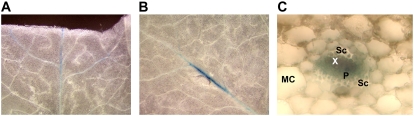
To identify another aminotransferase active in Met chain elongation, we reexamined the promoter activity of BCAT3 (At3g49680). A very weak activity of this promoter had previously been observed in the basal part of the leaf petiole directly at the site where the leaf was detached (Schuster and Binder, 2005). This observation suggested an induction of the promoter by wounding similar to the pattern found for BCAT4. To follow this issue, leaves of transgenic plants containing a BCAT3 promoter:GUS construct were wounded by removing the tip of a rosette leaf with scissors and by piercing the leaf blade with a needle. These treatments activated the BCAT3 promoter in leaf veins close to the site of physical treatment (Fig. 1, A and B) suggesting promoter activity in the vascular bundles, a pattern typical for genes and proteins involved in the formation of Met-derived glucosinolates (Chen et al., 2003; Grubb et al., 2004; Levy et al., 2005; Schuster and Binder, 2005; Schuster et al., 2006). A cross section through the leaf blade confirmed the BCAT3 promoter to be active in the phloem cells as had been observed for BCAT4. Furthermore BCAT3 is diurnally expressed in phase with BCAT4 and MAM1 as shown in previously published transcript profiling analyses (Bläsing et al., 2005; Supplemental Fig. S1). Thus the spatiotemporal expression pattern of BCAT3 identifies this protein as a strong candidate for an aminotransferase active in the Met chain elongation cycle.
Figure 1.
BCAT3 promoter activity is found in vascular bundles. Histochemical GUS staining of leaves from transgenic Arabidopsis plants containing a BCAT3 promoter:GUS construct. The leaves were wounded with scissors (A) or a pin (B), both inducing promoter activity in vascular bundles in areas close to the physical treatment. A cross section through a leaf vein reveals BCAT3 promoter activity in the phloem (C). MC, Mesophyll cell; Sc, sclerenchyma; X, xylem; and P, phloem.
BCAT3 Exhibits a Strong Substrate Preference for the Keto Acids of Leu, Ile, and Val
The biochemical abilities of BCAT3 were analyzed in vitro with the recombinant protein without the putative chloroplast targeting signal (amino acids 1–60; for details see “Materials and Methods”). Protein activities were assayed in a coupled enzyme test that had been applied in previous studies of BCATs from plants (Schadewaldt and Adelmeyer, 1996; Schuster and Binder, 2005; Schuster et al., 2006). To establish optimal ranges for substrate specificity tests, kinetics of this enzyme were followed with the keto acids of Val (3MOB), Leu (4MOP), Ile (3MOP), and Met (MTOB; Supplemental Figs. S2–S5). The analysis of substrate concentration and reaction velocities revealed that BCAT3 exhibits Michaelis-Menten kinetics with all tested compounds. Highest affinities were seen toward 4MOP (Km, 0.14 ± 0.04 mm) and 3MOP (Km, 0.14 ± 0.3 mm). Vmax was twice as high for 4MOP (27.42 ± 2.09 μmol (min mg)−1 as for 3MOP (13.33 ± 0.68 μmol [min mg]−1; Table I). A substantially lower affinity and maximal velocity was measured for 3MOB, the keto acid of Val (Km, 1.38 ± 0.10 mm; Vmax, 14.79 ± 0.35 μmol [min mg]−1). These values are in the range of those found for MTOB (Km, 1.92 ± 0.27 mm, Vmax, 21.01 ± 0.93 μmol [min mg]−1). Based on these results, substrate specificity tests were performed at two substrate concentrations, 0.1 and 2.0 mm.
Table I.
Km and Vmax values of recombinant BCAT3 with different substrates
KIC, Ketoisocaproate; KMV, ketomethylvalerate; KIV, ketoisovalerate.
| Substrate | Km | Vmax |
|---|---|---|
| mm | μmol (min mg)−1 | |
| 4MOP (KIC) | 0.14 ± 0.04 | 27.42 ± 2.09 |
| 3MOP (KMV) | 0.14 ± 0.03 | 13.33 ± 0.68 |
| 3MOB (KIV) | 1.38 ± 0.10 | 14.79 ± 0.35 |
| MTOB | 1.92 ± 0.27 | 21.01 ± 0.93 |
We used MTOB as standard substrate and the activity measured with this substrate was arbitrarily set to 100%. As expected from the kinetic data, highest activities were found with 4MOP (1,565%/353% at 0.1/2 mm substrate concentrations) and 3MOP (865%/328%), whereas the activity with 3MOB (182%/136%) was about 10 times lower compared with 4MOP. In contrast to these standard substrates for a BCAT, lower activities were found with keto acids from the Met chain elongation cycle, 5-methylthio-2-oxopentanoate (MTOP; 109%/112%), followed by MTOB (100%/100%) and 6-methylthio-2-oxohexanoate (MTOH; 62%/86%; Table II).
Table II.
Relative substrate specificities of recombinant BCAT3
| Substrate | Relative Activity
|
|
|---|---|---|
| 0.1 mm | 2 mm | |
| % | ||
| 4MOP (KIC) | 1,565 (± 4.4/3.3) | 353 (± 4.2/7.9) |
| 3MOP (KMV) | 875 (± 10.7/5.7) | 328 (± 8.6/4.8) |
| 3MOB (KIV) | 182 (± 1.4/1.4) | 136 (± 11.2/7.1) |
| MTOP | 109 (± 5.2/3.4) | 112 (± 4.4/6.2) |
| MTOB | 100 (± 4.5/5.6) | 100 (± 4.3/7.3) |
| MTOH | 62 (± 6.4/10) | 86 (± 4.2/2.4) |
No activities were detectable with a control lysate purified from Escherichia coli expressing an empty pET32a vector, demonstrating that the measured activities originate from the overexpressed BCAT3 and not from any copurified E. coli protein.
In summary, BCAT3 shows a strong preference for the standard substrates of a BCAT, but the enzyme also exhibits substantial activities with the keto acids of Met and Met derivatives. These activities suggest a function of BCAT3 in the biosynthesis of BCAAs, but do not exclude the involvement of this enzyme in the Met chain elongation cycle, as suggested by the gene expression data.
Distinct Changes in the Levels of Free Amino Acids and Glucosinolates Are Observed in a bcat3-1 T-DNA Insertion Mutant
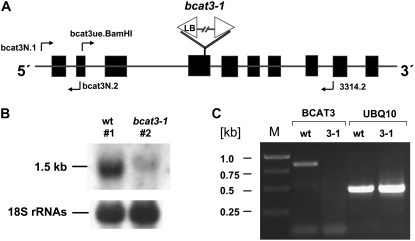
To gather more information about the particular role of BCAT3 in amino acid and/or glucosinolate biosynthesis in vivo, we analyzed amino acid and glucosinolate metabolites of a BCAT3 T-DNA insertion mutant obtained from the GABI-Kat collection (002A11; Rosso et al., 2003). The insertion of the T-DNA at the predicted location in exon 4 was confirmed by PCR and sequencing of a PCR product covering the T-DNA and the respective flanking sequence at the left border (Fig. 2A). Plants homozygous for the bcat3-1 allele and for the bcat3 wild-type allele (from the GABI-Kat collection) were further examined by northern-blot and reverse transcription (RT)-PCR analyses. In wild-type RNA a probe covering the 5′ terminal region of the BCAT3 cDNA detects an approximately 1.5-kb mRNA, whereas in the T-DNA insertion mutant only a very weak and slightly larger signal was detected (Fig. 2B). In addition, an RT-PCR analysis with primers bcat3ue.BamHI and 3314.2 flanking the T-DNA insertion was performed. As expected a 0.85-kb cDNA fragment was amplified from wild-type RNA, whereas no such product was obtained from RNA of the homozygous mutants demonstrating the T-DNA to cause a knockout (Fig. 2C). This was further confirmed by another RT-PCR covering the BCAT3 mRNA upstream of the insertion (data not shown).
Figure 2.
Characterization of a bcat3-1 T-DNA insertion mutant. A, The structure of the BCAT3 gene is depicted with exons indicated as black boxes. The T-DNA insertion is located in exon 4. Primers used for the generation of the probe used in the northern hybridization (bcat3N.1/bcat3N.2) or for cDNA analysis (bcat3ue.BamHI/3314.2) are indicated as bent arrows. B, Northern analysis of total RNA from a wild-type plant (no. 1) and a plant homozygous for bcat3-1 (no. 2), both isolated from the seed pool obtained from GABI-Kat. A 1.5-kb BCAT3 mRNA is detected in the wild-type RNA. Equal loading is inspected by a hybridization with oligonucleotide P18SrRNA (5′-AAGCATATGACTACTGGCAGG) detecting 18S rRNA (bottom). C, RT-PCR analysis of BCAT3 mRNA with primer pair bcat3ue.BamHI/3314.2 (BCAT3) flanking the T-DNA insertion. The expected fragment of 854 bp can only be amplified on cDNA from wild type (wt), but not from RNA isolated from the plants homozygous for bcat3-1 (3–1). A control PCR amplifying a 500-bp cDNA fragment from the ubiquitin 10 mRNA corroborated the integrity of the RNA and cDNA as well as the use of equal amounts of template (UBQ10). Lengths of DNA marker fragments (lane M) are given in kilobase pair.
From the progeny of both wild-type and mutant plants, levels of free amino acids were measured in rosette leaves of about 3-week-old plants and in seeds (data not shown). In the latter no significant differences were observed in the levels of free amino acids. This also holds true for most amino acids in rosette leaves with the exceptions of Val, Ser, and Thr, which are significantly reduced in the mutant by about 30%, 20%, and 10%, respectively (Table III).
Table III.
Significant differences in the levels of free amino acids found in rosette leaves from the wild type and the bcat3-1 knockout plants
Concentrations of amino acids were measured in at least three independent experiments and are given with the corresponding sd (±). According to t tests, the contents of these amino acids are significantly different (P < 0.01).
| Amino Acid | Amino Acid Content
|
||
|---|---|---|---|
| bcat3-1 | Col-0 Wild Type | t Test | |
| nmol/mg dry wt | |||
| Total amino acid | 147.92 ± 7.73 | 158.82 ± 8.13 | 0.02445 |
| Ser | 7.37 ± 0.52 | 9.21 ± 0.32 | 0.00000 |
| Thr | 5.70 ± 0.42 | 6.32 ± 0.31 | 0.00832 |
| Val | 0.67 ± 0.03 | 0.98 ± 0.07 | 0.00000 |
Because the recombinant enzyme also reacts with keto acids of Met and its derivatives, a glucosinolate profiling was done in leaves and seeds of wild-type and bcat3-1 knockout plants. In leaves of bcat3-1 plants the levels of total Met-derived glucosinolates were increased 1.2-fold (Table IV). The most substantial change was observed for 5-methylsulfinylpentylglucosinolate (5MSOP; 5.8-fold increase in the mutant), which accounts for the major part of the increase of Met-derived glucosinolates. Significant increases were also measured for 6-methylsulfinylhexylglucosinolate (6MSOH; 3-fold) and 7-methylsulfinylheptylglucosinolate (7MSOH; 2.4-fold). In addition, the Leu- or Val-derived aliphatic glucosinolates 4-methylpentylglucosinolate (4MP) and 5-methylhexylglucosinolate (5MH), usually barely detectable in ecotype Columbia of Arabidopsis (Col-0), were found in considerable quantities in rosette leaves in the mutant. Minor variations were seen in indole glucosinolates (Table IV).
Table IV.
Glucosinolate profile in rosette leaves of homozygous bcat3-1 plants
The wild type used in this analysis was selected from the seed probe provided by GABI-Kat for line 002A11. GS, Glucosinolates; Total Met GS, total Met-derived glucosinolates; 3MSOP, 3-methylsulfinylpropylglucosinolate; 8MSOO, 8-methylsulfinyloctylglucosinolate; 1MOI3M, 1-methoxyindol-3-ylmethylglucosinolate; 4MOI3M, 4-methoxyindol-3-ylmethylglucosinolate. For other abbreviations, see text.
| GS Type | Glucosinolate Content
|
||
|---|---|---|---|
| bcat3-1 | Col-0 Wild Type | t Test | |
| μmol/g dry wt | |||
| Total Met GS | 41.25 ± 5.57 | 33.93 ± 3.66 | 0.01321 |
| Total GS | 46.51 ± 6.13 | 37.38 ± 3.98 | 0.00626 |
| 3MSOP | 2.26 ± 0.33 | 2.87 ± 0.34 | 0.00542 |
| 4MTB | 0.05 ± 0.03 | 0.09 ± 0.02 | 0.01481 |
| 4MSOB | 26.51 ± 3.79 | 25.94 ± 3.02 | 0.76094 |
| 5MSOP | 6.97 ± 0.98 | 1.20 ± 0.12 | 0.00000 |
| 6MSOH | 0.46 ± 0.05 | 0.15 ± 0.02 | 0.00000 |
| 7MSOH | 1.34 ± 0.14 | 0.56 ± 0.08 | 0.00000 |
| 8MSOO | 3.67 ± 0.36 | 3.11 ± 0.47 | 0.02942 |
| 4MP | 1.32 ± 0.16 | 0.04 ± 0.01 | 0.00000 |
| 5MH | 0.54 ± 0.06 | 0.02 ± 0.00 | 0.00000 |
| I3M | 2.58 ± 0.34 | 2.23 ± 0.31 | 0.06553 |
| 1MOI3M | 0.54 ± 0.07 | 0.89 ± 0.25 | 0.00374 |
| 4MOI3M | 0.29 ± 0.03 | 0.26 ± 0.02 | 0.09043 |
In contrast to rosette leaves, no significant difference was observed in the total contents of Met-derived glucosinolates between wild-type and mutant seeds. However, besides minor statistically significant changes, substantial elevations were seen in the content of the pentylglucosinolates (5MTP; 2.8-fold increase) and 5MSOP (2.9-fold increase). Among the other glucosinolate types, indol-3-ylmethylglucosinolate (I3M) was reduced by 50% (Table V).
Table V.
Glucosinolate profile in seeds of the wild-type and the bcat3-1 knockout plants
3OHP, 3-Hydroxypropylglucosinolate; 8MTO 8-methylthiooctylglucosinolate. For other abbreviations, see text.
| GS Type | Glucosinolate Content
|
||
|---|---|---|---|
| bcat3-1 | Col-0 Wild Type | t Test | |
| μmol/g seeds | |||
| Total Met GS | 73.45 ± 6.79 | 75.66 ± 7.97 | 0.54308 |
| Total GS | 78.59 ± 6.89 | 85.84 ± 8.57 | 0.07038 |
| 3OHP | 0.53 ± 0.08 | 0.58 ± 0.13 | 0.36318 |
| 3BZO | 4.63 ± 0.43 | 5.37 ± 0.60 | 0.00984 |
| 4OHB | 2.85 ± 0.22 | 3.81 ± 0.59 | 0.00054 |
| 4MTB | 23.66 ± 3.43 | 27.87 ± 4.50 | 0.04432 |
| 4BZO | 14.31 ± 0.84 | 16.15 ± 0.52 | 0.00003 |
| 4MSOB | 0.71 ± 0.17 | 0.90 ± 0.32 | 0.14931 |
| 5MTP | 8.21 ± 0.76 | 2.95 ± 0.28 | 0.00000 |
| 5MSOP | 0.43 ± 0.07 | 0.15 ± 0.04 | 0.00000 |
| 7MTH | 5.43 ± 0.95 | 4.13 ± 0.78 | 0.00571 |
| 7MSOH | 1.23 ± 0.28 | 0.90 ± 0.25 | 0.01555 |
| 8MTO | 5.14 ± 0.72 | 5.88 ± 0.95 | 0.08754 |
| 8MSOO | 6.31 ± 0.82 | 6.96 ± 1.13 | 0.19237 |
| I3M | 5.14 ± 2.65 | 10.19 ± 1.18 | 0.00006 |
In summary, the metabolite profiling studies show that BCAT3 is involved in both the biosynthesis of Met-derived glucosinolates and the biosynthesis of BCAAs. Although the total amounts of amino acids and glucosinolates are more or less identical in both the wild-type and the bcat3-1 mutant, significant changes were found for individual amino acids and glucosinolate species. Because only a single knockout allele was investigated here, we also analyzed plants homozygous for bcat3-1 and the bcat4 wild-type allele, which were obtained in the F2 generation after crossings of bcat3-1 and bcat4-2 (see below). These plants correspond de facto to an F2 generation of an outcrossing of bcat3-1. These showed the same chemotype as described above, confirming the observed alterations in the metabolite profile to be the results of the BCAT3 knockout (data not shown). This is further supported by the chemotype of the bcat3-1/bcat4-2 double knockout plants (see below) and the characteristics of the recombinant enzyme.
Long-Term Feeding of BCAAs Triggers the Biosynthesis of 4MP and 5MH
So far our studies suggest that BCAT3 is active in biosyntheses of both BCAAs and Met-derived glucosinolates. The interdependence of the BCAA biosynthesis and the synthesis of aliphatic glucosinolates is also indicated by the occurrence of the unusual 4MP and 5MH in the bcat3-1 mutant. We speculated that the enhanced levels of these glucosinolates might be triggered by imbalances in BCAA metabolism, assuming that the formation of 4MP and 5MH contributes to homeostasis of this class of amino acids. Thus mutant plants were fed with BCAAs over different time periods to determine their effect on amino acid and glucosinolate composition.
Amino acid profiling 4.5 h after administration of 2 mm Val, Leu, or Ile clearly showed the uptake of the respective amino acid. In rosette leaves of both genotypes (wild type and bcat3-1) the given amino acid was elevated, although major differences were seen in the resulting variations (Supplemental Tables S1 and S2). Lowest increases (×2.6 in wild type/×3.6 in bcat3-1) are found after feeding of Val followed by Leu (×7.5/×8.2). The latter also leads to a slight decrease of free Val. Highest values were found after the addition of Ile (×14.7/×21.8) accompanied by a weak, significant increase in Thr (×1.3/×1.5). Almost no variation was seen in the glucosinolate profile with the exception of 1-methoxyindol-3-ylmethylglucosinolate, which is very moderately, but significantly increased upon feeding of Ile (data not shown).
A different pattern of changes is seen in the amino acid profile of leaves after repeated administration of BCAAs to 2-week-old wild-type and bcat3-1 plants, respectively (for details see “Materials and Methods”). In both genotypes feeding of Val and Leu provoked an increase of total amino acids, an effect not seen when Ile was added. In wild-type plants these increases are predominantly attributed to significantly elevated levels of Gln, but higher amounts are also observed for Ala, Asp, Asn, and Glu (Supplemental Table S3). A very similar profile is seen in the bcat3-1 mutant. However, compared with control plants no significant changes were seen in the Asp and Glu contents (Supplemental Table S4). Surprisingly in both genotypes no or only very moderate increases of the administered amino acids are seen.
When glucosinolates were analyzed in these plants, no significant changes were found for most of these secondary compounds (Supplemental Tables S5 and S6). However, in wild-type plants addition of Leu triggered the accumulation of 4MP and 5MH and all BCAAs moderately increased the levels of 7MSOH and 8MSOO. Similar alterations were found in mutant plants. An increase of 4MP and 5MH is seen upon feeding of Leu or Ile. Likewise the 8MSOO level is higher after addition of these amino acids.
We furthermore investigated the seeds of plants, which were grown for 3 weeks under normal conditions and which were then repeatedly fed with amino acids until seed set began (details are given in “Materials and Methods”). No substantial changes in amino acid profiles were observed in seeds of both wild-type and bcat3-1 plants with respect to control plants, which were treated identically without adding amino acids (data not shown). Likewise no substantial variations of standard glucosinolates were seen in these plants (data not shown). However, distinct changes were again observed in the levels of 4MP and 5MH (Table VI). Although no difference to control plants is detectable in 4MP levels after feeding of Val, the levels of this glucosinolate species increases slightly upon addition of Ile in wild-type (2.1-fold) and bcat3-1 plants (1.7-fold). Particularly strong increases are observed after supplementation of Leu. With respect to control plants, 4MP increased about 13.8- and 4.9-fold in wild type and the BCAT3 knockout, respectively. The increases of 4MP in wild type are much stronger, although they do not reach the absolute levels of bcat3-1 plants, which have higher amounts of these glucosinolate species per se (Table VI, left). A slightly different picture emerges for 5MH. Here administration of Ile has no effect, whereas the feeding of Val leads to increases of 5MH predominantly in bcat3-1 (5-fold). But again the strongest increases were seen after feeding of Leu, raising 5MH 25-fold in bcat3-1 and to 0.37 μmol/g seed from an undetectable amount in wild type. In contrast to 4MP, the increases of 5MH are stronger in bcat3-1 than in wild type (Table VI, right).
Table VI.
4MP and 5MH levels in seeds of wild-type and bcat3-1 plants grown in the presence of BCAAs
| Added Amino Acid | 4MP
|
5MH
|
||
|---|---|---|---|---|
| bcat3-1 | Wild Type | bcat3-1 | Wild Type | |
| μmol/g seeds | μmol/g seeds | |||
| Leu | 1.86 ± 0.26 | 1.10 ± 0.19 | 0.50 ± 0.06 | 0.37 ± 0.06 |
| Ile | 0.65 ± 0.12 | 0.17 ± 0.03 | 0.03 ± 0.03 | 0.02 ± 0.02 |
| Val | 0.43 ± 0.06 | 0.00 ± 0.00 | 0.10 ± 0.02 | 0.04 ± 0.02 |
| Control | 0.38 ± 0.04 | 0.08 ± 0.03 | 0.02 ± 0.03 | 0.00 ± 0.00 |
These experiments demonstrate that a long-term administration of Leu and to a lesser extent Ile and Val, results in the generation of the unusual glucosinolates 4MP and 5MH. This indicates that the generation of these glucosinolates may be a strategy to reduce long-term excess of Leu, and to a lesser extent Ile and Val.
Striking Changes in Amino Acid and Glucosinolate Profiles in the bcat3-1/bcat4-2 Double Knockout
Our data imply that besides the cytosolic BCAT4 the plastid-located BCAT3 is also involved in glucosinolate biosynthesis. This raises the question of whether these enzymes could at least partially complement each other. In particular, we speculated whether BCAT3 could be responsible for the remaining Met-derived glucosinolate biosynthesis in bcat4 knockout plants, where the total Met-derived glucosinolate content is reduced to 50% (Schuster et al., 2006). Thus plants homozygous for bcat3-1 and bcat4-2 alleles were crossed and individuals homozygous for both alleles were identified in the F3 progeny. Offspring of these plants were then subjected to glucosinolate and amino acid profiling (Tables VII–X).
Table VII.
Amino acid content in rosette leaves of a bcat3-1/bcat4-2 double knockout mutant
The wild-type plant used in this analysis was selected from the seed probe provided by GABI-Kat for line 002A11 (bcat3-1). Almost identical results were obtained with wild-type plants isolated from the seed pool obtained for line GABI-Kat 163D11 (bcat4-2; data not shown).
| Amino Acid | Amino Acid Content
|
||
|---|---|---|---|
| bcat3-1/bcat4-2 | Col-0 Wild Type | t Test | |
| nmol/mg dry wt | |||
| Total amino acid | 236.76 ± 20.88 | 270.04 ± 13.51 | 0.00832 |
| Ala | 9.16 ± 1.45 | 10.00 ± 0.81 | 0.24389 |
| Arg | 1.74 ± 0.46 | 0.82 ± 0.09 | 0.00072 |
| Asn | 10.08 ± 1.01 | 10.56 ± 0.86 | 0.39513 |
| Asp | 22.24 ± 1.41 | 18.88 ± 1.16 | 0.00115 |
| Gln | 113.12 ± 11.92 | 159.84 ± 10.23 | 0.00003 |
| Glu | 44.64 ± 3.14 | 43.68 ± 1.80 | 0.53032 |
| Gly | 5.48 ± 1.12 | 6.24 ± 1.08 | 0.25931 |
| His | 1.02 ± 0.26 | 0.40 ± 0.06 | 0.00020 |
| Ile | 0.36 ± 0.19 | 0.24 ± 0.00 | 0.14493 |
| Leu | 0.60 ± 0.11 | 0.36 ± 0.00 | 0.00027 |
| Lys | 0.82 ± 0.14 | 0.70 ± 0.09 | 0.10861 |
| Met | 3.04 ± 0.79 | 0.12 ± 0.00 | 0.00000 |
| Phe | 0.58 ± 0.09 | 0.38 ± 0.05 | 0.00076 |
| Ser | 13.76 ± 0.78 | 11.52 ± 0.61 | 0.00025 |
| SMM | 1.16 ± 0.33 | 0.00 ± 0.00 | 0.00001 |
| Thr | 7.48 ± 1.44 | 4.84 ± 0.51 | 0.00174 |
| Trp | 0.02 ± 0.05 | 0.00 ± 0.00 | 0.34089 |
| Tyr | 0.18 ± 0.10 | 0.14 ± 0.05 | 0.40103 |
| Val | 0.82 ± 0.09 | 1.04 ± 0.10 | 0.00235 |
Table VIII.
Amino acid content in seeds of the bcat3-1/bcat4-2 double knockout mutant
The wild-type plant used in this analysis was selected from the seed probe provided by GABI-Kat for line 002A11 (bcat3-1). Almost identical results were obtained with wild-type plants isolated from the seed pool obtained for line GABI-Kat 163D11 (bcat4-2; data not shown).
| Amino Acid | Amino Acid Content
|
||
|---|---|---|---|
| bcat3-1/bcat4-2 | Col-0 Wild Type | t Test | |
| μmol/g seeds | |||
| Total amino acid | 51.41 ± 6.09 | 13.13 ± 1.42 | 0.00000 |
| Ala | 1.44 ± 0.41 | 0.90 ± 0.51 | 0.03173 |
| Arg | 6.82 ± 1.48 | 0.68 ± 0.09 | 0.00000 |
| Asn | 5.89 ± 0.87 | 2.81 ± 0.32 | 0.00000 |
| Asp | 2.01 ± 0.26 | 1.34 ± 0.22 | 0.00006 |
| Gln | 2.38 ± 1.85 | 0.43 ± 0.21 | 0.01438 |
| Glu | 3.90 ± 0.68 | 4.46 ± 0.72 | 0.11948 |
| Gly | 1.40 ± 0.40 | 0.37 ± 0.11 | 0.00001 |
| His | 1.84 ± 0.38 | 0.10 ± 0.02 | 0.00000 |
| Ile | 3.64 ± 0.98 | 0.16 ± 0.03 | 0.00000 |
| Leu | 0.69 ± 0.16 | 0.09 ± 0.03 | 0.00000 |
| Met | 0.22 ± 0.06 | 0.05 ± 0.02 | 0.00001 |
| Phe | 0.31 ± 0.04 | 0.31 ± 0.05 | 0.99727 |
| Ser | 2.64 ± 0.49 | 0.52 ± 0.06 | 0.00000 |
| SMM | 15.57 ± 2.08 | 0.00 ± 0.00 | 0.00000 |
| Thr | 0.76 ± 0.44 | 0.25 ± 0.05 | 0.00871 |
| Trp | 0.44 ± 0.13 | 0.27 ± 0.05 | 0.00644 |
| Tyr | 0.18 ± 0.03 | 0.08 ± 0.03 | 0.00000 |
| Val | 1.29 ± 0.24 | 0.29 ± 0.05 | 0.00000 |
Table IX.
Glucosinolate content in rosette leaves of the bcat3-1/bcat4-2 double knockout mutant
The wild-type plant is identical to the one described in Table VII. 4OHI3M, 4-Hydroxy-indol-3ylmethylglucosinolate.
| GS Type | Glucosinolate Content
|
||
|---|---|---|---|
| bcat3-1/bcat4-2 | Col-0 Wild Type | t Test | |
| μmol/g dry wt | |||
| Total Met GS | 17.32 ± 3.61 | 39.38 ± 2.50 | 0.00000 |
| Total GS | 40.42 ± 9.52 | 46.61 ± 2.67 | 0.15375 |
| 3MSOP | 1.17 ± 0.27 | 3.48 ± 0.25 | 0.00000 |
| 4MTB | 0.59 ± 0.12 | 1.38 ± 0.21 | 0.00000 |
| 4MSOB | 11.69 ± 2.45 | 29.88 ± 1.81 | 0.00000 |
| 5MSOP | 3.01 ± 0.64 | 0.97 ± 0.06 | 0.00001 |
| 7MSOH | 0.09 ± 0.02 | 0.48 ± 0.04 | 0.00000 |
| 8MSOO | 0.65 ± 0.17 | 3.13 ± 0.22 | 0.00000 |
| 4MP | 3.91 ± 1.10 | 0.00 ± 0.00 | 0.00000 |
| 5MH | 8.76 ± 2.59 | 0.00 ± 0.00 | 0.00001 |
| I3M | 8.89 ± 1.82 | 5.28 ± 0.45 | 0.00066 |
| 1MOI3M | 0.92 ± 0.39 | 1.22 ± 0.24 | 0.13413 |
| 4MOI3M | 0.62 ± 0.11 | 0.73 ± 0.06 | 0.05430 |
| 4OHI3M | 0.12 ± 0.05 | 0.04 ± 0.03 | 0.00771 |
Table X.
Glucosinolate content in seeds of the bcat3-1/bcat4-2 double knockout mutant
The wild-type plant is identical to the one described in Table VII.
| GS Type | Glucosinolate Content
|
||
|---|---|---|---|
| bcat3-1/bcat4-2 | Col-0 Wild Type | t Test | |
| μmol/g seeds | |||
| Total Met GS | 23.51 ± 4.34 | 83.09 ± 8.92 | 0.00000 |
| Total | 29.70 ± 4.75 | 86.06 ± 8.83 | 0.00000 |
| 2MSOE | 1.53 ± 0.20 | 0.00 ± 0.00 | 0.00000 |
| 3OHP | 0.22 ± 0.07 | 0.85 ± 0.14 | 0.00000 |
| 3BZO | 0.54 ± 0.17 | 4.25 ± 0.49 | 0.00000 |
| 4OHB | 1.17 ± 0.31 | 5.12 ± 0.73 | 0.00000 |
| 4MTB | 3.30 ± 0.83 | 33.83 ± 4.57 | 0.00000 |
| 4BZO | 2.75 ± 0.68 | 14.08 ± 0.94 | 0.00000 |
| 4MSOB | 2.70 ± 0.72 | 1.83 ± 0.26 | 0.02288 |
| 5MTP | 5.00 ± 0.71 | 3.51 ± 0.46 | 0.00096 |
| 5BZO | 0.20 ± 0.04 | 0.14 ± 0.03 | 0.01079 |
| 5MSOP | 3.02 ± 0.51 | 0.27 ± 0.04 | 0.00000 |
| 6MSOH | 0.09 ± 0.01 | 0.12 ± 0.03 | 0.01310 |
| 7MTH | 0.38 ± 0.05 | 4.08 ± 0.73 | 0.00000 |
| 7MSOH | 0.11 ± 0.03 | 1.07 ± 0.26 | 0.00000 |
| 8MTO | 0.81 ± 0.16 | 5.94 ± 0.80 | 0.00000 |
| 8MSOO | 1.68 ± 0.23 | 7.90 ± 1.25 | 0.00000 |
| 5MH | 2.30 ± 0.37 | 0.02 ± 0.00 | 0.00000 |
| I3M | 3.71 ± 0.42 | 2.95 ± 0.12 | 0.00173 |
| 4MOI3M | 0.18 ± 0.04 | 0.10 ± 0.02 | 0.00149 |
Many significant changes are found in the amino acid profile in rosette leaves of the double knockout plants (Table VII). Besides several moderate increases the most striking elevations were observed for the levels of Met (25-fold) and for S-methyl-methionine (SMM), which is Met carrying a second methyl group on the sulfur atom. This amino acid is raised from undetectable amounts in wild type to 1.16 ± 0.33 nmol/mg dry weight in the double knockout mutant. In contrast, the level of Gln is lower in the double knockout mutant (72% of the wild-type level), which accounts for the lower amount of total amino acids in the mutant. In addition, Val is reduced as seen in bcat3-1 plants.
Severe changes were also found in the amino acid contents in seeds (Table VIII), where the amount of total amino acids is 4-fold elevated. With the exception of Glu and Phe, all amino acids measured are increased. The most dramatic rise is seen for SMM, which with 15.57 μmol/g seeds is the most abundant free amino acid in this tissue. Arg (10-fold), His (18.4-fold), and Ile (22.1-fold) are also highly increased.
Remarkable changes were also found in the levels of glucosinolates both in leaves and seeds (Tables IX and X). In both tissues the levels of total Met-derived glucosinolates were strongly reduced to 44% (leaves) and 28% (seeds) in comparison with wild type. In leaves almost all standard species of this class of glucosinolates were lowered, the sole exception being 5MSOP, which was increased 3-fold in the double knockout mutant. 4MP and 5MH were even more increased than in the bcat3-1 single knockout mutant. In the bcat3-1/bcat4-2 double knockout these compounds are among the most abundant aliphatic glucosinolates with a 5MH level reaching 75% of the most dominant leaf glucosinolate 4-methylsulfinylbutylglucosinolate (4MSOB). Much less fluctuation was seen for the indolic glucosinolates, with either slight decreases or up to 2.6-fold increases (Table IX).
Other alterations were found in seeds, where the main 4-methylthiobutylglucosinolate (4MTB) is severely reduced to about 10% of the wild-type level. In contrast to leaves, several standard glucosinolates are increased. For instance, the C5 group of glucosinolates derived from trihomo-Met is increased with the most remarkable elevation seen for 5MSOP (11.8-fold). In addition 2-methylsulfinylethylglucosinolate (2MSOE), which is directly synthesized from Met, and the BCAA-derived glucosinolate 5MH were measured at considerable amounts in this double knockout mutant. Both glucosinolate species are usually undetectable in seeds of Col-0 (Brown et al., 2003).
These data show that in the bcat3-1/bcat4-2 double knockout Met-derived glucosinolate levels are even more reduced than in the bcat4-2 single knockout mutant (Schuster et al., 2006). This strongly suggests that BCAT3 can act as a backup of BCAT4 by transaminating Met to MTOB and thus contributes to the overall biosynthesis of Met-derived aliphatic glucosinolates, which is probably localized in plastids.
DISCUSSION
BCAT3 Has a Dual Function in the Biosynthesis of BCAAs and the Met Chain Elongation
With the identification of the crucial roles of the methylthioalkylmalate (MAM) enzymes and of BCAT4 in the Met chain elongation pathway it became apparent that proteins active in this part of Met-derived aliphatic glucosinolate biosynthesis are encoded in the same gene families as enzymes of Leu biosynthesis (Kroymann et al., 2001; Textor et al., 2004; Schuster et al., 2006; Binder et al., 2007; Textor et al., 2007). This is particularly documented for the four members of the isopropylmalate synthase gene family, which encodes two “real” isopropylmalate synthases active in Leu biosynthesis (IPMS1, At1g18500 and IPMS2, At1g74040) and two MAM synthases (MAM1, At5g23010 and MAM3, At5g23020) catalyzing the condensation reactions in the Met chain elongation pathway (Kroymann et al., 2001; Field et al., 2004; Textor et al., 2004; de Kraker et al., 2007; Textor et al., 2007).
Now the investigation of BCAT3 demonstrates that this enzyme cannot be assigned exclusively to one of these pathways, but rather has a dual function in both primary and in secondary metabolism. Several arguments support the function of BCAT3 in the Met chain elongation pathway. First, the expression pattern of BCAT3 is very similar to MAM1 and BCAT4, two important enzymes of Met chain elongation pathway (Fig. 1; Supplemental Fig. S1). Second, the glucosinolate profile of the bcat3-1 mutant shows distinct alterations from that of the wild type (Tables IV and V). Third, in the bcat3-1/bcat4-2 double mutant (Tables IX and X), the knockout of BCAT3 has a striking additive effect on the reduction of total Met-derived glucosinolate content in comparison to the bcat4-1 or bcat4-2 single knockout mutants. Fourth, the BCAT3 protein exhibits substantial activity with substrates of the Met chain elongation pathway. These are in the range of the activity with the standard substrate 3MOB, the keto acid of Val. Fifth, BCAT3 and BCAT4 as well as other proteins active in Met chain elongation are under positive regulatory control of Myb28 (PMG1; Hirai et al., 2007).
We find similarly strong arguments for the function of BCAT3 in the final transamination steps of BCAA biosynthesis. First, the substrate specificity spectrum of recombinant BCAT3 shows a clear preference for 4MOP, 3MOP, and 3MOB, the standard substrates of BCATs involved in the formation of Leu, Ile, and Val (Tables I and II). Second, the enhanced levels of 4MP and 5MH in the bcat3-1 knockout mutant, both being usually present at very low levels in Arabidopsis Col-0, indicates an imbalance in BCAA biosynthesis in these plants (Table III). This conclusion is supported by the results of the feeding experiments performed in this investigation, which shows that a continuous excess of Leu and Ile triggers the generation of 4MP and 5MH. This may represent a mechanism for achieving homeostasis of these amino acids (Table VI). This result together with the reduced level of free Val in the bcat3-1 mutant (Table III) supports a role of BCAT3 in amino acid biosynthesis.
In summary these data show that this enzyme does not participate exclusively in one of the two pathways. BCAT3 is clearly active in both the BCAA and the Met-derived glucosinolate biosyntheses.
BCAT3 Is Important for the Generation of Val
Although the participation of BCAT3 in BCAA biosynthesis can be clearly documented, the assignment to the synthesis of a certain amino acid is difficult. Considering the results of the in vitro assays, the enzyme has the potential to catalyze transamination reactions forming all BCAAs. The lower affinity toward 3MOB in comparison with 4MOP may help assure that some 3MOB is available for the biosynthesis of Leu, rather than all of it being transaminated to Val. The enzymes of the first step of the Leu pathway, IPMS1 and IPMS2, which condense 3MOB with acetyl-CoA, have Km values for 3MOB of 0.3 and 0.28 mm, respectively, which are about 20% of that of BCAT3 for 3MOB (1.38 mm; de Kraker et al., 2007).
The reduction of Val in the bcat3-1 mutant suggests BCAT3 to be active in the formation of Val. This is supported by the appearance of the Val- or Leu-derived 4MP and 5MH glucosinolates in this mutant (Table IV) and in the feeding experiments (Table VI) that might be consequences of reduced Val formation according to the following scenario. The reduced rate of Val biosynthesis in the bcat3-1 mutant should lead to an enhanced synthesis of Leu. Because this amino acid is deleterious to plants at much lower levels than Ile and Val (Lee et al., 2007), it can be expected that plants would have a mechanism to reduce an excess of Leu. As a short-term response, feedback inhibition of both acetohydroxyacid synthase and isopropylmalate synthase by Leu could shut off or slow down biosynthesis of this amino acid. At the same time the inhibition of these enzymes also restricts the biosynthesis of Ile and particularly Val (Hagelstein and Schultz, 1993; Singh, 1999; Binder et al., 2007). Such a feedback effect is indeed seen after the addition of Leu, which leads to a reduction of Val (Supplemental Table S3). Consequently feedback inhibition by Leu, triggered either by the lack of BCAT3 activity or by ongoing administration of Leu, might cause a long-term depletion of Val and Ile.
Another mechanism for reducing excessive Leu is its conversion to glucosinolates. This might explain the occurrence of 4MP and 5MH in the bcat3-1 mutant and the accumulation of these glucosinolates in leaves and seeds after direct Leu feeding. With this interpretation, the occurrence of 4MP and 5MH after feeding Val and Ile might also be ascribed to increased Leu biosynthesis under these conditions.
In this context it is interesting to note that degradation of Leu in mitochondria might not be available as a means to avoid Leu excess in the BCAT3 mutant. The expression of enzymes involved in amino acid degradation in plants is induced principally by carbohydrate starvation (Binder et al., 2007), and so may not occur under the normal conditions used for Arabidopsis cultivation.
BCAT3 Is Responsible for the Transamination of Keto Acids to Met Derivatives
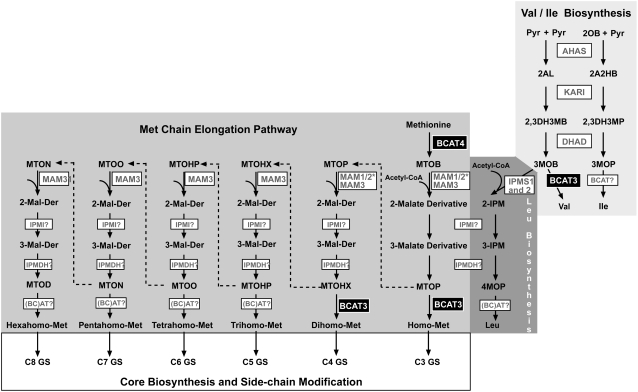
Our results clearly indicate the involvement of BCAT3 in the biosynthesis of Met-derived aliphatic glucosinolates. But which transamination reaction(s) of the chain elongation cycle is (are) catalyzed by this enzyme? In the bcat3-1 knockout mutant the total amount of aliphatic glucosinolates is either significantly increased by about 30% in rosette leaves or is unchanged in seeds. In both tissues of the mutant the pattern of aliphatic glucosinolates is altered with elevated levels of glucosinolate species with five or more carbon atoms. Glucosinolates with shorter side-chain lengths are uniformly (in seeds) or partially (leaves) reduced. These results indicate that BCAT3 is involved in the generation of Met derivatives extended by one or two methylene groups. The stepwise chain elongation of Met involves a three-part elongation cycle at the level of the respective keto acids (Graser et al., 2000). As elongated keto acids are formed, they can be transaminated to form chain-extended Met derivatives (which are then converted to glucosinolates), or proceed through additional rounds of elongation (Fig. 3). Hence an increase in the extent of elongation, as seen by the shift toward longer chain glucosinolates in the bcat3-1 mutant suggests a decreased rate of transamination of shorter keto acids (Table IV). This is consistent with preferential transamination of shorter keto acids by BCAT3 in wild-type plants.
Figure 3.
Model of the Met chain elongation and BCAA biosynthesis pathways. Enzymes are given in boxes. BCAT3 and BCAT4 are highlighted by black boxes. For clear distinction the Met chain elongation pathway is shaded in middle gray, the Leu biosynthesis pathway in dark gray, and the pathways leading to Val and Ile are highlighted in light gray. Abbreviations for enzymes not already defined in the text are as follows: AHAS, Acetohydroxyacid synthase; DHAD, dihydroxyacid dehydratase; IPMI, isopropylmalate isomerase; IPMDH, isopropylmalate dehydrogenase; KARI, ketolacid reductoisomerase; MAM1/2* indicates that either MAM1 (in Col-0) or MAM2 (in Landsberg erecta) catalyze condensation reactions leading to the preferential accumulation of C4 (in Col-0) and C3 (in Landsberg erecta) glucosinolates. Abbreviations for metabolites not defined in the text are: Pyr, pyruvate; 2OB, 2-oxobutyrate; 2AL, 2-acetolactate; 2A2HB, 2-aceto-2-hydroxybutyrate; 2,3DH3MB, 2,3-dihydroxy-3-methylbutyrate; 2,3DH3MP, 2,3-dihydroxy-3-methylpentanoate; IPM, isopropylmalate; MTOHX, 4-methylthio-2-oxohexanoate; MTOHP, 4-methylthio-2-oxoheptanoate; MTOO, 4-methylthio-2-oxooctanoate; MTON, 4-methylthio-2-oxononanoate; MTOD, 4-methylthio-2-oxodecanoate; Mal-Der, malate derivative.
The direct function of BCAT3 in transamination of intermediates of the Met chain elongation pathway was confirmed by in vitro assays. The recombinant enzyme converts MTOP to homo-Met and MTOH to dihomo-Met, whereas less activity of the enzyme is measured for 4-methylthio-2-oxohexanoate, which is converted to trihomo-Met (Table II). Thus the results of the in vitro substrate specificity tests are consistent with the in vivo profiling data.
Among the increased glucosinolates with longer side chains 5MTP and 5MSOP (both C5 glucosinolates) show the strongest increase whereas C6, C7, and C8 are only moderately increased. This might be explained by the preferential transamination of 7-methylthio-2-oxoheptanoate to trihomo-methionine by an additional BCAT. The presence of such an enzyme has to be postulated considering that there is still some residual biosynthesis of Met-derived glucosinolates in the bcat3-1/bcat4-2 double knockout mutant. Further studies will clarify this issue.
The functional analysis of the plastid-located BCAT3 and its assignment to the Met chain elongation pathway show that the transamination of at least certain keto acids, like MTOP and MTOH occurs in plastids, where MAM3 and most likely MAM1 are also located (Diebold et al., 2002; Textor et al., 2007). Thus these reactions are physically separated from the initial transamination of Met yielding MTOB by BCAT4 in the cytosol (Schuster et al., 2006). The biosynthesis of Met-derived aliphatic glucosinolates therefore requires at least two transport steps: the transport of MTOB into plastids, and the translocation of Met derivatives from these organelles back into the cytosol.
BCAT3 Partially Compensates for the Loss of BCAT4 Activity
The glucosinolate profile observed in the bcat3-1/bcat4-2 double knockout cannot be explained by the simple addition of the chemotypes of the single bcat3-1 (Tables IV and V) and bcat4-2 knockouts (Schuster et al., 2006). The additional reduction of total Met-derived glucosinolates from 50% of wild type in leaves and seeds of two bcat4-1 and bcat4-2 knockout mutants to 44% of wild type in leaves and 28% in seeds in bcat3-1/bcat4-2 double mutants suggests that BCAT3 might have a function equivalent to BCAT4, i.e. in catalyzing the formation of MTOB from Met. Although this function cannot be deduced from the results of the glucosinolate profiling of the bcat3-1 mutant, such a role of BCAT3 seems feasible considering the in vitro substrate specificity of this enzyme as shown here (Tables I and II).
This compensatory function of BCAT3 might only be obvious or even relevant in the BCAT4 knockout plants. A further complication is that BCAT3 is not the sole backup to BCAT4 as seen by the residual glucosinolate biosynthesis in the double mutant. Thus either another aminotransferase converts Met to MTOB (for instance BCAT2 or BCAT5), or perhaps MTOB or the chain elongated Met derivatives are recruited from other pathways.
Interdependence and Relationship of Amino Acid and Glucosinolate Metabolism in Arabidopsis
The close involvement of amino acid and glucosinolate metabolism had been indicated by the investigation of other Arabidopsis mutants, where alterations in the glucosinolate profile were found to have profound effects on amino acid composition (de Kraker et al., 2007; Field et al., 2004). The tight connection of glucosinolate and amino acid metabolism is also clear in this study particularly in the double knockout mutant, which shows unanticipated changes in both classes of metabolites, especially in seeds. One example is the accumulation of 2MSOE, a nonelongated glucosinolate directly derived from Met, which is not detectable in Col-0 wild type. The formation of this glucosinolate species presumably results from the severe reduction in the rate of Met chain elongation in the double mutant, which is accompanied by the accumulation of Met and SMM. Thus, the synthesis of this glucosinolate species can be viewed as a means of lowering the levels of free Met and SMM, in analogy to the synthesis of 4MP and 5MH as a means of responding to imbalances in levels of free BCAAs. And indeed, in the double knockout mutant, 4MP and 5MH accumulate to very high amounts in rosette leaves and 5MP is even detectable in seeds, whereas these abnormal glucosinolates are not detected in the bcat3-1 plants. This finding suggests that the double knockout mutant has to deal with even higher Leu levels than bcat3-1, which may mean that BCAT4 is also involved in Leu metabolism in addition to its function in Met elongation. Such a hypothesis is consistent with the observed substrate preference of this enzyme in vitro, which shows activity with Leu and its keto acid 4MOP, but not with Val, Ile, and the respective keto acids (Schuster et al., 2006). However, the potential role of BCAT4 in Leu metabolism is not obvious in the BCAT4 knockout plants, and so requires further investigation. In contrast, the data reported here unequivocally demonstrate the dual function of BCAT3 in amino acid and glucosinolate biosynthesis. Our findings show that in Leu biosynthesis and Met chain elongation pathway enzymes are not only encoded by members of the same gene families (Schuster et al., 2006; de Kraker et al., 2007), but one enzyme (BCAT3) acts in both pathways. Thus there is not necessarily a rigid barrier between primary and secondary metabolism in plants when it comes to enzyme sharing.
MATERIALS AND METHODS
Plant Material and Cultivation
Arabidopsis (Arabidopsis thaliana) plants ecotype Col-0 were grown under controlled conditions as recently described (Schuster et al., 2006). The genotypes of progeny of GABI-Kat line 002A11 were examined by PCR with BCAT3-specific primers 3314.1 (5′-GCTGGTGTACTCAACTATGG) and 3314.2 (5′-ACATTCCGTTCCTCCACCTG) and an oligonucleotide annealing in the T-DNA (LB-GabiKat, 5′-ATATTGACCATCATACTCATTGC). The corresponding allele was designated bcat3-1. Crossing of bcat3-1 and bcat4-2 plants was done following standard protocols (Weigel and Glazebrook, 2002). The genotype of double mutants was confirmed by respective PCRs as described above and previously (Schuster et al., 2006). In these crossing experiments, the F2 generation was also screened for plants homozygous for bcat3-1 and homozygous for the bcat4 wild-type allele. These plants were included in the metabolite profiling and showed a chemotype identical to the original bcat3-1 plants.
Enzyme Activity and Substrate Specificity Assays
For the generation of recombinant mature protein a BCAT3 cDNA was amplified with oligonucleotides bcat3.ue.BamHI (5′-TAGGATCCTGCAACGCTGTTTCGTCC) and bcat3ue.R (5′-TAGTCGACTTAACTAAGATTCACAG) on a previously established cDNA clone as DNA template (Diebold et al., 2002) using the Advantage 2 polymerase mix (CLONTECH Laboratories) supplied with the respective buffer system with the following conditions: 1 min at 94°C, 1 min at 50°C, and 1 min at 68°C. The amplified cDNA corresponds to 353 amino acids excluding the potential chloroplast targeting signal (60 amino acids). The cDNA fragment was digested with BamHI and SalI cloned into the respective site in pET32 overexpression vector (Novagen). After complete sequence analysis, overexpression and enrichment of recombinant BCAT3 on S-Tag agarose was done as described in the manufacturer's manual. For release from the S-Tag agarose recombinant BCAT3 was digested with recombinant enterokinase removing the S-Tag from the polypeptide and leaving a few additional amino acids at the N terminus. The amount of recombinant BCAT3 in the enriched fraction was determined as described previously (Schuster et al., 2006) and found to be at least 13.7%.
Kinetics and substrate specificity tests were done following established protocols (Schadewaldt and Adelmeyer, 1996; Schuster and Binder, 2005; Schuster et al., 2006). Calculations were done using the Origin 7.0 software (OriginLab). Substrates were purchased from Sigma-Aldrich or from Applichem.
Expression Analysis
Northern-blot and RT-PCR analyses as well as histochemical GUS staining were done as described before (Hull and Devic, 1995; Schuster and Binder, 2005; Schuster et al., 2006). The probe for the northern hybridization was amplified with primers bcat3N.1 (5′-GAGAGAAAGCCTCCGTCTCC) and bcat3N.2 (5′-GGAAGAAGAATGCTGAAG). RT-PCR analysis was done with Advantage 2 polymerase mix following the recommendations of the manufacturer (CLONTECH Laboratories). After cDNA synthesis initiated with oligo(dT) primer dTXSC, a PCR was done with primer pair bcat3ue.BamHI/3314.2. To check the integrity of the RNA and cDNA, respectively, a control reaction was done with primers UBQ10.H (5′-GATCTTTGCCGGAAAACAATTG)/UBQ10.R (5′-CGACTTGTCATTAGAAAGAAAGAG) amplifying a 500-bp fragment of the ubiquitin 10 gene.
Glucosinolate and Amino Acid Profiling
Glucosinolates were extracted and measured from rosette leaves of approximately 21-d-old plants and from mature seeds as described previously (Brown et al., 2003). Profiles of free amino acids were determined as described before (Roth, 1971; Sarwar and Botting, 1993; de Kraker et al., 2007).
Feeding of Amino Acids to Arabidopsis Plants
For determining glucosinolate and amino acid in seeds and leaves, single Arabidopsis plants (ecotype Col-0) were grown under normal conditions in pots.
For glucosinolate and amino acid profiling after short-term feeding, plants were grown under normal conditions for 3 weeks. Then 33 mL of a 2 mm amino acid solution in double distilled water was added to each pot and rosette leaves were harvested 4.5 h after administration.
To study the effects of repeated administration of BCAAs on amino acid and glucosinolate profiles, plants were grown for 2 weeks under normal conditions. Amino acid solutions as indicated above were given every second day until leaves were harvested when plants were 20-d-old. Harvesting was done 48 h after the final feeding.
For glucosinolate and amino acid profiling in seeds, feeding was started after 3 weeks of growth of the plants under normal conditions. The supplement was repeated 16 times until seed set about 3 weeks before harvest. Control plants were treated identically by adding water instead of amino acid solutions. Seeds were harvested from about 10-week-old plants. Glucosinolates and amino acids were profiled as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Time-resolved expression of BCAT3.
Supplemental Figure S2. Kinetic of BCAT3 with 3MOB.
Supplemental Figure S3. Kinetic of BCAT3 with 4MOP.
Supplemental Figure S4. Kinetic of BCAT3 with 3MOP.
Supplemental Figure S5. Kinetic of BCAT3 with MTOB.
Supplemental Table S1. Amino acid contents in leaves of Col-0 wild-type plants 4.5 h after administration of BCAAs.
Supplemental Table S2. Amino acid contents in leaves of bcat3-1 plants 4.5 h after administration of BCAAs.
Supplemental Table S3. Amino acid contents in leaves of Col-0 wild-type plants after repeated administrations of BCAAs.
Supplemental Table S4. Amino acid contents in leaves of bcat3-1 plants after repeated administrations of BCAAs.
Supplemental Table S5. Glucosinolate contents in leaves of wild-type Col-0 plants after repeated administrations of BCAAs.
Supplemental Table S6. Glucosinolate contents in leaves of bcat3-1 plants after repeated administrations of BCAAs.
Supplementary Material
Acknowledgments
We thank Conny Guha for the characterization of T-DNA insertion mutants. We are also very grateful to Carolin Müller for high quality cross sections.
This work was supported by a fellowship from the Landesgraduiertenförderungsgesetz des Landes Baden-Württemberg (T.K.), the Deutsche Forschungsgemeinschaft (Ge 1126/1–3 and Bi 590/9–1), the Max Planck Society, and a start-up grant from the Rudolph und Clothilde Eberhardt-Stiftung.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stefan Binder (stefan.binder@uni-ulm.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Binder S, Knill T, Schuster J (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129 68–78 [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481 [DOI] [PubMed] [Google Scholar]
- Chen S, Glawischnig E, Jorgensen K, Naur P, Jorgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33 923–937 [DOI] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143 970–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold R, Schuster J, Däschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Cardon G, Traka M, Botterman J, Vancanneyt G, Mithen R (2004) Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol 135 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser G, Schneider B, Oldham NJ, Gershenzon J (2000) The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch Biochem Biophys 378 411–419 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11 89–100 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Muller J, Masuno MN, Molinski TF, Abel S (2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40 893–908 [DOI] [PubMed] [Google Scholar]
- Hagelstein P, Schultz G (1993) Leucine synthesis in spinach chloroplasts: partial characterization of 2-isopropylmalate synthase. Biol Chem Hoppe Seyler 374 1105–1108 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57 303–333 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull GA, Devic M (1995) The beta-glucuronidase (gus) reporter gene system. Gene fusions, spectrophotometric, fluorometric, and histochemical detection. In H Jones, ed, Methods Mol Biol, Vol 49. Humana Press, Totowa, NJ, pp 125–141 [DOI] [PubMed]
- Keck AS, Finley JW (2004) Cruciferous vegetables: cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr Cancer Ther 3 5–12 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M (2007) AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J 50 305–319 [DOI] [PubMed] [Google Scholar]
- Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43 79–96 [DOI] [PubMed] [Google Scholar]
- Mehta PK, Christen P (2000) The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol 74 129–184 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Roth M (1971) Fluorescence reaction for amino acids. Anal Chem 43 880–882 [DOI] [PubMed] [Google Scholar]
- Sarwar G, Botting HG (1993) Evaluation of liquid chromatographic analysis of nutritionally important amino acids in food and physiological samples. J Chromatogr 615 1–22 [DOI] [PubMed] [Google Scholar]
- Schadewaldt P, Adelmeyer F (1996) Coupled enzymatic assay for estimation of branched-chain L-amino acid aminotransferase activity with 2-oxo acid substrates. Anal Biochem 238 65–71 [DOI] [PubMed] [Google Scholar]
- Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57 241–254 [DOI] [PubMed] [Google Scholar]
- Schuster J, Knill T, Reichelt H, Gershenzon J, Binder S (2006) Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In BK Singh, ed, Plant Amino Acids: Biochemistry and Biotechnology. Marcel Dekker, New York, pp 227–247
- Smith TK, Mithen R, Johnson IT (2003) Effects of Brassica vegetable juice on the induction of apoptosis and aberrant crypt foci in rat colonic mucosal crypts in vivo. Carcinogenesis 24 491–495 [DOI] [PubMed] [Google Scholar]
- Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA, Gershenzon J (2004) Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218 1026–1035 [DOI] [PubMed] [Google Scholar]
- Textor S, de Kraker J-W, Hause B, Gershenzon J, Tokuhisa JG (2007) MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol 144 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis, a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7 263–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.