Abstract
Plants damaged by insects can synthesize and release volatile chemicals that attract natural enemies of the herbivore. The maize (Zea mays subsp. mays) terpene synthase gene stc1 is part of that indirect defense response, being induced in seedling blades in response to herbivory by beet army worm. Many genes in maize are duplicated because of a past whole-genome duplication event, and several of these orthologs display different expression patterns. We report here the isolation and characterization of tps26 and confirm by homology and synteny criteria that it is the ortholog of stc1. Prior genetic analysis revealed that the stc1 function is not duplicated, raising the interesting question of how the two orthologs have become differentiated in their expression. tps26 encodes a 633-amino acid protein that is highly conserved with STC1. Like stc1, tps26 is induced by wounding, but in the roots and leaf sheath, instead of the blade, and not in response to beet army worm feeding. tps26 maps near a quantitative trait locus for Southwestern corn borer resistance, making it a plausible candidate gene for that quantitative trait locus. However, while possessing highly polymorphic tps26 alleles, the resistant and susceptible parents of the mapping population do not differ in levels of tps26 expression. Moreover, tps26 is not induced specifically by Southwestern corn borer feeding. Therefore, although they share a wounding response, the stc1 and tps26 maize orthologs differ in their tissue specificity and their induction by insect herbivores. The N termini of STC1 and TPS26 are predicted to encode plastid transit peptides; fusion proteins of green fluorescent protein to either N terminus localized to the plastid, confirming that prediction. The mature proteins, but not the respective complete proteins, were active and synthesized a blend of monoterpenes, indicating that they are monoterpene synthases. A gene closely related to stc1/tps26 is found in the sorghum (Sorghum spp.) genome at a location that is not orthologous with stc1. The possible origin of stc1-like genes is discussed.
Plants utilize remarkably sophisticated systems of direct and indirect defense against herbivores (Ryan and Jagendorf, 1995). In addition to direct defense systems, such as feeding deterrents and proteinase inhibitors, plants can employ indirect systems of defense by actively recruiting predators or parasitoids of the foraging herbivore (Turlings et al., 1990). For example, maize (Zea mays) plants being foraged upon by beet army worm (BAW) emit a blend of volatiles that selectively attract parasitoids of the caterpillar. Female parasitic wasps deposit their eggs in the caterpillars, which are eventually devoured by the emerging larvae (Turlings et al., 1993). Terpenoids are major components of the cocktails of volatiles released by plants under herbivore attack (Takabayashi and Dicke, 1996; Halitschke et al., 2001). It has been estimated that, in maize, terpene-mediated indirect defense may be more important than direct defense (Kollner et al., 2004a).
The key enzymes responsible for the biosynthesis of terpenoids are the terpene synthases or cyclases, which are encoded by genes often up-regulated by insect damage (e.g. Bohlmann et al., 1998; Crock et al., 1997). Many terpenoid synthase (tps) genes have been identified in both gymnosperms and angiosperms (Trapp and Croteau, 2001). Whole-plant genome sequencing projects have established that these genes occur in large families in Arabidopsis (Arabidopsis thaliana; Aubourg et al., 2002) and rice (Oryza sativa; Goff et al., 2002). Some tps genes are clustered and organized in tandem repeats of highly similar sequence, indicating that they represent paralogs of recent origin (Aubourg et al., 2002; Sakamoto et al., 2004). In maize, a terpene synthase gene (stc1) inducible by BAW injury was first identified by Shen et al. (2000), and several sesquiterpene synthase genes involved in the indirect defense response of maize against herbivores were subsequently isolated and extensively characterized by Degenhardt and coworkers (Schnee et al., 2002; Kollner et al., 2004b).
Maize is considered an ancestral allotetraploid (Gaut and Doebley, 1997) that arose from the hybridization of its two progenitors at least 4.8 million years ago (MYA; Swigonova et al., 2004). The paleopolyploid origin of maize presents us with the opportunity to investigate how the orthologs of genes involved in the indirect defense response have evolved within the same species. Toward that end, we have undertaken a characterization of tps26, the ortholog of the BAW-inducible stc1 gene located on chromosome 9. The tps26 gene lies in chromosome 6, in a region that is syntenic with a segment of the short arm of chromosome 9 (9S). We find that stc1 and tps26 differ in their tissue specificity; whereas stc1 is expressed in the leaf blade, tps26 is expressed in the leaf sheath and root. Thus, the two orthologs have evolved different patterns of expression since their estimated divergence from a common ancestor 11.9 MYA (Swigonova et al., 2004). The proteins encoded by stc1 and tps26 are highly conserved and carry predicted chloroplast transit peptides (CTP) at their N termini, as does the predicted sorghum (Sorghum spp.) ortholog. We confirm experimentally that these are functional CTPs and that the mature proteins possess predominantly monoterpene synthase activity when expressed in bacteria. tps26 is induced by wounding and maps close to a quantitative trait locus (QTL) for Southwestern corn borer (SWCB) resistance in tropical maize (Khairallah et al., 1998), observations that prompted us to investigate its possible role in this resistance. However, the lack of obvious expression differences between the parental tps26 alleles in the tropical inbred lines and the failure of SWCB damage to specifically induce tps26 do not support a role for tps26 as a candidate gene for this QTL.
RESULTS
Isolation of tps26 and Confirmation of Orthology to stc1
Under high stringency hybridization conditions, an stc1 probe detects a single band in wild-type genotypes but no band in the stc1 deletion sh-bz-x2 (Shen et al., 2000). However, under low stringency, the probe detects a second, weaker band in wild type that is also present in the deletion (data not shown). This observation suggests that there is an stc1 duplicate in the maize genome. To isolate it, PCR primers were designed based on stc1 sequences conserved among various terpene synthase genes.
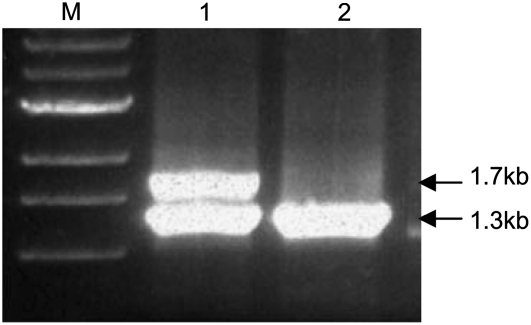
Genomic DNAs from wild-type line McC and the sh-bz-x2 deletion stock were used as templates for PCR amplification. These are near isogenic lines produced by introgression of a short segment of chromosome 9 from different stocks into the common genetic background of the inbred W22. Two PCR products, 1.7 kb and 1.3 kb in size, were amplified in wild type, but only the 1.3-kb fragment was amplified in sh-bz-x2 (Fig. 1). The latter fragment, considered to represent the amplification product of the duplicate gene, was cloned and sequenced. The sequence turned out to be identical to that of the terpene synthase cDNA clone tps26 isolated in one of our labs. Therefore, we designated the PCR clone tps26.
Figure 1.
Identification of tps26 by PCR in wild type and an stc1 deletion mutation. Genomic DNA from line McC (lane 1) and the stc1 deletion sh-bz-x2 (lane 2) was PCR amplified with primers from conserved parts of the stc1 coding region. The 1.3-kb fragment amplified in both lines is derived from the tps26 gene.
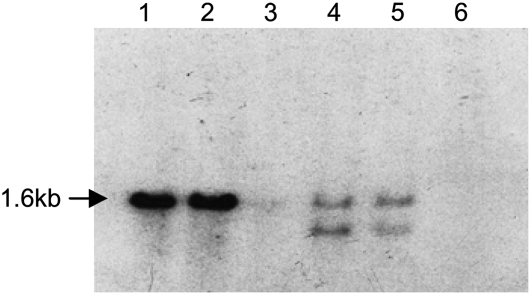
Many chromosomal segments are still duplicated in the maize genome (Gaut and Doebley, 1997). Because part of the long arm of chromosome 6 (6L) is syntenic with part of 9S, where stc1 resides (Helentjaris et al., 1988), tps26 could be located on chromosome 6. To test this, the cloned tps26 PCR fragment was used as probe to screen a series of oat-maize addition lines (OMALs) carrying single maize chromosomes (Ananiev et al., 1997). As can be seen in Figure 2, the tps26 probe detected a single band in lines McC and sh-bz-x2, indicating that there is only one copy of tps26 in W22 and that tps26 and stc1 do not cross hybridize under normal conditions. The probe detected two bands in the chromosome 6 OMAL and in the ‘Seneca 60’ maize parent of the addition lines, probably from a restriction polymorphism in the ‘Seneca 60’ tps26 gene, but no bands in the chromosome 9 OMAL or the oat (Avena sativa) parent. This result establishes that tps26 is located in chromosome 6.
Figure 2.
Mapping of tps26 to chromosome 6 by Southern blots of OMALs. Genomic DNA was digested with SacI and hybridized with the 1.3-kb tps26 PCR clone. Lane 1, McC; lane 2, sh-bz-x2; lane 3, Starter, oat parent of OMAL; lane 4, Seneca, maize parent of OMALs; lane 5, OMAL containing maize chromosome 6; lane 6, OMAL containing maize chromosome 9.
A more precise location of tps26 was determined with a set of recombinant inbred lines derived from the cross T232 × CM37 (Burr et al., 1988). In this population, tps26 mapped between STS markers umc65a and umc21, at a locus 4 cM proximal to pl1 on 6L. stc1 lies immediately adjacent to bz1 (Shen et al., 2000), which is 6 cM proximal to c1, so stc1 maps about 6 cM from c1 in 9S. Because pl1 and c1 are orthologs (Cone et al., 1993) and stc1 maps at about the same distance from c1 as tps26 does from pl1 and in the expected reverse orientation (Helentjaris et al., 1988), stc1 and tps26 lie in syntenic segments of the maize genome. Further evidence that the genes are orthologous comes from an analysis of maize bacterial artificial chromosome (BAC) sequences in the GenBank htgs database. The 6L BAC Z426F06 (AC152917) contains tps26 and a gene encoding a Cyt P450 that is 82% identical to one encoded by a cytP450 gene just proximal to stc1 (Fu et al., 2002). The two genes in the 6L BAC are in the same transcriptional orientation relative to each other, as are their cognates in 9S. Based on their high identity and synteny at the macro and micro levels, we conclude that tps26 is most likely the ortholog of stc1.
tps26 Expression Pattern
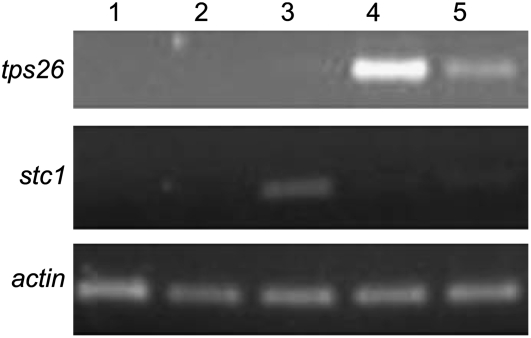
The specificity of tps26 expression was first determined in undamaged tissues of line McC. As can be seen in Figure 3, tps26 is expressed in seedling leaf sheaths and roots but not in the embryo, pollen, or seedling leaf blades, whereas stc1 is expressed exclusively in the leaf blades. Thus, although tps26 resembles stc1 in being expressed at the seedling stage of development, it differs from stc1 in being expressed in the sheaths and roots of seedlings rather than in the blades. This result indicates that the stc1 and tps26 orthologs are not performing redundant functions, as was anticipated from the prior observation that a specific terpenoid is missing in stc1 mutations (Shen et al., 2000).
Figure 3.
Expression of tps26 and stc1 orthologs in different maize tissues. RNA from mature embryos, pollen, and 2-week-old seedlings was analyzed by RT-PCR. Lane 1, Embryo; lane 2, pollen; lane 3, leaf blade; lane 4, root; lane 5, leaf sheath. The amplification of actin (maz95) is used as internal control.
Characterization of the tps26 Gene and Transcript
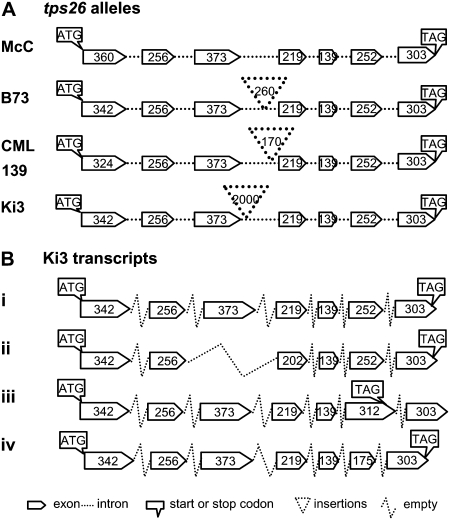
A 4.6-kb fragment containing the complete tps26 allele from McC was isolated by PCR and sequenced. A BLASTN homology search (Altschul et al., 1997) of the GenBank nr database identified maize stc1 alleles as having the highest similarity to the query (74% identity over the entire coding region). Analysis of the sequence by the gene-finding program Fgenesh (www.softberry.com) predicted that, like stc1, tps26 consisted of seven exons. This structure was confirmed experimentally after isolating the seedling tps26-McC cDNA by reverse transcription (RT)-PCR.
The predicted transcriptional start site was identified by 5′ RACE and verified with different primers based on the genomic sequence, and the 3′ end was identified by 3′ RACE. Three randomly selected cDNA clones were sequenced. The tps26-McC transcript has a 51-bp 5′ untranslated leader and a 201-bp 3′ untranslated region with a poly(A) tail 21 bp downstream of an AATAAA signal. As predicted, the tps26 mRNA is produced from the splicing of 6 introns (Fig. 4A), all of which have conserved GT and AG dinucleotides at their respective 5′ and 3′ splice sites. The 1902-bp open reading frame (ORF) in the cDNA would encode a 633-amino acid TPS26 protein.
Figure 4.
tps26 gene structure and sequence polymorphism. Exons are represented as pentagons and introns as dotted lines. The polymorphic insertions in intron 3 of several alleles are indicated as dotted triangles. Numbers inside of pentagons indicate exon sizes in base pairs. A, Gene structure in different maize inbreds: McC, CML139, B73, and Ki3. B, Different tps26 transcripts from inbred Ki3. i, Normal; ii, deletion of exon 3 and part of exon 4; iii, retention of 5′ part of intron 6; iv, deletion of part of exon 6.
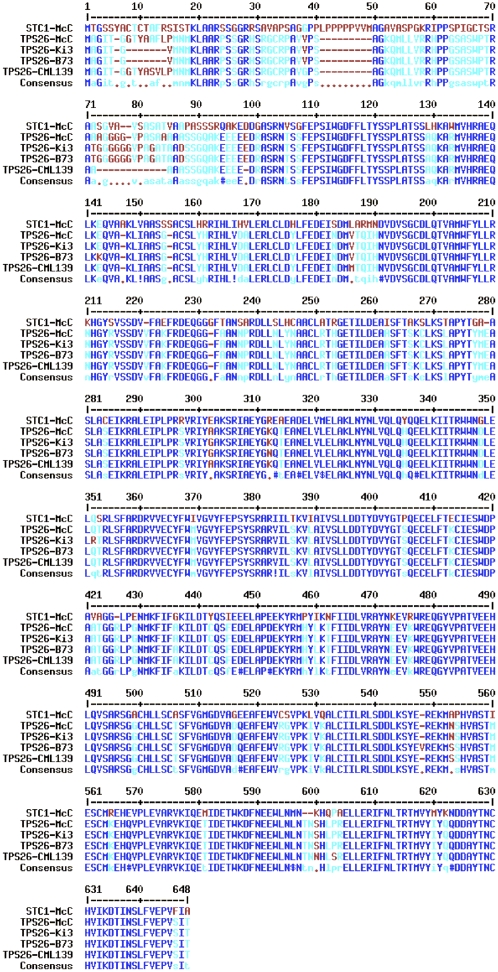
A comparison of the deduced protein sequence to other sequences in the database revealed that TPS26 was most similar to known terpene synthases from maize and other monocots. TPS26 contains the conserved DDxxD motif common to all terpenoid cyclases and shares 75% overall identity with its maize ortholog STC1 (Fig. 5), 42% to 46% identity with well-characterized sesquiterpene synthases from maize (TPS4 and TPS5 [Kollner et al., 2004b]; UMI2 or TPS6 [Basse, 2005]; and TPS10 [Schnee et al., 2006]), and 41% identity with a sesquiterpene synthase from Elaeis oleifera (Shah and Cha, 2000) and germacrene synthase from Zingiber officinale (Picaud et al., 2006). The deduced protein is predicted by the ChloroP program (http://www.cbs.dtu.dk/services/ChloroP/) to include a plastid-targeted transit peptide at the N terminus (Fig. 5). A similar structure was also identified at the STC1 n terminus (Shen et al., 2000), further supporting that the genes are orthologous.
Figure 5.
Alignment of TPS26 with STC1 by MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html). The proteins are highly conserved, except in the predicted plastid transit peptide (top row, 1–70), where substantial differences occur even among proteins encoded by different alleles. Blue denotes amino acid identity among all sequences, cyan denotes identity among three of four sequences, and brown denotes 50% or less identity. Missing amino acids in the N terminus are indicated with dashes.
Structure and Expression of tps26 Alleles in Different Maize Lines
The observation that stc1 and tps26 have largely nonoverlapping expression patterns coupled with the finding that stc1 is induced by BAW herbivory prompted us to investigate whether tps26 might be induced by a different insect pest. Interestingly, comprehensive studies on insect resistance in tropical maize at CIMMYT (Centro Internacional para el Mejoramiento de Maíz Y Trigo, Mexico) had mapped a QTL for SWCB resistance to a location in 6L close to where we placed tps26 (Khairallah et al., 1998), making tps26 a plausible candidate gene for that QTL. To test this possibility, we obtained from CIMMYT the two parental lines of the F2 population used in the mapping and proceeded to characterize the structure and levels of expression of their respective tps26 alleles.
The resistant line is CML139, a CIMMYT line, which, like other genotypes resistant to tropical borers, traces its origin to Caribbean germplasm, and the susceptible line is Ki3, which traces its origin to Suwan 1, a composite population from Thailand (Khairallah et al., 1998). tps26 genomic sequences were isolated from Ki3 and CML139 by PCR using primers based on the B73 and McC sequences. When regular PCR failed because of sequence divergence, inverse PCR was used to isolate the putative promoter regions. Total sequences of 7.1 and 8.6 kb were assembled from CML139 and Ki3, respectively, and aligned with the available genomic sequences of McC (4.6 kb) and B73 (10 kb).
The tps26 alleles show considerable interallelic polymorphisms in the promoter region and introns but high exon similarity (Fig. 4A). The 1.2-kb sequence immediately upstream of the translation start site in Ki3 and CML139 shares only 60% identity as a consequence of flanking retrotransposon polymorphisms (Fu and Dooner, 2002). The differences between the four alleles are most conspicuous in the third intron, the longest of the six. That intron measures 900 bp in McC, the only line that appears to lack an insertion of extraneous DNA. It is longer in the other lines, which carry insertions of different sizes and origins at different locations. A 260-bp sequence from a retroelement gag gene is inserted in the middle of the B73 intron, a 170-bp fragment of the chloroplast gene for ribosomal protein S2 is inserted about 100 bp upstream of exon 4 in CML139, and a 2-kb insertion with non-long terminal repeat (LTR) retrotransposon features is found 50 bp downstream of Ki3's exon 3. This insertion has a 17-bp AT-rich target site duplication and ends with a short poly(A) tail. At the nucleotide level, it has no homolog in GenBank but is predicted to encode a gag-pol polyprotein that is 53% identical to the putative maize protein AF466646. To investigate whether this insertion was unique to the tps26 allele of Ki3, the third tps26 intron of several lines was amplified by PCR and sequenced. The non-LTR retrotransposon was also found in A636 and I137TN, inbreds of unrelated origin, although it was absent in the majority of the lines examined (A188, B37, H99, W22, and W23). The TPS26 proteins encoded by the different alleles vary in length at the N terminus. For example, relative to the consensus sequence, Ki3 has a 7-amino acid deletion at position 8 and CML139 has a 12-amino acid deletion at position 73 (Fig. 5).
The presence of a large insertion in the tps26 third intron of Ki3 and other lines raises questions regarding the functionality of those tps26 alleles because the insertion of retroelements in introns has been shown to affect normal splicing in several wx mutants of maize (Varagona et al., 1992; Marillonnet and Wessler, 1997). To test the possible effect of the insertion on splicing of the tps26 transcript, full-length tps26 cDNAs from Ki3 and CML139 were isolated by RT-PCR and sequenced. Whereas only normal transcripts were present in CML139 (15/15), some abnormally spliced transcripts were found in Ki3 (3/20). The altered spliced products include: deletion of the third exon, deletion of the end of the sixth exon, or retention of part of the sixth intron (Fig. 4B). The exon deletions simply shorten the transcript, while retention of part of the sixth intron would result in premature termination. However, most Ki3 tps26 transcripts were normal, suggesting that the 2-kb insertion does not inactivate the tps26 gene.
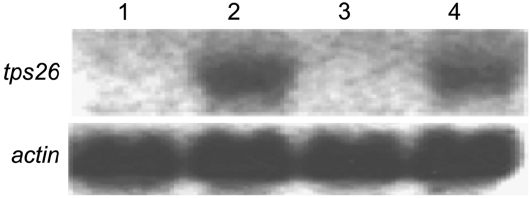
Total tps26 transcript levels in CML139 and Ki3 were compared by northern-blot analysis. RNA samples were collected separately from seedling leaf blades and sheaths of the two inbred lines. As can be seen in Figure 6, tps26 is expressed in sheaths but not blades, in agreement with the previous RT-PCR data, and the level of tps26 expression is similar in both lines, if not slightly higher in Ki3, the SWCB-susceptible line.
Figure 6.
tps26 expression in 2-week-old seedling leaves. Lane 1, Ki3 leaf blade; lane 2, Ki3 leaf sheath; lane 3, CML139 leaf blade; lane 4, CML139 leaf sheath. Twenty micrograms of total RNA was loaded per lane. The membrane was first hybridized to a tps26 probe, then rinsed with 0.1% boiling SDS to remove the tps26 signal and rehybridized to an actin probe.
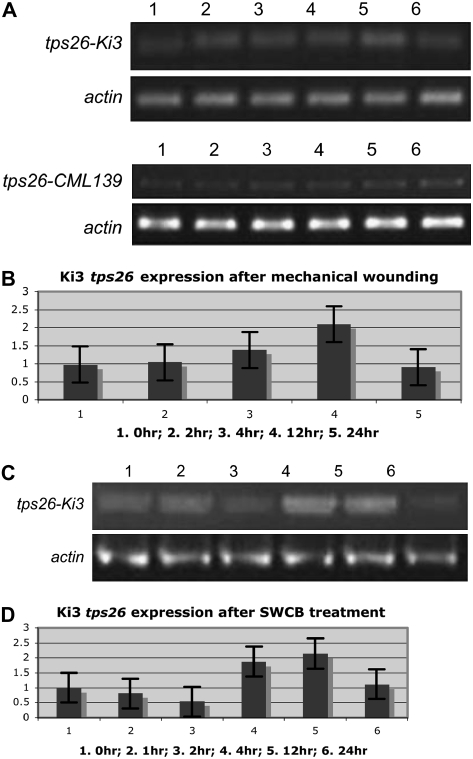
Shen et al. (2000) found that stc1 expression was strongly induced in seedlings wounded mechanically to mimic insect damage. To test whether tps26 was also induced by mechanical wounding, leaf sheaths of CML139 and Ki3 seedlings were punctured with a dissecting needle, RNA was extracted from materials collected at various times after wounding, and induction was quantified by RT-PCR and real-time PCR. As shown in Figure 7A, expression of tps26 was only slightly stimulated by wounding and was higher in Ki3 than CML139 at all times. To determine the extent of induction, expression levels were estimated by quantitative RT-PCR. Figure 7B shows the results for Ki3 RNA samples. A 2-fold maximum stimulation was detected 12 h after wounding, after which time tps26 transcript levels declined. A similar induction pattern was observed for CML139 (data not shown).
Figure 7.
Characterization of tps26 expression. A, RT-PCR of tps26 expression in mechanically wounded leaf sheaths. Sheaths from 2-week-old seedlings were punctured with a dissecting needle at several locations. Materials were collected at the following times after wounding: 1, 0 h; 2, 1 h; 3, 2 h; 4, 4 h; 5, 12 h; 6, 24 h. B, Quantitative PCR of tps26 induction in wounded leaf sheaths. Materials were collected at the following times after mechanical wounding: 1, 0 h; 2, 2 h; 3, 4 h; 4, 12 h; 5, 24 h. C, RT-PCR of tps26 expression in 3-week-old Ki3 sheaths after SWCB feeding. Materials were collected at the following times after feeding initiation: 1, 0 h; 2, 1 h; 3, 2 h; 4, 4 h; 5, 12 h; 6, 24 h. D, Quantitative PCR of tps26 expression in 3-week-old Ki3 sheaths after SWCB feeding. 1 to 6, As in C.
Induction of the stc1 gene by herbivore feeding is stronger and occurs earlier than by mechanical damage alone (Shen et al., 2000). To determine if the tps26 ortholog was induced specifically by SWCB, CML139 and Ki3 seedlings were exposed to foraging by SWCB larvae (see “Materials and Methods” for details). After being placed on the leaf whorl of 3-week-old seedlings, the larvae caused immediate and extensive damage to the youngest leaf in the center of the whorl, burrowed into the sheath, and either emerged from the damaged seedling by boring a hole in the sheath or continued burrowing into the roots. The damaged leaf blades and sheaths were collected separately at various times after treatment and tps26 induction was tested by RT-PCR and quantitative PCR. The results for Ki3 seedlings are shown in Figure 7, C and D. As with mechanical wounding, tps26 induction reached a 2-fold maximum 12 h after treatment. No increase over mechanical wounding was observed in CML139 seedlings either (data not shown), indicating that the tps26 gene is not induced specifically by SWCB feeding and is, therefore, unlikely to play a role in the different susceptibilities to SWCB of the two inbred lines.
Subcellular Localization of TPS26 and STC1
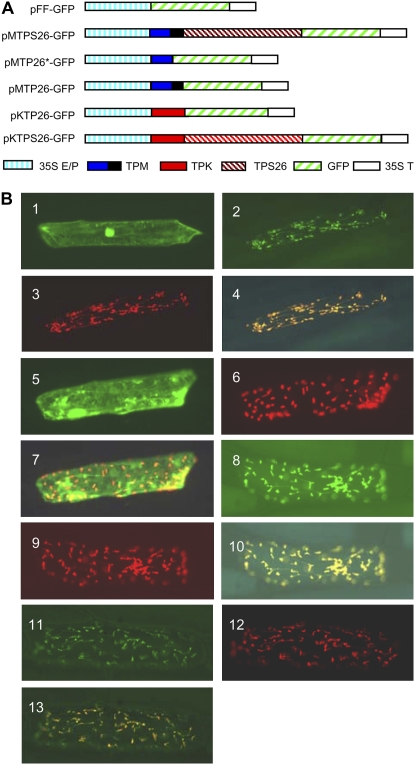
Both STC1 and TPS26 have predicted CTPs at their N termini. To test the plastid localization properties of these sequences, the putative transit peptides and full-length proteins of STC1 and TPS26 were fused C terminally to GFP and transformed into onion (Allium cepa) epidermal cells by particle bombardment. A fusion protein of acyl carrier protein (ACP), a soluble protein found in the plastid stroma, and DsRed was cotransformed as a positive control (Schnurr et al., 2002). Representative results are shown in Figure 8.
Figure 8.
Subcellular localization of TPS26. A, Schematic diagrams of the various GFP fusion constructs. 35S E/P, Cauliflower mosaic virus 35S enhancer-promoter; TPM, TPS26-McC transit peptide; TPK, TPS26-Ki3 transit peptide; TPS26, TPS26 protein; 35S T, cauliflower mosaic virus 35S terminator. Predicted transit peptides and full-length proteins were fused to the N terminus of GFP. B, Fluorescence visualization. Each of the constructs was cotransformed with ACP-DsRed into onion epidermal cells by particle bombardment. 1, pFF-GFP construct. 2 to 4, pMTPS26-GFP (fusion of the entire TPS26-McC protein to GFP) and ACP-DsRed; 2, GFP signal; 3, DsRed signal from same epidermal cell as 2; 4, merged images of 2 and 3. 5 to 7, pMTP26*-GFP (fusion of a 37-amino acid, nonfunctional transit peptide from TPS26-McC to GFP) and ACP-DsRed; 5, GFP signal; 6, DsRed signal; 7, merged images of 5 and 6. 8 to 10, pMTP26-GFP (fusion of a 60-amino acid, functional transit peptide from TPS26-McC to GFP) and ACP-DsRed; 8, GFP signal; 9, DsRed signal; 10, merged images of 8 and 9. 11 to 13, pKTP26-GFP (fusion of a 53-amino acid, functional transit peptide from TPS26-Ki3 to GFP) and ACP-DsRed; 11, GFP signal; 12, DsRed signal; 13, merged images of 11 and 12.
In control experiments with GFP alone (pFF-GFP), GFP diffused into the nucleus, as is commonly observed with this small protein (Fig. 8B, 1). A fusion of the entire TPS26-McC protein to GFP (pMTPS26-GFP) localized to the plastid (Fig. 8B, 2–4). Another one containing a predicted 37-amino acid transit peptide from TPS26-McC (pMTP2*-GFP) did not (Fig. 8B, 5–7), but extending the N-terminal sequence to 60 amino acids, the size of the predicted transit peptides in CML139 and Ki3, led to plastid localization of the fusion construct (pMTP2-GFP; Fig. 8B, 8–10). Because the TPS26 protein in Ki3 has a 7-amino acid deletion in the predicted transit peptide, it became of interest to test if the deletion affected the delivery of the protein into the plastid. GFP fusions containing either the predicted transit peptide (pKTP2-GFP) or the full-length protein of TPS26-Ki3 (pKTPS26-GFP) were constructed and transformed as described above. Both fusion proteins localized to the plastid (Fig. 8B, 11–13; data not shown), indicating that deletion of seven amino acids in the transit peptide does not affect the plastid localization of the TPS26 protein. GFP fusion proteins containing either the predicted transit peptide or the entire protein of STC1-McC also localized to the plastid (data not shown). Thus, as predicted from their sequence, the proteins encoded by both orthologs are plastid localized.
Heterologous Expression and Functional Characterization of TPS26 and STC1
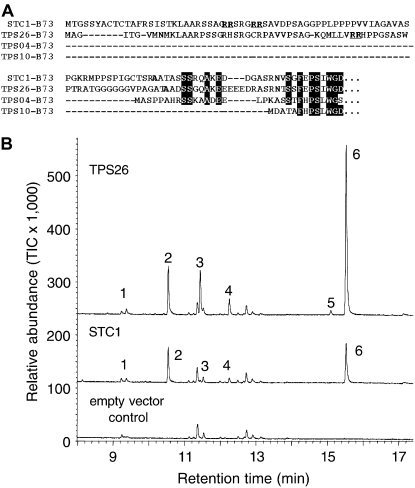
To identify the catalytic activity of the terpene synthases STC1 and TPS26, we cloned N-terminal truncations of the ORFs (Fig. 9A) into the expression vector pASK-IBA7 and expressed them in Escherichia coli. The partially purified recombinant enzymes were incubated with the potential substrates geranyl pyrophosphate (GPP) and farnesyl pyrophosphate (FPP). No enzymatic activity was observed with FPP, but both proteins were able to convert GPP into similar blends of monoterpene products (Fig. 9B). The major compound produced by both enzymes was identified as the oxygenated monoterpene α-terpineol. The minor monoterpene products of STC1 and TPS26 were the cyclic monoterpenes limonene, γ-terpinene, and terpinolene as well as the acyclic monoterpene myrcene. The absolute configuration of the limonene and α-terpineol products of STC1 and TPS26 by gas chromatography (GC) on a chiral column revealed that the (−)-enantiomer was formed almost exclusively in both cases (Supplemental Fig. S1).
Figure 9.
Functional characterization of TPS26 and STC1. A, Amino acid sequences of the N-terminal regions of maize terpene synthases. The N-terminal regions of STC1 and TPS26 were aligned with those of the sesquiterpene synthases TPS4 and TPS10. Amino acids identical to STC1 and TPS26 are marked by black boxes. The RR motifs in the transit peptides of STC1 and TPS26 are underlined and highlighted in bold. For overexpression in a bacterial system, the N-terminal ends of STC1 and TPS26 were truncated at the Ala residue highlighted in bold and cloned into the expression vector with an N-terminal strep-tag (not shown). B, Monoterpene products of TPS26 and STC1. The enzymes were expressed in E. coli, extracted, and incubated with the substrate GPP. The resulting terpene products were separated by GC-MS. The traces of the MS detector are shown for the active enzymes STC1 and TPS26 and for the products formed by an extract of E. coli transformed with the expression vector lacking a terpene synthase gene. The products were identified as β-myrcene (1), limonene (2), γ-terpinene (3), terpinolene (4), 4-terpineol (5), and α-terpineol (6) by comparison of their retention times and mass spectra to those of authentic standards.
To determine if major differences in monoterpene emission could be detected between wild-type and stc1 mutant seedlings, we re-examined the volatiles produced by 10-d-old seedlings under caterpillar feeding. We analyzed the emission of volatile terpenes from the lines Stc1-McC and of three isogenic lines carrying a transposon footprint within the stc1 coding region, which causes a frameshift and inactivates STC1 (Supplemental Fig. S2). Both the wild-type and frameshift mutation lines produced the herbivore-induced monoterpene alcohol linalool as well as the sesquiterpenes (E)-α-bergamotene and (E)-β-farnesene that are products of the terpene synthase TPS10 (Schnee et al., 2006). The main product of STC1, α-terpineol, and its minor products γ-terpinene and terpinolene are not emitted by either the wild type or frameshift mutant. Only traces of the minor products β-mycrene and limonene were detected but are unlikely to be formed by STC1 because they do not represent the monoterpene ratios found in vitro and are produced by all frameshift mutants. The absence of both olefinic and hydroxylated products in the volatile profile suggests that these products are not missing due to glucosylation in planta. Therefore, neither STC1 nor TPS26 appears to be active after induction by Spodoptera littoralis in the W22 background.
Possible Evolutionary Origin of stc1-Like Genes
STC1 and TPS26 carry a CTP at the N terminus, whereas none of their closest relatives, the rice predicted proteins EAZ22931 and EAY85744 or the maize sesquiterpene synthases TPS4, TPS5, and TPS10 (Kollner et al., 2004b), does. This observation suggests that stc1 and tps26 may represent a unique type of terpenoid synthase gene. To investigate whether this class of genes is found in grasses more closely related to maize than rice, we carried out a BLASTX search of the sorghum sequence database (http://www.phytozome.net) using the stc1 gene as query. The analysis uncovered a sorghum gene that encodes a protein highly identical (64%–66%) to STC1 and TPS26 (Supplemental Fig. S3) and with a predicted CTP of 62 amino acids at its N terminus. When tested against all GenBank sequences, the sorghum homolog formed a monophyletic clade with the maize STC1 and TPS26 proteins (Supplemental Fig. S4), suggesting that this unique class of tps gene arose in the maize-sorghum lineage after the divergence of their common ancestor from the rice progenitor.
DISCUSSION
We have isolated and characterized the maize tps26 gene, a close homolog of the stc1 gene that is induced by BAW herbivory and the elicitor volicitin (Shen et al., 2000). The two genes are very closely related, their predicted protein products sharing 75% overall amino acid identity. Maize is an ancestral allotetraploid (Gaut and Doebley, 1997) that is now approaching a diploid state by extensive gene loss (Ilic et al., 2003; Lai et al., 2004). Thus, the ortholog of any given maize gene may or may not have been retained today. For example, the adh1 region now contains only one duplicated pair of orthologs compared with the eight pairs that were present in the original hybrid progenitor (Ilic et al., 2003). Because of this selective retention of orthologs, it is not always possible to establish orthologous relationships between gene pairs in maize. We show here that, by macrosynteny (genetic maps) and microsynteny (BAC contigs) criteria, tps26 is the bona fide ortholog of stc1. tps26 maps to 6L, to a part of the long arm that is syntenic over several centimorgans with the part of 9S where stc1 resides (Helentjaris et al., 1988). In addition, the tps26 BAC also carries a cytP450 gene highly similar to one located just proximal to stc1 in the 9S BAC contig (Fu et al., 2002) and in the same transcriptional orientation relative to the stc1 gene. Thus, tps26 provides us with the opportunity to examine how orthologs of genes involved in indirect defense response may have evolved within the same species.
Like stc1, tps26 is expressed at the seedling stage of development. However, whereas stc1 is expressed in the leaf blade, tps26 is expressed in the leaf sheath and root, indicating that the genes are not performing redundant functions, as was already expected from the altered phenotype of the single stc1 mutants (Shen et al., 2000). Both stc1 and tps26 are induced by wounding, but the level of induction is much greater for stc1 than tps26 (30 times versus 2 times). Likewise, stc1 is induced by BAW foraging, whereas tps26 is not (Shen, 2001). Because of the close relationship between stc1 and tps26, we considered the possibility that tps26 was induced by a different insect pest. We focused on SWCB because a QTL for SWCB resistance in tropical maize had been mapped in the vicinity of the tps26 location in 6L (Khairallah et al., 1998). However, SWCB foraging failed to induce tps26 above the level induced by wounding alone. A comparison of the tps26 gene structure in several lines revealed the presence of a 2-kb non-LTR retrotransposon in the third intron of Ki3, the SWCB-susceptible line, but even though some Ki3 transcripts (approximately 15%) are abnormally spliced, most are normal, so the large insertion does not inactivate the tps26 gene. Lastly, the levels of tps26 expression did not differ in Ki3 and CML139, the two tropical inbred parents of the population used in the analysis of SWCB resistance. These data argue against the case that tps26 is a candidate gene for the mapped QTL. Instead, it may play a role in defense against subterranean herbivores or pathogens because its highest expression is in the root. The first case of a below-ground terpene defense signal produced by plant roots in response to herbivore attack was recently reported in maize (Rasmann et al., 2005).
Both STC1 and TPS26 proteins contain a predicted plastid transit peptide at their N terminus. The ability of the predicted CTPs from STC1 and TPS26 to transport proteins to the chloroplasts was confirmed experimentally by particle bombardment of the respective GFP fusions into onion epidermal cells. STC1 was deemed originally to be a sesquiterpene synthase on the basis of the chemical phenotype of stc1 mutants (Shen et al., 2000) and not on the basis of its in vitro product. The presence of a CTP was considered unusual because sesquiterpenes are synthesized in the cytosol via the mevalonic acid pathway, the plastids being the site of synthesis of monoterpenes and diterpenes via the methyl erythritol phosphate or mevalonic acid-independent pathway (Lange and Croteau, 1999). However, natural and engineered exceptions to this compartmentation do occur. The monoterpene synthase FvPINS from wild strawberry lacks a CTP and generates monoterpenes as sole products, evidence that monoterpene synthesis can occur in the cytosol (Aharoni et al., 2004). Conversely, by targeting a sesquiterpene synthase to the plastid, Wu et al. (2006) demonstrated synthesis of novel sesquiterpenes in plastids of transgenic tobacco. Terpene synthases capable of synthesizing sesquiterpenes and monoterpenes with different relative efficiencies have been reported in several plants (Crock et al., 1997; Colby et al., 1998; Aharoni et al., 2004; Kollner et al., 2004b). To determine the biochemical function of STC1 and TPS26, the mature proteins (i.e. proteins without the CTPs) were expressed in a bacterial system, purified, and tested for enzymatic activity with the substrates GPP and FPP. Both enzymes showed monoterpene synthase activity exclusively, producing six compounds, with α-terpineol and limonene as main products. The stereochemistry of the main products was determined on a chiral column.
Upon finding that STC1 has monoterpene synthase activity, we re-examined the chemical phenotype of caterpillar-foraged wild-type and stc1 mutant seedlings to determine if major differences in monoterpene emissions could be detected. However, while sesquiterpenes were present in the volatile collection, only traces of monoterpenes could be detected in either line (Supplemental Fig. S2). This could be a property of the W22 inbred background of the two genotypes, as other maize cultivars do produce mixtures of monoterpenes and sesquiterpenes in response to insect damage (Turlings et al., 1990). Thus, although the stc1 transcript is induced by herbivory and the recombinant enzyme displays monoterpene synthase activity, the role of this gene in plant defense remains conjectural. A possible explanation for the difference in sesquiterpene emissions between wild-type and stc1 mutant seedlings reported earlier (Shen et al., 2000) is that sesquiterpenes, the major compounds produced by maize seedlings under insect attack, are known to vary extensively in abundance with developmental stage (Kollner et al., 2004b), and the 10-d-old seedlings used in the earlier study may not have been at the same stage of development when harvested for the induction assay.
STC1 and TPS26 form a well-supported monophyletic clade with the maize sesquiterpene synthases TPS4, TPS5, and TPS10 (Schnee et al., 2002; Kollner et al., 2004b; Supplemental Fig. S4), suggesting that STC1 and TPS26 may represent an unusual type of terpene synthase not found in the few plant species whose genomes have been completely sequenced. To investigate if species closely related to maize have stc1-like genes, we conducted a search of the sorghum sequence database (http://www.phytozome.net) using either maize STC1 or TPS26 as query. This search revealed that sorghum carries a tps gene that is more closely related to the stc1 and tps26 genes of maize than to any other tps gene and that it, too, encodes a protein with a predicted CTP at its N terminus. Sorghum belongs to the Andropogoneae, the same tribe of the grass family as maize, and is a much closer relative of maize than rice is, having diverged from maize 11.9 MYA rather than 50 MYA. Thus, an stc1-like gene may have arisen in the common progenitor of maize and sorghum after its separation from the rice progenitor. It is interesting to note that the sorghum gene does not reside in a chromosomal region that is syntenic with either 6L or 9S in maize. Although the bz regions of maize and sorghum are largely colinear, sharing seven of eight genes, the stc1-like gene is the only one missing from the bz regions of Sorghum bicolor and Sorghum propinquum (Wang and Dooner, 2007), suggesting that it moved to its present location in either sorghum or maize after the sorghum-maize split 11.9 MYA (Swigonova et al., 2004). Similarly, the oat AsbAS1 gene, which encodes a triterpene synthase involved in the production of the antimicrobial avenacin, is clustered with other genes required for avenacin biosynthesis in a region of the genome not conserved in other cereals (Qi et al., 2004). This finding led the authors to suggest that such genes may have arisen by shuffling and accelerated evolution of existing genetic components. Intriguingly, a gene fragment encoding part of exon 4 of STC1/TPS26 is present in both S. bicolor and S. propinquum at the same position and orientation as the stc1 gene in maize 9S. A high fraction of the genes in the genome (1/7) have moved as single genes to new chromosomal locations in maize and sorghum in the 50 million years since the maize/sorghum-rice split (Lai et al., 2004). Apparently, the stc1/tps26 precursor in maize and sorghum was one such itinerant gene and, in its movement from one chromosomal location to another, may have fortuitously captured a CTP that redirected the protein to the plastid. Analysis of the structure and organization of stc1-like genes in other Andropogoneae, like Coix, may be informative. Regardless of their origin, it is clear that these maize orthologs have different expression profiles and are induced by different agents. Because this type of neofunctionalization after duplication is advantageous to the host plant (Lynch et al., 2001), one can similarly expect other retained maize orthologs involved in defense responses to have evolved unique tissue-specificity and/or inducibility of expression.
MATERIALS AND METHODS
Plant Genetic Stocks and Insect Stocks
The stc1-McC allele is carried in a fragment of chromosome 9 from a New England Flint stock that was introduced into the W22 inbred by repeated back-crossing. This subline of W22, identified as McC, carries, as expected, the tps26-W22 allele at the unlinked tps26 locus (formerly named stc2; Shen et al., 2000). sh-bz-x2 (Mottinger, 1970) is an x-ray-induced deletion of a 9S chromosomal segment that includes sh, bz, and other genes, such as stc1, located between them (Shen et al., 2000). This deletion was also introgressed into W22. Ki3 and CML139 are tropical inbreds kindly provided by David Hoisington and used by CIMMYT researchers as the parents of a mapping population for insect resistance QTLs (Khairallah et al., 1998). The OMALs were obtained from Ron Phillips and Howard Rines (Ananiev et al., 1997). The maize recombinant inbreds used to map tps26 were developed and provided by Ben Burr (Burr et al., 1988). Larvae of the SWCB Diatrea grandiosella Dyar. were obtained from Paul Williams at the U.S. Department of Agriculture-Agricultural Research Service Corn Host Plant Resistance Research Unit, Mississippi State University. Eggs of Spodoptera littoralis Boisd. were obtained from Aventis (Frankfurt).
Nucleic Acid Extraction and Hybridization
Genomic DNAs from seedlings and mature leaves were prepared by a urea-based method (Greene et al., 1994). A total of 10 to 15 μg genomic DNA was digested with various restriction enzymes (New England Biolabs), resolved on a 0.8% agarose gel in Tris acetate EDTA buffer, and transferred to a Hybond-N+ membrane according to the manufacturer's protocol (Amersham Biosciences). Radioactive probes were prepared with a Ready-to-go labeling kit (Amersham Biosciences). Membrane hybridization and washing conditions followed the recommendations of the manufacturer. Total RNA from different tissues previously frozen in liquid nitrogen was extracted by the Trizol method (Invitrogen). Separation of total RNA in agarose gels and blotting onto Hybond-N+ membranes were performed according to the manufacturer's instructions. Probe labeling and hybridization were as above.
PCR and Sequencing
PCR conditions followed an optimized protocol for maize (Zea mays) genomic DNA in the Dooner lab (Dooner and Martinez-Ferez, 1997). Genomic DNA was amplified with AmpliTaq (Perkin Elmer). PCR products were cloned into pGEM-T and pGEM-T Easy vectors (Promega) and transformed into DH5α competent cells. Plasmids were purified with Qiagen spin miniprep kit. For iPCR, 10 μg genomic DNA was completely digested by several restriction enzymes based on the known 5′-terminal sequences (500–1,000 bp). The digested DNA was extracted by phenol-chloroform, and 0.5 μg DNA were self-ligated in a total volume of 100 μL (4°C, overnight). Primer pairs residing in the 5′-terminal sequences were used to amplify the unknown region, and the amplification product was ligated to pGEM-T or pGEM-T easy vector for sequencing. The product overlapping with the known 5′-terminal sequence (more than 300 bp) was used for the assembly of genomic sequence. DNA sequencing was carried out in an ABI 3700 DNA Analyzer (Perkin Elmer) following the manufacturer's instructions. The following primer pairs were used to amplify the tps26 fragment used as probe: tps26F, ATGGAACCTCACAGGAGTGTGAGC (in exon 4); tps26R, CTGAGACCTGTAAGTGTTCCTCG (in exon 6). The length of the cDNA fragment is 279 bp and the length of the corresponding genomic region is 1.3 kb. All nucleotide sequences from this work have been deposited in GenBank under accession numbers EF-599327 to EF-599333.
cDNA Cloning, RACE, RT-PCR, Semiquantitative RT-PCR, and Quantitative PCR
In the artificial wounding experiments, RNA was extracted from 2-week-old plants growing in a growth chamber set at 25°C and a 16-h-light/8-h-dark cycle. In the SWCB feeding experiments, RNA was extracted from 3-week-old plants. Plants were dissected into roots, leaf blades, and sheaths for RNA extraction. Total RNA was used as a template for the downstream performance. cDNA was synthesized by the SuperScript first-strand synthesis system following essentially the instructions from Invitrogen. RACE analysis followed the manufacturer's instructions (Clontech). RT-PCR and semiquantitative RT-PCR were performed by FastStart High Fidelity PCR System from Hoffmann-La Roche. Quantitative PCR was carried out with iQ SYBR Supermix from Bio-Rad at the Rotergene 3000 (Corbett Research). PCR primers used in this study were as follows: for tps26 quantitative and semiquantitative RT-PCR, tps26realF1, CAGGTCTCAGCAAGAAGTGGT and tps26realR1, CATTCCAACAAACGATGTGC. For stc1 quantitative and semiquantitative RT-PCR, RTBstc1F, TTAGCGCCGGAAGAGAAATA and RTBstc1R, ACCTGCAACATCTCCCATTC. For the control of maize95 actin quantitative and semiquantitative RT-PCR, RTmaz95F, GAAGCACCTCTGAACCCAAA and RTmaz95R, GGCAGTCAGTCAGATCACGA. For the full-length cDNA of tps26, McCtps26-rtF1, AGAAGCCAAGAACGACCTCGCTGG and McCtps26-rtR1, AAGGATAGGGTTACCACGCCCAACA.
GFP and DsRed Fusion Protein Constructs
A red-shifted variant of wild-type GFP (Clontech) in plant expression vector pFF-GFP (kindly provided by Gregorio Segal) was used in this experiment. Using the tps26/stc1 cDNA clone as template, the DNA fragment of the full cDNA coding sequence and the putative transient peptides (TP) sequence were amplified with the Pfu enzyme (Stratagene), a high fidelity DNA polymerase. The PCR primers used in the amplification were: Tps26KpnF, ACAGGTACCTCGCTGGAAAATGGCAGG; TPtps26AgeR, TCTACCGGTTGCTTGCCGGCAGAGGGGACAA (for incomplete TP); TPTps26AgeR2, ATTACCGGTGCACTAGCTGGGACGC (for complete TP); tps26AgeR, GAGACCGGTGCTGTTGAAACAGGTCCAC (for full-length cDNA); Stc1KpnF, ACAGGTACCCCCCAAGCACACTGGCAAATGAC; TPstc1AgeR, TGAACCGGTGCCATGACAACTGGTGGTGG (for incomplete TP); TPstc1Age-R2, TACCGGTCTAGCAGAGACGGCGAC (for complete TP); and stc1AgeR, GAGACCGGTATGAAAACAGGCTCCACAAACAACGAG (for full-length cDNA). The PCR products were ligated to pGEM-T vector for sequencing to verify the sequence and then digested with restriction enzymes KpnI and AgeI. Gel-purified fragments were ligated in frame at the 5′ end of the GFP gene. The DsRed construct (Schnurr et al., 2002) was graciously supplied by Dr. Gert-Jan de Boer from the University of Amsterdam.
Particle Bombardment and GFP Transient Expression
Gold particle preparation and bombardment procedures followed the Iowa State University Plant Transformation Facility protocol (Register et al., 1994). Onion (Allium cepa) skins were cut into appropriate sizes and placed on supporting plates made with 0.8% agarose. In each shooting, 0.5 μg of plasmid was used. Plant samples were left at room temperature without light for 16 h, and then the epithelial layer was removed and mounted onto a slide with phosphate-buffered saline buffer for fluorescence microscopy observation (Ma and Dooner, 2004).
Protein Overexpression and Enzyme Assay
For expression in Escherichia coli, the ORFs of stc1 and tps26 were amplified with the primer pairs R26fwd (ATGGTAGGTCTCAGCGCATGGCCGCCACCGCTAGTAGT), R27rev (ATGGTAGGTCTCATATCAGGCTATGGAAACAGGCTCCACA) and BE12fwd (ATGGTAGGTCTCAGCGCATGGCCGCAGATTCTAGCGGG), BE9rev (ATGGTAGGTCTCATATCATGTTATTGAAACAGGCTCCACAAAC), respectively, from a cDNA made from herbivore-damaged B73 plants (Kollner et al., 2004b). The fragments were digested with BsaI and cloned into the expression vector pASK IBA7 with an N-terminal strep-tag (IBA). The constructs were introduced into the E. coli strain TOP10 (Invitrogen) and fully sequenced to avoid errors possibly introduced by DNA amplification. Liquid cultures of the bacteria harboring the expression constructs were grown at 37°C to an OD600 of 0.6. Expression was induced by addition of anhydrotetracycline (IBA) to a final concentration of 200 μg/L. After 20-h incubation at 18°C, the cells were collected by centrifugation and disrupted by a 4- × 30-s treatment with a sonicator (Bandelin UW2070) in chilled extraction buffer (50 mm Mopso, pH 7.0, with 5 mm MgCl2, 5 mm sodium ascorbate, 0.5 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, and 10% [v/v] glycerol). The cell fragments were removed by centrifugation at 14,000g and the supernatant was desalted into assay buffer (10 mm Mopso, pH 7.0, 1 mm dithiothreitol, 10% [v/v] glycerol) by passage through a Econopac 10DG column (Bio-Rad).
To determine the catalytic activity of the recombinant proteins, enzyme assays containing 20 μL of the bacterial extract and 80 μL of assay buffer with 10 μm substrate [GPP and (E,E)-FPP, respectively], 10 mm MgCl2, 0.2 mm NaWO4, and 0.1 mm NaF in a Teflon-sealed, screw-capped 1-mL GC glass vial were performed. The assays were overlaid with 100 μL of hexane and incubated for 1 h at 30°C. To extract the enzyme products, the assays were mixed for 60 s. The organic phase was then removed and analyzed by GC-mass spectrometry (MS). For chiral analysis of enzyme products, a 100-μL assay was performed as described above but without hexane overlay. A solid phase microextraction fiber consisting of 100 μm polydimethylsiloxane (Supelco) was placed into the headspace of the vial for 60 min incubation at 30°C. For analysis of the adsorbed reaction products, the solid phase microextraction fiber was directly inserted into the injector of the gas chromatograph.
GC
A Hewlett-Packard model 6890 gas chromatograph was employed with the carrier gas He at 1 mL min−1, splitless injection (injector temperature, 220°C; injection volume, 2 μL), a Chrompack CP-SIL-5 CB-MS column [(5%-phenyl)-methylpolysiloxane, 25-m × 0.25-mm i.d. × 0.25-μ film thickness; Varian], and a temperature program from 40°C (3-min hold) at 5°C min−1 to 240°C (3-min hold). The coupled mass spectrometer was a Hewlett-Packard model 5973 with a quadrupole mass selective detector with transfer line temperature, 230°C; source temperature, 230°C; quadrupole temperature, 150°C; ionization potential, 70 eV; and a scan range of 40 to 350 atomic mass units. The enantiomers of limonene and α-terpineol were separated and identified by GC-MS using a Hydrodex-β-3P column (2,6-di-O-methyl-3-O-pentyl, 25-m × 0.25-mm i.d.; Macherey-Nagel) and a temperature program from 45°C (1-min hold) at 100°C min−1 to 65°C further on at 1°C min−1 to 120°C. Products were identified by comparison of retention times and mass spectra with authentic reference compounds obtained from Fluka or Sigma.
Plant Volatile Collection and Analysis
Plants were grown in commercially available potting soil in a climate-controlled chamber with a 16-h photoperiod, 1 mmol (m2)−1 s−1 of photosynthetically active radiation, a temperature cycle of 22°C/18°C (day/night), and 65% relative humidity. Eggs of S. littoralis were reared on an artificial wheat germ diet (Heliothis mix, Stonefly Industries) for about 10 to 15 d at 22°C under an illumination of 750 μmol (m2)−1 s−1. For the herbivory treatments, three third-instar larvae were enclosed on the middle portion of each plant in a cage made out of two halves of a petri dish (9-cm diameter) with a circle cut out of each side and covered with gauze to allow for ventilation (Degenhardt and Gershenzon, 2000). An automated collection system (Analytical Research Systems) based on the design of Heath and Manukian (1994) was employed to analyze plant headspace volatiles. In brief, the aerial portion of a 10-d-old maize plant was placed in a large glass cylinder (50 cm × 20 cm) whose base was fitted with two adjustable blades that closed loosely around the stem. Air that had been passed through a charcoal-infused medium for purification and moistened to a relative humidity of 65% entered the chamber from above at a rate of 5 L min−1. After sweeping over the plant, the air exited the chamber through one of a series of collection traps, which were 150-mm × 5-mm diameter glass tubes containing 75 mg Super Q (80/100 mesh; Alltech) arrayed around the base of the chamber. Air was drawn through the traps at a rate of 1 L min−1 by an automated flow controller that could switch from one trap to the next at a designated time. The remaining air escaped through the opening around the stem, providing a positive pressure barrier against the entrance of ambient air. The entire volatile collection system was contained in a controlled environment chamber (Voetsch VB1014/S) set at 25°C and 75% relative humidity with a 16-h photoperiod and 750 μmol (m2)−1 s−1 of photosynthetically active radiation. All collections were performed between 9 am and 3 pm to avoid differences due to diurnal rhythms. After the 6-h collection period, the trap was rinsed with 0.2 mL dichloromethane and the sample was analyzed by GC-MS.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF-599327 to EF-599333.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Stereochemical analysis of limonene and α-terpineol produced by the enzymes STC1 and TPS26.
Supplemental Figure S2. Volatiles released by wild type (Stc1-McC) and stc1-s1.2 after herbivore attack.
Supplemental Figure S3. Sequences of maize STC1 and TPS26 and sorghum TPS were aligned by Clustal W.
Supplemental Figure S4. Phylogenetic tree of sesquiterpene synthases.
Supplementary Material
Acknowledgments
We thank Dr. Paul Williams and Dr. Frank Davis from the U.S. Department of Agriculture-Agricultural Research Service Corn Host Plant Resistance Research Unit at Mississippi State University for supplying us with SWCB and for advice on how to handle it, Dr. Gert-Jan de Boer for the DsRed construct, Dr. Gregorio Segal for the pFF-GFP construct, Janet McKim for assistance with fluorescent microscopy, and members of the Dooner and Degenhardt labs for helpful comments on the manuscript.
This work was supported by the National Science Foundation (grant no. IBN–02–35021 to H.K.D.), by Rutgers University (Busch-Waksman predoctoral fellowships to B.S. and Z.X.), by the German National Science Foundation (grant no. DE–837/2–2 to J.D. and T.G.K.), and by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hugo K. Dooner (dooner@waksman.rutgers.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev EV, Riera-Lizarazu O, Rines HW, Phillips RL (1997) Oat-maize chromosome addition lines: a new system for mapping the genome. Proc Natl Acad Sci USA 94 3524–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267 730–745 [DOI] [PubMed] [Google Scholar]
- Basse CW (2005) Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol 138 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann J, Crock J, Jetter R, Croteau R (1998) Terpenoid-based defenses in conifers: cDNA cloning, characterization, and functional expression of wound-inducible (E)-alpha-bisabolene synthase from grand fir (Abies grandis). Proc Natl Acad Sci USA 95 6756–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Burr FA, Thompson KH, Albertson MC, Stuber CW (1988) Gene mapping with recombinant inbreds in maize. Genetics 118 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R (1998) Germacrene C synthase from Lycopersicon esculentum cv. VFNT Cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc Natl Acad Sci USA 95 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crock J, Wildung M, Croteau R (1997) Isolation and bacterial expression of a sesquiterpene synthase cDNA clone from peppermint (Mentha x piperita, L.) that produces the aphid alarm pheromone (E)-beta-farnesene. Proc Natl Acad Sci USA 94 12833–12838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Gershenzon J (2000) Demonstration and characterization of (E)-nerolidol synthase from maize: a herbivory-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta 210 815–822 [DOI] [PubMed] [Google Scholar]
- Dooner HK, Martinez-Ferez IM (1997) Recombination occurs uniformly within the bronze locus, a meiotic recombination hotspot in the maize genome. Plant Cell 9 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Dooner HK (2002) Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA 99 9573–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Zheng Z, Dooner HK (2002) Recombination rates between adjacent genic and retrotransposon regions differ by two orders of magnitude. Proc Natl Acad Sci USA 99 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley J (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Greene B, Walko R, Hake S (1994) Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath B, Manukian A (1994) An automated system for use in collecting volatile chemicals released from plants. J Chem Ecol 18 1209–1226 [DOI] [PubMed] [Google Scholar]
- Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic K, SanMiguel PJ, Bennetzen JL (2003) A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc Natl Acad Sci USA 100 12265–12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairallah MM, Bohn M, Jiang C, Deutsch JA, Jewell DC, Mihm JA, Melchinger AE, Gonzalez-de-Leon D, Hoisington DA (1998) Molecular mapping of QTL for southwestern corn borer resistance, plant height and flowering in tropical maize. Plant Breed 117 309–318 [Google Scholar]
- Kollner TG, Schnee C, Gershenzon J, Degenhardt J (2004. a) The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distributions. Phytochemistry 65 1895–1902 [DOI] [PubMed] [Google Scholar]
- Kollner TG, Schnee C, Gershenzon J, Degenhardt J (2004. b) The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, et al (2004) Gene loss and movement in the maize genome. Genome Res 14 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Croteau R (1999) Isopentenyl diphosphate biosynthesis via a mevalonate-independent pathway: isopentenyl monophosphate kinase catalyzes the terminal enzymatic step. Proc Natl Acad Sci USA 96 13714–13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Dooner HK (2004) A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J 37 92–103 [DOI] [PubMed] [Google Scholar]
- Marillonnet S, Wessler SR (1997) Retrotransposon insertion into the maize waxy gene results in tissue-specific RNA processing. Plant Cell 9 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottinger JP (1970) The effects of X rays on the bronze and shrunken loci in maize. Genetics 64 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picaud S, Olsson ME, Brodelius M, Brodelius PE (2006) Cloning, expression, purification and characterization of recombinant (+)-germacrene D synthase from Zingiber officinale. Arch Biochem Biophys 452 17–28 [DOI] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S, Kollner TG, Degenhardt J, Hiltpold I, Toepfer S, Kuhlmann U, Gershenzon J, Turlings TCJ (2005) Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434 732–737 [DOI] [PubMed] [Google Scholar]
- Register JCI, Peterson DJ, Bell PJ, Bullock WP, Evans IJ, Frame B, Greenland AJ, Higgs NS, Jepson I, Jiao S (1994) Structure and function of selectable and non-selectable transgenes in maize after introduction by particle bombardment. Plant Mol Biol 25 951–961 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Jagendorf A (1995) Self defense by plants. Proc Natl Acad Sci USA 92 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Kollner TG, Gershenzon J, Degenhardt J (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-β-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol 130 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Kollner TG, Held M, Turlings TC, Gershenzon J, Degenhardt J (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA 103 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer GJ, Browse JA (2002) Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol 129 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah FH, Cha TS (2000) A mesocarp-and species-specific cDNA clone from oil palm encodes for sesquiterpene synthase. Plant Sci 154 153–160 [DOI] [PubMed] [Google Scholar]
- Shen B (2001) Transposon Ac-Facilitated Gene Search, Mutagenesis and Functional Characterization in Maize. Rutgers University, Piscataway, NJ
- Shen B, Zheng Z, Dooner HK (2000) A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: characterization of wild-type and mutant alleles. Proc Natl Acad Sci USA 97 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi J, Dicke M (1996) Plant-carnivore mutualism through herbivore-induced carnivore attractants. Trends Plant Sci 1 109–113 [Google Scholar]
- Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlings TCJ, McCall PJ, Alborn HT, Tumlinson JH (1993) An elicitor in caterpillar oral secretions that induces corn seedlings to emit chemical signals attractive to parasitic wasps. J Chem Ecol 19 411–425 [DOI] [PubMed] [Google Scholar]
- Turlings TCJ, Tumlinson JH, Lewis WJ (1990) Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250 1251–1253 [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Purugganan M, Wessler SR (1992) Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Dooner HK (2007) Variability of bz haplotypes in maize and its wild relatives (abstract no. P125). In 49th Annual Maize Genetics Conference Abstracts. Pheasant Run, St. Charles, IL, p 100
- Wu S, Schalk M, Clark A, Miles RB, Coates R, Chappell J (2006) Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol 24 1441–1447 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.