Abstract
Pathogens secrete effector molecules that facilitate the infection of their hosts. A number of effectors identified in plant pathogenic Phytophthora species possess N-terminal motifs (RXLR-dEER) required for targeting these effectors into host cells. Here, we bioinformatically identify >370 candidate effector genes in each of the genomes of P. sojae and P. ramorum. A single superfamily, termed avirulence homolog (Avh) genes, accounts for most of the effectors. The Avh proteins show extensive sequence divergence but are all related and likely evolved from a common ancestor by rapid duplication and divergence. More than half of the Avh proteins contain conserved C-terminal motifs (termed W, Y, and L) that are usually arranged as a module that can be repeated up to eight times. The Avh genes belong to the most rapidly evolving part of the genome, and they are nearly always located at synteny breakpoints. The superfamily includes all experimentally identified oomycete effector and avirulence genes, and its rapid pace of evolution is consistent with a role for Avh proteins in interaction with plant hosts.
Keywords: comparative genomics, gene family evolution, oomycete, avirulence genes, pathogenicity
Large repertoires of molecules, termed “effectors,” are used by microbial pathogens to promote effective colonization of their hosts. In some pathogens, a common mechanism deploys heterogeneous effectors to their site of action. For example, many bacterial effectors are delivered into host cells by a type III secretion system (1). Similarly, the malarial parasite, Plasmodium falciparum, translocates a set of heterogeneous effectors into human erythrocytes via a host targeting signal at the N termini of the effector proteins (2, 3).
The genus Phytophthora includes many destructive plant pathogens (4). For example, P. sojae, the soybean root rot pathogen, causes ≈$1–2 billion in losses per year worldwide. Another species, P. ramorum, is responsible for Sudden Oak Death and has destroyed many oak trees along the west coast of the United States (5). Phytophthora species are oomycetes. They belong to the lineage Stramenopiles that also includes diatoms and golden-brown algae (6). The phytopathogenicity of oomycetes evolved independently from pathogens in other lineages such as fungi, and Phytophthora genomes likely encode unique reservoirs of effectors.
Phytophthora species secrete many proteins with demonstrated or potential effector activity (7). Some effectors, called avirulence (Avr) proteins, display specific gene-for-gene interactions with host resistance proteins (8). Four oomycete Avr genes have been cloned, two from Phytophthora species and two from Hyaloperonospora parasitica (9–12). The encoded proteins share little sequence similarity except for two conserved motifs (RXLR and dEER) at the N terminus (10). These motifs have been shown to be required for Avr proteins to enter the host cell (ref. 13; D. Dou, S. D. Kale, and B.M.T., unpublished data). RXLR resembles the Pexel or VTF motif of Plasmodium effectors that is required to carry those effectors across the parasitiphorous vacuolar membrane into the cytoplasm of erythrocytes, suggesting a common origin for these effectors' transport motifs (2, 3). The RXLR and dEER motifs provide a powerful bioinformatics tool for identifying the effector reservoirs of oomycete pathogens. In this study, we have used recursive BLAST and Hidden Markov Model (HMM) searches to define and characterize the reservoir of P. sojae and P. ramorum RXLR-containing effectors from their genome sequences (14).
Results
P. sojae and P. ramorum Genomes Encode Large Numbers of Avirulence Homolog (Avh) Proteins.
Preliminary BLAST searches of the genomes with the Avr1b-1 gene revealed a diverse family of Avh genes with high levels of sequence divergence. No P. sojae Avh protein showed >30–50% sequence similarity to the most similar P. ramorum sequence, and this finding also was frequently true for the most similar P. sojae paralog. Using recursive BLAST searches, we discovered an initial 497 Avh sequences. HMM screens built on the initial 497 sequences discovered 258 more Avh sequences [supporting information (SI) Fig. 5]. The numbers must be regarded as approximate because they depended on the cutoffs used. The 755 sequences included 370 Avh genes in P. ramorum (PrAvh) and 385 in P. sojae (PsAvh). The search also revealed 68 pseudogenes or inactive alleles containing stop codons and frameshifts that were named P. sojae RXLRdEER Fragment (PsRF) and P. ramorum RXLRdEER Fragment (PrRF). In some cases, the distinction between genes and pseudogenes was unclear, particularly when the distance from the dEER motif to the C terminus was very short (<10 not allowed) or the distance from the N terminus to the RXLR motif was very long (<120 not allowed).
Of the Avh genes, 327 were missed from the automated genome annotation. The coding potential of these genes ranged from −3.3 to 12.3, with 95% >0; a value >0 suggests that the sequence is part of an authentic ORF (15). Nine sequences with a negative coding potential were excluded from the list of 755, but 11 genes were included because they had positive HMM scores in the RXLR-dEER domain, as well as significant homology to Avh genes with a positive coding potential. The P. sojae Avh genes had an average coding potential of 8.3, whereas noncoding regions had an average of −8.4 (SI Fig. 6). Because the Avh list was developed by using heuristic BLAST and HMM searches, the superfamily includes members with C-terminal homology that do not have a conventional RXLR string. Of the 755 Avh proteins, 552 have a perfect RXLR string, but the others have strings such as KXLR, RXLG, PXLR, and so on. Some of these variant RXLR sequences may still be functional cell-targeting motifs (D. Dou, S. D. Kale, and B.M.T., unpublished data), but some may represent pseudogenes or inactive alleles.
The Avh superfamily encompasses all 54 experimentally supported Phytophthora RXLR proteins and 30 close paralogs (13, 16).
Avh Proteins Dominate the RXLR Effector Reservoir in the Two Phytophthora Species.
To estimate the total number of RXLR effectors independently of the BLAST approach, we first identified all candidates (1,240) carrying the string RXLR in putative proteins with an N-terminal signal peptide (SP) obtained by translating the genome sequences in all reading frames. This number is similar to that of Win et al. (16). The string RXLR was allowed to occur between 30 and 60 amino acids after the SP cleavage site. We then permuted all of the putative proteins and reperformed the search to estimate the number of false positives. RXLR motifs were detected in 639 of the permuted proteins, indicating that about half of the 1,240 detected RXLR motifs could be expected by chance. When a control search was carried out with the string GXLG, the hits recovered from the permuted and nonpermuted proteins pool were similar (SI Fig. 7). Therefore, the nonrandom RXLR reservoir in the two species was estimated to be ≈600 proteins.
To identify the true RXLR effectors in the pool of 1,240, we constructed an HMM from all of the sequences in the pool, reasoning that the HMM would extract any signal present in the true members. The HMM was constructed from the 10 and 5 amino acid residues to the left and right of the string RXLR-(1–30aa)-[E D]R, respectively. Our rationale was that sequences flanking the RXLR string in known effectors are nonrandom (14) and, together with the dEER motif, have been demonstrated experimentally to be required for cell targeting in Phytophthora (D. Dou, S. D. Kale, and B.M.T., unpublished data) and Plasmodium (17). By using a cutoff score of 4.0, this HMM recovered 548 candidates from the unpermuted pool of 1,240, which is comparable to the number of true effectors estimated by permutation analysis. With a cutoff of 4.0, only 30 false positives were recovered from the permuted pool. Lowering the HMM cutoff to 3.0 retrieved 33 more hits from the permuted pool, but only 6 more from the unpermuted pool. The 548 candidates from the unpermuted pool include 533 Avh proteins. Because 30 of the 548 candidates are estimated to be false positives based on the search of the permuted pool, the Avh proteins could account for ≈100% of the true positives. These 533 Avh proteins account for 96.4% of the 552 Avh proteins that have perfect RXLR strings. Among all of the Avh candidates identified by the Blast and HMM searches, <5% contained RXLR after permutation, consistent with a low false positive rate in this subset.
Our set of 755 Avh proteins and the string search dataset of Bhattacharjee et al. (325 proteins) (17) have 227 proteins in common, whereas the RXLR string search set of Win et al. (16) has less than half overlapping with our set (SI Fig. 8). These two string search datasets (16, 17) contain 775 unique non-Avh proteins. Of these, 386 (50%) have low coding potential, whereas another 109 (14%) correspond to incorrect gene models in which the protein no longer qualified as an RXLR effector after the model was corrected. A further 269 (35%) scored negative with the HMM derived from the pool of 1,240 (SI Table 1), leaving 11 that pass our objective criteria. These 11 belong to the 15 non-Avh proteins detected in our HMM screen of the pool of 1,240 (the remaining 4 are fragments or reside inside other gene models and were discarded). The 11 were combined with our Avh list to create a consensus RXLR effector pool. Closer examination revealed that 4 of the 11 had significant matches to Avh proteins and so were reclassified as Avh. Four previously missed paralogs of the 11 also were added to the list, 3 of which matched Avh proteins (SI Fig. 8). The final RXLR-dEER effector consensus list is comprised of 770 proteins, 374 from P. ramorum and 396 from P. sojae. Of the 770, 762 are Avh proteins (370 PrAvh and 392 PsAvh) (SI Table 2).
Avh Proteins Are Related and Form a Superfamily.
As expected, because most Avh proteins were discovered by recurrent Blast searches, >90% of the Avh proteins could be grouped into one superfamily by the criteria that all members have at least one significant BLAST hit (E value <1e-5, identity >30%) to another Avh protein. Furthermore, the pairwise similarities among the superfamily members form a fully connected network. If the Avh sequences excluding the SP were permuted, <2% could be grouped, indicating that the pairwise similarities did not arise by chance. If the RXLR-dEER domains as well as the SPs were left unpermuted, 10% could be grouped, indicating that similarities in the RXLR-dEER region account for some of the family-wide similarity, but that the majority of the similarities lie within the C-terminal regions.
Based on shared sequence identity (proteins sharing BLASTP hits of E value <1e-8, identity >30%) and neighbor joining analysis, the Avh superfamily could be divided into 103 Avh groups (AGs), ranging in size from 2 to 42 members and one large group of 109 members (SI Fig. 9). The latter could be divided into subgroups based on higher sequence identity (sharing BLASTP hits of E value <1e-30, identity >40%) (SI Table 3). There were 66 singletons that did not belong to any of these groups, but were loosely associated with the superfamily because of a significant BLAST hit (E value <1e-2) to group members. They are unlikely to be spuriously identified because their average coding potential is the same as that of the superfamily.
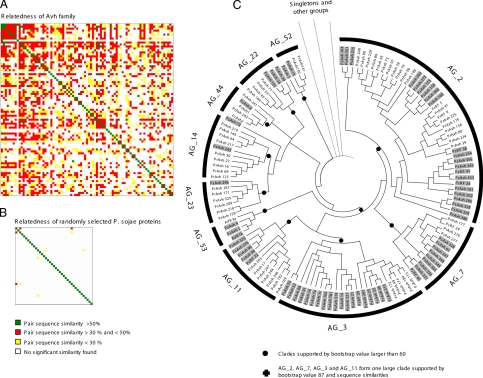
The relatedness of the superfamily can be illustrated by the pairwise similarity of the groups (Fig. 1A). By using the sequence identity of the most similar pair of proteins from any two groups to represent the similarity between the groups, every group was related to several other groups with an identity of >30%. The set of pairwise relationships defines a fully connected network, indicating that all of the sequences form a single superfamily. In contrast, when 1,000 random P. sojae proteins were grouped based on identity >30%, only 17 groups consisting of three to four members could be identified and only five pairwise similarities (Fig. 1B).
Fig. 1.
Relationship between Avh proteins. (A) Pairwise similarity of Avh groups (AGs). For every pair of groups, the two most similar Avh proteins were used for the sequence similarity analysis. The AGs from left to right and from top to bottom are AG_1_1 to AG_1_9, AG_2 to AG_78, and AG_83. (B) Pairwise similarity of randomly selected 1,000 P. sojae proteins. (C) Phylogram constructed with several AGs. The P. sojae Avh proteins are shaded in gray. The unrooted phylogram is based on NJ analysis. All major clades (indicated by black dots) are consistent with similarity groupings. Confidence of groupings was estimated by using 1,000 bootstrap replicates; numbers next to the branching point indicate the percentage of replicates supporting each branch. On the branch tip, the individual Avh proteins are indicated.
One example of the extended network of similarities across the superfamily is represented by the relationship between the P. sojae Avr protein, Avr1b-1 (9), and the P. infestans protein, ipiO1 (18). Both possess an RXLR-dEER domain, but exhibit <20% sequence identity. However, it is possible to identify a chain of seven other Avh proteins with pairwise relationships that connect Avr1b-1 and ipiO1, with each protein showing >25% identity to its neighbors in the chain (SI Fig. 10 and SI Table 4). A chain with slightly lower overall similarity also can be assembled by using only P. sojae Avh proteins (SI Fig. 11). When all of the Avh proteins were permuted excluding the SP and RXLR-dEER domain, no such chain between Avr1b-1 and ipiO1 could be assembled. Conversely, when the SP and RXLR-dEER domains were removed, the same chain could still be assembled, indicating that the similarities underlying the chain are located in the C termini.
A similar chain relationship can be found to connect every group containing more than three members (SI Fig. 12A), and all but six Avh proteins could be connected. Even these six proteins can still be connected, but at a lower threshold (E value <1). By contrast, when Avh proteins are permuted, the connectivity is almost totally lost (SI Fig. 12B). Furthermore, in 1,000 randomly selected P. sojae proteins, only five groups showed connection with a maximum three other groups (SI Fig. 12C).
To visualize the relationships within the Avh superfamily, similarity trees were constructed by NJ analysis with the large AG_2 and several smaller AGs (Fig. 1C). Because of the high sequence divergence, other methods like ML and Bayesian inference failed to resolve most clades. Hence, the phylogram in Fig. 1C does not show the true phylogenetic relationships, but rather represents the similarity of Avh proteins and visualizes groups within the superfamily. The four large AGs (2, 7, 3, and 11) are more closely related because they form a clade supported by high bootstrap values. These AGs contain members from both P. sojae and P. ramorum. However, within each group, often the major clades are specific to one species.
Conserved Motifs in the C Termini.
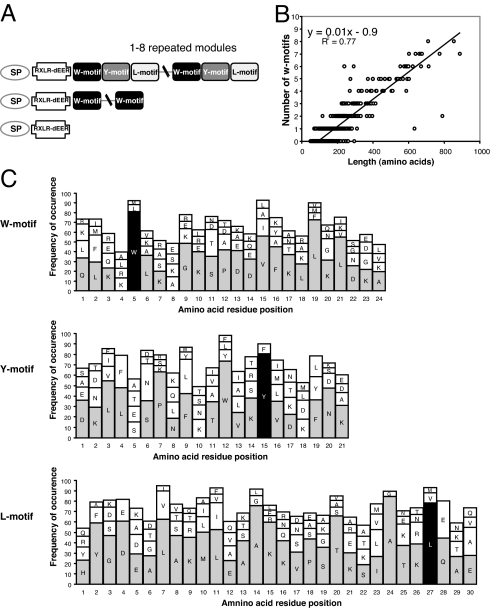
All members of the Avh superfamily lack significant sequence homology to known proteins in GenBank other than previously identified Avr proteins. To identify potential functional motifs in the Avh proteins, we used MEME to screen the C-terminal regions downstream of the RXLR-dEER motifs, where much of the sequence similarity of the superfamily resides. We identified three motifs from 21–30 residues in length, named the W, Y, and L motifs after highly conserved residues (Fig. 2 A and C and SI Fig. 13). The W motif occurs in 60% of all Avh members, whereas 30% of the members have Y and L motifs. The number of Y and L motifs is probably underestimated. For example, a variant Y motif occurs in the C terminus of Avr1b. The Y and L motifs occur after the W motif in 95% cases, forming a W–Y–L module.
Fig. 2.
Conserved C-terminal motifs. (A) Motifs in Avh proteins; 36% of the Avh proteins have a combination of W, Y, and L motifs, 22% have only W motifs, and the remaining 42% do not have identifiable W, Y, or L motifs. SP, signal peptide. (B) The correlation between the number of W motifs and the size of the protein. (C) The consensus sequence of W, Y, and L motifs. The size of the histograms indicates the frequency of the occurrence of the amino acid residue. The most abundant four residues are shown if their frequencies are >2%. The most frequent amino acid at each position is shaded if the frequency is >20%. The most abundant W, Y, and L residues are shaded in black.
Thus, 30% of the family members possess two to eight W–Y–L modules. The number of modules is correlated with the length of the proteins (Fig. 2B). Avh proteins with W–Y–L modules show a significantly higher coding potential value than members without the motifs (SI Fig. 5). Members of large groups often contain several modules. For example, AG_1_1 members typically have five to seven W motifs. Most groups include proteins with modules and proteins without detectable modules. However, some groups such as AG_3 have no detectable modules.
The Avh Gene Family Is One of the Most Rapidly Evolving Parts of the Genome.
The Avh superfamily is highly divergent between P. sojae and P. ramorum. Very few ortholog pairs can be found in the superfamily. The whole genome analysis showed that >55% of gene pairs in P. sojae and P. ramorum could be assigned as orthologs based on bidirectional best BLAST hits (14). However, only 84 pairs of Avh genes share bidirectional best BLAST hits, and only 34 pairs are located within regions of conserved synteny based on the Phylogenetically Inferred groups (PHIGs) analysis of the genome sequences (14). Two such ortholog pairs are shown in SI Fig. 14.
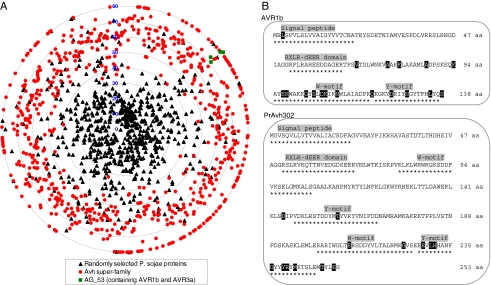
On average, the identity of the most similar paralogs of any Avh protein in the other species was only 31%. In contrast, the average sequence identity of a set of 1,000 randomly selected P. sojae proteins to the most similar P. ramorum protein was 70%. Fig. 3A shows the divergence (100% identity) of all of the Avh proteins plotted in comparison to the random subset. The vast majority (92%) of the Avh superfamily showed sequence divergence of >50%. The avirulence proteins Avr1b and Avr3a belong to the small group AG_53; all four AG_53 members showed sequence divergence of >70%.
Fig. 3.
High level of sequence divergence and polymorphisms. (A) Sequence divergence of Avh proteins. Sequence divergence is defined as one sequence identity. PsAvh and PrAvh proteins were compared against P. ramorum and P. sojae proteins, respectively. Each radius ranges from 0 (center) to 80 (outer circle) representing 100–20% (or less) identity. The distribution along each circumference is random. A set of 1,000 randomly selected P. sojae proteins was compared against P. ramorum. (B) Positively selected sites in Avh proteins. Codeml model 2a was used to detect positively selected sites (shaded in black). The motifs are indicated by asterisks underneath. The analysis was performed on alleles of Avr1b (9) and close paralogs of PrAvh302.
Rapid gene sequence divergence between species can be caused by various mechanisms, such as relaxation of selection, frequent gene duplication, rapid gene loss, and/or positive selection. Genes involved in an interaction between a host and a pathogen often show positive selection (19). To test for positive selection, Ka and Ks values were calculated (20, 21). However, the 18 syntenic ortholog pairs of Avh genes show a Ks value of >1, so their sequence divergence was too high to allow a reliable test for positive selection. Therefore, we tested for diversifying selection within 17 families of closely related paralogs, defined by the criterion of sequence identity >90% (SI Table 5). Positively selected residues were found in six groups, most of them located in the C termini of the proteins. One example is PrAvh302, which has four paralogs. All of the positively selected residues are located in the C terminus (Fig. 3B), and most are located within the boundaries of W and Y motifs. A second example is provided by the family formed by four alleles of Avr1b and the close paralog of Avr1b, Avh1 (9). This family showed an overall Ka/Ks value of 2.7, and there are 18 positively selected sites, most of them within the W and Y motifs (Fig. 3B).
The Genome Locations of Avh Family Members Are Associated with Frequent Genome Rearrangements.
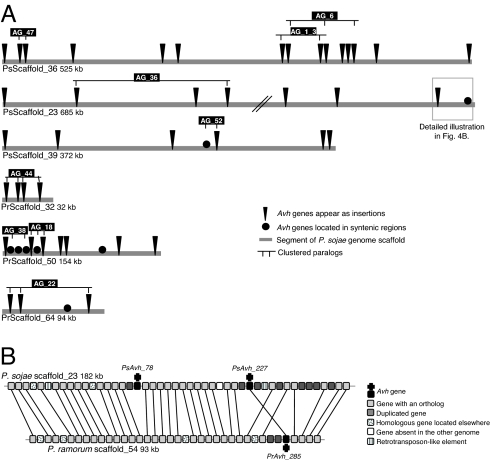
Despite the presence of very large numbers of Avh genes in the two genomes, presumably as a result of repeated gene duplications, there is only very limited clustering of the genes in the respective genomes (22). In P. sojae, the Avh genes are located on 107 scaffolds, whereas in P. ramorum, the Avh genes are located on 150 scaffolds. Several regions of both genomes show concentrations of Avh genes. However, the Avh genes typically are from diverse similarity groups. For example, in P. sojae, the largest Avh cluster spans one contig of 90 kb on scaffold 36; eight PsAvh genes belonging to six AGs are found in this contig. In P. ramorum, the largest cluster spans a region of 46 kb on scaffold 50, containing nine PrAvh genes that belong to six AGs (Fig. 4A). Conversely, Avh genes belonging to the same group do not form clusters of more than four members at an intergenic distance of 50 kb or less. Some small paralog gene clusters are found in AG_52, AG_44, and AG_22 (Fig. 4A). In contrast, some highly similar paralogs are widely separated. For example, Avh1, which encodes a protein 88% identical to Avr1b, is located 406 kb from Avr1b-1.
Fig. 4.
Synteny breakpoints at the locations of Avh genes. (A) Distribution of Avh genes represented as indels. Six scaffolds containing more than two PsAvh genes are shown. PsScaffold_36 and PrScaffold_50 contain the largest clusters of Avh genes in P. sojae and P. ramorum, respectively. Paralogs belonging to the same family are indicated by lines. (B) One deletion and one rearrangement of PsAvh genes in the P. sojae scaffold_23. Orthologs are connected by thin lines.
A comparison of the locations of Avh genes in the P. sojae and P. ramorum genomes reveals extensive rearrangements at the locations of these genes. In contrast to the majority of the genes in the two genomes, only ≈10% (34 pairs) of Avh genes can be found in regions showing synteny (SI Table 6 and SI Fig. 15) (14). Thus, the majority of Avh genes are located at nonconserved genomic locations (Fig. 4). Even for the 34 pairs with conserved locations, reversals of gene orientation and local changes of gene order are frequent (Fig. 4B).
Discussion
In this study, we have identified, by two independent methods, and carefully curated, a very large superfamily of RXLR-dEER effectors encoded in two sequenced Phytophthora genomes. These effectors are highly diverse in their sequences; a single BLAST search recovers only 5–10% of the superfamily members. However, the extended relatedness of these diverse proteins can be demonstrated by iterative sequence similarity searches and the identification of a network of similarity relationships that spans 90% of the superfamily members. This extended similarity suggests that a single gene within a common ancestor of the two species has spawned hundreds of highly divergent, fast-evolving genes within each pathogen genome.
The Avh superfamily displays high sequence divergence, minimal paralog clustering, and frequent rearrangements. One can speculate that the pressures molding this highly fluid effectome are counteracting selections for increased gene number in the pathogens to promote virulence and, at the same time, the need to evade host surveillance by resistance genes that may cause the pathogen to become avirulent on particular hosts. Similar to our findings, Win et al. (16) described the rapid divergence of C termini of closely related paralogs of Phytophthora RXLR genes. Rapid coevolution of pathogen-avirulence and host-resistance genes has been described in the differential recognition of Hyaloperonospora parasitica ATR1NdWsB alleles by RPP1 genes in Arabidopsis (10) and in the recognition of flax rust AvrL567 genes by alleles of the flax L resistance gene (23). Rapid sequence divergence in newly formed Avh genes would enable the number of effector genes in the pathogen to increase while minimizing the likelihood of host recognition. The Avh effectors presumably have a function that contributes to virulence that is reflected in the W, Y, and L motifs found in many of the Avh proteins; the presence of these motifs likely results from some degree of purifying selection. Selection within plant hosts has presumably favored resistance gene products that can detect sequences within the more conserved W, Y, and L motifs. Therefore, it is not surprising to find strong evidence for host-driven positive selection within these motifs. Selection against these motifs as a result of host recognition also would lead to loss of recognizable W, Y, and L motifs, resulting in the population of Avh genes lacking these motifs. Furthermore, as the number of Avh genes becomes larger and the net contribution of each gene to virulence becomes smaller, it is expected that the effect of purifying selection on maintaining the motifs would become smaller, and increasing numbers of Avh proteins would be lost from the functional effectome because of mutations in the W, Y, and/or L motifs. Consistent with this hypothesis, the average coding potential of the non-W–Y–L Avh genes was found to be lower than that of the W–Y–L-containing Avh genes. It also is possible that some non-W–Y–L Avh genes have acquired new functions that do not require W, Y, or L motifs; these genes would be retained in the effectome by purifying selection, but would not retain recognizable W, Y, or L motifs. Consistent with this hypothesis, there are many non-W–Y–L Avh genes that show conservation between P. sojae and P. ramorum.
Frequent duplications are presumably responsible for the expansion of this family. Genes derived from recent local duplication events as a result of illegitimate recombination are often physically clustered (24). In the case of the Avh superfamily, however, extensive clustering is not found, suggesting that newly formed genes are rapidly dispersed to other loci in the genome. The frequent genomic rearrangements associated with nearly all Avh loci suggest that these genes are located within highly fluid regions of the genome (25). In Phytophthora genomes, retrotransposons flank Avh genes significantly more frequently than average genes (R.H.Y.J. and M. C. Zody, unpublished data). The rapid dispersal of new Avh genes would limit the homogenization that occurs among tandemly arrayed genes as a result of gene conversion and illegitimate recombination (26). This dispersal would facilitate rapid divergence among the newly formed genes, driven by host-imposed positive selection.
In the pathogenic bacterium Xanthomonas, the AvrBs3 effector family consists of ≈40 highly similar proteins, which differ mainly in an internal repeat domain (27). Recombination within the repeat generates effectors with unique virulence specificities (28). In the case of Phytophthora effectors, new specificities of effectors most likely are generated via gene duplication and rapid divergence. Considering that oomycetes have acquired pathogenicity independently from fungi and bacteria, expansion of the Avh family should have contributed greatly to the evolution of Phytophthora pathogenicity.
Materials and Methods
Gene Mining and Motif Finding.
For BLAST searches, we used the NCBI BLAST and Standalone-BLAST Version 2.2.3 (29). The protein sequence Avr1b (AAR05402) was used as the initial query to search the total predicted protein set of P. sojae and P. ramorum (14) by BLASTP, as well as the genome scaffold sequences by TBLASTN. Significant hits (E value <1e-5) were manually checked for the presence of an obvious RXLR-dEER domain and an N-terminal signal peptide. New Avh proteins identified by the BLAST searches were used to repeat the process until no new candidates could be discovered. By using the program HMMER 2.3.2 (http://hmmer.janelia.org/) (15), two HMMs were built from the full set of candidates, one using the RXLR motif and 10 amino acids on the left side and the other using the complete RXLR-dEER domains, with the variable spacing arbitrarily placed in between. The RXLR-dEER domain is defined as the occurrence of the string RXLR, together with the trailing acidic motif (containing >10% D or E residues). To increase the sensitivity of a database search, the model was calibrated by hmmcalibrate to give an empirical E value calculation according to the HMM model, as suggested by the program instructions. The whole predicted protein set, as well as all six-frame translations from the complete genome sequences of P. sojae and P. ramorum (14), were used for screening. Proteins with a significant HMM score (E value <0.05) were considered as candidates and were manually examined.
To search for motifs, the program MEME (30, 31) was used to search the C-terminal sequences of the collected Avh proteins, from the end of the dEER motif to the C terminus of each protein. Limits on the allowed motif lengths were a minimum of 6 amino acid residues and a maximum of 100 residues. The resultant motifs were used to build a HMM model with HMMER 2.3.2 (15) that was used to rescreen all of the Avh proteins. Avh proteins with a positive HMM score were considered as motif-containing Avh sequences and were used to build an updated HMM model. The process was repeated until no new motif-containing Avh sequences could be found. When the C-terminal sequences of the Avh proteins were permuted (amino acid residue order was randomly shuffled), the MEME search did not yield any conserved motifs. Furthermore, none of the permuted sequences yielded a significant hit with any HMM models discovered from the C termini of the nonpermuted Avh protein sequences.
Sequence Analysis of Avh Genes and Avh Proteins.
Coding potential was calculated (32) from the average value derived from a window of 300 bp sliding 1 bp at a time using the codon usage frequencies derived from the total predicted gene sets of P. sojae and P. ramorum.
Tests for purifying or diversifying selection were performed with the codeml program in the PAMLv3.14 package (20, 21). Models M0, M1a, M2a, M7, and M8 were used for the analysis. Positively selected amino acid sites were assigned based on a probability >95% with Bayes empirical Bayes statistics (33) in model M2a.
Syntenic regions between P. sojae and P. ramorum were found by using PHIGs (14, 34). For the Avh gene ortholog assignment, genes sharing best bidirectional BLAST hits were considered to be candidate ortholog pairs. If such gene pairs were found to be located in regions showing conserved synteny, they were confirmed as orthologs.
Phylogeny Reconstruction and Sequence Grouping.
Multiple sequence alignment was performed by ClustalX 1.8 (35). Molecular Evolutionary Genetic Analysis 3 (36) was used for Neighbor-Joining (NJ) analysis. Poisson correction was chosen as the distance parameter. The inferred phylogeny was tested by 1,000 bootstrap replicates.
Avh proteins were initially grouped based on BLASTP similarity. Proteins having a BLASTP hit (E value <1e-8, sequence identity >30%) to any other Avh proteins were grouped together. In AG_1, sequences sharing identity of >40% to any other Avh protein within AG_1 were defined as subgroups. For several large groups, NJ trees were constructed to check whether the grouping was consistent with well supported phylogenetic clades (Fig. 1C).
Supplementary Material
Acknowledgments.
We thank Pieter van Poppel, Klaas Bouwmeester, Rob Weide, Hans-Peter Versluis, and Daolong Dou for encouraging discussions. This work was supported by Netherlands Genomics Initiative Fellowship 050-72-404 (to R.H.Y.J.); the Centre for BioSystems Genomics, which is part of Netherlands Organization for Scientific Research; National Research Initiative of the United States Deparment of Agriculture Cooperative State Research, Education and Extension Service Grants 2001-35319-14251, 2002-35600-12747, 2007-35600-18530, and 2007-35319-18100 (to B.M.T.); U.S. National Science Foundation Grants MCB-0242131 and MCB-0731969; and the Virginia Bioinformatics Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The amino acid and DNA sequences of the effector candidate genes have been deposited in the Virginia Bioinformatics Institute Microbial Database, vmd.vbi.vt.edu (accession nos. 158991–159443 for P. sojae and 97196–97586 for P. ramarum), and in GenBank as part of the P. sojae and P. ramorum whole-genome shotgun projects (accession nos. AAQY01000000 for P. sojae and AAQX01000000 for P. ramorum). Virginia Bioinformatics Institute Microbial Database accession nos. of individual genes are listed in SI Table 2.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709303105/DC1.
References
- 1.Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 2002;10:462–469. doi: 10.1016/s0966-842x(02)02451-4. [DOI] [PubMed] [Google Scholar]
- 2.Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 3.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 4.Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St. Paul, MN: Am Phytopathol Soc; 1996. [Google Scholar]
- 5.Rizzo DM, Garbelotto M, Hansen EM. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu Rev Phytopathol. 2005;43:309–335. doi: 10.1146/annurev.phyto.42.040803.140418. [DOI] [PubMed] [Google Scholar]
- 6.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 7.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 8.Flor HH. Inheritance of pathogenicity in a cross between physiologic races 22 and 24 of Melampsora lini. Phytopathology. 1942;32:653–669. [Google Scholar]
- 9.Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- 10.Rehmany AP, et al. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong MR, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen RL, et al. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 13.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 14.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 15.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 16.Win J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharjee S, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieterse CMJ, et al. Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene. 1994;138:67–77. doi: 10.1016/0378-1119(94)90784-6. [DOI] [PubMed] [Google Scholar]
- 19.Stahl EA, Bishop JG. Plant pathogen arms races at the molecular level. Curr Opi Plant Biol. 2000;3:299–304. doi: 10.1016/s1369-5266(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. An integrated BAC, genome sequence physical map of Phytophthora sojae. Mol Plant-Microbe Interact. 2006;19:1302–1310. doi: 10.1094/MPMI-19-1302. [DOI] [PubMed] [Google Scholar]
- 23.Ellis JG, Dodds PN, Lawrence GJ. The role of secreted proteins in diseases of plants caused by rust, powdery mildew and smut fungi. Curr Opi Microbiol. 2007;10:326–331. doi: 10.1016/j.mib.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 25.Jiang RHY, Tyler BM, Govers F. Comparative analysis of Phytophthora genes encoding secreted proteins reveals conserved synteny and lineage-specific gene duplications and deletions. Mol Plant-Microbe Interact. 2006;19:1311–1321. doi: 10.1094/MPMI-19-1311. [DOI] [PubMed] [Google Scholar]
- 26.Reams AB, Neidle EL. Selection for gene clustering by tandem duplication. Annu Rev Microbiol. 2004;58:119–142. doi: 10.1146/annurev.micro.58.030603.123806. [DOI] [PubMed] [Google Scholar]
- 27.Lahaye T, Bonas U. Molecular secrets of bacterial type III effector proteins. Trends Plants Sci. 2001;6:479–485. doi: 10.1016/s1360-1385(01)02083-0. [DOI] [PubMed] [Google Scholar]
- 28.Schornack S, Meyer A, Romer P, Jordan T, Lahaye T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Gribskov M. Score distributions for simultaneous matching to multiple motifs. J Comput Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- 31.Bailey TL, Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 32.Tripathy S, Pandey VN, Fang B, Salas F, Tyler BM. VMD: a community annotation database for oomycetes and microbial genomes. Nucleic Acids Res. 2006;34:D379–D381. doi: 10.1093/nar/gkj042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 34.Dehal PS, Boore JL. A phylogenomic gene cluster resource: The Phylogenetically Inferred Groups (PhIGs) database. BMC Bioinformatics. 2006;7:201. doi: 10.1186/1471-2105-7-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.