Abstract
Root endosymbioses vitally contribute to plant nutrition and fitness worldwide. Nitrogen-fixing root nodulation, confined to four plant orders, encompasses two distinct types of associations, the interaction of legumes (Fabales) with rhizobia bacteria and actinorhizal symbioses, where the bacterial symbionts are actinomycetes of the genus Frankia. Although several genetic components of the host–symbiont interaction have been identified in legumes, the genetic basis of actinorhiza formation is unknown. Here, we show that the receptor-like kinase gene SymRK, which is required for nodulation in legumes, is also necessary for actinorhiza formation in the tree Casuarina glauca. This indicates that both types of nodulation symbiosis share genetic components. Like several other legume genes involved in the interaction with rhizobia, SymRK is also required for the interaction with arbuscular mycorrhiza (AM) fungi. We show that SymRK is involved in AM formation in C. glauca as well and can restore both nodulation and AM symbioses in a Lotus japonicus symrk mutant. Taken together, our results demonstrate that SymRK functions as a vital component of the genetic basis for both plant–fungal and plant–bacterial endosymbioses and is conserved between legumes and actinorhiza-forming Fagales.
Keywords: actinorhizal symbioses, Casuarina glauca, mycorrhizae, signaling
Root endosymbioses are associations between plants and soil microorganisms involving intracellular accommodation of microbes within host cells. The most widespread of these associations is arbuscular mycorrhiza (AM), which is formed by the majority of land plants with fungi belonging to the phylum Glomeromycota (1). In contrast, nitrogen-fixing nodulation symbioses of plant roots and bacteria are restricted to four orders of eurosid dicots (2). Actinorhiza, formed by members of the Fagales, Rosales, and Cucurbitales with Gram-positive Frankia bacteria, differs from the interaction of legumes with Gram-negative rhizobia in several morphological and cytological aspects (3). Although these differences suggest independent regulatory mechanisms, the close relatedness of nodulating lineages indicates a common evolutionary basis of root nodulation symbioses (2). In the legume–rhizobia interaction, among the key factors mediating recognition between the plant and the bacteria are Nod factors (NFs). NFs are bacterial lipochitooligosaccharides with an N-acetylglucosamine backbone (4). The perception of NFs induces a series of responses in host roots, including ion flux changes and membrane depolarization, rhythmic calcium oscillations in and around the nucleus (calcium spiking), cytoskeletal modifications and root hair curling, and activation of cortical cell divisions (5). Extensive mutant screenings performed in legumes led to the identification of several loci involved in this signaling cascade, and recently most of the corresponding genes were identified by map-based approaches (6). In Lotus japonicus, two genes, NFR1 and NFR5 encoding receptor-like serine/threonine kinases with LysM domains, are assumed to be involved in NF perception, because the corresponding mutants are impaired in the earliest NF responses (7). Several downstream components of the NF signaling cascade, including the leucine-rich-repeat receptor kinase gene L. japonicus SymRK (DMI2/NORK in Medicago truncatula and M. sativa, respectively) (8, 9), are dually involved in AM and nodulation symbiosis. SymRK is likely active near the junction of fungal and rhizobial signaling cascades (8). This makes it a particularly interesting candidate for studying a possible role of legume symbiosis genes in Casuarina glauca, which similarly forms AM, but in contrast to legumes interacts not with rhizobia but with Frankia bacteria to form actinorhiza.
In actinorhizal symbioses, very little is known about signaling mechanisms involved in plant-bacteria recognition. Analyses of the genome of three Frankia strains (10), the biochemical characterization of a Frankia root hair-deforming factor whose chemical structure is unknown (11), and the failure of Frankia DNA to complement rhizobial nod gene mutants (12) suggest that Frankia symbiotic signals are structurally different from rhizobial NFs. No plant genes involved in the perception and transduction of Frankia symbiotic signals have been identified to date, mostly due to the lack of genetic tools in actinorhiza-forming plants. Here, we isolate CgSymRK, a predicted SymRK gene from the actinorhizal tree C. glauca, and analyze its role in root endosymbioses. Our results reveal that SymRK is required for both AM and actinorhiza formation in C. glauca, indicating shared genetic mechanisms between fungal and bacterial root endosymbioses in C. glauca and legumes.
Results
Isolation of C. glauca SymRK.
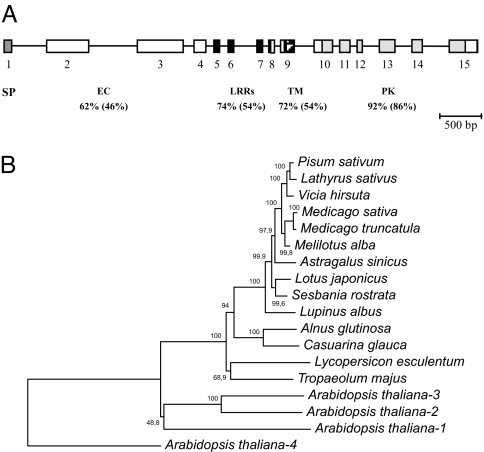
A C. glauca SymRK candidate, CgSymRK, was isolated by using a degenerate priming approach based on similarity with legume SymRK sequences. The gene is 7,280 bp long and contains 15 putative exons, encompassing a 2,829-bp coding sequence. Intron positions and phases are identical to SymRK genes of L. japonicus and other legumes, including Medicago truncatula, Pisum sativum, and Sesbania rostrata. The predicted protein of 941 aa contains an N-terminal region of unknown function, three leucine-rich repeat motifs, a transmembrane region, and a serine/threonine protein kinase (Fig. 1A). The SYMRK kinase domain is highly conserved between legumes and actinorhizal plants. However, SYMRK extracellular regions are conserved between the two actinorhizal plants C. glauca and A. glutinosa but highly variable between legumes and actinorhizal plants (data not shown). Both actinorhizal proteins cluster together in a phylogenetic distance tree in the same subgroup as the legume SYMRK (Fig. 1B). Southern blot experiments suggested that only one SymRK gene exists in C. glauca (data not shown). In C. glauca roots infected with Frankia, real-time expression analysis revealed very little change in CgSymRK transcript abundance within 2 weeks after inoculation [supporting information (SI) Fig. 5A]. However, we cannot rule out that localized changes in CgSymrk expression might occur. CgSymRK expression was three times higher in 3-week-old nodules than in uninoculated roots (SI Fig. 5B).
Fig. 1.
C. glauca SymRK gene. (A) Genomic structure of CgSymRK with indicated predicted protein domains. Exons are depicted as boxes, introns as a black line. SP, predicted signal peptide; EC, extracellular domain; LRR leucine-rich repeat motifs; TM, transmembrane domain; PK, protein kinase domain. Percentages of similarity and identity between CgSYMRK and LjSYMRK are indicated below each predicted protein domain. (B) Distance tree of predicted SYMRK protein sequences based on a CLUSTALW alignement. Numbers above the branches represent the percentages of 1,000 bootstrap replications.
CgSymRK Is Necessary for Actinorhizal and AM Symbioses in C. glauca.
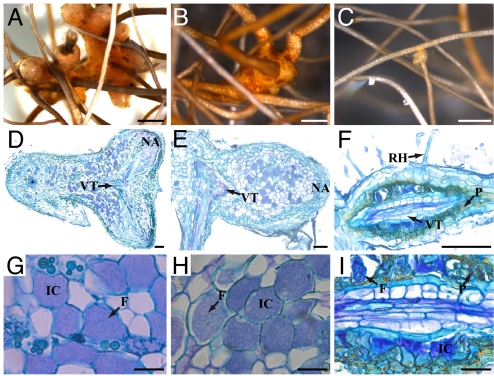
To investigate the role of SymRK in root endosymbioses in C. glauca, we reduced CgSymRK expression levels in Agrobacterium-induced hairy roots by using RNAi. In parallel, control plants bearing nontransgenic and hairy roots transformed with a control vector comprising the GFP reporter gene but lacking the RNAi cassette were analyzed. A total of 78 RNAi composite plants and 48 transgenic control composite plants showing high GFP fluorescence in hairy roots were analyzed in two independent experiments. At 3 weeks after inoculation, plants transformed with the control vector began to develop nodules that were similar in size and shape to those produced on nontransgenic roots (Fig. 2 A and B). As observed in ref. 13, nodulation of transgenic control hairy roots was reduced compared with nontransgenic controls. In CgSymRK RNAi roots, the frequency of nodulated root systems was 50% less than in transgenic control root systems (SI Table 1). Nodulated RNAi roots showed strong alterations in nodule development compared with control roots. We observed a gradient of phenotypes ranging from aborted prenodules (Fig. 2 C and F) to nodules usually consisting of one thin lobe, whereas mature nodules in transgenic and nontransgenic control roots were multilobed (Fig. 2 A and B and D and E). On CgSymRK RNAi root nodules, the nodular roots, which are unbranched roots exhibiting negative geotropism growing at the apex of each nodule lobe, often behaved like adventitious roots exhibiting normal root growth and branching (data not shown). These aberrant nodules and nodular roots were never seen on transgenic or nontransgenic control roots. Histological analysis of 10 aberrant symbiotic structures of CgSymRK RNAi roots revealed an accumulation of phenolic compounds (Fig. 2F) and the presence of small infected cells in the cortex (Fig. 2I) contrasting with the hypertrophied infected cells observed in nontransgenic and transgenic control nodules (Fig. 2 G and H). We tested the ability of CgSymRK RNAi nodules to fix nitrogen via acetylene reduction activity (ARA) assays. CgSymRK RNAi nodules exhibited a quasinull ARA compared with transgenic control nodules (SI Fig. 6). To test the efficacy of CgSymRK knockdown in RNAi roots, CgSymRK expression was tested by quantitative RT-PCR (qPCR) in subcultivates of five CgSymRK RNAi roots. A 52–76% reduction of CgSymRK mRNA levels was observed in RNAi roots compared with transgenic control roots (SI Fig. 7). Taken together, our results indicate that a reduction in CgSymRK expression results in severe impairment in actinorhiza formation and symbiotic nitrogen fixation.
Fig. 2.
Knockdown phenotype of CgSymRK after Frankia inoculation. (A) Nontransgenic nodule consisting of multiple lobes 10 weeks postinoculation (wpi). A nodular root develops at the apex of each nodule lobe. (B) Nodule on a hairy root transformed with a control vector at 10 wpi. Nodule morphology is similar to wild-type nodules. (C) Nodule-like structure formed on CgSymRK knockdown (RNAi) roots 10 wpi. Nodule lobes are small and do not branch to form a multilobed structure. (D and E) Sections of wild-type and transgenic control nodules. Each nodule lobe exhibits a central vascular bundle and cortical parenchyma infected with Frankia. (F) Section of a nodule-like structure observed on an RNAi plant showing few small infected cells and abnormal accumulation of polyphenols in the endodermis. (G) Closeup of area in D, showing both infected and uninfected cortical cells. Infected cells are hypertrophied and filled with Frankia. (H) Closeup of area in E. As in nontransgenic nodules, hypertrophied cortical cells are filled with Frankia. (I) Closeup of area in F. Infected cells are few and small compared with cells in nontransgenic and transgenic control nodules. IC, infected cell with Frankia; RN, root nodule; NA, nodule apex; VT, vascular tissue; P, polyphenol droplets; RH, root hair. [Scale bars: 1 mm (A–C); 100 μm (D–F); 25 μm (G–I).]
In legumes, SymRK has been shown to play also a crucial role in the establishment of AM symbiosis (14). To investigate whether CgSymRK is also involved in AM formation in C. glauca, RNAi hairy roots plants were generated and cultivated in pots containing Glomus intraradices inoculum. Plants were harvested after 4 or 8 weeks, and GFP fluorescence was checked in transformed roots. Seventeen control and 21 RNAi plants showing GFP fluorescence and six nontransformed root systems were subjected to AM analysis. In nontransgenic control plants, fungal structures such as intraradical hyphae, vesicles, and arbuscules (SI Figs. 8 and 9 A and B) were observed at high frequencies ranging from 22% to 52% total root length colonization. Similar structures were observed at relatively high frequencies in roots of most transgenic control plants, 4 or 8 weeks after inoculation (SI Figs. 8 and 9 C and D). The slight reduction of colonization compared with nontransgenic control roots might be linked to modifications of hormonal balance. In contrast, most plants transformed with the CgSymRK RNAi construct showed very weak levels of AM colonization, and four composite plants showed a complete absence of intraradical structures 4 weeks after inoculation (SI Fig. 8). The absence of intracellular colonization was not due to an absence of inoculum, because extraradical hyphae were very often observed. Some RNAi roots showed extensive development of extraradical mycelium, usually growing along the epidermal cells and forming appressoria, which were frequently associated with abnormal, swollen hyphal structures (SI Fig. 9 E and F). Most fungal penetration attempts aborted, resulting in very low levels of intraradical colonization (SI Fig. 8). However, on the rare occasions where penetration succeeded, intraradical hyphae, arbuscules, and vesicles morphologically similar to those found in transgenic and nontransgenic control roots were observed (SI Fig. 9 G and H). Compared with control roots, colonized patches were generally smaller, spreading over few cells near the entry point and never succeeding in colonizing the whole root. These results indicate that CgSymRK knockdown strongly affects early steps of the AM interaction, especially fungal penetration into the root cortex, thereby revealing a conservation of SymRK function in AM between legumes and C. glauca.
CgSymRK Can Restore Root Endosymbioses in a Legume symrk Mutant.
To test whether CgSymRK can function in root endosymbioses in a legume, we introduced its coding sequence linked to the L. japonicus SymRK promoter region into Agrobacterium rhizogenes-induced roots of L. japonicus symrk-10 (15) mutants. Interaction with AM fungi is usually aborted in L. japonicus symrk mutants at the epidermal level (14), with few hyphae invading the root cortex and no arbuscules developing within 3–6 weeks of exposure to fungal inoculum.
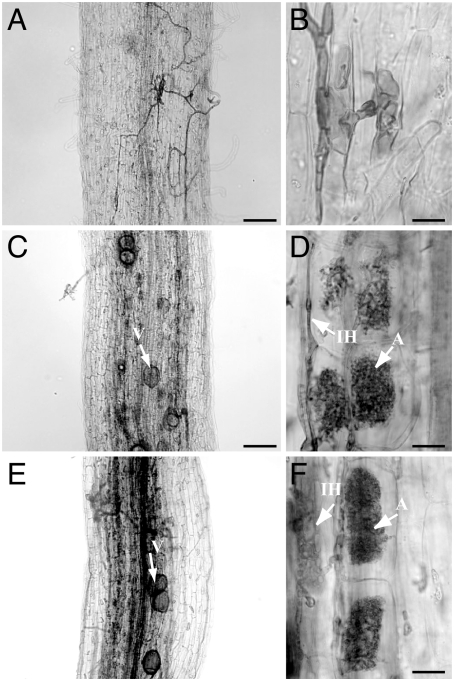
Similarly, after 3 weeks of cocultivation with G. intraradices, symrk-10 roots transformed with a vector lacking a SymRK expression cassette (control vector) formed no AM, and typical hyphal swellings formed in epidermal cells indicating abortion of fungal infections (SI Table 2; Fig. 3 A and B). In contrast, wild-type (Fig. 3 C and D) and symrk-10 (Fig. 3 E and F) plants transformed with CgSymRK developed AM (SI Table 2), involving the formation of wild-type-like cortical arbuscules (Fig. 3F) and infection sites in the complemented mutants. Similar results were obtained with wild-type and symrk-10 mutant plants transformed with an LjSymRK expression cassette controlled by the same promoter region (SI Table 2). These results demonstrate that CgSymRK can complement the mycorrhization defect of L. japonicus symrk mutants.
Fig. 3.
AM formation in L. japonicus symrk-10 mutants complemented with the CgSymRK coding sequence under control of the LjSymRK promoter, after 3 weeks of cocultivation with G. intraradices. Cleared roots with fungal structures are stained with acidic ink. (A and B) symrk-10 roots transformed with a control vector. (A) Noncolonized root with extraradical mycelium and aborted infection structure (arrow). (B) Fungal appressorium and entry point associated with aborted infection structure within host epidermal cell. (C and D) Wild-type and (E and F) symrk-10 roots transformed with CgSymRK linked to the LjSymRK promoter. Fungal hyphae grow through epidermis and exodermis and form arbuscules and vesicles in the inner root cortex. A, arbuscule; IH, intraradical hyphae; V, vesicle. [Scale bars: 100 μm (A, C, and E); 20 μm (B, D, and F).]
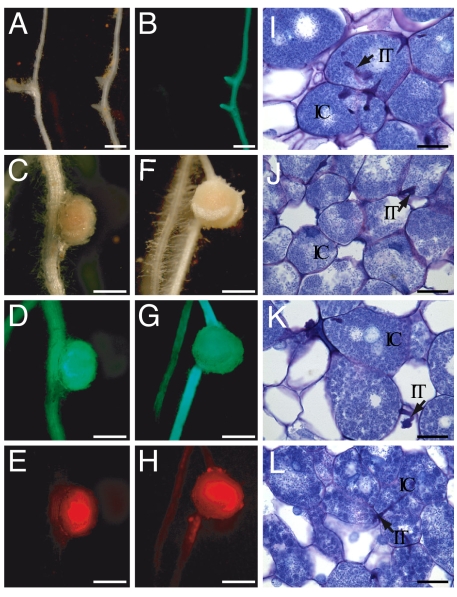
Wild-type L. japonicus plants respond to inoculation with their rhizobial symbiont Mesorhizobium loti by root hair curling, infection thread formation, and nodule development. L. japonicus symrk mutants, in contrast, show no normal curling reaction of root hairs, and bacteria are unable to induce infection thread or nodule formation (8). This was equally the case in symrk-10 roots transformed with a control vector (SI Table 2; Fig. 4 A and B), whereas L. japonicus wild-type and symrk-10 roots carrying CgSymRK formed wild-type-like infection threads and nodules (Table 2; Fig. 4 C–E and F-H, respectively). These exhibited bacteria-filled and noninfected host cells (Fig. 4 I and J). L. japonicus wild-type and symrk-10 roots transformed with a L. japonicus SymRK expression cassette equally formed infection threads and infected nodules (SI Table 2; Fig. 4 K and L, respectively). The observation that SymRK from the actinorhiza-forming plant C. glauca can restore not only the interaction with AM fungi but also with M. loti bacteria in L. japonicus symrk mutants indicates that this gene is highly conserved in its function in both AM and nodulation symbioses, whereas the specificity of recognition of bacterial partners is SymRK-independent.
Fig. 4.
Nodulation in L. japonicus symrk-10 mutants complemented with the CgSymRK coding sequence under control of the LjSymRK promoter, 8 weeks after inoculation with M. loti MAFF expressing DsReD. Transgenic roots carried an sGFP reporter gene. (A, C, and F) Roots and nodules under white light. (B, D, and G) Transgenic roots and nodules showing GFP fluorescence. (E and H) Red fluorescence of bacterial DsRED. (A and B) symrk-10 root transformed with the control vector, showing no nodules. (C–E) Transgenic wild-type root carrying the CgSymRK coding sequence. Nodules contain DsReD-expressing bacteria (E). (F–H) symrk-10 mutant root transformed with the CgSymRK coding sequence, carrying wild-type-like nodules. (I–L) Semithin sections of nodules stained with toluidine blue. (I and J) Nodules on symrk-10 mutant and wild-type roots complemented with the CgSymRK coding sequence, respectively. (K and L) Nodules on symrk-10 mutant and wild-type roots complemented with the LjSymRK coding sequence, respectively. Infection threads (IT) are contained within bacteria-infected cells (IC). [Scale bars: 500 μm (A–H); 25 μm (I–L).]
Discussion
There are three major types of root endosymbioses in angiosperms. These include the arbuscular mycorrhiza symbiosis with fungi and nitrogen-fixing root nodulation of legumes and actinorhiza-forming plants. In recent years, there has been a tremendous increase in knowledge of the molecular mechanisms responsible for NF perception and signal transduction in legumes (6). Genetic approaches in model legumes led to the identification of several components and the definition of a signaling cascade (5). Part of this signaling cascade is also involved in transduction of the symbiotic signal in AM symbioses (5). This gave rise to the hypothesis that the evolutionarily recent legume–rhizobia symbiosis reuses some of the molecular mechanisms of the more ancient AM symbiosis (16). This common signaling pathway includes the receptor kinase SymRK/DMI2. So far, nothing is known about the symbiotic signals and their perception during actinorhizal symbioses. Available data indicate only that the Frankia symbiotic signal is likely chemically different from NFs (10–12). Here, we report the isolation and characterization of CgSymRK, a SymRK/DMI2 homolog from the actinorhizal tree C. glauca. Our data demonstrate that CgSymRK is functionally equivalent to LjSymRK in symbiosis formation in L. japonicus. In our experience, both AM and nodulation symbiosis formation in hairy roots of L. japonicus can vary in efficiency, particularly in complemented mutant tissue. Despite the differences in numbers of rescued root systems between the symbiosis types (CgSymRK) and constructs, phenotypic analyses clearly suggest that both symbiosis types can be fully supported by CgSymRK in L. japonicus.
Moreover, we were able to show that CgSymRK is necessary for functional symbiosis with Frankia. We therefore conclude that CgSymRK is probably a component of the signaling pathway involved in the perception and the transduction of yet-unknown Frankia factors. As in legumes (17, 18) CgSymRK expression level remained constant during root infection and increased in mature nodules compared with noninoculated roots. The reduction in the number of nodulated plants obtained by RNAi is less pronounced than the one obtained in legumes (18, 19); however, it clearly indicates that CgSymRK is involved during the early stages of Frankia root hair infection. A second symbiotic defect was observed downstream of this infection with striking differences in the nodule morphology and tissue organization relative to the control. CgSymRK RNAi hairy roots mostly developed small nonfixing nodule-like structures. Light microscopy revealed that the nodule apical meristem was absent, and we did not observe the gradient of infection and differentiation in the cortex that is present in transgenic and nontransgenic control nodules. Cortical cells also seemed to be less infected, and infected cells were smaller than those of control nodules. In addition, we observed the formation of dense deposits of polyphenols in CgSymRK RNAi nodules. These data suggest that the loss of CgSymRK function also affects C. glauca–Frankia symbiotic interaction after bacterial penetration. This is consistent with qPCR results that indicate an enhancement of CgSymRK expression in mature nodules.
We also analyzed the role of CgSymRK in the G. intraradices–C. glauca interaction. Hairy roots of C. glauca carrying the CgSymRK RNAi construct were able to form arbuscules and vesicles morphologically similar to those found in control plants, suggesting that CgSymRK is not involved in the formation of these late symbiotic structures. However, most RNAi plants showed a significant decrease in fungal colonization. At the root surface, hyphae developed abundant appressoria, but these colonization attempts rarely succeeded, pointing to a role of CgSymRK during hyphal penetration. Similar results were shown for L. japonicus symrk (14) and M. truncatula dmi2 mutants (20, 21). This work report a role of SymRK in AM symbiosis formation in a nonlegume plant.
In summary, our data indicate that SymRK is involved in the symbiotic signal transduction pathway leading to actinorhizal symbioses. Our results demonstrate that, in C. glauca as in legumes, SymRK is involved in the establishment of both nitrogen-fixing nodule and AM symbioses, thus supporting the hypothesis that signaling genes have been recruited from the more ancient AM symbiosis during the evolution of nitrogen-fixing symbioses. It will now be essential to compare signal transduction pathways involved in endosymbiotic accommodation of AM fungi, rhizobia, and Frankia to develop strategies for the transfer of nodulation to nonnodulated plants.
Materials and Methods
Plant, Bacterial, and Fungal Material.
C. glauca seeds were provided by Carter Seeds and grown as described in ref. 22. L. japonicus ecotype B-129 Gifu and L. japonicus symrk mutant symrk-10 from the same ecotype (15) were grown for transformation as described in ref. 23. C. glauca and L. japonicus plants were transformed with A. rhizogenes strains A4RS (24) and AR1193 (25). For nodulation phenotyping of C. glauca, plants were inoculated with Frankia strain CcI3. C. glauca mycorrhization experiments were performed in pots containing an autoclaved mixture of quartz sand and soil (4:1). Plants were transferred from in vitro cultures and grown for 4–8 weeks in a growth chamber and watered with a modified Hoagland solution (22) containing 10 μM phosphate. G. intraradices inoculum was prepared by extracting spores from in vitro cultures of G. intraradices (26). One Petri dish showing extensive sporulation was kept at 4°C for at least 3 weeks and used to inoculate 2 liters of sand:soil mixture. For nodulation phenotyping of L. japonicus, composite plants were grown in plastic pots with 300 ml of Seramis substrate (Mars) and 150 ml of FP medium (27) and inoculated with Mesorhizobium loti strain MAFF expressing DsReD (M. Hayashi, personal communication). To test for AM formation, plants were cocultivated with G. intraradices BEG195 in chive nurse pots as described (14) and harvested after 3 weeks. Transgenic roots were selected via GFP fluorescence and stained with acidic ink as described (14) for visualization of fungal structures.
Identification and Cloning of CgSymRK Sequences and Phylogenetic Analysis.
Amplification of CgSymRK was conducted on a cDNA library prepared from C. glauca uninfected roots by using the degenerated primers SymRKdeg-5 (5′-CCAAGACATGAATGGTCTCTGGTNGARTGGGC-3′) and SymRKdeg-3 (5′-GAATCCATAGATCTCATATATTCAGAAGCRTTRTTYTC-3′). The amplified fragment was cloned into a pGEM-T easy vector (Promega) and sequenced. cDNA fragments were obtained by RACE-PCR on a root cDNA library by using the Marathon cDNA amplification kit (Clontech), and the CDS was amplified by using primers CgSymRKATG (5′-ATGATGGAGGGATTGCATAAT-3′) and CgSymRKSTOP (5′-TCCTCCACAGCCAAGATAA -3′). The CgSymRK genomic sequence was obtained by using a Genome Walker kit (Clontech) and cloned in a pGEM-T easy vector. For the phylogenetic analysis, sequences (with GenBank accession numbers in parentheses) from Alnus glutinosa (62946487), Sesbania rostrata (56412259), Melilotus alba (21698802), Pisum sativum (21698794), Lathyrus sativus (89213719), Vicia hirsuta (21698800), Medicago truncatula (21698783), Lotus japonicus (21622628), Medicago sativa (21698781), Lupinus albus (62946493), Tropaeolum majus (62946489), Astragalus sinicus (61723807), and Lycopersicon esculentum (62944413) were retrieved from GenBank via BLASTP search performed with the L. japonicus-predicted SYMRK sequence. Sequences of SYMRK homologs in Arabidopsis thaliana (−: At5g48740; −2: At2g37050; −3 At1g67720) described in ref. 28 were also included. A. thaliana (−: At3g25560), an NSP-interacting kinase (29), was used to root the tree. The alignment was performed by using the CLUSTALW and Neighbor-Joining algorithms in the CLC-Free Workbench 4 software package (CLC bio).
Agrobacterium rhizogenes-Mediated Transformation, RNAi and Complementation.
C. glauca and L. japonicus hairy root transformation was performed by following standard procedures (13, 23). To produce the knockdown construct, 365 bp corresponding to the kinase domain of CgSymRK sequence were amplified from genomic DNA by using CgRNAi-5 (5′-GGGAGCTGGAGGATGCTTTGA-3′) and CgRNAi-3 (5′-TAAGTAGTAGTAGGTGGGGAGATTATTC-3′) primers containing 5′ XhoI or BamHI restriction sites for CgRNAI-5 and KpnI or ClaI for CgRNAI-3. Amplified fragments (XhoI-CgRNAi-KpnI and BamHI-CgRNAi-ClaI) were then cloned into pKannibal (30) downstream of the CaMV 35S promoter, and the RNAi cassette was then cloned into the pHKN29 binary vector (31). This vector also contains the GFP gene under the control of the CaMV 35S promoter. For functional complementation, the CgSymRK coding sequence was fused to LjSymRK promoter (2,415-bp) and terminator (315-bp) regions and transferred into the pHKN29 binary vector. For complementation of L. japonicus symrk mutants with LjSymRK, the binary vector pCAMBIA 1302 was equipped with 4,970 bp of the LjSymRK promoter region fused to the LjSymRK coding sequence and a 285-bp NOS terminator fragment amplified from pJawohl8 RNAi [kind gift of P. Schulze-Lefert (Max Planck Institute, Cologne, Germany)] by using primers TNOS-5 5′-AATAAACCTAGGATCAGCTTGCATGCCGGTCG-3′ and TNOS-3 5′-AAATAAGTCGACCTAGAGTCAAGCAGATCGTTCAAAC-3′.
qPCR and Acetylene Reduction Assay.
Total RNA was extracted by using the RNeasy Plant Mini Kit (Qiagen). RNAs were quantified with Quant-iT Ribogreen RNA Reagent (Invitrogen). One hundred nanograms of total RNA was reverse-transcribed by using SuperScriptIII H− reverse transcriptase (Invitrogen) and oligo(dT)12–18. qPCR was performed by using the FullVelocity SYBR Green QPCR Master Mix (Stratagene). The primers used were qCgSymRKFor1 5′-GCAGGAGGTAGCAGTGAAGGTTC-3′ and qCgSymRKRev2 5′-GCGATCTTGAAGCGAGCCATTAG-3′. The FullVelocity cycling PCR program on an MX 3005P (Stratagene) was as follows: 1 cycle at 95°C for 5 min, 40 cycles at 95°C for 10 s and 60°C for 30 s, ended by 1 cycle at 95°C for 1 min, 60°C for 30 s, and 95°C for 30 s. Reactions were performed in triplicate, and the comparative threshold-cycle method was used to quantify CgSymRK expression (32). The results were standardized with CgUbi expression levels (32). The acetylene reduction assay was performed according to ref. 33.
Histochemical Analysis and Microscopy.
C. glauca nodules were fixed as described in ref. 34. L. japonicus nodules were fixed in a solution containing 50% EtOH, 4% formaldehyde, and 5% acetic acid and dehydrated in 70% EtOH. Samples were embedded in Technovit 7100 resin (Heraeus Kulzer), and thin sections (6 μm) were cut with a microtome (Microm HM355S), stained with toluidine blue (0.01%), and mounted in Clearium Mountant (Surgipath). Visualization of AM in L. japonicus was performed as described in ref. 14. To visualize AM in C. glauca, roots were cleared for 1–2 days in 10% KOH at 90°C, rinsed, and stained for at least 1 h with a 0.05% trypan blue/5% acetic acid solution at 60°C and destained in water as described (35). Samples were viewed under a DMRB microscope (Leica). Colonization was assessed with a microscope by using the gridline intersect method (35) on at least 100 intersections per sample.
Supplementary Material
Acknowledgments.
We thank Dr. G. Bécard (Unité Mixte de Recherche 5546, Toulouse) for providing the G. intraradices starting cultures, Dr. H. Kouchi (National Institute of Agrobiological Sciences, Ibaraki, Japan) for providing pHKN29, Dr. A. Galiana [Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), Montpellier, France] for help with the ARA experiment, and M. Collin and V. Vaissayre (IRD) for technical assistance in cytology and the in vitro culture. Financial support was provided by IRD, the Center for French–Bavarian University Cooperation (CCUFB/BFHZ) Project, the PROCOPE (EGIDE/DAAD, French–German Bilateral Cooperation) project, and the Agence Nationale de la Recherche Project NewNod (ANR-06-BLAN-0095).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU294188 (CgSymRK genomic) and EU273286 (CgSymRK CDS)].
See Commentary on page 4537.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710618105/DC1.
References
- 1.Schüssler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res. 2001;105:1413–1421. [Google Scholar]
- 2.Soltis DE, et al. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawlowski K, Bisseling T. Rhizobial and actinorhizal symbioses: What are the shared features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Haeze W, Holsters M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology. 2002;12:79R–105R. doi: 10.1093/glycob/12.6.79r. [DOI] [PubMed] [Google Scholar]
- 5.Oldroyd GE, Downie JA. Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol. 2006;9:351–357. doi: 10.1016/j.pbi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 8.Stracke S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 9.Endre G, et al. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- 10.Normand P, et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceremonie H, Debelle F, Fernandez MP. Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Can J Bot. 1999;77:1293–1301. [Google Scholar]
- 12.Ceremonie H, Cournoyer B, Maillet F, Normand P, Fernandez MP. Genetic complementation of rhizobial nod mutants with Frankia DNA: artifact or reality? Mol Gen Genet. 1998;260:115–119. doi: 10.1007/s004380050877. [DOI] [PubMed] [Google Scholar]
- 13.Diouf D, et al. Hairy root nodulation of Casuarina glauca: a system for the study of symbiotic gene expression in actinorhizal tree. Mol Plant–Microb Interact. 1995;8:532–537. doi: 10.1094/mpmi-8-0532. [DOI] [PubMed] [Google Scholar]
- 14.Demchenko K, Winzer T, Stougaard J, Parniske M, Pawlowski K. Distinct roles of Lotus japonicus SYMRK and SYM 15 in root colonization and arbuscule formation. New Phytol. 2004;163:381–392. doi: 10.1111/j.1469-8137.2004.01123.x. [DOI] [PubMed] [Google Scholar]
- 15.Perry JA, et al. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kistner C, Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends Plants Sci. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- 17.Bersoult A, et al. Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol Plant–Microbe Interact. 2005;18:869–876. doi: 10.1094/MPMI-18-0869. [DOI] [PubMed] [Google Scholar]
- 18.Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M. SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA. 2005;102:10369–10374. doi: 10.1073/pnas.0504250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limpens E, et al. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA. 2005;102:10375–10380. doi: 10.1073/pnas.0504284102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morandi D, Prado E, Sagan M, Duc G. Characterisation of new symbiotic Medicago truncatula (Gaertn.) mutants, and phenotypic or genotypic complementary information on previously described mutants. Mycorrhiza. 2005;15:283–289. doi: 10.1007/s00572-004-0331-4. [DOI] [PubMed] [Google Scholar]
- 21.Calantzis C, Morandi D, Arnould C, Gianinazzi-Pearson V. Cellular interactions between G. mosseae and a Myc(-) dmi2 mutant in Medicago truncatula. Symbiosis. 2001;30:97–108. [Google Scholar]
- 22.Franche C, et al. Genetic transformation of the actinorhizal tree Allocasuarina verticillata by Agrobacterium tumefaciens. Plant J. 1997;11:897–904. [Google Scholar]
- 23.Diaz G, Gronlund M. In: Lotus japonicus Handbook. Marquez AJ, editor. New York: Springer; 2005. pp. 261–277. [Google Scholar]
- 24.Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid. 1986;16:124–134. doi: 10.1016/0147-619x(86)90071-5. [DOI] [PubMed] [Google Scholar]
- 25.Stougaard J, Abildsten D, Marcker KA. The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet. 1987;207:251–255. [Google Scholar]
- 26.St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res. 1996;100:328–332. [Google Scholar]
- 27.Fahraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Riely BK, Burns NJ, Ane JM. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontes EPB, Santos AA, Luz DF, Waclawovsky AJ, Chory J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004;18:2545–2556. doi: 10.1101/gad.1245904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wesley SV, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai H, Kouchi H. Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant–Microbe Interact. 2003;16:663–668. doi: 10.1094/MPMI.2003.16.8.663. [DOI] [PubMed] [Google Scholar]
- 32.Hocher V, et al. Expressed sequence-tag analysis in Casuarina glauca actinorhizal nodule and root. New Phytol. 2006;169:681–688. doi: 10.1111/j.1469-8137.2006.01644.x. [DOI] [PubMed] [Google Scholar]
- 33.Hardy RW, Holsten RD, Jackson EK, Burns RC. The Acetylene-Ethylene Assay for N (2) Fixation: Laboratory and Field Evaluation. Plant Physiol. 1968;43:1185–1207. doi: 10.1104/pp.43.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svistoonoff S, et al. cg12 expression is specifically linked to infection of root hairs and cortical cells during Casuarina glauca and Allocasuarina verticillata actinorhizal nodule development. Mol Plant–Microbe Interact. 2003;16:600–607. doi: 10.1094/MPMI.2003.16.7.600. [DOI] [PubMed] [Google Scholar]
- 35.Brundrett MC, Piché Y, Peterson RL. A new method for observing the morphology of vesicular-arbuscular mycorrhizae. Can J Bot. 1984;62:2128–2134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.