Abstract
In contrast to other cereals, typical barley cultivars have caryopses with adhering hulls at maturity, known as covered (hulled) barley. However, a few barley cultivars are a free-threshing variant called naked (hulless) barley. The covered/naked caryopsis is controlled by a single locus (nud) on chromosome arm 7HL. On the basis of positional cloning, we concluded that an ethylene response factor (ERF) family transcription factor gene controls the covered/naked caryopsis phenotype. This conclusion was validated by (i) fixation of the 17-kb deletion harboring the ERF gene among all 100 naked cultivars studied; (ii) two x-ray-induced nud alleles with a DNA lesion at a different site, each affecting the putative functional motif; and (iii) gene expression strictly localized to the testa. Available results indicate the monophyletic origin of naked barley. The Nud gene has homology to the Arabidopsis WIN1/SHN1 transcription factor gene, whose deduced function is control of a lipid biosynthesis pathway. Staining with a lipophilic dye (Sudan black B) detected a lipid layer on the pericarp epidermis only in covered barley. We infer that, in covered barley, the contact of the caryopsis surface, overlaid with lipids to the inner side of the hull, generates organ adhesion.
Keywords: caryopsis, domestication, epidermis, ethylene response factor, grass
Barley (Hordeum vulgare L.) is the world's fourth most important cereal crop behind wheat, rice, and maize. A particular botanical feature of domesticated barley is that most cultivars have covered (hulled) caryopses in which the hull (outer lemma and inner palea) is firmly adherent to the pericarp epidermis at maturity; but a few cultivars are of a free-threshing variant called naked (hulless) barley (Fig. 1). No other Poaceae (grass) family crops show such hull-caryopsis adhesion. Both caryopsis types of barley have agronomic value and are used for different purposes. Covered barley is mainly used as an animal feed and for brewing. The hull of covered barley protects embryos from damage during mechanical harvest, and it also provides a filtration medium in separation of fermentable extract (wort) during malt processing (1). In contrast, naked barley is preferred for human food, because extensive pearling to remove the hull is unnecessary. Now that healthy effects of the soluble-fiber-rich barley products have been officially approved (2, 3), consumers' current interest in nutrition might boost the status of barley as human food.
Fig. 1.
Morphology of covered (Nud) and naked (nud) barley represented by the Bowman isogenic lines. (A) Mature caryopses of covered (left two) and naked (right two) barley. In each pair, dorsal (Left) and ventral (Right) sides are shown. (Scale bar: 5 mm.) (B) Part of a mature spike of covered (Left) and naked (Right) barley. (Scale bar: 1 cm.)
Easy processing of edible part can be a primary character of selection during domestication of a food crop (4, 5). Consequently, the naked caryopsis is considered a key domestication character in barley (5–8). The wild progenitor of barley, H. vulgare subsp. spontaneum, has covered grains. The covered grain is therefore considered adaptive in the wild: the hulls protect the caryopses from various biotic and abiotic stresses, and the awn attached to the distal end of the lemma facilitates seed dispersal and burial (9). According to archeological evidence (4), the earliest domestication event in barley was acquisition of nonbrittle rachises at ≈10,500 to ≈9,500 years before present (yBP), followed by the appearance of six-rowed spikes at ≈8,800 to ≈8,000 yBP. Then, naked barley appeared at ≈8,000 yBP. Naked barley, mostly with six-rowed spikes, has been excavated from many Neolithic agricultural settlement sites in the Near East and western India (4, 8). It is believed that quick spread of naked barley happened in ancient times, reaching areas in Turkey, western and northern Europe, and Scandinavia and throughout Asia and Ethiopia (6). Although naked barley is today distributed worldwide, it is frequent in East Asia, especially in the highlands of Nepal and Tibet. Its earlier maturity than that of wheat fostered its adaptation to highlands, where the growing season is short; naked barley became a staple food there. About the origin of naked barley, different interpretations (mono versus multiple origins) have been proposed (6, 10). Because the easily noticeable naked caryopsis trait can be subject to human selection, the current distribution of naked barley is unlikely to reflect its geographic origin accurately (5). Our work in ref. 11, based on molecular variation of a marker tightly linked to the nud locus, suggested the monophyletic origin of naked barley, but the issue remains unsolved yet. Recent comprehensive molecular evolutionary studies on the barley crop as a whole favor the interpretation of multiple domestication events at different locations (12, 13).
The covered/naked caryopsis in barley is controlled by a single locus (nud, for nudum) located on chromosome arm 7HL (14); the covered caryopsis allele (Nud) is dominant over the naked one (nud). Harlan (15) reported that, in covered barley, a sticky adhesive substance appears 10 days after flowering on the caryopsis surface and that the substance is produced by the caryopsis, not by the hull. Transmission electron microscopy showed that a cementing substance is secreted from the pericarp epidermis 2 days after flowering. Its thickness increases during grain development, but its chemical composition remains unknown (1). We have been attempting map-based isolation of the Nud/nud gene (16, 17). The present study reports molecular cloning of the Nud/nud gene. We also performed histochemical and biochemical analyses to elucidate the mechanisms controlling the covered/naked caryopsis trait. Finally, on the basis of the molecular variation of the Nud/nud gene itself, the issue of the origin of naked barley is revisited.
Results
Positional Cloning of Nud.
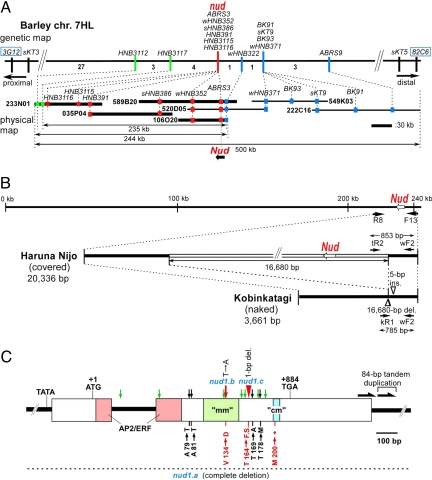
We screened 2,828 progeny segregating for the trait from two cross combinations, using flanking markers developed in ref. 17, and nud was delimited to a 0.64 cM interval between markers sKT3 and sKT9 (Fig. 2A). We then exploited barley/rice microsynteny for further localization. Integration of the flanking markers into a high-density barley expressed sequence tag (EST) map (18) selected two barley ESTs flanking the nud locus (accession nos. BJ462032 for marker 3G12 and AV935407 for marker 82C6). BLASTN analysis identified their respective homologous rice ESTs (accession nos. AK068856 and AK070667) 370 kb apart on rice chromosome arm 6L. Two rice genes (accession nos. AK061163 and AK121264) within the collinear region were successfully used as vehicles to develop closer barley markers (ABRS3 and ABRS9). Starting with markers sKT9 and ABRS3, we screened the bacterial artificial chromosome (BAC) library of the covered barley cultivar Haruna Nijo (19). Seven rounds of chromosome walks selected 20 BAC clones [supporting information (SI) Table 1], and a 500 kb-contig spanning the nud locus was constructed (see SI Table 2 for markers used for BAC contig assembly). An ≈235-kb region cosegregated with nud. In the physical map, the nud locus was covered completely with four overlapping BAC clones (Fig. 2A). Annotation of the sequenced BAC contig showed two predicted genes. Approximately 50% of the contig sequence was classified as repetitive elements (SI Table 3). An ethylene response factor (ERF) family transcription factor was the only gene that lies in the region delimited by the genetic and physical mapping and, therefore, is considered as a Nud candidate gene.
Fig. 2.
Positional cloning of naked caryopsis gene (nud) in barley. (A) High-resolution genetic map and physical map. Thick lines show four BAC clones sequenced. The recombinant numbers are shown below the genetic map. (B) Structural differences near the nud locus between covered barley (Haruna Nijo) and naked barley (Kobinkatagi). Black arrows indicate the appropriate location of PCR primers. (C) Structure of the Haruna Nijo Nud gene (standard) and nucleotide changes found in two radiation-induced naked mutant alleles, nud1.b (red arrow) and nud1.c (red arrowhead). Boxes indicate exons, and the black bar between the boxes shows an intron. Deduced functionally important domain/motif(s) are colored. The motif names, “mm” (middle motif) and “cm” (C-terminal motif), follow Aharoni et al. (21). The asterisk indicates a stop codon resulting from the frame shift (F.S.). Natural allelic variations found among 33 covered lines are also indicated by green arrows (synonymous substitutions) and black arrows (nonsynonymous substitutions).
To isolate the candidate gene from naked cultivars, we attempted PCR-amplification, using the primer pair ABRS3 shown in SI Table 2. However, no fragment amplified in any naked cultivar tested. Similarly, all other PCR primer pairs designed for every 2-kb interval in the region between 10.8 kb upstream and 2.8 kb downstream of the ERF gene failed amplification specifically in naked cultivars. A long PCR was attempted with a primer pair HNB32C2 F13-R8 (Fig. 2B and SI Table 4). A 3.6-kb fragment was amplified from naked cultivars, whereas the control PCR, using DNA of BAC HNB 106O20 as a template, amplified the expected ≈20-kb band. Sequencing of the 3.6-kb fragment obtained from two naked lines [Kobinkatagi (a Japanese landrace) and nud-Bowman (an isogenic line carrying the nud allele in the genetic background of the covered cultivar Bowman)] revealed a deletion of 16,680 bp relative to the corresponding region of the Haruna Nijo BAC contig sequence (Fig. 2B). The 16,680-bp deletion (hereafter called the 17-kb deletion) included the entire ERF gene. Thus, the gene structure analysis of naked cultivars supports the candidacy of the ERF gene.
Sequence Analysis of Induced Naked Mutants.
We then analyzed the candidate gene sequences in two x-ray-induced naked mutants (20). We had confirmed their allelism to the nud locus through crossing experiments. Sequence analysis showed that each of the two induced naked mutants carried a different single base mutation in the putative functional motif of the ERF gene, but their wild-type varieties (Haisa's and Ackermann's Donaria, respectively) had a nucleotide sequence that is identical to that of Haruna Nijo (Fig. 2C and SI Fig. 5). Mut.4129 had a T to A nucleotide substitution in the second exon that resulted in a valine (V) to aspartic acid (D) change at position 134. Mut.3041/6a had a 1-bp deletion in the second exon that caused a frame shift and generated a premature stop codon resulting in a C-terminally truncated 199-amino acid protein (Fig. 2C and SI Fig. 6). The sequence analysis of the induced mutants validated that the ERF gene comprises the nud locus, and, therefore, we will refer to the gene as nud. We designated the null (deletion) allele in naked cultivars as nud1.a, and the induced naked mutant alleles as nud1.b (Mut.4129) and nud1.c (Mut.3041/6a), respectively, according to Franckowiack and Konishi (14).
Natural Variation of the nud Locus.
Six additional naked cultivars of diverse origin [one Iranian, two Turkish, two Ethiopian, and one Nepalese] were directly sequenced for the ≈1.4-kb region encompassing the deletion point, using primer pairs shown in SI Table 4; they all shared an identical nucleotide sequence with the two standard naked lines. We developed a simple PCR assay to determine the presence or absence of the 17-kb deletion (Fig. 2B, SI Table 4, and SI Methods). A survey of 259 world barley lines selected by Taketa et al. (11) revealed that all 100 naked cultivars shared the 17-kb deletion (nud1.a allele) and that none of 159 covered lines had the deletion. These results indicate the monophyletic origin of naked barley.
Natural variation of Nud was studied in 33 (12 domesticated and 11 wild) covered lines of diverse origin. Sequencing of an ≈1.7-kb region of Nud, including 5′-noncoding and 3′-noncoding regions, detected various types of nucleotide polymorphisms at 16 sites relative to the standard sequence of Haruna Nijo (SI Table 5). In the coding region, 11 types of single nucleotide substitution were detected; all were in the second exon. All four nonsynonymous substitutions were located outside the AP2/ERF domain, “mm” or “cm” motifs (Fig. 2C). Nucleotide changes were also observed in the first intron and 5′-noncoding and 3′-noncoding regions. Variable microsatellites were also found. In the 3′-noncoding region, an 84-bp tandem duplication was detected in most domesticated lines, but no wild lines sequenced carried it. Taken together, our survey of natural allelic variation supported the identity of the nud gene.
Characterization of Nud.
The Nud gene consists of two exons and one intron, and the ORF encodes a deduced protein of 227 aa (Fig. 2C and SI Fig. 5). The deduced amino acid sequence of Nud is 59% and 74% identical to the Arabidopsis WAX INDUCER1/SHINE1 (WIN1/SHN1) protein (21, 22) and a rice putative ERF protein on chromosome 6 (Os06ERF), respectively (SI Fig. 6). Both rice homologs had an intron in the AP2/ERF domain positioned 83 bp from the start codon, as is observed also for Nud, whereas three Arabidopsis homologs (WIN1/SHN1, SHN2, and SHN3) all contained a single intron positioned 3 bp upstream at 80 bp from the start codon (21).
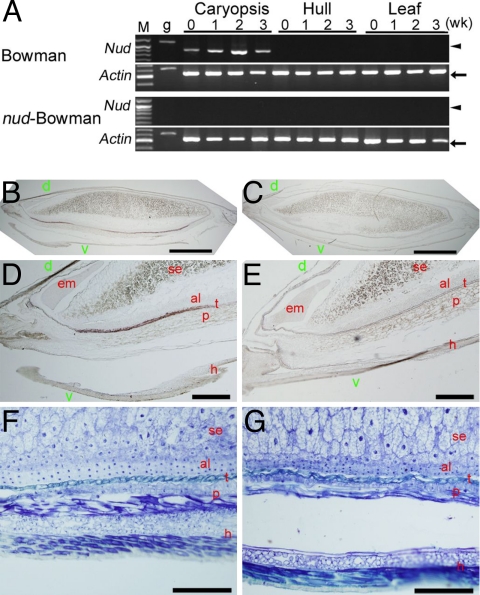
Isogenic lines of the covered cultivar Bowman and its naked version (nud-Bowman) were used in the following analyses (for details, see SI Methods). RT-PCR showed that, in Bowman, Nud was expressed only in the caryopsis with a peak at two weeks after anthesis; no expression was detected in hulls or leaves (Fig. 3A). In nud-Bowman, no expression was detected in any tissue examined. To know the spatial expression, RNA in situ hybridization using the antisense probe was performed. Results showed that, in Bowman, Nud was expressed predominantly in the testa of the ventral side, and very weakly on the dorsal side (Fig. 3 B–F). In nud-Bowman, no signal above background was detected (Fig. 3 C–G). These results revealed that Nud was expressed in the tissues where adherence occurs.
Fig. 3.
Expression patterns of Nud in the Bowman isogenic lines. (A) RT-PCR analysis of Nud expression. Lane M is a 100-bp ladder marker (arrowheads and arrows indicate 1,000 bp and 500 bp, respectively). Lane g represents amplicons using barley genomic DNA as the template. Numbers indicate weeks (wk) after anthesis. (B–E) RNA in situ hybridization with the antisense probe against longitudinal sections of 2-week-old caryopsis. (B and D) Entire caryopsis and close-up of the micropylar end of Bowman caryopsis. (C and E) Corresponding areas of nud-Bowman. (F and G) Toluidine blue O stained ventral-side caryopsis section in Bowman (F) and nud-Bowman (G). d, dorsal side; v, ventral side; em, embryo; se, starchy endosperm; al, aleurone layers; t, testa; p, pericarp; h, hull (palea). (Scale bars: B and C, 1 mm; D and E, 500 μm; F and G, 200 μm.)
Histochemical Analyses.
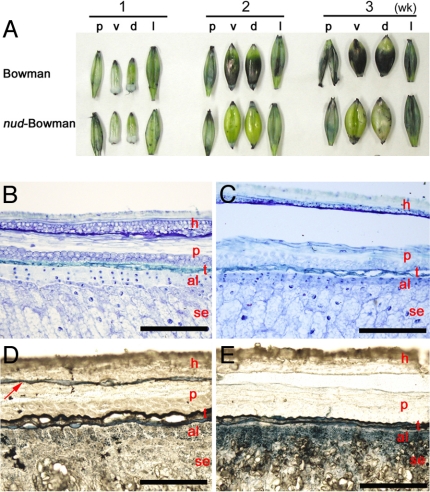
When WIN1/SHN1 was overexpressed in Arabidopsis, shiny and twisted leaf phenotype appeared. Leaves and flowers accumulated different compositions and increased amounts of wax and cutin on the epidermis (21–23). Wax and cutin are two main components of the cuticle (24). Biosyntheses of wax and cutin initially share a common lipid pathway starting from the same fatty acid precursors but later diverged into distinct pathways of fatty acid modification (25). To determine whether developing barley caryopses accumulate lipids on their surface, we attempted staining with the lipophilic dye Sudan black B (Fig. 4A). In Bowman, 1-week-old dehulled caryopses were not stained, but 2- and 3-week-old dehulled caryopses showed strong staining on their surface except for areas over the embryo. Caryopses of nud-Bowman were not stained at any stage studied. In barley grains, the pericarp and testa lie between the hull and the outermost aleurone layer. The pericarp consists of several layers of cells including parenchyma cells and cross cells. Inside the pericarp is the testa, consisting of two layers of cells (26). Sudan black B staining of longitudinal sections from 2-week-old caryopses detected a clear lipid layer on the pericarp epidermis of Bowman only (Figs. 4 B–E). In both genotypes, the inner sides of the lemma and palea were weakly stained. Similarly, 2-week-old dehulled caryopses of the two induced naked mutants were not stained with Sudan black B, but those of their original varieties were heavily stained (data not shown). Therefore, the presence or absence of the lipid layer on the pericarp epidermis is a critical difference that separates covered and naked barley.
Fig. 4.
Caryopses stained with a lipophilic dye Sudan black B. (A) Hull and dehulled caryopses 1–3 weeks after anthesis. p, palea; v, ventral side of caryopsis; d, dorsal side of caryopsis; l, lemma. Toluidine blue O (B and C) and Sudan black B (D and E) staining of the dorsal-side caryopsis section. (B and D) Bowman. (C and E) nud-Bowman. Red letters are the same as in Fig. 3, except that h indicates lemma. The arrow indicates the lipid layer found only in Bowman. (Scale bars: 200 μm.)
Pericarp Permeability Analyses.
In Arabidopsis WIN1/SHN1 overexpression plants, both chlorophyll elution and water loss were enhanced (21). We conducted similar experiments, using dehulled caryopses 2 weeks after anthesis. Both chlorophyll elution and water loss through the pericarp epidermis were faster in Bowman than in nud-Bowman (SI Fig. 7), suggesting that the pericarp epidermis in covered barley has enhanced permeability.
Lipid Analyses.
TLC of surface lipids revealed clear differences between caryopses and hulls in each Bowman isogenic line. However, neither hull lipids nor caryopsis lipids exhibited apparent differences between the Bowman isogenic lines (SI Fig. 8). We presume that lipids on the dehulled-caryopsis surface of covered barley are resistant to the extraction protocol used here, because the caryopses after the extraction treatment were still strongly stained with Sudan black B.
Discussion
Using positional cloning, we identified an ERF family transcription factor gene as Nud in barley. This conclusion was validated by (i) annotation of the candidate region delimited by high-resolution genetic and physical mapping, (ii) fixation of the 17-kb deletion harboring the ERF gene among all 100 naked cultivars studied, (iii) discovery of a nonsynonymous substitution(s) affecting the putative functional motif of the ERF gene in two x-ray induced nud alleles, and (iv) testa-specific gene expression. As discussed later, information from Arabidopsis WIN1/SHN1 can help explain the drastic morphological change from covered to naked caryopses in barley as a result of recessive nud mutations.
Function of the Barley Nud Gene.
The Arabidopsis genome includes 122 ERF family transcription factors (27). Characterization of the limited members suggests their important roles in plant morphogenesis and stress responses (27–30). The ERF family is classified into 10 subfamilies, and WIN1/SHN1 belongs to subfamily V (27). Functions of Arabidopsis WIN1/SHN1 are being investigated intensively. To date, 35S:WIN1/SHN1 overexpression lines showed altered composition and increased accumulation of epidermal lipids, both in leaves and flowers, whereas the WIN1/SHN1 RNAi lines with incomplete gene silencing exhibited opposite effects on lipids (21–23). Strangely, WIN1/SHN1 overexpression lines showed a shiny leaf phenotype despite an increased lipid deposition on the epidermis. This conflict was ascribed to altered compositions of epidermal cuticle (21, 23). However, promoter:GUS reporter experiments showed that, in normal plants, the WIN1/SHN1 gene is predominantly expressed in areas of cell separation such as abscission and dehiscence zones. These observations suggest that the WIN1/SHN1 gene plays important dual roles in the regulation of lipid biosynthesis pathways (23) and in control of adequate tissue separation during development (21).
By analogy to the deduced functions of the Arabidopsis WIN1/SHN1 summarized above, the barley Nud gene probably regulates composition of lipids deposited on the pericarp epidermis (Fig. 4A). Hydrophobic lipids are synthesized in plastids within a cell. They need to move both intracellularly and intercellularly and finally be secreted to the epidermis through mediation of various transfer systems (31). Pighin et al. (32) showed that cuticular lipid export from Arabidopsis stem epidermis requires an adenosine triphosphate binding cassette transporter. The discrepancy between the site of Nud transcription in the testa and the heavy lipid accumulation on the pericarp surface might be explained as follows: Nud transcription factor protein might activate production of special lipids in the testa, so either final or intermediate lipids produced there are transported through the pericarp layers and are secreted out of the pericarp epidermis.
The epidermal lipids in plants are considered to serve as a repellent, providing a waterproof protective cover and preventing abnormal tissue adhesion during development (24). In covered barley, however, the epidermal lipid layer on the caryopsis appears to exert opposite effects. In naked barley, the lack of the lipid layer probably blocks adhesion, thereby rendering free-threshing caryopses. Organ fusion mutants showing abnormal adhesion between epidermal cell walls have been reported in various plants. Well known examples are the maize adherent 1 (ad1), showing fusion of a younger leaf to an older one (33), and the Arabidopsis fiddlehead (fdh) with irregular fusion in leaves and floral organs (34). Most organ fusion mutants are accompanied by enhanced chlorophyll permeability and altered epicuticular wax compositions (for a review, see ref. 35). We observed similar symptoms in covered barley caryopses (Fig. 4A and SI Fig. 7). We surmise that the hull-caryopsis adhesion in covered barley might be caused by mechanisms similar to those speculated to exist in organ fusion mutants.
Advantages of Barley in Lipid Regulation Studies.
In barley, >1,580 eceriferum (cer) mutants with different degrees of reduced wax in different parts, are classified into 79 complementation groups, 21 of which are assigned to a specific chromosome (36, 37). Three barley cer mutants (cer-zv, cer-yl, and cer-ym) show weak hull-caryopsis adhesion and reduced wax in all aerial parts except the nodes; they show poor growth and reduced fertility under dry conditions (38). These cer mutants, which are nonallelic to nud, had caryopses that were unstained with Sudan black B (data not shown). Thus, they also suggest a link between epidermal lipids and hull-caryopsis adhesion.
From Arabidopsis studies, the AP2/ERF domain is thought to function as DNA binding motifs (39), but functional importance of the “mm” and “cm” motifs in the WIN/SHN transcription factor genes has not been clarified because of unavailability of mutants and functional redundancy among three homologs. Availability of nud mutants and tissue-specific gene expression unlike to the ubiquitously expressed Arabidopsis homolog might make barley an ideal system for further functional analyses of the WIN/SHN transcription factors. The present study provides the first evidence for functional importance of the highly conserved valine residue (position 134) in the “mm” motif of this ERF transcription factor regulating lipid biosynthesis pathways.
Implications in Crop Domestication.
As described above, in barley, some potential loci expressing the naked caryopsis phenotype exist. However, only nud has important agricultural value without pleiotropic deleterious effects on other agronomic characters, being the sole target of selection for this trait during the domestication of barley. The absence of deleterious effects associated with nud mutation is probably attributable to its strict control of gene expression localized to the testa. The present nud locus analyses reinforce our previous interpretation of the monophyletic origin of domesticated naked barley based on a tight-linked marker (11). It is surprising that only a single fortuitous mutation event of the complete deletion of the entire Nud gene was selected by early cautious farmers and spread worldwide, because recent molecular evolutionary studies (12, 13) favored the interpretation of multiple domestication events for the barley crop as a whole. The monophyletic origin of the naked caryopsis trait sharply contrasts with the multiple origins of six-rowed spike in barley (40).
Domestication genes for efficient recovery of edible parts have been cloned in some cereal crops. In the domestication of maize from the wild progenitor teosinte, tga1 (teosinte glume architecture 1) mutation in a squamosa-promoter binding protein family transcription factor gene resulted in exposed kernels on the cob (41). In rice, two genes controlling seed shattering were found to encode transcription factors controlling abscission layer formation, but they belong to different families: sh4 (shattering 4) is a Myb3 family (42) and qSH1 is a BEL1-type homeobox gene (43). In macaroni and bread wheats, the Q gene on chromosome 5A was identified to encode an AP2 family transcription factor (44). The dominant Q gene in wheat renders a free-threshing trait by reducing the tenacity of glumes and fragility of rachises, whereas the recessive q allele shows tough glumes and fragile rachises (also called “hulled” wheat). In barley, recessive nud mutations express naked caryopsis. It should be emphasized that the meaning of the term “hulled” is completely different between wheat and barley, because “hulled” wheat never shows hull-caryopsis adhesion. The present example of the nud gene in barley might provide further support to the important roles of mutations in transcription factors in crop domestication as proposed by Doebley (45).
The cloning of Nud in barley might help confer the naked caryopsis trait to more distantly related wild species of the genus Hordeum or other wild grasses with covered grain and agricultural potential. Comparative Poaceae-wide analyses of Nud homologs and possible Nud target genes are in progress to elucidate how barley, alone among the grass species that have been successfully domesticated by humans, acquired the hulled caryopsis as the major type.
Materials and Methods
Plant Materials and Genetic Mapping.
Genetic mapping was performed in 2,828 plants segregating for nud from two populations. For analysis of the allelic variation of Nud, two induced naked mutants (20) and 33 lines listed in SI Table 5 were used. For functional analyses, Bowman and its isogenic naked line (abbreviated as nud-Bowman) were used (for details, see SI Methods).
BAC Contig Development.
BAC clones near the nud locus were selected from the Haruna Nijo BAC library (19). Chromosome walking was repeated until a recombinant on the proximal side was identified (see SI Methods and SI Table 1).
BAC Sequencing and Annotation.
Four BAC clones were shotgun-sequenced. Annotation analyses were carried out as described in SI Methods.
Long PCR and RACE.
For determination of the deletion in naked cultivars, long PCR amplification was attempted. The full-length cDNA sequence of the Nud gene was determined by 3′ and 5′ RACE (see SI Table 4 and SI Methods).
Sequence Analysis of Nud Alleles.
Allelic variation of the approximately 1.7-kb Nud region was studied as described in SI Methods.
Phylogenetic Analysis.
We constructed a phylogenetic tree based on the deduced amino acid sequences of Nud and its homologs from Arabidopsis and rice, using the neighbor-joining method provided with CLUSTAL W (46).
Histochemical Analysis.
We performed sectioning of fixed grains and staining of sections or dehulled caryopses with Toluidine blue O or Sudan black B as described in SI Methods.
Chlorophyll Leaching and Water-Loss Analyses.
Chlorophyll leaching and water-loss analyses were performed according to Ahanori et al. (21) with modifications described in SI Methods.
Surface Lipid Analysis.
Lipids were separated by using TLC according to Tsukagoshi et al. (47) and were visualized using detection methods described in SI Fig. 8. Methods of surface lipid extraction are described in SI Methods
RT-PCR and RNA in Situ Hybridization.
We performed RT-PCR and RNA in situ hybridization as described in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank T. Awayama, E. Myoraku, H.T. Mai, and former students for technical assistance; P. Heslop-Harrison for critical reading; K. Murai, E. Himi, and H. Tamura for technical advice; U. Lundqvist (Nordic Gene Bank, Alnarp, Sweden). for cer mutants; and M. Ichii and S. Tajima for encouragement. The resources were provided by the National Bioresource Project-Barley, Japan. This work was supported by a Core Research for Evolutional Science and Technology grant, Green Techno Project Grant GD3002, Ministry of Education, Culture, Sports, Science and Technology Grant 18580006, and Kagawa University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AP009567 (BAC contig)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711034105/DC1.
References
- 1.Gaines RL, Bechtel DB, Pomeranz Y. A microscopic study on the development of a layer in barley that causes hull-caryopsis adherence. Cereal Chem. 1985;62:35–40. [Google Scholar]
- 2.Newman CW, Newman RK. A brief history of barley foods. Cereal Foods World. 2006;51:4–7. [Google Scholar]
- 3.Pins JJ, Kaur H. A review of the effects of barley β-glucan on cardiovascular and diabetic risk. Cereal Foods World. 2006;51:8–11. [Google Scholar]
- 4.Zohary D, Hopf M. Domestication of Plants in the Old World. New York: Oxford Univ Press; 2000. [Google Scholar]
- 5.Staudt G. The origin of cultivated barleys: A discussion. Economic Bot. 1961;15:203–212. [Google Scholar]
- 6.Helbeck H. Domestication of food plants in the Old World. Science. 1959;130:365–372. doi: 10.1126/science.130.3372.365. [DOI] [PubMed] [Google Scholar]
- 7.Harlan JR. In: Evolution of Crop Plants. 2nd Ed. Smartt J, Simmonds NW, editors. London: Longman; 1995. pp. 140–147. [Google Scholar]
- 8.Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the Near East. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 9.Harlan JR. Barley: Origin, Botany, Culture, Winterhardiness, Genetics, Utilization, Pests. Agriculture Handbook No. 338. Washington, DC: U.S. Department of Agriculture; 1968. pp. 9–31. [Google Scholar]
- 10.Takahashi R, Yamamoto J. Studies on the classification and the geographical distribution of the Japanese barley varieties. XIII. Covered/naked caryopsis character. Nogaku Kenkyu. 1950;39:32–36. [Google Scholar]
- 11.Taketa S, et al. Monophyletic origin of naked barley inferred from molecular analyses of a marker closely linked to the naked caryopsis gene (nud). Theor Appl Genet. 2004;108:1236–1242. doi: 10.1007/s00122-003-1560-1. [DOI] [PubMed] [Google Scholar]
- 12.Morrell PL, Clegg MT. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA. 2007;104:3289–3294. doi: 10.1073/pnas.0611377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saisho D, Purugganan MD. Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics. 2007;177:1765–1776. doi: 10.1534/genetics.107.079491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franckowiack JD, Konishi T. Naked caryopsis. Barley Genet Newsletter. 1997;26:51–52. [Google Scholar]
- 15.Harlan HV. Daily development of kernels of Hannchan barley from flowering to maturity at Aberdeen, Idaho. J Agric Res. 1920;19:393–429. [Google Scholar]
- 16.Kikuchi S, Taketa S, Ichii M, Kawasaki S. Efficient fine mapping of the naked caryopsis gene (nud) by HEGS (high-efficiency genome scanning)/AFLP in barley. Theor Appl Genet. 2003;108:73–78. doi: 10.1007/s00122-003-1413-y. [DOI] [PubMed] [Google Scholar]
- 17.Taketa S, Awayama T, Amano S, Sakurai Y, Ichii M. High-resolution mapping of the nud locus controlling the naked caryopsis in barley. Plant Breed. 2006;125:337–342. [Google Scholar]
- 18.Sato K, Nankaku N, Motoi Y, Takeda K. A large scale mapping of ESTs on barley genome.. Proc 9th Int Barley Genet Symp; Brno, Czech Republic: 2004. pp. 79–85. [Google Scholar]
- 19.Saisho D, Myoraku E, Kawasaki S, Sato K, Takeda K. Construction and characterization of a bacterial artificial chromosome (BAC) library from the Japanese malting barley “Haruna Nijo.”. Breed Sci. 2007;57:29–38. [Google Scholar]
- 20.Scholz F. Mutationsversuche an Kulturpflanzen. IV. Über den züchterischen Wert zweier röntgeninduzierter nacktkörniger Gerstenmutanten. Kulturpflanze. 1955;3:69–89. [Google Scholar]
- 21.Aharoni A, et al. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannangara R, et al. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell. 2007;19:1278–1294. doi: 10.1105/tpc.106.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieber P, et al. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell. 2000;12:721–738. doi: 10.1105/tpc.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman PL, Palmer GH. The structure of the pericarp and testa of barley. J Inst Brew. 1984;90:88–94. [Google Scholar]
- 27.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcriptional factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 29.Sakuma Y, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 30.Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog in Lipid Res. 2003;42:51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 32.Pighin JA, et al. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- 33.Sinha N, Lynch M. Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta. 1998;206:184–195. [Google Scholar]
- 34.Yephremov A, et al. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lolle SJ, Pruitt RE. Epidermal cell interactions: a case for local talk. Trend in Plant Sci. 1999;4:14–20. doi: 10.1016/s1360-1385(98)01353-3. [DOI] [PubMed] [Google Scholar]
- 36.Lundqvist U, Lundqvist A. Mutagen specificity in barley for 1580 eceriferum mutants localized to 79 loci. Hereditas. 1988;108:1–12. [Google Scholar]
- 37.von Wettstein-Knowles P. Munck L, editor. Barley Genetics VI vol II. 1992:753–771. [Google Scholar]
- 38.Lundqvist U, Franckowiack JD. In: Diversity in Barley (Hordeum vulgare) von Bothmer R, Knüpffer H, van Hintum T, Sato K, editors. Amsterdam: Elsevier; 2003. pp. 77–96. [Google Scholar]
- 39.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a β-sheet revealed the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 43.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 44.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doebley J. Unfallen grains: How ancient farmers turned weeds into crops. Science. 2006;312:1318–1319. doi: 10.1126/science.1128836. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukagoshi H, Morinaka A, Nakamura K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA. 2007;104:2543–2547. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.