Abstract
We report that aminoacylation of minimal RNA helical substrates is enhanced by mismatched or unpaired nucleotides at the first position in the helix. Previously, we demonstrated that the class I methionyl-tRNA synthetase aminoacylates RNA microhelices based on the acceptor stem of initiator and elongator tRNAs with greatly reduced efficiency relative to full-length tRNA substrates. The cocrystal structure of the class I glutaminyl-tRNA synthetase with tRNAGln revealed an uncoupling of the first (1⋅72) base pair of tRNAGln, and tRNAMet was proposed by others to have a similar base-pair uncoupling when bound to methionyl-tRNA synthetase. Because the anticodon is important for efficient charging of methionine tRNA, we thought that 1⋅72 distortion is probably effected by the synthetase–anticodon interaction. Small RNA substrates (minihelices, microhelices, and duplexes) are devoid of the anticodon triplet and may, therefore, be inefficiently aminoacylated because of a lack of anticodon-triggered acceptor stem distortion. To test this hypothesis, we constructed microhelices that vary in their ability to form a 1⋅72 base pair. The results of kinetic assays show that microhelix aminoacylation is activated by destabilization of this terminal base pair. The largest effect is seen when one of the two nucleotides of the pair is completely deleted. Activation of aminoacylation is also seen with the analogous deletion in a minihelix substrate for the closely related isoleucine enzyme. Thus, for at least the methionine and isoleucine systems, a built-in helix destabilization compensates in part for the lack of presumptive anticodon-induced acceptor stem distortion.
Aminoacyl-tRNA synthetases (RSs) form an isofunctional family of enzymes that catalyze the addition of amino acids to tRNA molecules in a strict one-to-one relationship that establishes the genetic code. Although the genetic code is based on tRNA anticodons, many synthetases efficiently aminoacylate small RNA substrates made up of the amino acid acceptor stems of their cognate tRNAs (1–7). These functional substrates, lacking the tRNA anticodons, reveal an operational RNA code for aminoacylation based on the sequence and/or structure of the acceptor stem minihelices (8).
Methionyl-tRNA synthetase (MetRS) aminoacylates both initiator (tRNAfMet) and elongator (tRNAMet) molecules and contacts with the CAU anticodon are critical for catalysis (9, 10). Small RNA substrates based on the acceptor stems of these tRNAs are methionylated in a sequence-specific manner but with catalytic efficiencies decreased approximately six orders of magnitude relative to full-length tRNA (3, 11, 12). The low rate of mini- and microhelix aminoacylation appears to be due to a difficulty in forming the transition state rather than to a binding defect (12).
The 20 RSs can be partitioned into two 10-enzyme classes, each based on sequence similarities and structural motifs (13, 14). The only crystal structure of a class I RS complexed with its cognate tRNA is that of Escherichia coli GlnRS with tRNAGln (15, 16). Unusual features of the tRNA structure in the complex include the uncoupling of the U1⋅A72 base pair in the first position of the acceptor stem helix and distortion of the adjacent 3′-CCA end of the molecule toward the active site of the enzyme. Similarities between the structures of the GlnRS-tRNAGln complex and uncomplexed class I E. coli MetRS (17) led to the construction of a model for the MetRS-tRNAMet cognate pair in which tRNAMet has an orientation like that of tRNAGln in its complex, including the disrupted first base pair (18). The orientation of tRNAMet in this model complex is consistent with previous biochemical and genetic studies that identified specific enzyme–tRNA contacts (19–21).
The mechanism or “trigger” for local acceptor helix destabilization in a synthetase–tRNA complex is not known. For many class I enzymes, the anticodon is important for efficient aminoacylation of the full tRNA substrate. A previous analysis (12) evaluated the relative contribution of the free energy of binding of the microhelix and of the anticodon stem–loop structure to MetRS. This analysis established that the sum of the binding energies of the two components (anticodon stem–loop and acceptor stem domains) exceeded that of the full tRNA by 7 kcal (1 kcal = 4.18 kJ). The excess free energy was proposed to be used at least in part to compensate for the cost of straining the tRNA, such as a distortion of the 3′ end so that it fits into the active site pocket.
In this scenario, the low efficiency of aminoacylation of microhelix substrates for MetRS would be due in part to the inability to induce the active conformation of the 3′ end of the substrate. This inability might be compensated by creating a structure that could more easily pass into or mimic the transition state. We imagined that introduction of mismatches at the first position of the helix might be beneficial, provided that such mismatches did not remove an essential enzyme–tRNA contact at this point in the helix. Because tRNAMet and tRNAfMet differ in sequence at the first position, we reasoned that an essential contact may not occur at this location. Thus, the possibilities for activation of microhelix aminoacylation by manipulations of the first position of the helix seemed plausible.
MATERIALS AND METHODS
RNA Substrates.
RNA molecules (microhelices and duplex strands) were chemically synthesized on a Gene Assembler Special (Pharmacia) using N-benzyl- and N-isobutyl-protected or N-phenoxyacetyl-protected 2′-t-butyl-dimethylsilyl-ribonucleoside 3′-cyanoethyl phosphoramidites from Chem Genes (Waltham, MA). Synthesis, deprotection, and purification of benzyl- or isobutyl-protected RNAs were as described by Musier-Forsyth and coworkers (22). The N-PAC protected RNAs were deprotected with 1:1 ammonia:methylamine and triethylamine trihyrofluoride according to the protocol of Wincott et al. (23). Deprotected RNAs were purified by electrophoresis on denaturing 16% polyacrylamide gels.
Construction of (His)6-Tagged MetRS.
An active monomer fragment of E. coli MetRS [consisting of the N-terminal 547 residues (24)] was constructed with an N-terminal six-histidine tag. Complementary oligonucleotides encoding the sequence MRGSHHHHHHSSGST were inserted between the Bsu36I and SalI restriction sites of the plasmid pJB104. This plasmid is a derivative of pBlueScript KS(+) (Strategene) (25). (Further details are available upon request from the authors.) The resulting plasmid pRA101, encoding 6H-MetRS 547, was transformed into E. coli TG1 (26). The overexpressed 6H-MetRS 547 enzyme was purified by nickel nitrilotriacetic acid-agarose affinity chromatography according to the manufacturer’s protocol (Qiagen, Chatsworth, CA). We established that fusion of an N-terminal six-histidine tag to monomeric MetRS had no effect on the charging of tRNAfMet or of microhelices, relative to the activity of the unmodified protein. All data reported herein were obtained with the (His)6-tagged enzyme.
Aminoacylation Assays.
MicrohelixMet and duplexMet aminoacylation assays were carried out at 25°C in 20 mM Hepes (pH 7.5), 100 μM Na2EDTA, 150 mM NH4Cl, 4 mM ATP, 10 mM MgCl2, 100 μM methionine, and [35S]methionine (NEN-Dupont; 50 μCi/ml; 1 Ci = 37 GBq). RNAs were annealed before use by heating at 80°C followed by slow cooling to room temperature in the presence of 1 mM MgCl2. MetRS was used at 10 μM and RNA substrate concentrations were typically 200 μM. Reaction aliquots were quenched by spotting on Whatman filters soaked in 5% trichloroacetic acid and 1 mM methionine. The filters were washed for five 10-min periods in ice-cold 5% trichloroacetic acid and 1 mM methionine before liquid scintillation counting. The amount of filter-bound [35S]methionine from an aminoacylation reaction in the absence of RNA [presumably reflecting the “self-methionylation” activity of MetRS (27)] was subtracted from the reported aminoacylation values. Self-methionylation of MetRS represented approximately 20% of wild-type microhelixMet counts and 1–2% of Δ1-C72 microhelixMet counts. Aminoacylation of minihelixIle and Δ1-U72 minihelixIle was carried out at 25°C in 20 mM Hepes (pH 7.5), 100 μM Na2EDTA, 150 mM NH4Cl, 2 mM ATP, 20 mM MgCl2, 21 μM isoleucine, [3H]isoleucine (Amersham; 100 μCi/ml), and 100 μM minihelix. E. coli IleRS was purified as described (28) and used at a concentration of 5 μM.
Acid gel analyses of aminoacyl-microhelixMet variants were carried out as described (11) with minor alterations. Reactions were carried out at 25°C in 20 mM Hepes (pH 7.5), 100 μM Na2EDTA, 150 mM NH4Cl, 4 mM ATP, 10 mM MgCl2, 20 μM methionine, [35S]methionine (900 μCi/ml), 300 μM microhelix substrate, and 10 μM MetRS. The reactions were quenched after 15 min in 8 M urea, 50 mM sodium acetate (pH 5.2), 0.025% xylene cyanol, and 0.025% bromophenol blue running buffer and heated at 50°C for 10 min before loading onto a 12% polyacrylamide (19:1 acrylamide/N,N′-methylenebisacrylamide) gel made up in 8 M urea/10 mM sodium acetate, pH 5.2. After electrophoresis, the gel was soaked in 10% methanol/12% acetic acid for 15 min and dried under vacuum. The [35S]Met-containing microhelices were visualized by analysis with a PhosphorImager (Molecular Dynamics).
RESULTS
Oligonucleotide Substrates for Aminoacylation.
Twelve substrates that were tested for aminoacylation by MetRS are shown in Fig. 1. These substrates are based on the sequences of tRNAMet and tRNAfMet. For example, microhelixMet and microhelixfMet recapitulate the acceptor stem helix and TΨC loop of the respective tRNAs. In addition to microhelix substrates, we synthesized RNA duplexes that represent the 12 bp of the acceptor-TΨC stem of full-length tRNAMet. The duplexes are constructed as single strands that are then hybridized together. Compared with microhelices, the duplex substrates offer the technical convenience and efficiency of enabling us to hybridize one strand to several complementary strands that differ by a single nucleotide. We saw little if any difference between the efficiency of charging of the two types of substrates (see below and data not shown) and for that reason used them interchangeably.
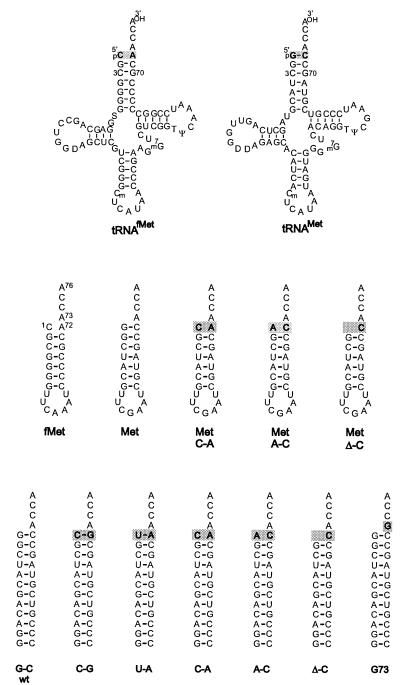
Figure 1.
Substrates for E. coli MetRS. (Top) Sequences of E. coli elongator and initiator tRNAs. The base pairs at the first position of the acceptor stems are shaded. (Middle) Microhelix substrates that were assayed. Wild-type microhelixfMet and microhelixMet substrates and variants of microhelixMet are shown, with substitutions of the first base pair being shaded. (Bottom) DuplexMet substrates, with base substitutions shaded.
Collectively, nine substitutions at the first base pair (G1⋅C72) and discriminator base (A73) were made within the microhelixMet and duplexMet frameworks (Fig. 1 Middle and Bottom). These substitutions create new base pairs at the first (1⋅72) position of the helix and also introduce mismatches and a deletion. In addition, microhelixMet and microhelixfMet have a natural variation at the first position of the helix (G⋅C versus C⋅A). As stated, this variation suggested that the identity of the nucleotides at the first position is not essential. In contrast, the adjacent G2⋅C71 and C3⋅G70 base pairs are conserved. These considerations suggested that we might not be able to test the effect of helix destabilizations beyond the first base pair, without the potential for disruption of a direct synthetase–RNA contact at one or both of the 2⋅71 and 3⋅70 base-pair positions. It is perhaps noteworthy that, in the complex of GlnRS with tRNAGln, it is only the first pair of the helix that is disrupted.
Specificity of Aminoacylation.
We confirmed the earlier report (3) that microhelixMet is a substrate for methionylation by MetRS. (Similar results were obtained with the duplexMet substrate.) At longer reaction times (60 min), 1–2% of microhelixMet is aminoacylated. Because the rate of aminoacylation is so slow, deacylation reactions compete with charging, so that the “plateau level” of acylation is severely limited (29). In spite of this limitation, we observed that the initial rates of charging were linearly dependent on enzyme concentration. Thus, the initial rates can be used to obtain an apparent kcat/Km.
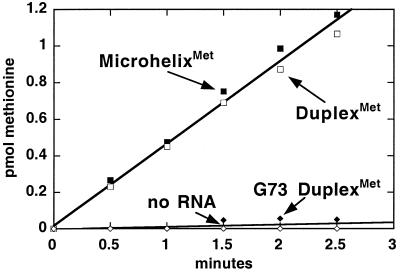
To verify that the low aminoacylation rate resulted from a specific interaction between the enzyme and microhelix, we tested a substrate with a G73 substitution. The same G73 substitution in the full tRNA is known to attenuate the charging of that substrate (30, 31). We could detect no aminoacylation of G73 duplexMet (Fig. 2). Thus, the full tRNA and the oligonucleotide substrate have a similar sensitivity to the G73 substitution.
Figure 2.
Aminoacylation of microhelix and duplex substrates of MetRS. The substitution of the A73 discriminator base abolishes aminoacylation.
Activation of Aminoacylation by Localized Helix Melting.
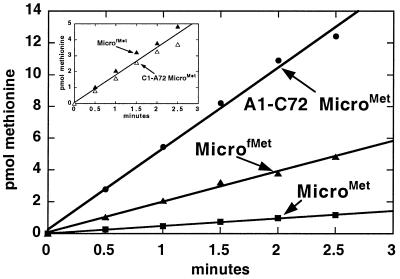
A comparison of microhelixMet and microhelixfMet charging efficiencies demonstrated that the initiator-type microhelix with the mismatched 1⋅72 pair had a significantly higher rate of aminoacylation (Fig. 3). This result is consistent with the lack of direct base recognition at the 1⋅72 position. It is also consistent with the hypothesis that local helix melting at the 1⋅72 base pair is needed for more efficient aminoacylation. However, microhelixMet and microhelixfMet also differ in sequence at the 4⋅69, 5⋅68, and 6⋅67 base-pair positions and the enhanced charging of the latter substrate could be due to more favorable synthetase interactions at one or more of these positions. To determine the contribution of the mismatch at the first helix position to microhelixfMet charging, the C1⋅A72 pair was substituted into the context of microhelixMet. Microhelices with the C⋅A pair at the first position are aminoacylated at a higher rate (3- to 4-fold) than the wild-type microhelixMet, whether the other base pairs of the acceptor stem recapitulate the elongator or initiator tRNA sequences (Fig. 3 Inset). Thus, the higher rate of charging of microhelixfMet compared with microhelixMet may be entirely due to the mismatched 1⋅72 pair.
Figure 3.
Enhancement of microhelixMet aminoacylation by 1⋅72 base-pair mismatches.
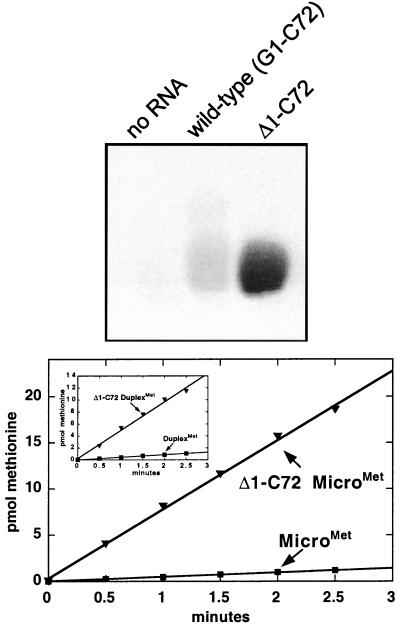
To see whether the activation of charging with the C⋅A mismatch was specific to that combination of bases at the 1⋅72 base-pair position, we also tested the A1⋅C72 transversion, in the context of microhelixMet. Interestingly, this substrate is even more active, with a rate of aminoacylation that is about 10-fold higher than that of the wild-type microhelixMet (Fig. 3). Thus, the enhancement of acylation is not specific to the C·A replacement and, instead, appears to be related to having a mismatched base pair. [Possibly the higher rate of aminoacylation in the presence of the A⋅C base pair relative to the C⋅A pair is due to a disruption of purine–purine stacking interactions between A72 and A73. The resulting flexibility could facilitate the proposed hairpin conformation of the 3′ end of the substrate (18).] With this in mind, a microhelixMet variant lacking the G1 base was synthesized to test a substrate completely unable to form a base pair at the first helix position. This Δ1⋅C72 microhelixMet is a substantially better substrate than wild-type microhelixMet. This can be seen by direct side-by-side visualization of the intense band corresponding to [35S]methionylated Δ1⋅C72 microhelixMet compared with charged microhelixMet on an acidic polyacrylamide gel (Fig. 4 Upper). A study of the time course showed that the initial rate of Δ1⋅C72 microhelixMet charging is 16-fold higher than that of wild-type microhelixMet (Fig. 4 Lower). A similar rate increase is seen on methionylation of the duplexMet substrate lacking the G1 nucleotide (Fig. 4 Lower Inset).
Figure 4.
Enhancement of aminoacylation by terminal nucleotide deletion. (Upper) Acid gel analysis of microhelix aminoacylation showing the enhanced aminoacylation of the Δ1⋅C72 microhelixMet substrate with [35S]methionine after a 15-min reaction. (Lower) Time course of aminoacylation of microhelixMet and Δ1⋅C72 microhelixMet. The enhanced aminoacylation conferred by the Δ1⋅C72 substitution is also seen with the duplexMet substrate (Inset).
We also tested other substitutions at the end of the acceptor helix, by using duplexMet as the context for those changes. None of these substitutions enhanced the charging in the way seen with the mismatches or deletion described above. Replacing the wild-type G1⋅C72 pair with the weaker U⋅A base pair enhanced aminoacylation by 2- to 3-fold. A similar effect was seen with the C⋅G substitution (data not shown). These results further support the idea that direct base recognition does not occur at the 1⋅72 base-pair position in this system. It is also possible that the small effects in charging rates seen with the U1⋅A72 and C1⋅G72 substitutions could be related to a minor perturbation of specific stacking energies at the end of the helix. For example, the C1⋅G72 substitution in duplexMet removes the strong G1⋅G2 stacking interaction (32, 33), resulting in a less stable helix as evidenced by the lower melting temperature Tm of a C1⋅G72 versus G1⋅C72 duplexAla substrate (34). DuplexAla has the same A73 and G2⋅C71 stacking partners as duplexMet for the 1⋅72 base-pair position.
Activation of Minihelix Charging by the Related IleRS.
IleRS and MetRS are part of a subclass of more closely related synthetases that also includes the leucine, cysteine, and valine enzymes (35, 36). The close similarity of the two enzymes made possible a reasonably accurate model of IleRS [before the determination of its three-dimensional structure (37)], which led to studies showing close parallels in their respective modes of tRNA recognition (38). Like MetRS, aminoacylation by the isoleucine enzyme is greatly enhanced by interactions with the anticodon. Consequently, the rate of charging of minihelices based on the acceptor stem of tRNAIle is greatly reduced (39, 40). In addition, some data suggested that the nature of the 1⋅72 base pair of the acceptor stem was not essential for charging with isoleucine (40, 41). Thus, we wondered whether the close relatedness of the isoleucine and methionine systems extended to the way they formed the transition state with the 3′ end of the tRNA. If so, then minihelix aminoacylation might be activated by the same sort of localized helix destabilization observed in this example with microhelixMet.
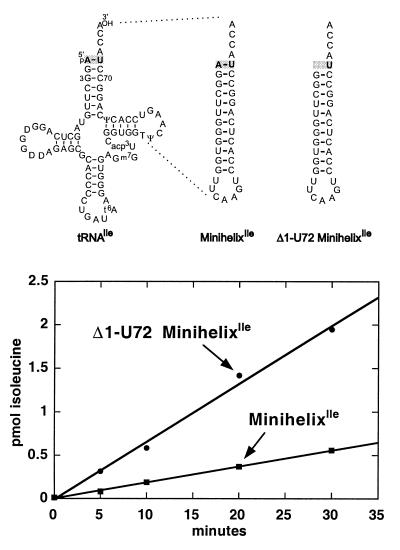
To investigate this possibility, we constructed two minihelices (Fig. 5 Upper). One was based on the acceptor-TψC stem–loop structure of tRNAIle, where the first base pair of the helix is A1⋅U72. The second minihelix was identical, except that A1 was removed to give the Δ1⋅U72 construction. The linear time course of isoleucine incorporation into the minihelices was significantly higher (3.5-fold) with the Δ1·U72 construction (Fig. 5 Lower). Although the activation is less than the 16-fold seen with the ΔG1⋅C72 deletion in the methionine system, the qualitatively similar behavior of the Δ1⋅U72 minihelixIle is consistent with related mechanisms of transition state formation that underlie both systems.
Figure 5.
Enhancement of minihelixIle aminoacylation by deletion of nucleotide A1. (Upper) MinihelixIle recapitulates the acceptor stem and TΨC loop of tRNAIle. Δ1⋅U72 minihelixIle is lacking the 5′ terminal adenosine. (Lower) Time course of aminoacylation of minhelixIle and Δ1⋅U72 minihelixIle.
CONCLUSION
Previous work suggested that a “fold-back” structure was important for the formylase that catalyzes attachment of a formyl group to the charged initiator tRNAfMet, and gave evidence for this sort of structure in a microhelix variant (30, 42). Thus, the fold-back conformation may be of particular significance for tRNAfMet where two enzymes appear to require flexibility of the 3′ end for their respective transition states.
While RNA microhelix and duplex substrates for aminoacylation have been demonstrated for 11 RSs, this work investigates the effects of helix-destabilizing substitutions at the 1⋅72 base-pair position in an RNA oligonucleotide substrate for a class I enzyme. The most common base pair closing the acceptor stem helix of prokaryotic tRNAs is G1⋅C72. Nearly all tRNAs aminoacylated by class II enzymes contain a G1⋅C72 pair, but about 50% of the tRNAs for class I enzymes have other nucleotides at these positions (43). A previous analysis (44) suggested that this class-specific conservation reflected the use of the G1⋅C72 base pair for specific functional contacts by at least some class II enzymes.
For example, replacement of the G1⋅C72 base pair in a duplexAla substrate (for the class II E. coli AlaRS) with a C1⋅C72 pair severely reduced aminoacylation, but a C1⋅G72 or G1⋅G72 pair eliminated aminoacylation (44). In the class II E. coli HisRS system, tRNAHis contains a unique G−1⋅C73 base pair at the first position of the acceptor helix. This base pair is essential for aminoacylation of a microhelix with histidine (45). Thus, the severely negative effect of any manipulation of the first base pair in microhelixAla and microhelixHis is in striking contrast to that seen in the microhelixMet and minihelixIle systems.
The difference between MetRS (and IleRS) and AlaRS (and HisRS) in their response to nucleotide replacements at the first position of the helix is paralleled in the crystal structures of the related synthetase–tRNA complexes. In the class I GlnRS–tRNAGln structure (15), the distorted acceptor stem is bound by the protein domain (connective polypeptide or CP1) inserted between halves of the dinucleotide (Rossmann) binding fold. This distortion directs the amino acid acceptor end into the active site of the enzyme. Protein contacts responsible for discrimination are made on the minor groove side of the acceptor stem. In contrast, in the structure of the class II AspRS–tRNAAsp complex (46, 47), the enzyme approaches the acceptor stem from the major groove side of the helix. Specific contacts are made between residues of conserved motif 2 and the major groove of the acceptor stem, including the U1⋅A72 base pair. In further contrast to complexed tRNAGln, the single-stranded acceptor end of tRNAAsp maintains a helical path into the active site. Therefore, in at least some class I enzymes, acceptor stem helix distortion is necessary to properly orient the amino acid attachment site in the active site. For the class II AspRS, and probably for AlaRS and HisRS, no helix destabilization appears necessary to place the 3′ end of the RNA into the active site. For certain other class II enzymes, localized helix distortion during aminoacylation remains an open possibility (48, 49).
Localized helix destabilization as a mechanism to activate aminoacylation may be specific to a particular subgroup of class I enzymes and not to the entire class. For example, TyrRSs from bacteria to humans aminoacylate microhelix substrates based on the acceptor stems of the tRNATyr. Unlike MetRS, where there is no requirement for a specific base pair, the G1⋅C72 pair (found in prokaryotic tRNATyrs) or the C1⋅G72 pair (found in eukaryotes) is critical for aminoacylation (50, 51). Direct recognition of these base pairs by a small peptide within the CP1 insertion of TyrRSs is responsible for the specificity. Thus, when this peptide element from CP1 was transplanted from the E. coli to the human enzyme, the microhelix charging specificity was switched from C1⋅G72 to G1⋅C72. The opposite change in specificity was achieved by transplantation of the same peptide from the human to the bacterial enzyme (51). Although TyrRS is a class I enzyme, it and TrpRS form a structural subclass distinct from that of MetRS or IleRS (52, 53). We suspect that localized helix melting does not play the role in tRNATyr that it does in tRNAMet.
For all synthetase–tRNA complexes, the acceptor stem is a major locus of interactions. For many of them, a second set of interactions also occurs at the anticodon. We imagine that the activity-enhancing distortion of the acceptor stem, suggested by this work and by the crystal structure of GlnRS with tRNAGln, is promoted by the interaction of the synthetase with the anticodon triplet. However, this effect appears to require continuity of the tRNA structure. For example, the anticodon is important for highly efficient aminoacylation of tRNAVal with ValRS (9, 54). The rate of charging of minihelices with valine is greatly reduced compared with that of the full tRNA. But only a small stimulation of the aminoacylation of minihelices with valine is seen when the anticodon stem–loop structure of tRNAVal is added to the minihelix substrate (2). Only weak stimulation of minihelix charging is also seen when the anticodon stem–loop structure is added to minihelixIle (40). Similarly, we added the tRNAMet anticodon stem–loop structure, which associates strongly with the enzyme (55), to our microhelix aminoacylation system. This addition had no effect on the efficiency of aminoacylation of either microhelixMet or Δ1⋅C72 microhelixMet (data not shown). Thus, in the closely related isoleucine, methionine, and valine systems, communication between distal parts of a continuous tRNA structure appears essential for substrate activation. The importance of “long-range” communication within the tRNAIle structure was also suggested by recent studies of a tRNA-dependent editing reaction (56).
If, in the full tRNA, localized helix melting is promoted through a conformational change initiated by a synthetase–anticodon contact, then it is not obvious whether base-pair-destabilizing substitutions at the 1⋅72 pair would enhance or diminish charging of tRNA. On the one hand, a built-in mismatch at the 1⋅72 pair should make it easier to distort the 3′ end of the tRNA. On the other hand, any disturbance of the region around the 1⋅72 position could adversely affect the kinetic pathway for the specific conformational change at the 3′ end that is promoted by interactions at the anticodon. In studies with tRNAMet, tRNAfMet, tRNAGln, and tRNAVal, substitutions at the 1⋅72 pair generally diminish or have little effect on aminoacylation efficiency (30, 31, 44, 57, 58). Any effects are typically not large and are consistent with the 1⋅72 position not being used for essential functional contacts by the enzyme.
The substrates for MetRS tested herein that contained a mismatch or deletion at the first helix position were significantly enhanced in aminoacylation compared with wild-type microhelixMet. Thus, the engineered destabilization of the first base pair has reduced the apparent free energy of activation, possibly by making it easier to recreate the conformation of the acceptor stem of tRNAMet during catalysis. With the most active oligonucleotide substrate—the Δ1⋅C72 microhelixMet variant—the apparent free energy of transition state formation is lowered by about 1.6 kcal/mol. (In the isoleucine system, the Δ1⋅U72 deletion lowered the apparent transition state barrier by about 0.7 kcal/mol.) Thus, the free energy contribution of the “conformational effect” in micro- and minihelices is comparable to that of some of the individual functional contacts between a synthetase and specific atomic groups in the acceptor stem helix (34, 59).
The activation of microhelix aminoacylation seen herein only partially compensates for the loss of interactions with other parts of the tRNA (such as the anticodon). A far greater compensation might be achieved with a microhelix substrate whose 3′ end was specifically bent like that seen in bound tRNAGln (15). A substrate of this sort would have a much lower entropic barrier to formation of the presumed transition state than the helix-destablized substrates studied herein.
Acknowledgments
We thank Prof. Karin Musier-Forsyth for comments on the manuscript and Dr. Lluís Ribas de Pouplana for helpful discussions. This work was supported by Grant GM15539 from the National Institutes of Health. R.W.A. is a National Institutes of Health Postdoctoral Fellow. B.E.N. is a Howard Hughes Medical Institute Predoctoral Fellow.
ABBREVIATION
- RS
aminoacyl-tRNA synthetase
References
- 1. Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 2.Frugier M, Florentz C, Giegé R. Proc Natl Acad Sci USA. 1992;89:3990–3994. doi: 10.1073/pnas.89.9.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinis S A, Schimmel P. Proc Natl Acad Sci USA. 1992;89:65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright D J, Martinis S A, Jahn M, Söll D, Schimmel P. Biochimie. 1993;75:1041–1049. doi: 10.1016/0300-9084(93)90003-b. [DOI] [PubMed] [Google Scholar]
- 5.Frugier M, Florentz C, Giegé R. EMBO J. 1994;13:2219–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamann C S, Hou Y M. Biochemistry. 1995;34:6527–6532. doi: 10.1021/bi00019a034. [DOI] [PubMed] [Google Scholar]
- 7.Saks M E, Sampson J R. EMBO J. 1996;15:2843–2849. [PMC free article] [PubMed] [Google Scholar]
- 8.Schimmel P, Ribas de Pouplana L. Cell. 1995;81:983–986. doi: 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- 9.Schulman L H, Pelka H. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- 10.Kim H Y, Pelka H, Brunie S, Schulman L H. Biochemistry. 1993;32:10506–10511. doi: 10.1021/bi00090a029. [DOI] [PubMed] [Google Scholar]
- 11.Martinis S A, Schimmel P. J Biol Chem. 1993;268:6069–6072. [PubMed] [Google Scholar]
- 12.Gale A J, Shi J P, Schimmel P. Biochemistry. 1996;35:608–615. doi: 10.1021/bi9520904. [DOI] [PubMed] [Google Scholar]
- 13.Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. Nature (London) 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 14.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 15.Rould M A, Perona J J, Söll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 16.Rould M A, Perona J J, Steitz T A. Nature (London) 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 17.Brunie S, Zelwer C, Risler J L. J Mol Biol. 1990;216:411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- 18.Perona J J, Rould M A, Steitz T A, Risler J L, Zelwer C, Brunie S. Proc Natl Acad Sci USA. 1991;88:2903–2907. doi: 10.1073/pnas.88.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon O, Schulman L H. Biochemistry. 1987;26:5416–5422. doi: 10.1021/bi00391a030. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh G, Pelka H, Schulman L H. Biochemistry. 1990;29:2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- 21.Meinnel T, Mechulam Y, Le Corre D, Panvert M, Blanquet S, Fayat G. Proc Natl Acad Sci USA. 1991;88:291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Proc Natl Acad Sci USA. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellot P, Mechulam Y, Le Corre D, Blanquet S, Fayat G. J Mol Biol. 1989;208:429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 25.Burbaum J J, Schimmel P. Biochemistry. 1991;30:319–324. doi: 10.1021/bi00216a002. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Gillet S, Hountondji C, Schmitter J M, Blanquet S. Protein Sci. 1997;6:2426–2435. doi: 10.1002/pro.5560061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard A, Shiba K, Schimmel P. Proc Natl Acad Sci USA. 1992;89:9964–9968. doi: 10.1073/pnas.89.20.9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnet J, Ebel J P. Eur J Biochem. 1972;31:335–344. doi: 10.1111/j.1432-1033.1972.tb02538.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee C P, Dyson M R, Mandal N, Varshney U, Bahramian B, RajBhandary U L. Proc Natl Acad Sci USA. 1992;89:9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meinnel T, Mechulam Y, Lazennec C, Blanquet S, Fayat G. J Mol Biol. 1993;229:26–36. doi: 10.1006/jmbi.1993.1005. [DOI] [PubMed] [Google Scholar]
- 32.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers M H, Neilson T, Turner D H. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto N, Kierzek R, Turner D H. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 34.Beuning P J, Gulotta M, Musier-Forsyth K. J Am Chem Soc. 1997;119:8397–8402. [Google Scholar]
- 35.Starzyk R M, Webster T A, Schimmel P. Science. 1987;237:1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- 36.Hou Y M, Shiba K, Mottes C, Schimmel P. Proc Natl Acad Sci USA. 1991;88:976–980. doi: 10.1073/pnas.88.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nureki O, Vassylyev D G, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson T L, Schimmel P, Yokoyama S. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 38.Auld D S, Schmimmel P. EMBO J. 1996;15:1142–1148. [PMC free article] [PubMed] [Google Scholar]
- 39.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Nature (London) 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 40.Nureki O, Niimi T, Muto Y, Kanno H, Kohno T, Muramatsu T, Kawai G, Miyazawa T, Giegé R, Florentz C, Yokoyama S. In: The Translational Apparatus. Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. New York: Plenum; 1993. pp. 59–66. [Google Scholar]
- 41.Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giegé R, Yokoyama S. J Mol Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 42.Puglisi E V, Puglisi J D, Williamson J R, RajBhandary U L. Proc Natl Acad Sci USA. 1994;91:11467–11471. doi: 10.1073/pnas.91.24.11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinberg S, Misch A, Sprinzl M. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Kessler J, Peterson R, Musier-Forsyth K. Biochemistry. 1995;34:9795–9800. doi: 10.1021/bi00030a017. [DOI] [PubMed] [Google Scholar]
- 45.Francklyn C, Shi J P, Schimmel P. Science. 1992;255:1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- 46.Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry J C, Moras D. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 47.Cavarelli J, Rees B, Ruff M, Thierry J C, Moras D. Nature (London) 1993;362:181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Peterson R, Kessler J, Musier-Forsyth K. Nucleic Acids Res. 1995;23:165–169. doi: 10.1093/nar/23.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzger A U, Heckl M, Willbold D, Breitschopf K, RajBhandary U L, Rösch P, Gross H J. Nucleic Acids Res. 1997;25:4551–4556. doi: 10.1093/nar/25.22.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinn C L, Tao N, Schimmel P. Biochemistry. 1995;34:12489–12495. doi: 10.1021/bi00039a001. [DOI] [PubMed] [Google Scholar]
- 51.Wakasugi K, Quinn C L, Tao N, Schimmel P. EMBO J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter C W. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 53.Doublie S, Bricogne G, Gilmore C, Carter C W., Jr Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 54.Chambers R W, Aoyagi S, Furukawa Y, Zawadzka H, Bhanot O S. J Biol Chem. 1973;248:5549–5551. [PubMed] [Google Scholar]
- 55.Gale A J, Schimmel P. Biochemistry. 1995;34:8896–8903. doi: 10.1021/bi00027a042. [DOI] [PubMed] [Google Scholar]
- 56.Hale S P, Auld D S, Schmidt E, Schimmel P. Science. 1997;276:1250–1252. doi: 10.1126/science.276.5316.1250. [DOI] [PubMed] [Google Scholar]
- 57.Jahn M, Rogers M J, Söll D. Nature (London) 1991;352:258–60. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- 58.Liu M, Chu W C, Liu J C H, Horowitz J. Nucleic Acids Res. 1997;25:4883–4890. doi: 10.1093/nar/25.24.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musier-Forsyth K, Schimmel P. Nature (London) 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]