Abstract
Despite the fact that important genetic diseases are caused by mutant mitochondrial ribosomes, the molecular mechanisms by which such ribosomes result in a clinical phenotype remain largely unknown. The absence of experimental models for mitochondrial diseases has also prevented the rational search for therapeutic interventions. Here, we report on the construction of bacterial hybrid ribosomes that contain various versions of the mitochondrial decoding region of ribosomal RNA. We show that the pathogenic mutations A1555G and C1494T decrease the accuracy of translation and render the ribosomal decoding site hypersusceptible to aminoglycoside antibiotics. This finding suggests misreading of the genetic code as an important molecular mechanism in disease pathogenesis.
Keywords: decoding, mitochondria, mutant rRNA, ribosomes, disease
Genes encoded by mitochondrial DNA (mtDNA) have been linked to a variety of diseases (1, 2). One of the most common phenotypes associated with mitochondrial diseases is sensorineural deafness; corresponding mitochondrial mutations localize to tRNA and rRNA genes (2, 3). In particular, the single-nucleotide alteration A1555G has been identified as a major source of nonsyndromic deafness (4) and has since been found in patients from all ethnic backgrounds and geographic origins. More recently, a C1494T point mutation has been associated with nonsyndromic deafness (5); C1494 forms a noncanonical RNA base pair with the adenine encoded by 12S rRNA position 1555 (Fig. 1). Both mutations locate to the penultimate helix of mitochondrial 12S rRNA, which is a component of the aminoacyl-tRNA acceptor site (A site), a region of the small subunit rRNA essential for mRNA decoding (6). In bacteria, the A site is a target for aminoglycoside antibiotics, compounds that are compromised by a substantial degree of ototoxicity (7). Presumably, drug toxicity relates, at least in part, to limited selectivity between the bacterial and the mitochondrial A site and to the mechanism of drug action, i.e., miscoding (8). Interestingly, and in addition to congenital deafness, the A1555G and the C1494T mutations render affected individuals highly susceptible to aminoglycoside-induced deafness (4, 5).
Fig. 1.
Secondary-structure comparison of decoding-site rRNA sequences in the small ribosomal subunit. (A) Decoding region of 16S rRNA helix 44 in wild-type ribosomes of M. smegmatis; rRNA nucleotides are numbered according to the bacterial nomenclature, i.e., to homologous E. coli 16S rRNA positions. (B) Homologous 12S rRNA sequence in human mitochondrial ribosomes; rRNA residues are numbered according to the mitochondrial nomenclature: mtDNA 1494, 1555, and 1556 correspond to E. coli 16S rRNA 1410, 1490, and 1491. (C–E) Decoding site rRNA of human–bacterial hybrid ribosomes. The transplanted helix is boxed, and nucleotide positions depicted in blue represent residues that are specific for human mitochondrial rRNA. Mutations that are associated with congenital deafness, mtDNA position 1555A to G (D), and 1494C to U (E) are highlighted in red.
By itself, the A1555G mutation produces a clinical phenotype that may range from severe congenital deafness through moderate progressive hearing loss of later onset to normal hearing (3). Evidence has accumulated that the nuclear background plays an important role in the phenotypic manifestation of nonsyndromic deafness associated with mitochondrial rRNA mutations (9, 10), although cochlear alterations have been demonstrated to be present in symptomatic and asymptomatic carriers of the A1555G mutation (11). Biochemical investigations of deafness-associated mtDNA mutations have so far relied on the characterization of transmitochondrial cell lines, constructed by transferring mitochondria from cell lines of affected patients into the human cell line p°206 lacking mtDNA (9, 10). Nonetheless, the mechanistic link between mitochondrial mutation and disease has remained largely elusive and is still to be established, mainly because of the absence of suitable experimental models. Mice models for mitochondrial diseases have been limited to nuclear-encoded proteins, as the technical hurdles to create animal models of pathogenic mtDNA mutations remain formidable (12). In the absence of experimental models for ribosomes of higher eukaryotes, a spontaneous yeast mutant with a mitochondrial rRNA C1477-to-G alteration conferring resistance to paromomycin has been used as a model for the human mitochondrial A1555G deafness mutation (13). However, this yeast mutant does not show a discernible phenotype in translation (13, 14), and yeast mitochondrial rRNA position 1477 corresponds to human mitochondrial rRNA position 1493 rather than 1494 [see supporting information (SI) Fig. 5 and compare with Fig. 1]. We wanted to test whether the prokaryotic A site in the bacterial ribosome can be modified so as to serve as a model for the A site of mitochondrial ribosomes, as it would allow the study of the pathogenesis of mitochondrial rRNA-associated deafness mutations.
Results and Discussion
Much of the small subunit A-site rRNA, i.e., the corresponding regions of helices 18, 34, and 44 of 16S rRNA, is highly conserved in structure and nucleotide sequence between bacteria and mitochondria (15). Toward this end, we constructed bacterial hybrid ribosomes comprising the mitochondrial homologue of bacterial helix 44 (H44) of the 16S RNA decoding region by using a single-rRNA allelic derivative of Mycobacterium smegmatis and a previously described mutagenesis strategy (16). After several attempts to transfer parts of the mitochondrial H44 into M. smegmatis ribosomes, we were eventually able to generate recombinant strains with homogenous populations of hybrid ribosomes in which the central 34-nt part of rRNA helix 44 is identical to that of the human mitochondrial decoding site (Fig. 1). We next constructed recombinants where the bacterial H44 is replaced with mitochondrial deafness alleles corresponding to mtDNA mutations A1555G and C1494T. The resulting mutant mitochondrial hybrid ribosomes differ from the wild-type mitochondrial hybrid only in nucleotide position 1555 or 1494, corresponding to Escherichia coli positions 1490 and 1410 (Fig. 1).
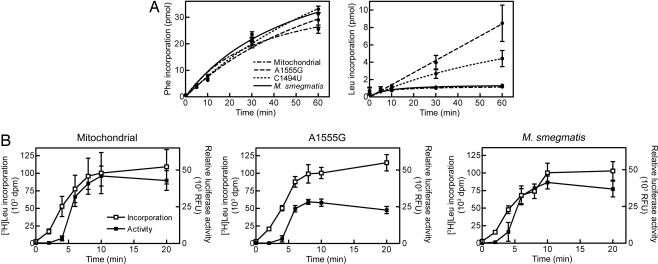
Because both mitochondrial rRNA mutations localize within the ribosomal decoding site, we hypothesized that they may affect the rate or accuracy of protein synthesis. For a biochemical analysis of the deafness mutations and their effect on mRNA decoding and polypeptide synthesis, 70S hybrid ribosomes were purified and studied in a cell-free translation assay with an AUG(UUU)12 mRNA as template. This template allows the determination of both the rate of amino acid incorporation and the frequency of codon misreading; codon ambiguity between leucine and phenylalanine is at the third codon position, UUU/UUC for phenylalanine versus UUA/UUG for leucine. This assay with its artificial mRNA has an intrinsically high error rate (17) and should thus provide a sensitive measure for studying misreading in wild-type compared with mitochondrial hybrid ribosomes. Translation of AUG(UUU)12 into oligopeptides composed of the cognate amino acid phenylalanine was comparable in hybrid ribosomes carrying either the mitochondrial wild-type or the A1555G- or C1494U-mutated decoding site (Fig. 2A and Table 1). In contrast, the accuracy of mRNA translation was significantly reduced in the hybrid ribosomes carrying a deafness mutation. Misincorporation of the near-cognate amino acid leucine by AUG(UUU)12-directed synthesis was increased 4-fold in the C1494U mutant as compared with wild-type mitochondrial and bacterial ribosomes (Fig. 2A and Table 1). Hybrid ribosomes carrying the A1555G mutation showed a 7-fold increase in leucine misincorporation relative to the wild-type mitochondrial hybrid. In absolute numbers, the A1555G and C1494U mutant A sites accommodated two leucine tRNAs per 7 and 15 phenylalanine tRNAs, respectively. Thus, although both deafness mutations result in substantial codon misreading, the error frequency is more pronounced in the A1555G mutant.
Fig. 2.
Rate and accuracy of mRNA translation in wild-type and mutant mitochondrial hybrid ribosomes. (A) AUG(UUU)12 mRNA-directed incorporation of phenylalanine and leucine into oligopeptides synthesized by ribosomes containing either a wild-type mitochondrial, mutant A1555G, or mutant C1494U decoding site (mean ± SD; n = 3). M. smegmatis wild-type ribosomes are included for comparison. For leucine per phenylalanine ratios, see Table 1. (B) Relative bioluminescence in luciferase synthesis assays (■) and total leucine incorporation (□) by mitochondrial hybrid and mutant A1555G ribosomes (mean ± SD; n = 3). Bacterial wild-type ribosomes are included for comparison. For luciferase activity per amino acid incorporation ratios, see Table 2.
Table 1.
Capacity and accuracy of amino acid incorporation
| Decoding site | AUG(UUU)12-directed amino acid incorporation |
||
|---|---|---|---|
| Phe, pmol | Leu, pmol | Leu/Phe | |
| M. smegmatis | 31 | 1.3 | 0.04 |
| Mitochondrial | 25 | 1.2 | 0.05 |
| A1555G | 29 | 8.5 | 0.29 |
| C1494U | 33 | 4.4 | 0.13 |
| C1556G | 29 | 0.7 | 0.03 |
The codon ambiguity exploited in the AUG(UUU)12 assay is located at the third position. Distinction of the third position is relevant for mixed codon boxes, where the third codon base is important for discriminating between the correct cognate or wobble codons, and the near-cognate codons, e.g., UUU/C for Phe versus UUA/G for Leu, CAU/C for His versus CAA/G for Gln, AAU/C for Asn versus AAA/G for Lys, GAU/C for Asp versus GAA/G for Glu (18). To determine ribosome fidelity in translation of a natural mRNA, we used a luciferase-encoding mRNA to analyze synthesis of luciferase based on a coupled transcription–translation assay. The functional enzymatic activity of firefly luciferase [bioluminescence measured as relative fluorescence units (RFU)] was determined and related to the amount of amino acids incorporated (measured as [3H]leucine incorporation). Compared with wild-type bacterial and mitochondrial hybrid ribosomes, the mutant A1555G hybrid ribosomes incorporated similar amounts of amino acids in quantitative terms. This observation indicates that the A1555G mutation does not grossly affect ribosome mechanics nor blocks a rate-limiting step in polypeptide synthesis. However, in qualitative terms, the mutant produced ≈50% less functional enzymatic activity per amino acid incorporated (Fig. 2B and Table 2). This finding would also be compatible with miscoding, as amino acid misincorporation during the synthesis of luciferase (550 aa in length) would be expected to reduce the enzymatic activity (19), thereby diminishing the level of luminescence without grossly affecting the overall amount of protein synthesized. To distinguish misincorporation from premature termination as cause of decreased luciferase activity, we analyzed the synthesized luciferase by SDS/PAGE and autoradiography. Mutant and wild-type mitochondrial hybrid ribosomes produced the same level of full-length protein (SI Fig. 6).
Table 2.
Ribosomal accuracy in luciferase synthesis
| Decoding site | Relative luciferase activity per [3H]leucine incorporation, RFU/dpm |
||
|---|---|---|---|
| 10 min | 20 min | 30 min | |
| M. smegmatis | 0.29 | 0.22 | 0.25 |
| Mitochondrial | 0.24 | 0.20 | 0.29 |
| A1555G | 0.11 | 0.12 | 0.13 |
To study the effect of aminoglycosides on translational fidelity of the deafness alleles, we determined tobramycin-induced misincorporation of leucine in the AUG(UUU)12-mRNA translation assay. Fig. 3 shows the dose-dependent exacerbation of translation infidelity in mutant mitochondrial hybrid ribosomes. Wild-type mitochondrial ribosomes are affected only slightly at drug concentrations >50 μM. The intrinsic infidelity of the A1555G and C1494U mutants, however, is further aggravated by drug concentrations of as little as 2 μM.
Fig. 3.
Aminoglycoside-induced exacerbation of translation infidelity. Dose–response curves show tobramycin-induced misincorporation of leucine per phenylalanine by wild-type, A1555G, and C1494U mitochondrial hybrid ribosomes after 60 min incubation with an AUG(UUU)12 mRNA template (mean ± SD; n = 3).
Most nucleotides in 16S rRNA helices 18, 34, and 44 that form the small subunit decoding site are universally conserved, although subtle but significant variations have evolved in helix 44. Although the hybrid ribosomes in this study do not carry a complete mitochondrial A site, transplanting the mitochondrial version of helix 44 revealed that, in comparison to the wild-type mitochondrial helix, the disease-associated mutation resulted in significant misreading. How to explain that the deafness mutations C1494U and A1555G result in mistranslation? Kinetic models for tRNA selection suggest that the ribosome discriminates between cognate and noncognate tRNAs by induced fit (20). At the structural level, decoding is linked to conformational changes, in which A1492, A1493, and G530 (E. coli numbering) interact intimately with the minor groove of the first two codon-anticodon base pairs, thus allowing for discrimination between cognate and noncognate tRNAs (21) and to initiate transition of the ribosome from an open to a closed form (22). The internal, asymmetric loop containing the bulged-out nucleotides A1492 and A1493 is closed by nucleotides of the lower stem of helix 44 (Fig. 1A). Compared with the bacterial A site, where the loop is closed by a Watson–Crick C1409–G1491 interaction, the internal loop in the mitochondrial A site is able to adopt an extended conformation becoming closed further down the helix by a C1495–G1554 base pair (Fig. 1B). Both the C1494U and the A1555G mutation replace the noncanonical C1494·A1555 interaction by a Watson–Crick base pairing, thus shortening the extended conformation of the mitochondrial loop. To investigate a possible effect of internal loop closure on mitochondrial decoding accuracy, we introduced a C1493–G1556 base pair to the mitochondrial hybrid (SI Fig. 7B). In contrast to a C1494–G1555 interaction, introduction of the C1493–G1556 base pair did not result in miscoding (Table 1 and SI Fig. 7). This finding indicates that closing the bulge does by itself not affect the error rate of mRNA decoding.
The base-pairing pattern of the mitochondrial A1555G or C1494U decoding site resembles that of the bacterial A site carrying a G1491C mutation. In both sites, the C·C opposition at position 1409–1491 (mitochondrial 1493–1556) is followed by a Watson–Crick base pair 1410–1490 (mitochondrial 1494–1555, as depicted in SI Fig. 8A). To test whether this more general pattern accounts for the translation phenotype observed in A1555G and C1494U mutant mitochondrial hybrid ribosomes, we determined the translation fidelity of purified G1491C bacterial ribosomes. We found that the bacterial G1491C decoding site shows no significant misreading, but instead exhibits a translation fidelity that compares well with that of wild-type bacterial and wild-type mitochondrial decoding sites (SI Fig. 8B). This observation indicates that translation fidelity also depends on base-pair interactions within the lower stem of helix 44, which is part of the entire mitochondrial region transplanted, and which is significantly different from its bacterial counterpart (Fig. 1 and SI Fig. 8A).
For studying a mechanism for mutation-induced miscoding at the structural level, we made two assumptions: first, the conformation of the decoding region in the native and mutated mitochondrial ribosomes is identical to that of its bacterial counterpart, except for the consequences of known sequence differences; second, the mutations influence their immediate vicinity. For the human cytosolic A site, two theoretical possibilities for a noncanonical C·A base pair have been described based on RNA oligonucleotides: one with a protonated adenine shifted to the minor groove side, the other with a nonprotonated adenine shifted to the major groove side (23). The former is practically superimposable on the Thermus thermophilus structure (24, 25) and can explain how codon–anticodon interactions are monitored to ensure translation fidelity. For the latter, no biological correlate has been provided. Modeling the mitochondrial decoding site on the Thermus structure revealed that the C1494·A1555 interaction is established by a slight rotation of A1555 toward the outline of helix 44 (Fig. 4). This motion is accompanied by the rotation of C1494 into the helix to create an acceptable donor–acceptor relationship between the adenine and the cytosine. This rotation would increase the surface area between H44 and H27. By introducing a Watson–Crick base pairing, as in C1494–A1555G and C1494U–A1555, nucleotide 1555 will be rotated back into H44, thus reducing the contact between H44 and H27. Destabilizing the interaction between H44 and H27 could make a relative movement between H27 and H44 easier. Such a movement is likely to be part of the induced conformational change required in decoding (22, 26), and lowering the energetic penalty for the change would allow near-cognate tRNAs to be accepted more easily. According to this interpretation, rotation of residue 1555 should be affected by the relative strength of the 1494–1555 interaction: compared with a U–A interaction, a C–G pair would limit the possibility of nucleotide 1555 to escape its displacement imposed by base-pairing with nucleotide 1494; the presence of three as compared with two H bonds provides additional binding energy and makes G–C base pairs intrinsically more stable than A–U pairs. This prediction is in line with the finding that the A1555G mutation results in a more pronounced misreading than the C1494U mutation.
Fig. 4.
Structural interpretation of mutation A1555G and C1494U. (A) The interface of 16S rRNA helices 44 and 27 in the crystal structure of T. thermophilus ribosomes (24), showing the bacterial G1410–C1490 base pair, which corresponds to the homologous C1494·A1555 interaction in human mitochondrial 12S rRNA. (B) Enlarged view into the vicinity of C1490. The arrow indicates the rotation that C1490 has to undergo (Upper) to create a noncanonical C·A interaction present in mitochondrial ribosomes (Lower). This rotation increases the interface area between helix 44 and helix 27. Note that owing to the position of nucleotide 1556, the mutation C1556G (E. coli G1491) should have no effect on the interface area.
Simple single-cell prokaryotic organisms are apparently able to tolerate significant misreading (27, 28), as also evidenced by the viability of the A1555G and C1494U hybrid mutants. However, the situation in complex multicellular eukaryotes and in highly specialized tissues is notably different. Misfolded proteins have been implicated in a variety of diseases (29), in particular involving cell types that have limited ability to regenerate by cell division. At least two mechanisms may account for the disease phenotype conferred by mistranslation: dysfunctional proteins and misfolded protein response (30). The 13 proteins encoded by human mtDNA and translated by mitochondrial ribosomes are subunits of the respiratory chain and oxidative phosphorylation pathway at the inner mitochondrial membrane. Lymphoblastoid cell lines with mutations in mitochondrial 12S rRNA have been reported to show decreased rates of oxygen consumption (9, 10). In cell lines of patients with the A1555G mutation no alterations of mitochondrial translation products at the level of electrophoretic mobility were found (31). This observation is compatible with our luciferase data in the mitochondrial hybrid ribosomes, which demonstrate that mutation-mediated misreading manifests primarily at the level of functional activity of the translated protein by missense rather than by nonsense decoding (SI Fig. 6). Increased mistranslation of mitochondrial genes might also account for a misfolded protein response to result in the cochlear alterations observed in symptomatic and asymptomatic carriers of the A1555G mutation (11).
Identification of misreading as key mechanism in pathogenesis also allows us to integrate the role of the nuclear background in disease manifestation and address the association of the A1555G and C1494U mutations with aminoglycoside-induced deafness. The nuclear-encoded proteins thus far linked to the expression of the deafness phenotype are involved in mitochondrial tRNA or rRNA modifications (32–34). On the basis of a misreading-prone ribosome, any change in translational efficiency, regardless of its specific mechanism of action, would further impair mitoribosome function, to aggravate translational dysfunction beyond the threshold required to result in the phenotypic expression of severe hearing loss that is associated with mutations A1555G and C1494U. Likewise, the misreading phenotype of the A1555G and C1494U mutant mitochondrial ribosomes is significantly exacerbated by aminoglycoside antibiotics.
Although the basis for the tissue specificity of disease expression is unclear, the biochemical phenotype of the mitochondrial deafness alleles, as revealed by the study of hybrid ribosomes, is identical to that of aminoglycoside action, i.e., increased mistranslation. Together with the dose-related, irreversible ototoxicity of aminoglycosides, which presumably at least in part reflects drug-mediated dysfunction of the mitochondrial ribosome (8), we conclude that the findings we observe are significant in understanding the mechanisms of mitochondrial deafness. We can presently not exclude that, in addition to mistranslation, other aspects of ribosome function may result in pathogenic properties of the mutant. Although additional effects may be involved, our results provide a mechanistic link between mtDNA deafness mutations and pathogenesis. The experimental model developed here may help in the identification of prokaryotic homologues of nuclear genes modifying phenotypic expression of the disease and in the design of RNA-specific agonists for treatment. As a further outcome of our work, we note that identification of error-prone protein synthesis in mitochondria with an A1555G mutation also allows testing of the long-standing hypothesis of mitochondrial dysfunction and aging (35).
Materials and Methods
Construction of Mutant Strains with Hybrid Ribosomes.
The recently described strain M. smegmatis ΔrrnB (16) was used for all genetic manipulations. Site-directed mutagenesis of its single rRNA operon was performed by PCR mutagenesis with hybrid rDNA oligonucleotides comprising the wild-type or mutant mitochondrial helix 44 decoding site sequence. The resulting hybrid gene fragment was cloned into an integration-proficient plasmid used to transform M. smegmatis ΔrrnB. Transformants were selected on LB agar plates containing 20 μg/ml paromomycin for gene replacement by homologous recombination. After several attempts to introduce various parts of the mitochondrial H44 into bacterial ribosomes by site-directed mutagenesis, we were finally able to generate recombinant M. smegmatis cells where the central 34-nt part of the bacterial H44 had been replaced by its mitochondrial counterpart. Successful replacement of the bacterial decoding site sequence with the mitochondrial sequence was controlled by sequence analysis of the chromosomal rrnA locus. Transplanting the mitochondrial decoding sites into the bacterial ribosome affected the generation times of the resulting M. smegmatis mutants slightly (mitochondrial hybrid 5.1 h; C1494U 4.9 h; A1555G 6.0 h; as compared with M. smegmatis ΔrrnB 3.7 h).
Isolation and Purification of Ribosomes.
Ribosomes were isolated from bacterial cell pellets as described (36). For further fractionation, isolated ribosomes were resuspended in overlay buffer [20 mM Tris·HCl (pH 7.4), 60 mM NH4Cl, 5.25 mM MgCl2, 0.25 mM EDTA, 3 mM 2-mercaptoethanol, and 5% sucrose] loaded on a sucrose gradient (10–40% sucrose in overlay buffer) and centrifuged in a Beckman Ti 15 rotor at 28,000 rpm for 18 h. Gradient fractions were collected by unloading the zonal rotor with 50% sucrose in overlay buffer. The 70S ribosome-enriched fractions were pooled according to the absorption profile at 260 nm, and applied to centrifugation at 180,000 × g for 20 h. The final ribosome pellets were resuspended in buffer A [50 mM Tris·HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2], incubated for 30 min at 4°C, dispensed into aliquots, and stored at −80°C after shock freezing in liquid nitrogen. 70S ribosome concentrations were determined by absorption measurements on the basis of 23 pmol per A260 unit. Integrity and functional activity of purified 70S ribosomes was determined by analytical ultracentrifugation and by assessing their capacity to form initiation complexes, as described (36).
Cell-Free AUG(UUU)12 Translation Assays.
Cell-free translation reactions in buffer A (pH 7.5) were prepared on ice and contained M. smegmatis tRNAbulk (0.5 mg/ml), amino acids mixture (30 μM each) lacking phenylalanine and/or leucine, 10% (vol/vol) S100 extract, energy mix (1 mM DTT, 1 mM GTP, 4 mM ATP, and 5 mM phosphoenolpyruvate), pyruvate kinase (0.1 mg/ml), and polyamines (2 mM spermidine and 8 mM putrescine). The reaction mixture was preincubated with 30 μM [14C]phenylalanine (110 mCi/mmol) and/or 30 μM [3H]leucine (500 mCi/mmol) at 37°C for 15 min. The translation reaction was started by addition of ribosomes to a final concentration of 0.25 μM and AUG(UUU)12-mRNA [5′-GCGGCAAGGAGGUAAAUAAUG(UUU)12UAAGCAGG-3′, obtained from Dharmacon] to 1 μM; in experiments studying tobramycin-induced misreading, the drug was added simultaneously. After incubation at 37°C for the times indicated, the reaction was stopped by addition of KOH to 0.5 M and subsequent hydrolysis at 37°C for 30 min. Synthesized polypeptides were precipitated with 200 μl of 5% trichloroacetic acid (TCA) for 10 min on ice and bound to filters. Filter-bound polypeptides were washed with cold 30% 2-propanol, dissolved in 10 ml of scintillation mixture, and quantified.
Cell-Free Luciferase Translation Assays.
Purified 70S hybrid ribosomes were used in a coupled transcription-translation reaction of firefly luciferase (plasmid pBESTluc; Promega). A typical reaction (30 μl volume) contained 0.25 μM 70S ribosomes, 600 ng DNA, 40% (vol/vol) of M. smegmatis S100 extract, 100 μM amino acid mixture minus leucine, 1.7 μM [3H]leucine (59 Ci/mmol), and RNasin (40 units; Promega). rNTPs, tRNAs, and energy were supplied by the addition of commercial S30 Premix Without Amino Acids (Promega). The reaction mixture was incubated at 37°C and stopped on ice. Fifteen microliters of the reaction volume was added to 100 μl of luciferase assay substrate (Promega), and the bioluminescence was measured in a luminometer (Bio-Tek Instruments; FL ×800). In the remaining 15 μl of reaction volume, polypeptides were hydrolyzed, precipitated, applied to a GF/A filter (Whatman), and washed three times with 3 ml of ice-cold 5% TCA. Filters were air-dried for 10 min, and [3H]leucine incorporation was quantified in 10 ml of scintillation mixture.
Structural Modeling.
Modeling the mitochondrial decoding site on the Thermus thermophilus structure (24) was done manually, according to the following procedure: (i) substituting the bacterial residues G1410 with C and C1490 with A according to the homologous positions in mitochondrial ribosomes (C1494·A1555), using the backbone conformation as a guide; (ii) fitting A1555 into the helix so that a noncanonical C·A base pair could be formed; and (iii) combining attempts to obtain a relevant H bond with estimating (by actual measurements) the deviation of the new structure from a perfect helix and searching for structural elements that can be exploited for the extra stabilization needed for the slightly shifted-rotated region of the helix. In addition to the slight rotation, a small shift was included in the modeling; the latter, however, is of negligible magnitude at the resolution level of the structure.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Raz Zarivach, Anat Bashan, and Ada Yonath for valuable advice on structural interpretations, Marina Rodnina (Universität Witten/Herdecke, Witten, Germany) for generously providing [3H]fMet-tRNAfMet; and Tanja Janušić for expert technical assistance. This work was supported by grants from the Swiss National Science Foundation (to E.C.B.) and the Bonizzi-Theler-Stiftung (to S.N.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707265105/DC1.
References
- 1.Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 2.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mutat. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 4.Prezant TR, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and nonsyndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 7.Begg EJ, Barclay ML. Aminoglycosides: 50 years on. Br J Clin Pharmacol. 1995;39:597–603. [PMC free article] [PubMed] [Google Scholar]
- 8.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, et al. Functional characterization of the mitochondrial 12S rRNA C1494T mutation associated with aminoglycoside-induced and nonsyndromic hearing loss. Nucleic Acids Res. 2005;33:1132–1139. doi: 10.1093/nar/gki262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bravo O, Ballana E, Estivill X. Cochlear alterations in deaf and unaffected subjects carrying the deafness-associated A1555G mutation in the mitochondrial 12S rRNA gene. Biochem Biophys Res Commun. 2006;344:511–516. doi: 10.1016/j.bbrc.2006.03.143. [DOI] [PubMed] [Google Scholar]
- 12.Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat Genet. 2001;27:191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Li R, Lin X, Guan MX. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J Biol Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 14.Colby G, Wu M, Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 15.Gutell RR. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbie SN, et al. A genetic model to investigate structural drug–target interactions at the ribosomal decoding site. Biochimie. 2006;88:1033–1043. doi: 10.1016/j.biochi.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Jelenc PC, Kurland CG. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agris PF. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- 20.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 21.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 22.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 23.Kondo J, Urzhumtsev A, Westhof E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006;34:676–685. doi: 10.1093/nar/gkj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3-Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 25.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 26.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurland CG, Hughes D, Ehrenberg M. In: Escherichia coli and Salomonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, et al., editors. Vol 1. Washington, DC: Am Soc Microbiol; 1996. pp. 979–1004. [Google Scholar]
- 28.Bacher JM, de Crecy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci USA. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 30.Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Annu Rev Genomics Hum Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- 31.Guan MX, Fischel-Ghodsian N, Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- 32.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 33.Bykhovskaya Y, et al. Phenotype of nonsyndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol Genet Metab. 2004;83:199–206. doi: 10.1016/j.ymgme.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Guan MX, et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hipkiss AR. Errors, mitochondrial dysfunction and ageing. Biogerontology. 2003;4:397–400. doi: 10.1023/B:BGEN.0000006560.84660.fe. [DOI] [PubMed] [Google Scholar]
- 36.Hobbie SN, et al. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 2007;35:6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.