Abstract
DNA barcoding is a technique in which species identification is performed by using DNA sequences from a small fragment of the genome, with the aim of contributing to a wide range of ecological and conservation studies in which traditional taxonomic identification is not practical. DNA barcoding is well established in animals, but there is not yet any universally accepted barcode for plants. Here, we undertook intensive field collections in two biodiversity hotspots (Mesoamerica and southern Africa). Using >1,600 samples, we compared eight potential barcodes. Going beyond previous plant studies, we assessed to what extent a “DNA barcoding gap” is present between intra- and interspecific variations, using multiple accessions per species. Given its adequate rate of variation, easy amplification, and alignment, we identified a portion of the plastid matK gene as a universal DNA barcode for flowering plants. Critically, we further demonstrate the applicability of DNA barcoding for biodiversity inventories. In addition, analyzing >1,000 species of Mesoamerican orchids, DNA barcoding with matK alone reveals cryptic species and proves useful in identifying species listed in Convention on International Trade of Endangered Species (CITES) appendixes.
Keywords: CITES, Kruger National Park, Mesoamerica
DNA barcoding is a diagnostic technique for species identification, using a short, standardized DNA region, i.e., the “DNA barcode” (www.barcoding.si.edu). It is, however, challenging to find a suitable genomic region for DNA barcoding a wide range of taxa. Indeed, for DNA barcoding to work, sequence variation must be high enough between species so that they can be discriminated from one another; however, it must be low enough within species that a clear threshold between intra- and interspecific genetic variations can be defined. Although the use of DNA barcoding for identification and taxonomy has been controversial (1, 2), a growing scientific community has embraced DNA barcoding as a practical tool for biodiversity studies, for example to facilitate inventories of very diverse but taxonomically poorly known regions (3–6). DNA barcoding, using the mitochondrial coxI gene (COI) (7–10), is now well established for animals, but the quest for a universal DNA barcode in plants is still disputed (11, 12).
Kress et al. (13) proposed originally that the trnH-psbA plastid region would be a suitable universal barcode for land plants. Concurrently, the newly established “plant working group” from the consortium for the barcoding of life tested a series of other genomic regions at first disregarding trnH-psbA because of its complex molecular evolution (14). It was also proposed that, because the plastid genome is evolving so slowly relative to other genomes, more than one barcode may be necessary to provide enough variation for this technique to work (15–17). However, several competing proposals have so far been put forward, which need thorough evaluations. Kress and Erickson (16) proposed to combine the original trnH-psbA barcode from Kress et al. (13) with rbcL, following analyses from Newmaster et al. (17). By contrast, Chase et al. (15) proposed either to combine rpoc1, rpoB, and matK or rpoc1, matK, and trnH-psbA, whereas Taberlet et al. (18) suggested the trnL intron as a suitable plant barcode. Furthermore, tests of potential DNA barcodes have been based on a taxonomic coverage approach, necessarily encompassing just a few representatives from a wide range of distantly related groups of land plants (13, 15–17). However, the critical test of evaluating the applicability of DNA barcoding for biodiversity inventories in species-rich geographic areas has been lacking.
Here, we focus on two biodiversity hotspots (19, 20), Mesoamerica and Maputaland–Pondoland–Albany in southern Africa, in which we analyze >1,600 plant specimens. We test eight potential DNA barcodes, six of which were made publicly available at the plant working group's website [www.kew.org/barcoding (15)], whereas a further two were proposed by Kress and Erickson (16). Our study sites have been chosen for their exceptional plant diversity and contrasting habitats. Costa Rica comprises tropical forests and has one of the richest orchid floras in the world. Although there is a well developed network of protected areas in Costa Rica, the orchid flora remains under constant threat from deforestation and illegal trade. Orchids are also well known to be difficult to identify, particularly when they are sterile, which makes them an ideal model group in which to test DNA barcoding techniques. In southern Africa, we have undertaken our study in the Kruger National Park (KNP), one of the largest protected areas in the world. The KNP is renowned for its large game animals but less for its flora, which is under continuous pressure from mega-herbivores. Home to ≈600 species of trees and shrubs (21), the KNP area has the highest tree diversity of any of the world's temperate regions.
During 2005–2007, we conducted extensive fieldwork to collect samples for this study. We used several metrics to evaluate the various potential barcoding regions. Intra- and interspecific genetic divergences were assessed by using pairwise calculations (22). Statistical tests were used to compare divergences. Phylogenetic analyses were performed to look for species monophyly. Genetic clustering algorithms (23, 24) were applied to test whether the coalescent process in a given barcode matched species delimitation.
Results and Discussion
PCRs were generally successful with all potential barcodes, except ndhJ and ycf5, which did not amplify efficiently in orchids. It is known that rbcL is not variable enough in orchids (25), so we did not sequence this gene in this group. The rbcL and trnH-psbA regions did not amplify in the achlorophyllous Hydnora johanis but amplified in other parasitic plants. A portion of the matK exon amplified easily by using primers 390F and 1326R from Cuénoud et al. (26). Alignment of sequences was straightforward, except for trnH-psbA that required the addition of several gaps. In orchids and amaryllids, we also found that trnH-psbA hosts a well conserved exon, which corresponds to an extra copy of the rps19 gene (14).
We assessed genetic divergences within and between species, using various metrics (22). We comment here on calculations, using the best-fit models for each barcode (Table 1). For comparison purposes with other studies, we also provide as SI the results based on other distances [supporting information (SI) Tables 6 and 7]. A suitable barcode must exhibit high interspecific but low intraspecific divergence. Here, the highest interspecific divergence is provided by trnH-psbA (KNP and combined datasets; Table 1). The next most variable barcode at interspecific level is matK for all datasets. Three different metrics were used to characterize intraspecific divergence: (i) average of all pairwise distances between all individuals sampled within those species that had at least two representatives; (ii) “mean theta,” with theta being the average pairwise distances calculated for each species that have more than one representative, thereby eliminating biases associated with uneven sampling among taxa; and (iii) average coalescent depth, i.e., the maximum distance from tips of a node linking all sampled extend members of a species, “book-ending” intraspecific variability (see also SI Table 8). The results from these calculations of intraspecific differences do not show a clear pattern. In orchids, the barcodes exhibiting the lowest intraspecific divergence are rpoC1 (average mean divergence), accD/matK (mean theta) and matK (coalescent depth). In the KNP, the lowest intraspecific divergence is provided by ndhJ with all three metrics. Wilcoxon signed rank tests on combined data show that trnH-psbA is the most variable barcode at interspecific level, followed by matK (Table 2). At intraspecific level, Wilcoxon signed rank tests show rpoC1 and accD having the lowest level of divergence, whereas the highest is provided by trnH-psbA (Table 3). Based on these results alone, it is difficult to decide on which barcode is the most suited for plants.
Table 1.
Measures of inter- and intraspecific divergences for eight potential barcodes sampled in Costa Rica and in the KNP of South Africa
| Dataset | Mean | Potential Barcode |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| trnH-psbA | matK | ycf5 | rbcL | rpoB | ndhJ | accD | rpoc1 | ||
| All interspecific distances | 0.0271 ± 0.0258 | 0.013 ± 0.0126 | 0.0104 ± 0.0092 | 0.0082 ± 0.0066 | 0.0061 ± 0.006 | 0.0029 ± 0.004 | 0.0033 ± 0.0037 | 0.0019 ± 0.0033 | |
| All intraspecific distances | 0.0012 ± 0.0021 | 0.0016 ± 0.0057 | 0.0002 ± 0.001 | 0.0005 ± 0.001 | 0.0007 ± 0.0021 | 0 | 0.0005 ± 0.0017 | 0.0002 ± 0.0009 | |
| Theta | 0.0015 ± 0.0033 | 0.0012 ± 0.0012 | 0.0005 ± 0.0017 | 0.0003 ± 0.0006 | 0.0004 ± 0.0013 | 0.00003 ± 0.0001 | 0.0004 ± 0.0013 | 0.0001 ± 0.0005 | |
| Coalescent depth | 0.0024 ± 0.0049 | 0.0017 ± 0.0017 | 0.0009 ± 0.0027 | 0.0004 ± 0.001 | 0.0007 ± 0.002 | 0.00009 ± 0.0005 | 0.0008 ± 0.0025 | 0.0002 ± 0.0008 | |
| Costa Rican | All interspecific distances | 0.0082 ± 0.0069 | 0.0079 ± 0.0086 | — | — | 0.0067 ± 0.0086 | 0.0163 ± 0.0211 | 0.0071 ± 0.0069 | 0.0022 ± 0.003 |
| All intraspecific distances | 0.0033 ± 0.0034 | 0.0016 ± 0.0022 | — | — | 0.0038 ± 0.0096 | 0.0077 ± 0.0146 | 0.0017 ± 0.0038 | 0.0014 ± 0.002 | |
| Theta | 0.0024 ± 0.0025 | 0.001 ± 0.001 | — | — | 0.0067 ± 0.0133 | 0.0132 ± 0.0138 | 0.001 ± 0.0019 | 0.0015 ± 0.0018 | |
| Coalescent depth | 0.0034 ± 0.0037 | 0.0015 ± 0.0015 | — | — | 0.0081 ± 0.0146 | 0.0174 ± 0.02 | 0.0018 ± 0.0035 | 0.0021 ± 0.0027 | |
| Combined | All interspecific distances | 0.0236 ± 0.0246 | 0.0121 ± 0.0121 | 0.0104 ± 0.0092 | 0.0082 ± 0.0066 | 0.0062 ± 0.0065 | 0.0046 ± 0.0095 | 0.0039 ± 0.0047 | 0.002 ± 0.0033 |
| All intraspecific distances | 0.0023 ± 0.0031 | 0.0016 ± 0.0042 | 0.0002 ± 0.001 | 0.0005 ± 0.001 | 0.0023 ± 0.0071 | 0.0037 ± 0.0107 | 0.0012 ± 0.003 | 0.0008 ± 0.0017 | |
| Theta | 0.0018 ± 0.0031 | 0.001 ± 0.001 | 0.0005 ± 0.0017 | 0.0003 ± 0.0007 | 0.0021 ± 0.0074 | 0.003 ± 0.0084 | 0.0006 ± 0.0015 | 0.0005 ± 0.0012 | |
| Coalescent depth | 0.0027 ± 0.0046 | 0.0014 ± 0.0014 | 0.0009 ± 0.0027 | 0.0004 ± 0.001 | 0.0027 ± 0.0082 | 0.0039 ± 0.0116 | 0.0011 ± 0.0028 | 0.0007 ± 0.0017 | |
Table 2.
Wilcoxon signed rank tests of inter-specific divergence among loci
| W+ | W− | Relative Ranks, n, P value | Result |
|---|---|---|---|
| trnH-psbA | matK | W+ = 21,089, W− = 3,001, n = 219, P ≤ 5.886 × 10−22 | trnH-psbA ≫ matK |
| trnH-psbA | ycf5 | W+ = 13,226, W− = 802, n = 167, P ≤ 3.276 × 10−23 | trnH-psbA ≫ ycf5 |
| trnH-psbA | rbcL | W+ = 15,878, W− = 1,327, n = 185, P ≤ 2.001 × 10−23 | trnH-psbA ≫ rbcL |
| trnH-psbA | rpoB | W+ = 23,403, W− = 2,022, n = 225, P ≤ 7.967 × 10−28 | trnH-psbA ≫ rpoB |
| trnH-psbA | ndhJ | W+ = 20,363, W− = 2,642, n = 214, P ≤ 1.546 × 10−22 | trnH-psbA ≫ ndhJ |
| trnH-psbA | rpoc1 | W+ = 23,709, W− = 162, n = 218, P ≤ 1.55 × 10−36 | trnH-psbA ≫ rpoc1 |
| trnH-psbA | accD | W+ = 23,669, W− = 1,756, n = 225, P ≤ 3.828 × 10−29 | trnH-psbA ≫ accD |
| matK | ycf5 | W+ = 6,833, W− = 6,862, n = 165, P ≤ 0.9818 | matK = ycf5 |
| matK | rbcL | W+ = 12,312, W− = 4,893, n = 185, P ≤ 3.673 × 10−7 | matK > rbcL |
| matK | rpoB | W+ = 21,020, W− = 3,290, n = 220, P ≤ 6.803 × 10−21 | matK > rpoB |
| matK | ndhJ | W+ = 19,554, W− = 2,812, n = 211, P ≤ 4.287 × 10−21 | matK > ndhJ |
| matK | rpoc1 | W+ = 24,054, W− = 477, n = 221, P ≤ 3.17 × 10−35 | matK > rpoc1 |
| matK | accD | W+ = 22,666, W− = 2,087, n = 222, P ≤ 6.824 × 10−27 | matK > accD |
| rbcL | ycf5 | W+ = 4,564, W− = 10,487, n = 173, P ≤ 7.186 × 10−6 | rbcL < ycf5 |
| rbcL | rpoB | W+ = 11,985, W− = 5,220, n = 185, P ≤ 3.536 × 10−6 | rbcL > rpoB |
| rbcL | ndhJ | W+ = 14,475, W− = 576, n = 173, P ≤ 6.202 × 10−26 | rbcL > ndhJ |
| rbcL | rpoc1 | W+ = 14,908, W− = 143, n = 173, P ≤ 4.702 × 10−29 | rbcL > rpoc1 |
| rbcL | accD | W+ = 15,215, W− = 1,438, n = 182, P ≤ 3.803 × 10−22 | rbcL > accD |
| ycf5 | rpoB | W+ = 10,796, W− = 2,899, n = 165, P ≤ 1.338 × 10−10 | ycf5 > rpoB |
| ycf5 | ndhJ | W+ = 11,259, W− = 987, n = 156, P ≤ 1.037 × 10−19 | ycf5 > ndhJ |
| ycf5 | rpoc1 | W+ = 11,952, W− = 294, n = 156, P ≤ 6.297 × 10−25 | ycf5 > rpoc1 |
| ycf5 | accD | W+ = 10,755, W− = 1,026, n = 153, P ≤ 8.128 × 10−19 | ycf5 > accD |
| rpoB | ndhJ | W+ = 11,709, W− = 6,057, n = 188, P ≤ 0.0001556 | rpoB > ndhJ |
| rpoB | rpoc1 | W+ = 16,047, W− = 3,456, n = 197, P ≤ 3.984 × 10−15 | rpoB > rpoc1 |
| rpoB | accD | W+ = 11,614, W− = 5,777, n = 186, P ≤ 7.227 × 10−5 | rpoB > accD |
| ndhJ | rpoc1 | W+ = 11,859, W− = 5,346, n = 185, P ≤ 8.037 × 10−6 | ndhJ > rpoc1 |
| ndhJ | accD | W+ = 7,469, W− = 6,392, n = 166, P ≤ 0.3857 | ndhJ = accD |
| rpoc1 | accD | W+ = 3,891, W− = 14,064, n = 189, P ≤ 1.447 × 10−11 | rpoc1 < accD |
Table 3.
Wilcoxon signed rank tests of intraspecific difference among loci
| W+ | W− | Relative Ranks, n, P value | Result |
|---|---|---|---|
| trnH-psbA | matK | W+ = 1,949, W− = 826, n = 74, P ≤ 0.002509 | trnH-psbA > matK |
| trnH-psbA | ycf5 | W+ = 327, W− = 108, n = 29, P ≤ 0.01843 | trnH-psbA > ycf5 |
| trnH-psbA | rbcL | W+ = 436, W− = 92, n = 32, P ≤ 0.001342 | trnH-psbA > rbcL |
| trnH-psbA | rpoB | W+ = 1,113, W− = 483, n = 56, P ≤ 0.01031 | trnH-psbA > rpoB |
| trnH-psbA | ndhJ | W+ = 973, W− = 567, n = 55, P ≤ 0.08976 | trnH-psbA ≥ ndhJ |
| trnH-psbA | rpoc1 | W+ = 1,596, W− = 234, n = 60, P ≤ 5.464 × 10−7 | trnH-psbA > rpoc1 |
| trnH-psbA | accD | W+ = 1,579, W− = 437, n = 63, P ≤ 9.399 × 10−5 | trnH-psbA > accD |
| matK | ycf5 | W+ = 260, W− = 175, n = 29, P ≤ 0.3638 | matK = ycf5 |
| matK | rbcL | W+ = 299, W− = 197, n = 31, P ≤ 0.3224 | matK = rbcL |
| matK | rpoB | W+ = 695, W− = 790, n = 54, P ≤ 0.6857 | matK = rpoB |
| matK | ndhJ | W+ = 585, W− = 640, n = 49, P ≤ 0.7883 | matK = ndhJ |
| matK | rpoc1 | W+ = 1,220, W− = 491, n = 58, P ≤ 0.004829 | matK > rpoc1 |
| matK | accD | W+ = 1,059, W− = 594, n = 57, P ≤ 0.06529 | matK > accD |
| rbcL | ycf5 | W+ = 66, W− = 124, n = 19, P ≤ 0.2579 | rbcL = ycf5 |
| rbcL | rpoB | W+ = 104, W− = 127, n = 21, P ≤ 0.7022 | rbcL = rpoB |
| rbcL | ndhJ | W+ = 96, W− = 9, n = 14, P ≤ 0.004028 | rbcL > ndhJ |
| rbcL | rpoc1 | W+ = 98, W− = 22, n = 15, P ≤ 0.03015 | rbcL > rpoc1 |
| rbcL | accD | W+ = 66, W− = 105, n = 18, P ≤ 0.4171 | rbcL = accD |
| ycf5 | rpoB | W+ = 94, W− = 96, n = 19, P ≤ 0.9843 | ycf5 = rpoB |
| ycf5 | ndhJ | W+ = 44, W− = 1, n = 9, P ≤ 0.007812 | ycf5 > ndhJ |
| ycf5 | rpoc1 | W+ = 68, W− = 10, n = 12, P ≤ 0.021 | ycf5 > rpoc1 |
| ycf5 | accD | W+ = 46, W− = 59, n = 14, P ≤ 0.7148 | ycf5 = accD |
| rpoB | ndhJ | W+ = 297, W− = 406, n = 37, P ≤ 0.4153 | rpoB = ndhJ |
| rpoB | rpoc1 | W+ = 496, W− = 207, n = 37, P ≤ 0.02982 | rpoB > rpoc1 |
| rpoB | accD | W+ = 465, W− = 438, n = 42, P ≤ 0.8709 | rpoB = accD |
| ndhJ | rpoc1 | W+ = 243, W− = 82, n = 25, P ≤ 0.03135 | ndhJ > rpoc1 |
| ndhJ | accD | W+ = 322, W− = 174, n = 31, P ≤ 0.1498 | ndhJ = accD |
| rpoc1 | accD | W+ = 276, W− = 427, n = 37, P ≤ 0.2579 | rpoc1 = accD |
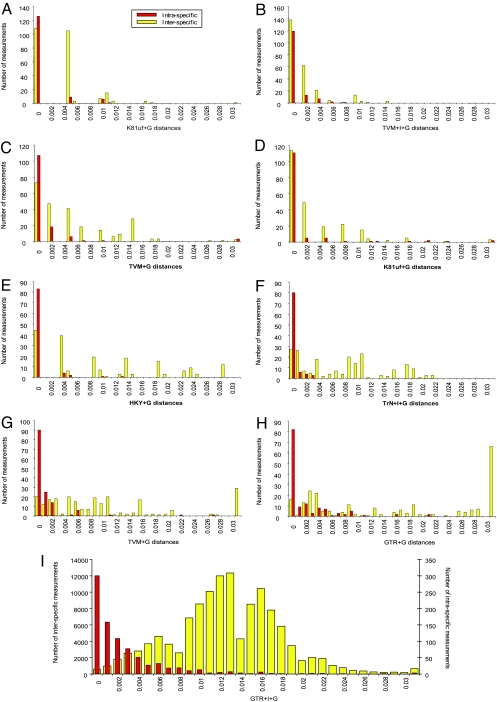
Ideally, barcodes must exhibit a “barcoding gap” between inter- versus intraspecific divergences (22). To evaluate whether such a gap is present, we looked at the distribution of divergences in classes of 0.001 distance units (Fig. 1). Median and Wilcoxon two-sample tests were significant in each case, i.e., the distribution and mean of intraspecific differences were lower than that of interspecific divergences, with the highest significances found for matK (Wilcoxon two-sample test, P < 0.0001), followed by trnH-psbA (Wilcoxon two-samples test, P < 0.0001; SI Table 9). We did not find, however, any large barcoding gap typical of cox1 in animals (22), although with matK in the Mesoamerican orchids matrix the distributions of intra- versus interspecific divergence are relatively well separated (Fig. 1I).
Fig. 1.
Relative distribution of interspecific divergence between con-generic species (yellow) and intraspecific distances (with best fit-model; red) for eight loci. (A) accD. (B) rpoC1. (C) rpoB. (D) ndhJ. (E) ycf5. (F) rbcL. (G) matK reduced matrix. (H) trnH-psbA. (I) matK expanded Mesoamerican orchids matrix. x axis, increments of 0.001; y axis, number of occurrences. Barcoding gaps were assessed with Median tests and Wilcoxon two-samples tests, and all were highly significant (P < 0.0001).
We evaluated for each barcode whether species are recovered as monophyletic, using phylogenetic techniques and bootstrap resampling. We compared the performance of potential barcodes in recovering species as monophyletic, using maximum parsimony (MP), likelihood, Bayesian, and distance methods. The trnH-psbA and matK barcodes both recovered the highest value of species monophyly [highest score with unweighted pair group method with arithmetic mean (UPGMA), 90.9%; Table 4]. These two barcodes also recovered the highest percentage of species monophyly with other tree building techniques than UPGMA but with lower percentages (Table 4). When we combined trnH-psbA with matK, the percentage of species monophyly did not increase notably, except with MP (+7%). Similarly when all barcodes were combined, the percentage of monophyly did not show much increase (93.1% recovered). Combining all potential barcodes did not provide 100% of species monophyly, and for example Faurea (Proteaceae), Ficus glumosa, and Ficus abutilifolia (Moraceae) were always polyphyletic, and the multiple accessions of the palm Hyphaene coriacea and orchid Prosthechea radiata did not cluster as single species.
Table 4.
Proportion (%) of monophyletic species recovered with different phylogenetic techniques and loci
| Dataset | UPGMA | NJ | MP | ML | BI |
|---|---|---|---|---|---|
| accD | 56.8 (36.3) | 45.4 (29.5) | 29.5 (27.2) | 31.8 (29.5) | 29.5 (29.5) |
| rpoc1 | 63.6 (38.6) | 40.9 (27.2) | 34 (29.5) | 34 (29.5) | 31.8 (34) |
| ndhJ | 63.4 (39) | 51.2 (36.5) | 39 (26.8) | 34.1 (26.8) | 34.1 (29.2) |
| ycf5 | 80 (66.6) | 60 (43.3) | 50 (46.6) | 53.3 (46.6) | 53.3 (53.3) |
| rpoB | 72.7 (56.8) | 61.3 (50) | 54.5 (50) | 59 (50) | 56.8 (54.5) |
| rbcL | 87.5 (78.1) | 65.6 (75) | 68.7 (68.7) | 71.8 (68.7) | 71.8 (71.8) |
| trnH-psbA | 90.6 (65.1) | 53.4 (32.5) | 76.7 (62.7) | 72 (60.4) | 69.7 (69.7) |
| matK | 90.6 (76.7) | 79 (76.7) | 79 (79) | 79 (79) | 79 (79) |
| matK + trnH-psbA | 90.9 (86.3) | 79.5 (70.4) | 86.3 (81.8) | 81.8 (75) | 81.3 (79.5) |
| All barcodes combined | 93.1 (84) | 72.7 (77.2) | 88.6 (86.3) | 88.6 (86.3) | 88.6 (88.6) |
Proportions supported by posterior probabilities or bootstrap >50% are in brackets.
Finally, we used coalescence analyses to compare the branching patterns along trees and identify distinct genetic clusters (24). The highest number of independent clusters was found by using UPGMA with matK (SI Fig. 2), followed by rpoB and trnH-psbA (Table 5). With matK, 41 clusters were identified, of which 30 fully correspond to previously recognized taxonomic species, 4 partially matched taxonomic species (i.e., failed to group all representatives into a single cluster), and 7 mixed species together (Table 5 and SI Fig. 2). With rpoB, 36 clusters were identified, of which 20 fully correspond to taxonomic species; whereas with trnH-psbA, 34 clusters were identified, which a slightly higher proportion corresponding to previously recognized species (i.e., 19 clusters).
Table 5.
Coalescence analyses indicating the number of independent genetic clusters and their correspondence with taxonomically recognized species
| Dataset | No. of Genetic Clusters | Full match | Partial match | No match | |
|---|---|---|---|---|---|
| UPGMA | Combined | 31 | 23 | 3 | 5 |
| matK + trnHpsbA | 33 | 12 | 19 | 2 | |
| accD | 33 | 17 | 3 | 13 | |
| matK | 41 | 30 | 4 | 7 | |
| ndhJ | 28 | 15 | 3 | 10 | |
| rbcL | 28 | 20 | 5 | 3 | |
| rpoB | 36 | 20 | 5 | 11 | |
| rpoC1 | 29 | 13 | 3 | 13 | |
| trnH-psbA | 34 | 19 | 12 | 3 | |
| ycf5 | 16 | 7 | 0 | 9 | |
| MP branch lengths plus NPRS | Combined | 8 | 3 | 4 | 1 |
| matK + trnHpsbA | 11 | 7 | 3 | 1 | |
| accD | 20 | 13 | 3 | 4 | |
| matK | 20 | 16 | 1 | 3 | |
| ndhJ | 20 | 16 | 1 | 3 | |
| rbcL | 21 | 17 | 3 | 1 | |
| rpoB | 17 | 14 | 0 | 3 | |
| rpoC1 | 16 | 10 | 0 | 6 | |
| trnH-psbA | 16 | 13 | 3 | 0 | |
| ycf5 | 15 | 10 | 3 | 2 | |
| ML plus NPRS | Combined | 32 | 26 | 2 | 4 |
| matK + trnHpsbA | 11 | 9 | 1 | 1 | |
| accD | 20 | 13 | 3 | 4 | |
| matK | 23 | 17 | 2 | 4 | |
| ndhJ | 16 | 9 | 2 | 5 | |
| rbcL | 20 | 16 | 3 | 1 | |
| rpoB | 19 | 14 | 1 | 4 | |
| rpoC1 | 18 | 10 | 1 | 7 | |
| trnH-psbA | 16 | 15 | 1 | 0 | |
| ycf5 | 14 | 9 | 3 | 2 |
Altogether, our results indicate that either matK or trnH-psbA are the most suitable regions for plant DNA barcoding. In this sense, we agree with previous relatively small-scale studies that focused on the nutmeg family [Myristicaceae (27)] or the 50-acre forest of the New York Botanical Garden (28). Because several matK sequences were already available in GenBank for orchids, we expanded our Costa Rican sampling and compiled a large matrix for Mesoamerican species, assembling 1,566 DNA sequences. Coalescence analyses from the UPGMA tree identified 212 genetic clusters, of which 86 fully matched previously recognized species and a further 25 partially matched taxonomic species (SI Fig. 3). An examination of these clusters reveals cryptic species, which need further taxonomic work. For example, we sequenced four accessions of Lycaste tricolor (Klotszch.) Rchb. (numbered 841, 840, 838, and 1011 in SI Table 10). Lycaste cf. tricolor 1011 does not cluster with the other three accessions and taxonomists had indeed suspected it could be another, separate, species. Lycaste cf. tricolor 1011 grows on the Pacific slopes of Costa Rica, whereas the other three representatives (i.e., the “typical” L. tricolor) grow on the Atlantic side. There are also discrete morphological differences. The pollinarium of Lycaste cf. tricolor 1011, like all other representatives of that species in the Pacific slope, have long stipes, a twisted column and a hairy anther cap, whereas the typical tricolor have short stipes, a straight column and a smooth anther cap (SI Fig. 4). These differences in column are probably also involved in reproductive isolation whereby L. cf. tricolor 1011 would deposit pollinia on the shoulder of the pollinating bees and the “true” tricolor, with their straight column, would deposit pollinia on the back of the bees.
Our sampling is more comprehensive than previous studies on DNA barcoding in plants. Kress et al. (13) used 19 species with duplicates/triplicates and a further 83 species with only one representative per species. Kress and Erickson (16) used 48 pairs of species, each represented by one sample. Cowan et al. (29) and Chase et al. (15) report that the plant working group started with 96 pairs of taxa but narrowed it down to fewer species. Cameron used 343 species from within the botanical garden in New York (28). We used here 86 species in which all barcodes were tested and a further 1,036 orchid species in the dataset restricted to matK. Because the assessment of intraspecific variability is crucial for deciding on a suitable barcode, we included 44 species in which there were at least two and up to seven representatives per species. Our results are robust and all point toward the same pair of loci. Given that the second half (5′ end) of the matK exon is easy to amplify and align, we propose that matK is used as a preferred universal DNA barcode for flowering plants. The trnH-psbA region performs nearly equally well, although its pattern of molecular evolution is complex. Therefore, we propose that trnH-psbA is used as either an alternative to matK or a complementary barcode to matK. When combined, these loci achieve only moderate improvement, as shown by our analyses of recovering species monophyly.
The use of matK as a barcode has been criticized mainly because no universal primers were available (15), hence it had the lowest amplification success in Kress and Erickson (16). However, we found that primers 390F and 1326R from Cuénoud et al. (26) amplify the same region with a 100% success. The use of trnH-psbA has been criticized because of the difficulty in the alignment due to extensive length variation and because certain species host a pseudogene (15). Although in certain cases trnH-psbA might indeed be problematic, we found here that it was one of the most useful regions across a wide range of angiosperms.
Using matK alone or in combination with trnH-psbA, our tests of monophyly reach >90% of correct species identification. If our sampling was restricted to sister species rather than natural geographic assemblages of species, we may have found this value to drop. However, our samples do include very closely related species, given that Costa Rica and southern Africa both have experienced extensive rapid radiations (30, 31).
Apart from combining matK and trnH-psbA, we found that adding the other barcodes did not improve species identification by >3% and therefore was not worth pursuing if one balances gains in identification versus sequencing efforts. It is possible that some regions yet untested here may be useful as a complementary barcode, and we await further studies. Alternatively, we may need to accept that no more than ≈90% of species will be identified with universal plastid barcodes and that those difficult lineages will need “case-by-case” analyses, using, for example, nuclear population genetic markers and taking advantage of recent developments in DNA sequencing technology (32).
Our results differ from the proposal of Kress and Erickson (16) in the sense that we advocate matK rather than rbcL, although we agree with the utility of trnH-psbA. As explained above, the amplification of matK is not problematic, as Kress and Erickson thought before, and the pattern of variation in its second half (5′ end) is particularly appropriate for its use as a DNA barcode, as exemplified by our large-scale analysis in orchids. The matK gene also presents another advantage: its first half (3′ end) was useful to reconstruct the phylogeny of angiosperms (33), and therefore the complete sequence of this gene can be used as dual barcode-phylogenetic marker. The matK gene has an unusual mode and tempo of evolution; it is the only putative chloroplast-encoded group II intron maturase, and its function relates to the regulation of plant development. Analyses of the expression of this gene suggested that “genetic buffers” are in operation and constrain its evolution, which may explain why relatively low intraspecific but high interspecific variation is found and therefore why it fits DNA barcoding purposes so well. We disagree with Kondo et al. (34), who argued that matK on its own was not useful for species identification, but their study focused exclusively on species of liquorices in the legume family. We also disagree with the proposal of Chase et al. (15), because we found that neither rpoC1 nor rpoB were performing well as a barcode (Tables 1–5). These two loci amplify easily in non-angiosperms (15), but we found that they were too conserved in angiosperms. It might in fact not be so important to design primer pairs or barcodes that work universally from ferns, mosses, to seed plants. Several of the DNA barcoding applications (e.g., rapid inventories for conservation) may not need to identify non-seed plants at the species level, and alternatively if this was required then moss- and fern-specific primers or barcodes could be used in complement to seed plant barcodes. In the meantime, we propose that DNA barcoding with matK is used on a large scale.
DNA barcoding with matK alone (or matK plus trnH-psbA combined) has the potential to speed up the exploration and preservation of plant life on Earth by facilitating considerably biodiversity inventories beyond South Africa and Costa Rica. In addition, new methods are now being developed in which DNA barcoding data can be used in conservation (35). As an example, we illustrate how customs officers could use DNA barcoding to identify plant fragments from species in which trade is controlled by the Convention on International Trade of Endangered Species (CITES). All orchids are in Appendix 2 of CITES [i.e., a special permit is required for their trade (www.cites.org)], but a few species, such as the lady's slipper orchids in Mesoamerica (genus Phragmipedium), are so threatened in the wild that their trade is prohibited altogether (i.e., they are listed in Appendix 1 of CITES). We included in our large matK matrix one sequence of Phragmipedium as a reference and ran a UPGMA analysis with all 1,500+ orchids with 10 additional Phragmipedium sequences representing another seven species (GenBank accession nos. AY918826–31, AJ581442, AY557204). All species of Phragmipedium clustered together correctly. This means that in our theoretical case, using our proposed DNA barcode, the custom services would have positively identified species from CITES Appendix 1 (i.e., the lady's slipper orchids) from species in Appendix 2 (i.e., the other orchids) and those not listed by CITES (here, the species from the KNP).
To ensure even longer-term benefit of the DNA barcoding efforts, it is also essential to put in place DNA banking strategies (36) so that complementary barcodes to the ones identified here can be produced in the future. More importantly, if DNA barcoding is to achieve its goals, it must urgently become available to countries rich in biodiversity but poor in resources through efficient capacity building and judicious funding programs.
Methods
Sampling.
In total, 1,667 taxa were sampled (SI Table 10). In the KNP, we collected 101 specimens of trees, shrubs, and achlorophyllous parasites, including 32 species in which we have more than one representative per species. The first dataset of Costa Rican orchids comprises 71 specimens representing 48 species in which 12 have more than one representative per species. A second orchid dataset was assembled with matK only, but with a much increased taxon sampling with a total of 1,566 specimens representing 1,084 species from Mesoamerica in which 295 have at least two representatives.
DNA Sequencing.
Total DNA was extracted by using the method of Doyle and Doyle (37). We amplified and sequenced accD, rpoC1, rpoB, ndhJ, matK, and ycf5, following guidelines from the plant working group. For matK, additional primers 390F and 1326R (26) were used. Primers trnHf and psbA3′f were used for trnH-psbA (13). For the first half of the rbcL exon, primers 1F and 724R were used following Kress et al. (13). DNA sequences were aligned in PAUP4b10 (38).
Genetic and Phylogenetic Analyses.
Inter- and intraspecific genetic divergences were calculated following Meyer and Paulay (22). Pairwise distances were calculated with PAUP4b10 (38) and the best-fitting model as given by applying MODELTEST 3.7 (39). Wilcoxon signed rank tests were performed to compare intra- and interspecific variability for every pairs of barcodes following Kress and Erickson (16). We evaluated DNA barcoding gaps by comparing the distribution of intra- versus interspecific divergences (22). To evaluate whether species were recovered as monophyletic with each barcode, we used standard phylogenetic techniques: MP, maximum likelihood (ML), neighbor joining (NJ), and UPGMA with PAUP4b10 (38). Bayesian statistical inferences (BI) were performed with MrBayes software, Version 3.1.2 (40). The parsimony analysis of the large matK matrix of Mesoamerican orchids was performed by using the parsimony ratchet method (41). We identified genetic clusters by coalescence analyses, using methods developed by Pons et al. (23) and Fontaneto et al. (24). Details are available from the corresponding author upon request.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the KNP, South African National Parks, H. Eckhardt, I. Smit, G. Zambatis, T. Khoza, Ministerio de Ambiente y Energía, and Sistema Nacional de Areas de Conservación, for granting access to the park and sharing data; M. Chase, R. Cowan, M. Powell, H. van Niekerk, two anonymous reviewers, and the editor for comments; and T. Rikombe, R. Bryden, T. Mhlongo, H. van der Bank for fieldwork. This work was supported by the South African National Research Foundation, the University of Johannesburg, the United Kingdom Darwin Initiative, The Royal Society (U.K.), and the European Commission (HOTSPOTS Consortium).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU254252–EU254410 and EU213263–EU214530.
See Commentary on page 2761.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709936105/DC1.
References
- 1.Ebach MC, Holdrege C. Nature. 2005;434:697. doi: 10.1038/434697b. [DOI] [PubMed] [Google Scholar]
- 2.Will KW, Mishler BD, Wheeler QD. Syst Biol. 2005;54:844–851. doi: 10.1080/10635150500354878. [DOI] [PubMed] [Google Scholar]
- 3.Blaxter ML. Philos Trans R Soc London Ser B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajibabaei M, de Waard JR, Ivanova NV, Ratnasingham S, Dooph RT, Kirk SL, Mackie PM, Hebert PDN. Philos Trans R Soc London Ser B. 2005;360:1959–1967. doi: 10.1098/rstb.2005.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janzen DH. Philos Trans R Soc London Ser B. 2004;359:731–732. doi: 10.1098/rstb.2003.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janzen DH, Hajibabaei M, Burns JM, Hallwachs W, Remigio E, Hebert PDN. Philos Trans R Soc London Ser B. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert PDN, Cywinska A, Ball SL, De Waard JR. Proc R Soc Biol Sci Ser B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith MA, Fisher BL, Hebert PDN. Philos Trans R Soc London Ser B. 2005;360:1825–1834. doi: 10.1098/rstb.2005.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vences M, Thomas M, Bonett RM, Vieites DR. Philos Trans R Soc London Ser B. 2005;360:1859–1868. doi: 10.1098/rstb.2005.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. Philos Trans R Soc London Ser B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubinoff D, Cameron S, Will K. Trends Ecol Evol. 2006;21:1–2. doi: 10.1016/j.tree.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Pennisi E. Science. 2007;318:190–191. doi: 10.1126/science.318.5848.190. [DOI] [PubMed] [Google Scholar]
- 13.Kress JW, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Proc Natl Acad Sci USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, Cheng CH, Lin CY, Liu SM, Chang CC, et al. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 15.Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, Petersen G, Seberg O, Jorgsensen T, Cameron KM, Carine M, et al. Taxon. 2007;56:295–299. [Google Scholar]
- 16.Kress WJ, Erickson DL. PLoS One. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newmaster SG, Fazekas AJ, Ragupathy S. Can J Bot. 2006;84:335–341. [Google Scholar]
- 18.Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E. Nucleic Acids Res. 2007;35:e1–e8. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 20.Myers N. Bioscience. 2003;53:916–917. [Google Scholar]
- 21.van der Schijff HP. Publikasies van die Universiteit van Pretoria, Nuwe reeks. 1969;53:1–100. [Google Scholar]
- 22.Meyer CP, Paulay G. PLoS Biol. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. Syst Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- 24.Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. PloS Biol. 2007;5:914–921. doi: 10.1371/journal.pbio.0050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron KM, Chase MW, Whitten WM, Kores PJ, Jarrell DC, Albert VA, Yukawa T, Hills HG, Goldman DH. Am J Bot. 1999;86:208–224. [PubMed] [Google Scholar]
- 26.Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW. Am J Bot. 2002;89:132–144. doi: 10.3732/ajb.89.1.132. [DOI] [PubMed] [Google Scholar]
- 27.Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J. Mol Ecol Notes. 2008 doi: 10.1111/j.1471-8286.2007.02002.x. [DOI] [PubMed] [Google Scholar]
- 28.Cameron K. The Botanical Society of America. Botany and Plant Biology 2007 Joint Congress. Chicago: The Botanical Society of America; 2007. [Google Scholar]
- 29.Cowan RS, Chase MW, Kress JW, Savolainen V. Taxon. 2006;55:611–616. [Google Scholar]
- 30.Linder HP. Biol Rev. 2003;78:597–638. doi: 10.1017/s1464793103006171. [DOI] [PubMed] [Google Scholar]
- 31.Gravendeel B, Smithson A, Slik FJW, Schuiteman A. Philos Trans R Soc London Ser B. 2004;359:1523–1535. doi: 10.1098/rstb.2004.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilu K, Borsch T, Müller K, Soltis DE, Soltis PS, Savolainen V, Chase MW, Alice L, Evans R, Sauquet H, et al. Am J Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- 34.Kondo K, Shiba M, Yamaji H, Morota T, Zhengmin C, Huixia P, Shoyama Y. Biol Pharm Bull. 2007;30:1497–1502. doi: 10.1248/bpb.30.1497. [DOI] [PubMed] [Google Scholar]
- 35.Faith DP, Baker A. Evol Bioinf Online. 2006;2:70–77. [Google Scholar]
- 36.Savolainen V, Reeves G. Science. 2004;304:1445. doi: 10.1126/science.304.5676.1445b. [DOI] [PubMed] [Google Scholar]
- 37.Doyle JJ, Doyle JL. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 38.Swofford DL. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (* and other methods) Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 39.Posada D. Nucleic Acids Res. 2006;34:W700–W703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. Bioinformatics (Oxford) 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Nixon KC. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.