Abstract
Multigene families encoding class XI myosins are conserved in higher plants, however, little information is available on specific functions of these ubiquitous molecular motors. We isolated gene knockout mutants for all 13 class XI myosins present in Arabidopsis (Arabidopsis thaliana) genome. Inactivation of 11 myosin genes resulted in no discernible phenotypes under the normal growth conditions. In contrast, the knockouts of the remaining two myosin genes, XI-2 (formerly MYA2) and XI-K, exhibited similar defects in root hair elongation suggesting that the myosin-driven motility plays a significant role in a polar tip growth. Strikingly, inactivation of each of these myosins also reduced trafficking of Golgi stacks, peroxisomes, and mitochondria in root hairs and in leaf epidermal cells. These results indicate that myosins XI-K and XI-2 play major and overlapping roles in the cell dynamics in Arabidopsis and highlight the redundant nature of myosin function in plants.
Myosins are signature molecular motors of eukaryotes that are involved in a broad spectrum of actin cytoskeleton-associated types of cellular dynamics (Vale, 2003). Comparative genomics revealed that myosins are conserved throughout the eukaryotic domain of life (Richards and Cavalier-Smith, 2005; Foth et al., 2006). Land plants possess two myosin classes, XI and VIII, each of which is evolutionary, related to animal and fungal class V myosins (Desnos et al., 2007), suggesting their origin was from a common ancestor that antedated the divergence of Plantae and Opisthokonts (Foth et al., 2006). Subsequent evolution of myosins was dominated by gene duplication and diversification that resulted in the presence of more than 10 myosin genes in all plant genomes sequenced so far (see accompanying article Avisar et al., 2008). In particular, Arabidopsis (Arabidopsis thaliana) encodes 13 class XI and four class VIII myosins (Reddy and Day, 2001). The studies that mostly involved cytoskeletal inhibitors have demonstrated the principal role of actomyosin motilty in plant cell dynamics including organelle trafficking, remodeling, and inheritance (Boevink et al., 1998; Nebenfuhr et al., 1999; Sheahan et al., 2004; Kim et al., 2005; Runions et al., 2006). Some of the class XI myosins were found in association with organelles suggesting their involvement in organelle transport (Wang and Pesacreta, 2004; Hashimoto et al., 2005; Li and Nebenfuhr, 2007; Reisen and Hanson, 2007). However, information on functional profiles of individual myosin motors is very limited. Two recent publications have demonstrated the role of Arabidopsis myosin XI-K in root hair growth (Ojangu et al., 2007), and implicated rice (Oryza sativa) myosin XI-B in pollen development (Jiang et al., 2007).
Here we screen the gene knockouts of all 13 class XI myosins of Arabidopsis to show that, in addition to myosin XI-K, myosin XI-2 (MYA2) is also required for root hair development. Furthermore, we demonstrate that each of these two highly expressed myosins functions in the rapid movement of the Golgi stacks, peroxisomes, and mitochondria in roots and leaves. Interestingly, inactivation of the genes encoding the most closely related paralogs of myosins XI-K and XI-2 and myosins XI-1 (MYA1) and XI-B, respectively, did not impair root hair growth or organelle trafficking. These results indicate that evolution of myosins in plants combines opposing tendencies of functional specialization and functional redundancy.
RESULTS
Isolation of the Homozygous Knockout Lines
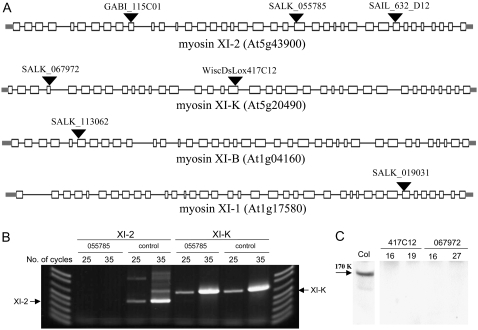
Sixteen homozygous lines in which each of the 13 class XI myosin genes of Arabidopsis was inactivated by T-DNA insertion were obtained and the exact localization of the insert was determined by sequencing (Fig. 1A; data not shown). Furthermore, inactivation of the target genes was demonstrated using semiquantitative reverse transcription (RT)-PCR (e.g. Fig. 1B). The corresponding mRNAs were undetectable by RT-PCR analysis, therefore confirming complete abolishment of myosin expression for each of the 16 knockout lines. To ensure that the observed phenotype is attributed solely to the inactivation of the myosin XI-K locus, line SALK_067972 was further backcrossed twice to the wild-type plant, and the homozygous progeny was selected and used for the experiments described below. In addition, two independent lines for myosin XI-K and three for myosin XI-2 genes were selected for further experiments to ensure that the observed phenotypes were due to the T-DNA insertion in the corresponding locus rather than to secondary site mutations. For both of the obtained independent myosin XI-K gene knockout lines, the lack of an expressed protein was confirmed using a polyclonal antibody specific to this myosin (Fig. 1C).
Figure 1.
A, Diagrams of the four Arabidopsis myosin class XI genes with the positions of the T-DNA insertions. White boxes represent exons, black lines represent introns, and gray bars correspond to the 5′ and 3′ untranslated regions. Designations of each line are shown above the corresponding T-DNA insertion sites (black triangles). Sizes of exons and introns are drawn to scale using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn). The diagram corresponding to the gene At5g20490 (myosin XI-K) was modified to accommodate corrections in the exon structure (Ojangu et al., 2007). B, Semiquantitative RT-PCR analysis of the insertion line SALK_055785 in which the myosin XI-2 gene was inactivated. Primers complementary to sites flanking the insertion site in the myosin XI-2 mRNA (lines under XI-2 in the image) or complementary to two regions within the myosin XI-K mRNA (lines under XI-K) were used. In the control, the RNA used for analysis was isolated from the parental Columbia line. The expected position of the DNA amplification products for each mRNA is shown by an arrow. C, Immunoblot analysis of the protein extracts from the control (Col in the image) and myosin XI-K knockout plants using specific polyclonal antibody. Samples from two plants for each line are shown.
Inactivation of Myosins XI-2 and XI-K Induces Similar Defects in Root Hair Growth
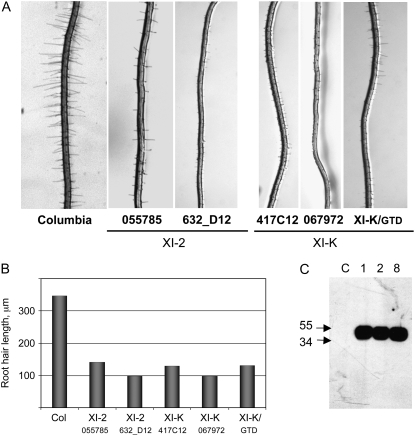
Inspection of the insertional lines in which each of the 13 class XI myosin genes present in Arabidopsis genome was inactivated (Fig. 1; data not shown) revealed no detectable developmental defects in the aerial organs of plants grown under normal conditions. Therefore, the seeds of each line were planted to vertical plates to screen for root morphology. Although the overall root sizes and shapes appeared normal in all knockout lines, four lines exhibited obvious defects in the elongation of the root hairs (Fig. 2A). These lines were knockouts of two class XI myosin genes, namely, XI-2 and XI-K. Quantification of the root hair length for each of these lines revealed a dramatic reduction in the mean root hair length that varied from 28% to 40% of that in the parental Columbia line (Fig. 2B). Comparative analysis showed that the differences in the root hair length between each of these four knockout lines and the wild-type line were statistically significant with P < 0.001 for all lines.
Figure 2.
Root hair development in the knockout mutants and transgenic plants expressing a dominant negative mutant of the myosin XI-K. A, Typical images of the roots for the control (Columbia) and four knockout lines are shown as indicated. XI-K/GTD is a transgenic line expressing GTD of the myosin XI-K. B, Analysis of the mean root hair length for the lines shown (A). C, Immunoblot analysis of the HA epitope-tagged XI-K/GTD expression using HA-specific monoclonal antibody. Samples were from a control plant (C in the image) and plants representing three independent transgenic lines (1, 2, and 8). Positions of two size markers (molecular mass in kilodaltons) are shown by arrows.
Interestingly, analysis of the light-regulated chloroplast relocalization performed as described in an accompanying work by Avisar et al. (2008) revealed that none of the myosin gene knockouts exhibited observable defects in chloroplast movement (data not shown). This result suggested that several myosin motors may have overlapping functions in chloroplast movement. Alternatively, it is possible that although chloroplast movement requires intact actin cytoskeleton (Paves and Truve, 2007), it does not involve myosins.
To confirm the results of the gene knockout experiments by an independent approach and to examine the usefulness of the dominant negative inhibition of the myosin function that we used for Nicotiana benthamiana in an accompanying article (Avisar et al., 2008) for Arabidopsis, we generated several transgenic lines that expressed the cargo-binding, globular tale domain (GTD) of myosin XI-K. The hemagglutinin (HA)-epitope tagging of this domain was used to select transgenic lines that exhibited the highest levels of GTD expression (Fig. 2C). Because the mean root hair length in these selected lines was only 38% of that in the control (Fig. 2A, far right), very similar to that found for myosin XI-K knockout mutants, these data validated the use of the dominant negative inhibition approach in Arabidopsis.
Given that the principal function of myosin motors is physical translocation of various cargoes, our findings suggest that such translocation powered by myosins XI-2 and XI-K is required for the rapid polarized growth of the root hairs.
Myosins XI-2 and XI-K Are Required for the Rapid Organelle Transport in Root Hairs and Leaves
What are the cargoes transported by the myosins XI-2 and XI-K within the growing root hairs? To address this question, we studied trafficking of Golgi stacks and peroxisomes in the gene knockout lines that were transformed to express the fluorophore-tagged, organelle-specific reporters (Fig. 3, A–C). The mitochondrial transport was examined using a vital fluorescent dye Rhodamine 123 (Fig. 3E). Organelle trafficking in the live root hairs was visualized by confocal microscopy and the resulting movies were used for computer-assisted organelle tracking and velocity measurements.
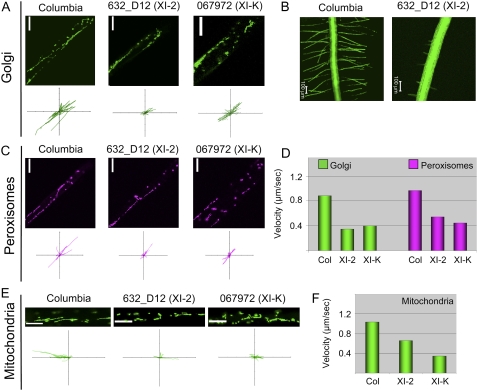
Figure 3.
Roles of myosins XI-2 and XI-K in organelle trafficking in root hairs. A, C and E, Representative images of the indicated organelles (top rows) and paths of individual organelles plotted relative to a common origin (bottom rows; each axis is 100 μm). B, Images of the roots of parental and knockout lines transformed with the Golgi-specific GFP reporter. D, Mean velocities of the Golgi stacks and peroxisomes. F, Mean velocities of mitochondria.
As expected, the predominant pattern of Golgi movement was along the root hair longitudinal axis (Fig. 3A). It should be noted, however, that not infrequently the neighboring individual Golgi stacks were moving in the opposite directions or with the drastically distinct velocities (data not shown). This pattern and a mean Golgi velocity of approximately 1 μm/s were very similar to those recently described for Arabidopsis root hairs (Campanoni et al., 2007). It seems, therefore, that Golgi stacks move independently of each other rather than in a uniform stream-like pattern.
Strikingly, we found that inactivation of either myosin XI-2 or myosin XI-K resulted in an approximately 2-fold reduction in the mean velocity of the Golgi stacks and peroxisomes (Fig. 3D). In the case of mitochondria, the myosin XI-K knockout line exhibited more than 3-fold lower velocity than that in the wild type, whereas a less dramatic, approximately 1.5-fold velocity reduction was observed in the myosin XI-2 knockout line (Fig. 3F). In all cases, differences between the organelle velocities in knockout lines versus the parental Columbia line were statistically significant with P < 0.001. Taken together, these observations demonstrate that myosins XI-K and XI-2 each make a significant contribution into the rapid transport of Golgi stacks, peroxisomes, and mitochondria in the root hairs. This, however, does not necessarily imply that the defects in root hair growth seen in the corresponding knockout lines can be directly attributed to the slower organelle movement.
To determine whether or not myosins XI-2 and XI-K are required for rapid organelle trafficking in organs other than root hairs, we examined motility of Golgi stacks and peroxisomes in the elongated epidermal cells found along the central vein on the leaf underside (Fig. 4B, central area). These elongated cells are most amenable to observing and measuring organelle movement in leaves. Interestingly, the mean velocity of these organelles in leaf cells was at least 30% greater than that in the root hairs (compare with Figs. 3D and 4D). Inactivation of either myosin XI-2 or myosin XI-K resulted in an approximately 3- to approximately 5-fold reduction of the mean velocity of Golgi stacks and peroxisomes (Fig. 4, A, C, and D), and also a stronger effect compared with that in the root hairs. The dominant negative inhibition of myosin XI-K closely mimicked the effect of the gene knockout, once again confirming the utility of this approach for the study of myosin function in plants (Fig. 4, A and D).
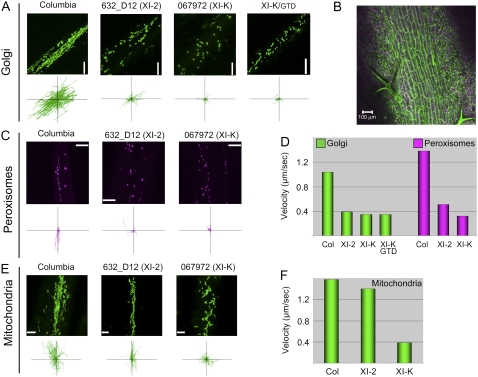
Figure 4.
Roles of myosins XI-2 and XI-K in organelle trafficking in leaves. A, C, and E, Representative images of the indicated organelles (top rows) and paths of individual organelles plotted relative to a common origin (bottom rows; each axis is 100 μm). B, Image of the leaf vein in the Columbia line transformed with the Golgi-specific GFP reporter showing a file of elongated epidermal cells used for organelle tracking. D, Mean velocities of the Golgi stacks and peroxisomes. F, Mean velocities of mitochondria.
The mean velocity of mitochondria in the leaf cells was approximately 40% greater than that in the root hairs (compare with Figs. 3F and 4F). Interestingly, inactivation of the two myosin genes had distinct effects on the translocation of mitochondria: the myosin XI-2 gene knockout line showed only a moderate, although statistically significant (P = 0.045) reduction in the velocity of this organelle, whereas the myosin XI-K knockout line exhibited a drastic, 3.5-fold velocity reduction (Fig. 4, E and F).
Collectively, these results demonstrated that two class XI myosins, XI-2 and XI-K, make comparable contributions to rapid trafficking of Golgi stacks and peroxisomes in the Arabidopsis roots and leaves. It seems, however, that myosin XI-K plays a more significant role in the translocation of mitochondria than myosin XI-2 and that this difference is more pronounced in leaves than in root hairs.
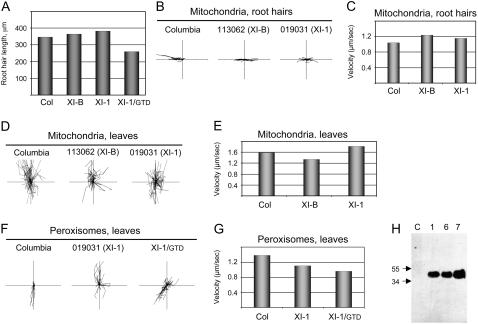
Paralogous Myosins XI-B and XI-1 Are Not Essential for Organelle Transport and Root Hair Growth
Phylogenetic analysis of plant myosins presented in Figure 1A of an accompanying article (Avisar et al., 2008) shows that Arabidopsis myosins XI-2 and XI-K each possess closely related paralogs, myosins XI-B and XI-1, respectively, suggesting that the myosins in each paralogous pair may perform similar functions. To test this possibility, we examined root hair growth and trafficking of the mitochondria and peroxisomes in the knockout lines with inactivated myosins XI-B or XI-1. Our analyses revealed no defects in root hair growth in either of the mutant lines (Fig. 5A). Examination of mitochondria in the root hairs (Fig. 5, B and C) or leaf epidermal cells (Fig. 5, D and E) showed no significant changes in the mean organelle velocities due to mutational inactivation of either of two myosin paralogs. Only a modest reduction of the peroxisome velocity in leaf epidermal cells was observed in the myosin XI-1 knockout line compared with that in the parental Columbia line (Fig. 5, F and G).
Figure 5.
Myosins XI-B and XI-1 do not play significant roles in root hair development and organelle movement. A, Analysis of the mean root hair length for the indicated gene knockout mutant lines and transgenic line that expresses XI-1/GTD. B, D and F, Paths of individual organelles plotted relative to a common origin (each axis is 100 μm). C and E, Mean velocities of mitochondria in root hairs and leaf epidermal cells, respectively. G, Mean velocities of the peroxisomes in leaf epidermal cells. H, Immunoblot analysis of the HA epitope-tagged XI-1/GTD expression using HA-specific monoclonal antibody. Samples were from a control plant (C, Columbia in the image) and plants representing three independent transgenic lines (1, 6, and 7). Positions of two size markers (molecular mass in kilodaltons) are shown by arrows.
It was also found that transgenic expression of the myosin XI-1/GTD (Fig. 5H) had only moderate negative effects on the root hair growth (Fig. 5A) or on peroxisome trafficking (Fig. 5, F and G). Taken together, these observations indicate that myosins XI-B and XI-1 might play only a relatively minor role in the root hair development and organelle translocation in the presence of intact paralogous myosins XI-2 and XI-K.
DISCUSSION
It is well established that actin cytoskeleton dynamics plays a paramount role in the polarized growth of the root hair cells (Hepler et al., 2001; Carol and Dolan, 2002; Smith and Oppenheimer, 2005; Staiger and Blanchoin, 2006). However, there is only limited insight into the significance of the myosin-powered motility of vesicles and organelles for the elongation of the root hairs. In fact, only one class XI Arabidopsis myosin, XI-K, was recently implicated in this process (Ojangu et al., 2007), and even in this study, the potential role of this myosin in organelle movement has not been addressed.
Here we describe systematic screening of all 13 class XI myosins of Arabidopsis for their potential functions in the root hair development. Using gene knockout and dominant negative inhibition approaches, we reveal that two class XI myosins, XI-2 and XI-K, are essential for the normal elongation of the root hairs. Inactivation of each of these myosins in four distinct insertion lines (Fig. 1) results in very similar phenotypes with a mean root hair length of approximately one-third that in the parental Columbia line (Fig. 2). In addition, we investigated the roles of myosins XI-2 and XI-K, and their most closely related paralogs, myosins XI-B and XI-1, in the trafficking of Golgi stacks, peroxisomes, and mitochondria in the root hairs. Conspicuously, we found that myosins XI-2 and XI-K are each required for the rapid movement of all three organelles (Fig. 3). Inactivation of the corresponding genes reduced the mean velocity of organelles from 1.5- to 3-fold depending on the organelle or myosin identity. In contrast, inactivation of the closely related, paralogous myosins XI-B and XI-1, had no detectable effects on either root hair growth or organelle motility (Fig. 5); additional work is needed to identify functions of these myosins.
The observed correlation in the roles of myosins XI-2 and XI-K in root hair growth and organelle trafficking suggests that the latter might be functionally required for the former. Indeed, the rapid movement of the Golgi stacks, peroxisomes, and mitochondria in a “reverse fountain” manner (Hepler et al., 2001) may be needed to enhance protein secretion and turnover of metabolites and energy in the growing root hair tip. It also seems possible that the vesicular trafficking that delivers building materials for the plasma membrane and cell wall expansion in the tip is powered by myosins XI-K and XI-2. Finally, the possibility that these myosins are also involved in microfilament remodeling or even more general processes required for maintenance of cell metabolism cannot be ruled out. However, because interference with cargo binding by myosin XI-K using transgenic expression of the cognate GTD mimics the phenotype of XI-K gene knockout in defective root hair growth and slow organelle movement (Figs. 2 and 3), both of these effects are likely due to the lack of the proper cargo translocation in both knockout and dominant negative inhibition approaches used to determine myosin XI-K function.
Recent insight into the mechanistic parallelism in the development of the root hairs and pollen (Hepler et al., 2001; Cole and Fowler, 2006) implies potential involvement of myosins in the polarized growth of the pollen tubes. However, relatively low levels of myosins XI-2 and XI-K in pollen (the myosin transcription data were found at the Weigel World Web site; http://www.weigelworld.org/resources) make these myosins unlikely candidates for the major role in this process. In contrast, myosins XI-A, XI-B, XI-C, XI-D, and XI-J are almost exclusively expressed in pollen and therefore are much better candidates for direct involvement in pollen tube growth. Therefore, it seems likely that the closest paralog of myosin XI-2, myosin XI-B, has evolved to mediate organelle and vesicle transport in the pollen tubes.
We found that myosins XI-2 and XI-K are required for the rapid organelle trafficking not only in root hairs, but also in the leaves (Fig. 4). This conclusion resonates with the relatively high levels of these myosins in leaves (http://www.weigelworld.org/resources). In fact, myosins XI-2, XI-K, and XI-1 are the most abundant myosins in the entire Arabidopsis plants. Surprisingly, this abundance and major roles played by myosins XI-2 and XI-K in organelle movement do not translate into substantial defects in the development of leaves, stems, or flowers in the corresponding gene knockout lines. It appears that the reduction in the leaf organelle velocity in these lines is not critical for leaf development under normal growth conditions. Therefore, perhaps due to their rapid elongation, root hairs are a more sensitive indicator for the defects in organelle trafficking than the leaf cells.
The lack of major developmental defects in two myosin XI-2 knockout lines characterized here and in an additional line described elsewhere (Hashimoto et al., 2005) is in a stark contrast to the earlier article that reported dwarf growth and flower sterility in a single knockout line (Holweg and Nick, 2004). The reason for this severe phenotype could be the secondary site mutation(s).
Rapid organelle trafficking is a hallmark of plant cell physiology that is traditionally referred to as “cytoplasmic streaming” (Hepler et al., 2001; Smith and Oppenheimer, 2005; Taiz and Ziegler, 2006; Shimmen, 2007). However, early studies on Golgi trafficking (Boevink et al., 1998; Nebenfuhr et al., 1999) demonstrated that individual stacks are moved independently of each other by the actomyosin motility system rather than being passively carried along by indiscriminate cytosol flow (Nebenfuhr and Staehelin, 2001). Our analyses of the trafficking of Golgi, peroxisomes, and mitochondria in N. benthamiana (Avisar et al., 2008) stressed the need to revise the concept of cytoplasmic streaming. The patterns of organelle movement in Arabidopsis (e.g. Fig. 4) are also incompatible with passive “floating with the flow” and further support introduction of a mechanistically more relevant concept of active organelle translocation defined by the actomyosin transport network.
On a broader scale, this work is relevant to the problem of the multigene families' evolution in eukaryotes: to what extent the lineage-specific expansion of these families is due to adaptation as opposed to stochastic gene birth and death processes (Lynch, 2007)? The myosin gene families provide an excellent model to address this problem because they are nearly ubiquitous in eukaryotes. Our phylogenetic analysis (Avisar et al., 2008) suggested that the common ancestor of higher plants possessed five class XI myosins. This number has more than doubled in Arabidopsis, the plant that encodes more class XI myosins than the other three plants with completely sequenced genomes. The functional analysis of the Arabidopsis class XI myosins described here revealed a rather complex picture. The lack of major developmental defects in 11 myosin gene knockouts attests to a redundant nature of myosin functions in plants. The functional profiles of myosins XI-2 and XI-K that belong to distinct paralogous families largely overlap: both myosins are involved in the organelle transport and in root hair growth. The only functional specialization apparent between these Arabidopsis myosins is a somewhat larger contribution of myosin XI-K in the movement of mitochondria, an effect that is even more pronounced in N. benthamiana (Avisar et al., 2008). More extensive characterization of the myosin functions is required to determine how many and which myosins are essential for plant development.
MATERIALS AND METHODS
T-DNA Insertion Mutants
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia T-DNA insertion lines SALK_055785 and SAIL_632_D12 (At5g43900; myosin XI-2), SALK_067972 and WiscDsLox417C12 (At5g20490; myosin XI-K), SALK_113062 (At1g04160; myosin XI-B), SALK_019031 (At1g17580; myosin XI-1), SALK_082078 (At2g33240; myosin XI-D), SALK_072023 (At1g54560; myosin XI-E), SALK_018032 (At2g20290; myosin XI-G), SAIL_365_D03 (At4g28710; myosin XI-H), SALK_082443 (At4g33200; myosin XI-I), and SALK_063159 (At3g58160; myosin XI-J) were acquired from the Arabidopsis Biological Resource Center (Alonso et al., 2003). Lines GABI_115C01 (At5g43900; myosin XI-2), GABI_622E02 (At1g04600; myosin XI-A), GABI_262B03 (At1g08730; myosin XI-C), and GABI_070F03 (At2g31900; myosin XI-F) were obtained from the European Arabidopsis Stock Center. All lines were selfed and screened for homozygosity by PCR using primers flanking putative insertion sites in combination with the T-DNA specific primers. The PCR products were sequenced to determine the exact positions of the T-DNA insertions some of which are shown in Figure 1. Total RNA was purified from pooled vegetative and reproductive organs of each of the selected homozygous lines using RNeasy Plant Mini Kit (QIAGEN). Semiquantitative RT-PCR with the sets of primes flanking the T-DNA insertion sites was carried out to confirm disruption of the correct mRNA sequences.
Rabbit polyclonal antiserum against synthetic oligopeptide AFSEAEARNSELATELENA-TRKAD corresponding to the amino acid residues 936 to 959 of the deduced sequence of the Arabidopsis myosin XI-K was custom-made by Genemed Synthesis and used for immunoblot analysis at 1:5,000 dilution.
Root Hair Morphology
Seeds were surface sterilized and grown on vertical plates containing 0.5× Murashige and Skoog medium, 5 mm MES, pH 5.8, 1% Suc, and 0.6% Phytogel under a 16-h light/8-h dark cycle. Root hair phenotypes of the 5-d-old seedlings were photographed using a Leica MZ6 stereozoom microscope equipped with a charge-coupled device camera and measured using the Image-Pro (Media Cybernetics) software. At least 200 root hairs from four or more individual plants were measured to determine the mean length for each line shown in Figures 2 and 5.
Arabidopsis Transformation
A Golgi-specific reporter was obtained by fusing rat α-2,6-sialyltransferase (Saint-Jore et al., 2002) with yellow fluorescent protein, while fusion between mCherry and a signal peptide of the pumpkin (Cucurbita mixta) hydroxypyruvate reductase (Mano et al., 2002) was used to tag the peroxisomes. Each of these cDNAs was cloned into binary vector pMDC32 (Curtis and Grossniklaus, 2003). The resulting plasmids were mobilized into Agrobacterium tumefaciens strain GV3101 and used to transform homozygous myosin gene knockout plant lines SAIL_632_D12 and SALK_067972 by floral dipping. Seeds harvested from treated flowers were surface sterilized and grown under 16-h light/8-h dark cycles at 22°C. Plant lines exhibiting hygromycin resistance were selected and grown in soil. The gene knockout/transgenic lines expressing organelle markers were selected using epifluorescent microscopy. At least two, but normally four to six, independent transgenic lines were analyzed for each experimental design. Analogous protocol was used to generate double transgenic lines expressing either myosin XI-K/GTD and Golgi marker or myosin XI-1/GTD and peroxisome marker. Each of the GTD domains was tagged with the triple HA epitope at the N terminus. The borders of GTD domains were from amino acid residue 1,100 to the C terminus for myosin XI-K (predicted molecular mass approximately 54 kD) and from residue 1,082 to the C terminus for myosin XI-1 (predicted molecular mass approximately 53 kD). The expression of GTD in transgenic lines was assayed by immunoblotting using anti-HA monoclonal antibody (Roche) in 1:3,000 dilution. At least three independent transgenic lines were used for analyses (Figs. 2C and 5H).
Organelle Trafficking
For visualization of live mitochondria in root hairs, seeds were germinated and grown for 5 d on the same medium supplemented with 50 nm of Rhodamine 123 (Invitrogen). A series of eight consecutive images was acquired using a 510 Meta (Zeiss) confocal microscope and was used to measure the organelle velocities. Samples were excited using Argon laser at 488 nm, emission signal was collected through a band-pass 505- to 530-nm filter. The Golgi stacks and peroxisomes were observed using the following configurations of excitation and emission filters, respectively: 488 and 508 nm for GFP, 513 and 527 nm for yellow fluorescent protein, and 587 and 610 nm for mCherry. For time-lapse experiments, the consecutive images were taken at 1-s intervals for mitochondria or 2 s for Golgi and peroxisomes. For the root hair and leaf epidermis observations, more than 150 and 300 individual organelles, respectively, were traced. Tracking and measurements of velocities of individual organelles was performed using the Volocity3.7.0 Classification software (Improvision; Image Processing and Vision Company). Statistical analysis of the data was done using t test and Excel software. Additional details of organelle trafficking analyses are provided in an accompanying work (Avisar et al., 2008).
Acknowledgments
We are grateful to Eugene Koonin for critical reading of the manuscript, to Amit Gal-On for kindly providing lab space to D.A., and to Maria Ivanchenko and Rex Cole for their technical help. The authors acknowledge the confocal microscopy facility of the Oregon State University Center for Genome Research and Biocomputing.
This work was supported in part by a grant from the National Institutes of Health (GM053190 to V.V.D.), and by a Vaadia-Binational Agricultural Research and Development Postdoctoral Fellowship (award no. F1–354–2004 to D.A.) from the U.S.-Israel Binational Agricultural Research and Development Fund. The publication was made possible in part by support from the National Institutes of Health (grant no. 1S10RR107903–01).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Valerian V. Dolja (doljav@science.oregonstate.edu).
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV (2008) Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka KJ, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15 441–447 [DOI] [PubMed] [Google Scholar]
- Campanoni P, Sutter JU, Davis CS, Littlejohn GR, Blatt MR (2007) A generalized method for transfecting root epidermis uncovers endosomal dynamics in Arabidopsis root hairs. Plant J 51 322–330 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Dolan L (2002) Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci 357 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9 579–588 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos C, Huet S, Darchen F (2007) ‘Should I stay or should I go?’: myosin V function in organelle trafficking. Biol Cell 99 411–423 [DOI] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D (2006) New insights into myosin evolution and classification. Proc Natl Acad Sci USA 103 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Igarashi H, Mano S, Nishimura M, Shimmen T, Yokota E (2005) Peroxisomal localization of a myosin XI isoform in Arabidopsis thaliana. Plant Cell Physiol 46 782–789 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17 159–187 [DOI] [PubMed] [Google Scholar]
- Holweg C, Nick P (2004) Arabidopsis myosin XI mutant is defective in organelle movement and polar auxin transport. Proc Natl Acad Sci USA 101 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jiang SY, Cai M, Ramachandran S (2007) Oriza sativa myosin XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol 304 579–592 [DOI] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I (2005) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Nebenfuhr A (2007) Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. J Biol Chem 282 20593–20602 [DOI] [PubMed] [Google Scholar]
- Lynch M (2007) The origins of genome architecture. Sinauer Assoc., Inc., Sunderland, MA
- Mano S, Nakamori C, Hayashi M, Kato A, Kondo M, Nishimura M (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol 43 331–341 [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin L (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A, Staehelin LA (2001) Mobile factories: Golgi dynamics in plant cells. Trends Plant Sci 6 160–167 [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Jarve K, Paves H, Truve E (2007) Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230 193–202 [DOI] [PubMed] [Google Scholar]
- Paves H, Truve E (2007) Myosin inhibitors block accumulation movement of chloroplastsin Arabidopsis thaliana leaf cells. Protoplasma 230 165–169 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Day IS (2001) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol 2 RESEARCH0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen D, Hanson MR (2007) Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol 7 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436 1113–1118 [DOI] [PubMed] [Google Scholar]
- Runions J, Brach T, Kuhner S, Hawes C (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57 43–50 [DOI] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29 661–678 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37 379–390 [DOI] [PubMed] [Google Scholar]
- Shimmen T (2007) The sliding theory of cytoplasmic streaming: fifty years of progress. J Plant Res 120 31–43 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21 271–295 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L (2006) Actin dynamics: old friends with new stories. Curr Opin Plant Biol 9 554–562 [DOI] [PubMed] [Google Scholar]
- Taiz L, Ziegler E (2006) Plant Physiology, Ed 4. Sinauer Associates, Sunderland, MA
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112 467–480 [DOI] [PubMed] [Google Scholar]
- Wang Z, Pesacreta TC (2004) A subclass of myosin XI is associated with mitochondria, plastids, and the molecular chaperone subunit TCP-1 in maize. Cell Motil Cytoskeleton 57 218–232 [DOI] [PubMed] [Google Scholar]