Abstract
Retroposition is increasingly recognized as an important mechanism for the acquisition of new genes. We show that a glycogen synthase kinase-3 gene, shaggy (sgg), retroposed at least 50 MYA in the Drosophila genus to generate a new gene, mojoless (mjl). We have extensively analyzed the function of mjl and examined its functional divergence from the parental gene sgg in Drosophila melanogaster. Unlike Sgg, which is expressed in many tissues of both sexes, Mjl is expressed specifically in the male germ line, where it is required for male germ line survival. Our analysis indicates that mjl has acquired a specific function in the maintenance of male germ line viability. However, it has not completely lost its ancestral biochemical function and can partially compensate for loss of the parental gene sgg when ectopically expressed in somatic cells. We postulate that mjl has undergone functional diversification and is now under stabilizing selection in the Drosophila genus.
Keywords: germ cell, male sterile, gene duplication, GSK-3, adaptive evolution, retroposition, male germ line
Introduction
New genes arise from exon shuffling (Long, Deutsch, et al. 2003), gene splits and joins (Wang et al. 2004), or duplication (Li et al. 2005) followed by diversification of function (Long, Betran, et al. 2003). Duplication can occur at the DNA level or via an RNA intermediate. RNA-mediated gene duplication, or retroposition, occurs by a retrovirus-like integration mechanism. Relatively recent retropositions can be identified based on simple criteria (Betran and Long 2002): retroposed genes show strong sequence homology with a parent gene in the genome, the coding sequence of the newly retroposed gene is not interrupted by introns (due to the mechanism of transmission), the PolyA tract from the mRNA intermediate is encoded in the genome, and the terminal repeats associated with the integration event are evident. These newly created genes have been under intense study, but older retroposed genes have not yet been extensively characterized. One might expect that some of the features of retroposed genes, such as PolyA tracts, would be unconstrained by evolution and eventually be lost. Such genes might ultimately acquire introns and become more difficult to distinguish from genes arising from strictly DNA-mediated duplication (Betran and Long 2002). Retroposed genes have several different fates: one member of the duplicated pair can become nonfunctional and silenced (nonfunctionalization), there could be a partitioning of the ancestral function between the 2 copies (subfunctionalization), or 1 of the copies could acquire new functions (neofunctionalization) (Lynch and Conery 2000).
We have identified a new retroposed glycogen synthase kinase-3 (GSK-3)–encoding gene, derived from one of the alternatively spliced mRNAs from the shaggy (sgg) locus. In Drosophila melanogaster, sgg locus is on the X chromosome and the retroposed duplicate, and mojoless (mjl) is on the third chromosome. We show that this retroposition event occurred near the root of the Drosophila genus and that sgg and mjl have diverged rapidly.
This gene pair is of particular interest because of the highly pleiotropic function of the parental sgg gene. GSK-3 proteins are kinases essential for metabolic regulation, cell fate determination, stem cell maintenance, and nuclear import in a wide range of eukaryotes (Jope and Johnson 2004). Sgg, the Drosophila homolog of GSK-3β is probably best known for its role as the key negative regulator of the Wingless (wg) signaling pathway, but is also intimately involved in diverse developmental processes, such as establishment of anterior-posterior polarity in the embryo, patterning of the embryonic cuticle as well as various adult structures (wing, eye, haltere etc.), and regulating circadian rhythms (Siegfried et al. 1994; Stanewsky 2002). Sgg is maternally loaded and has been detected ubiquitously throughout development (Bourouis et al. 1990). In addition, it has multiple splice variants ranging in size from 10 to 110 kilo basepair (kb) (Bourouis et al. 1990). These encode for at least 7 predicted protein isoforms (FlyBase 2003). Multiple isoforms of sgg mRNA show no obvious temporal pattern of expression and in situ hybridization using common probes show uniform expression in embryos (Bourouis et al. 1990; Siegfried et al. 1990).
On the other hand, mjl is intronless and it encodes for a single transcript with highly specific expression in the male germ line. Altered expression patterns are expected to promote neofunctionalization, and indeed, mjl knockdown results in male sterility due to ablation of the male germ line. The loss of mjl causes a germ line-specific defect with no obvious effects in the soma. This indicates that the retrocopy has now acquired a critical novel function, which probably explains why it has been retained in the genome. However, this neofunctionalization is not complete. Ectopically Mjl was capable of partially rescuing an sgg mutant. These results are consistent with retention of ancestral biochemical function (as a gene family member), but evolution of a new spatial expression pattern along with a specific novel role.
Methods
Fly Stocks and Crosses
All flies were raised and maintained on standard corn-meal media (Tucson Drosophila Stock Center, Tucson, AZ) at 25 °C, except the ones used for heat-shock regimens and the dsRNA-mediated interference (RNAi) studies, as noted. Enhancer trap 3914 expresses LacZ in all the somatic cells of the gonad (Asaoka et al. 1998). The nos-GAL4 line drives expression of Upstream Activating Sequence (UAS) transgenes in germ lines of both males and females (Schulz et al. 2004). The hsp-70-GAL4 line drives expression of UAS transgenes ubiquitously following heat shock (FlyBase 2003). The armadillo (arm)-GAL4 line was used to induce expression of transgenes is cells where Sgg is required in embryos (FlyBase 2003).
The coding region of mjl was polymerase chain reaction (PCR) amplified from expressed sequence tag (EST) clone GH16447 (Celniker et al. 2002) with primers designed to include ~120 bp upstream of the start of transcription (forward primer [FP]: 5′-CAG TTC CAC ACG CAT ACG CAC-3′; reverse primer [RP]: 5′-GTG CGG TTC AAA TCA AGA CC-3′). The 1.8-kb fragment was cloned into the pUAST vector (Brand and Perrimon 1993) to create the UAS-mjl transgene. To generate the UAS-mjl-RNAi transgene, two 700-bp fragments encoding the C-terminal region of mjl were PCR amplified (fragment 1: FP 5′-GCGAATTCGATCGTCAAGGTTATGGGCACTC-3′; RP 5′-GCACTAGTGCTATTGCACTGCCCACCATCAT-3′; fragment 2: FP 5′-GCGGTACCGATCGTCAAGGTTATGGGCACTC-3′; RP 5′-GCACTAGTGCTATTGCACTGCCCACCATCAT-3′). The fragments were then cloned into a modified pUAST vector (Han K, personal communication) containing a 500-bp heterologous intron inserted between the insertion sites. Transgenic lines were generated by P element–mediated transformation using the stock y w;Δ2–3 Sb/TM6, Ubx (Robertson et al. 1988). Eight and 10 independent lines of insertion were established for the UAS-mjl and UAS-mjlRNAi transgenes, respectively.
Phylogenetic Analysis
Sequence alignments were performed using RevTrans 1.4 (Wernersson and Pedersen 2003), which aligns coding sequence in translated amino acid space. Alignments were visualized using Jalview (Clamp et al. 2004). Ka/Ks was calculated pairwise using the Nei-Gojobori method (Nei and Gojobori 1986) and the modified Nei-Gojobori method (Zhang et al. 1998) in MEGA3.1 with Jukes-Cantor correction and transition/transversion R value of 2. For comparison, Ka/Ks was also calculated with a maximum likelihood approach using the yn00 program within the phylogenetic analysis by maximum likelihood package, implementing the method developed by Yang and Nielsen (2000). Phylogenetic trees were constructed using the minimum evolution method in MEGA3.1, and significance was measured via bootstrapping with 1,000 replicates (Kumar et al. 2004) and Neighbor-Joining to draw the trees, with the same substitution model as the modified Nei-Gojobori calculation.
Antibody Generation
To minimize cross-reaction of the antibody with Sgg, the divergent C-terminal region of Mjl was fused to bacterial glutathione-S-transferase (GST) and the recombinant protein was purified and then used to raise antibodies. A 763-bp C-terminal region fragment was amplified by PCR (FP: 5′-GACGAATTCATGTTATGGGCACT-3′; RP: 5′-GCACTGGAGTTAGTTTTCATCCTC-3′). The fragment was cloned into the pGEX-4T-2 vector (Amersham-Pharmacia, Piscataway, NJ) to produce an in-frame GST fusion protein. The protein was purified on a GST affinity column, using the manufacturer’s protocol (Amersham-Pharmacia), and injected into rabbits (Pocono Rabbit Farm, Canadensis, PA). The antibody was affinity purified against the fusion protein (Brent et al. 1987).
Immunoblots
Standard molecular biology techniques were used for protein transfer onto nitrocellulose membranes. Samples were homogenized in 50 μl of 2× Gel buffer (8 M urea, 2 M thio-urea, 3% sodium dodecyl sulfate (SDS), 75 mM dithiothreitol, 25 mM Tris pH 6.8, 0.1% bromophenol blue) using a motorized homogenizer. Samples were spun for 2 min in a microfuge to pellet cell debris and loaded onto a 12% SDS-polyacrylamide gel. Protein transfer was confirmed by Ponceau-S staining. Blots were blocked with 5% milk in Tris-buffered saline (TBS) (20 mM Tris, 150 mM NaCl) for 2–3 h at room temperature on a rocker, incubated with rabbit polyclonal anti-Mjl antibody overnight at 4 °C in 5% milk in TBS-Tween (20 mM Tris, 150 mM NaCl, 0.1% Tween) at a final concentration of 1:2000, washed, incubated in horseradish peroxidase-conjugated goat anti-rabbit antibody for 2–3 h at room temperature (Jackson Immuno Research Laboratories Inc., West Grove, PA), and developed using a commercial Enhanced chemiluminescence kit (Amersham-Pharmacia).
Rescue of sgg Mutant Embryos
Virgin females of the genotype sggM11-1 FRT101/FM7; arm-GAL4/arm-GAL4 were mated to ovoD1 FRT101/Y; FLP recombine (FLP)38/FLP38 males. The progeny of this cross were heat shocked at 37 °C for 1 h during the crawling third instar larval stage to induce FLP-directed recombination (Siegfried et al. 1992). Virgin females containing mosaic germ lines as well as 1 copy of the arm-GAL4 chromosome were mated to homozygous UAS-mjl14B, UAS-sggWDT7A, or Oregon-R males. Embryo sorting and cuticle preparations were performed (Steitz et al. 1998). Embryos were scored as (+/−) for rescue, based on presence or absence of denticle bands. Five hundred embryos were screened for each genotype. Embryos were viewed and photographed in dark field.
RNAi Treatments
Virgin nos-GAL4 females were crossed to various RNAi transgenic males and maintained at 29 °C (GAL4 is more effective at higher temperature). The progeny of the crosses were collected and newly eclosed males and females were individually mated to their respective wild-type counterparts. The crosses were maintained at 29 °C for up to 10 days and vials were examined for evidence of larvae. As a control, the experiment was repeated with virgin hsp-70-GAL4 females instead of nos-GAL4 females. The progenies were heat shocked at 38 °C for 15 min a day. The mating experiments were repeated at 25 °C, as above.
Immunofluorescence Staining of Testes
Testes from newly eclosed males (<24 h) were dissected and fixed (Fabrizio et al. 2003). Samples were blocked in 3% bovine serum albumin in phosphate buffered saline with 1% Triton X-100 overnight at 4 °C. Antibodies were used at the following concentrations: rabbit polyclonal anti-Mjl antibody at 1:20; chicken anti-Vasa at 1:1,000 (Burnett and Howard 2003). Goat anti-rabbit Alexa 488 and anti-chicken Alexa 555 (Invitrogen Molecular Probes, Carlsbad, CA) secondary antibodies were used at a final concentration of 1:250. After the final wash, testes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (50 μg/ml) and mounted (50% glycerol with 1,4-diazabicyclo[2.2.2]octane), and images were captured on a LSM510 confocal laser scanning microscope (Ziess, Thornwood, NY).
Results
mjl Arose in Drosophila by Retroposition
The mjl (CG11338) locus is on the third chromosome and encodes a novel GSK-3 protein sharing 53% amino acid identity with Sgg, the well-studied X-linked D. melanogaster GSK-3-encoding gene (fig. 1A). This gene was previously named gasket for GSK-3 based on its sequence homology (FlyBase 2003). We have subsequently renamed the gene mjl based on its loss-of-function phenotype, which is male sterility.
Fig. 1.
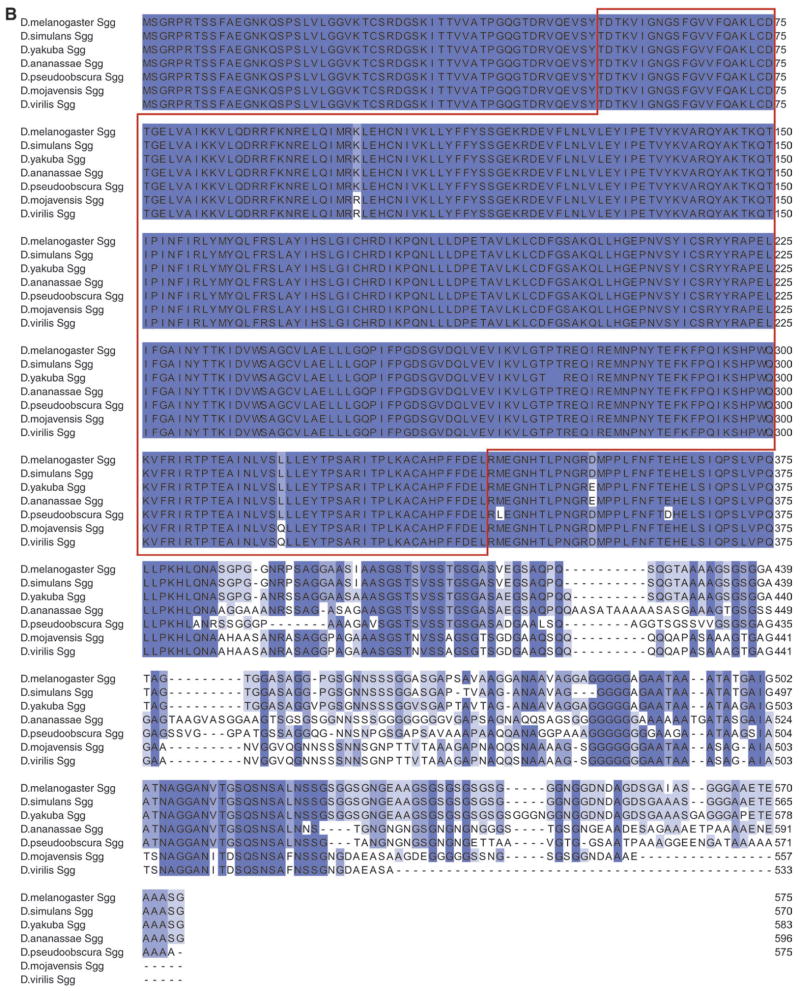
Mjl is a unique GSK-3 family member related to Sgg. (A) Sequence alignment between Sgg and Mjl proteins from Drosophila melanogaster. Identical residues are shaded blue. The red box marks the canonical kinase domain and the green box marks the divergent C-terminal region used to generate anti-Mjl antibody. (B) Phylogenetic tree of Drosophila Sgg and Mjl proteins generated using MEGA3.1 software. The Sgg proteins all cluster together as do the Mjl proteins, indicating that they are distinct from one another. (C) Model depicting retroposition of sgg-RH mRNA from X to an autosome (the third chromosome in D. melanogaster) giving rise to mjl.
The greatest level of conservation between Sgg and Mjl is within the canonical kinase domain (69% identity, outlined in red box in fig. 1A) and the least conserved region is the C-terminal region (11% similarity, outlined in green box in fig. 1A). Mjl and Sgg are the only predicted GSK-3 proteins we found encoded in the D. melanogaster genome (Adams et al. 2000; Celniker et al. 2002; FlyBase 2003). The mjl transcript is intronless, unlike sgg, which has 9 introns, indicating that it arose by retroposition of sgg. To determine which of the 11 sgg mRNA isoforms might have given rise to mjl, we examined the regions specific to those transcripts and found that mjl is most similar to sgg-RH. This suggests that an ancient version of sgg-RH mRNA retroposed to give rise to mjl (fig. 1C). There is no evidence of a PolyA tract or short repeats in mjl, implying that it is not a newly retroposed gene.
In order to investigate how long ago in the history of the genus mjl retroposed, we examined the genomic sequence of newly sequenced Drosophila species. We found that mjl was present in each species examined (fig. 1B; D. melanogaster, Drosophila sechellia, Drosophila mauritiana, Drosophila simulans, Drosophila yakuba, Drosophila ananassae, Drosophila pseudoobscura, Drosophila mojavensis, and Drosophila virilis). Interestingly, we have been unable to identify a homologue of mjl in the genomes of non-Drosophila insects (Anopheles gambiae, Apis mellifera, and Tribolium castaneum) or indeed any other published genomes. We also examined the linkage of sgg and mjl in the various species, and where available, the data indicate that sgg is X linked, whereas mjl is on the third chromosome (Muller element E). Collectively, these results demonstrate that mjl arose as a retroposed sgg, an event that was probably Drosophila specific. Based on estimates of molecular clocks, we can date this event to at least 50 MYA (Russo et al. 1995; Tamura et al. 2004).
Mjl Evolution
We examined the sequence conservation of sgg and mjl within the genus Drosophila. The predicted Sgg proteins are more conserved as a group than the predicted Mjl proteins, especially at their N termini (fig. 2). Residues that are crucial for functioning of the kinase domain, however, are well conserved within both groups of orthologs. This suggests that the sgg genes are under greater evolutionary constraint on their divergence. The observation that the N-terminal region outside the canonical kinase domain is strictly conserved within the predicted Sgg proteins and not within the Mjl proteins suggests that this domain may not be integral to function in the Mjl proteins. The C-terminal regions of both predicted Sgg and Mjl proteins show poor conservation indicating that this region is experiencing the least functional constraint within both groups of proteins (fig. 2).
Fig. 2.
Alignments of Mjl and Sgg orthologs. Sequence alignment of various Drosophila (A) Mjl and (B) Sgg proteins generated using ClustalW. Residues in blue are identical and the red box outlines the canonical kinase domain.
We next examined nonsynonymous and synonymous substitution rates (Ka/Ks) among orthologs of sgg and mjl (table 1). Maximum likelihood methods for comparing the rate of substitution at synonymous sites in a gene sequence (Ks) and comparing it with the rate of substitution at non-synonymous sites (Ka) are effective approaches to characterizing evolutionary forces (Nei and Kumar 2000; Hurst 2002). Orthologs of mjl have experienced on average a greater number of nonsynonymous substitutions per site than sgg (table 1). The average synonymous substitution rate (Ks) is much higher for mjl orthologs than for sgg orthologs (table 1). Most saliently, the fact that the Ka/Ks ratio is much less than 0.5 for both genes (table 1) implies that both genes are under strong selective constraints and therefore are functional.
Table 1.
Estimation of Codon Substitution Events in mjl and sgg
| mjl
|
sgg
|
|||||
|---|---|---|---|---|---|---|
| Ka | Ks | Ka/Ks | Ka | Ks | Ka/Ks | |
| PAMLa | 0.19 | 3.51 | 0.06 | 0.09 | 0.61 | 0.14 |
| PAMLb | 0.19 | 1.51 | 0.13 | 0.08 | 0.60 | 0.14 |
| MEGA | 0.22 | 0.84 | 0.24 | 0.09 | 0.46 | 0.19 |
Note.—PAML, phylogenetic analysis by maximum likelihood.
Yang-Nielsen method.
Nei-Gojobori method.
Mjl Has Retained GSK-3 Family Function
The greatest level of conservation between the Mjl and Sgg proteins is within the canonical GSK-3 kinase domain (69% identity, outlined in red in fig. 1A). All the residues crucial for GSK-3 function, as deduced from the mammalian crystal structure studies, are conserved in Mjl, such as R96, R180, and K205, which form a positively charged pocket to attract the primed substrate; Y216 within the activation loop whose phosphorylation increases catalytic activity >200-fold; R96 whose phosphorylation is important to align the (β-strand domain and the α-helical domain; S9 whose phosphorylation inhibits GSK-3 activity; and K85, which forms a salt bridge with E97 at the active site (Dajani et al. 2001; ter Haar et al. 2001). This indicates that the canonical kinase domain in the duplicate gene copy, mjl, is under strong selective pressure in order to retain its biochemical activity. However, the other regulatory domains within the coding sequence and perhaps upstream and downstream sequences are not subject to the same level of conservation, suggesting that mjl has started functionally diverging from sgg in its ability to recognize and bind to substrates, a key step in neofunctionalization (Zhang et al. 2004).
Sequence conservation suggests that the kinase domain is the most constrained, but does this conservation mean that Mjl and Sgg have the potential to recognize some of the same substrates? If Mjl and Sgg share biochemical activities, then ectopic expression of mjl should mimic sgg overexpression. Previous work has shown that overexpression of sgg along the presumptive wing margin disrupts the endogenous wg signaling pathway (Steitz et al. 1998).
Overexpression of sgg in the wing imaginal disc results in a loss of bristles and marginal tissue in the adult wing (compare panels A and B in fig. 3). When mjl is similarly expressed there is a comparable disruption of the wing margin (fig. 3C). The observation that ectopic expression of sgg and mjl result in similar phenotypes suggests that Mjl is also capable of interacting with and disrupting the endogenous wg signaling pathway. To confirm this hypothesis, we modulated the levels of 2 known components of the wg signaling pathway, Drosophila Axin (dAxin) and Arm. Expressing mjl in the wing imaginal disc in the presence of an extra copy of arm (fig. 3D) or a single copy of dAxin (fig. 3E) suppressed the wing phenotype. Similar types of suppression are seen with overexpression of sgg and modulation of gene dosages of either arm or d-Axin (Steitz 2000). Therefore, we conclude that Mjl is capable of interacting with the canonical wg signaling pathway. These suggest that any neofunctionalization of Mjl protein function is subtle or that any dramatic subfunctionalization does not include portions of the proteins involved in wg function.
Fig. 3.

Ectopic Mjl shows Sgg-like genetic interactions. Wings from adults of the following genotypes are shown: (A) Wild type, (B) vg-GAL4/UAS-sggWDT7A, (C) vg-GAL4/UAS-mjl14B, (D) +/DpYarmYD35; vg-GAL4, UAS-mjl, (E) vg-GAL4, UAS-mjl; dAxinl[3] SO44230/+,and (F) Vg-GAL4, UAS-mjl14B/UAS-mjlRNAi10.2. Ectopic expression of mjl in the wing margin results in a notched wing phenotype, which is similar to the phenotype seen with sgg overexpression. This phenotype can be suppressed by the reduction of dAxin levels (dAxin heterozygous) or by the increase of Arm levels (DpYarm duplication of arm on the Y), indicating that Mjl interacts with components of the wg pathway. Simultaneous expression of UAS-mjlRNAi10.2 and UAS-mjl suppresses the dominant phenotype associated with expression of UAS-mjl alone indicating the RNAi transgene is functional. (G) Cartoon depiction of the destruction complex: In the absence of a wg signal, Arm (yellow circle) is phosphorylated and targeted for degradation by a collection of proteins that comprise the “destruction complex.” These proteins include Sgg (red box), dAxin (blue box), and antigen-presenting cells (green box). Once Arm is phosphorylated, it is targeted for degradation via the ubiquitin-proteosome pathway. In the presence of wg signal, Arm escapes phosphorylation translocates into the nucleus where it activates wg target genes.
We also addressed the question of whether Mjl and Sgg have overlapping biological activity by attempting to rescue sgg mutant embryos with ectopic expression of mjl. Because sgg is a maternal effect gene, mutant embryos were derived from mothers with homozygous mutant germ lines or germ line clone females (see Methods). Mutant embryos are not viable and exhibit a distinct perturbation of segmentation that is revealed in a loss of all denticles or a “naked” phenotype (fig. 4, compare panels A and B). Zygotic expression of sgg in these embryos can partially restore segmentation (fig. 4C) (Steitz et al. 1998). In a similar fashion, ectopic expression of mjl also partially restores segmentation (fig. 4D). Again, this suggests that any differences in Mjl and Sgg protein function in this spatiotemporal context are subtle. This does not mean that there is no evidence of functional divergence. Indeed, the extent of sgg mutant rescue by the mjl transgene is less than the rescue by the sgg transgene. A rescue of 43% was seen for the sgg transgene as opposed to 10% for the mjl transgene (fig. 4E). These results are consistent with functional divergence, but could also be due to technical factors, such as degree of overexpression and position effects, which are inherent in all transgenic analysis.
Fig. 4.
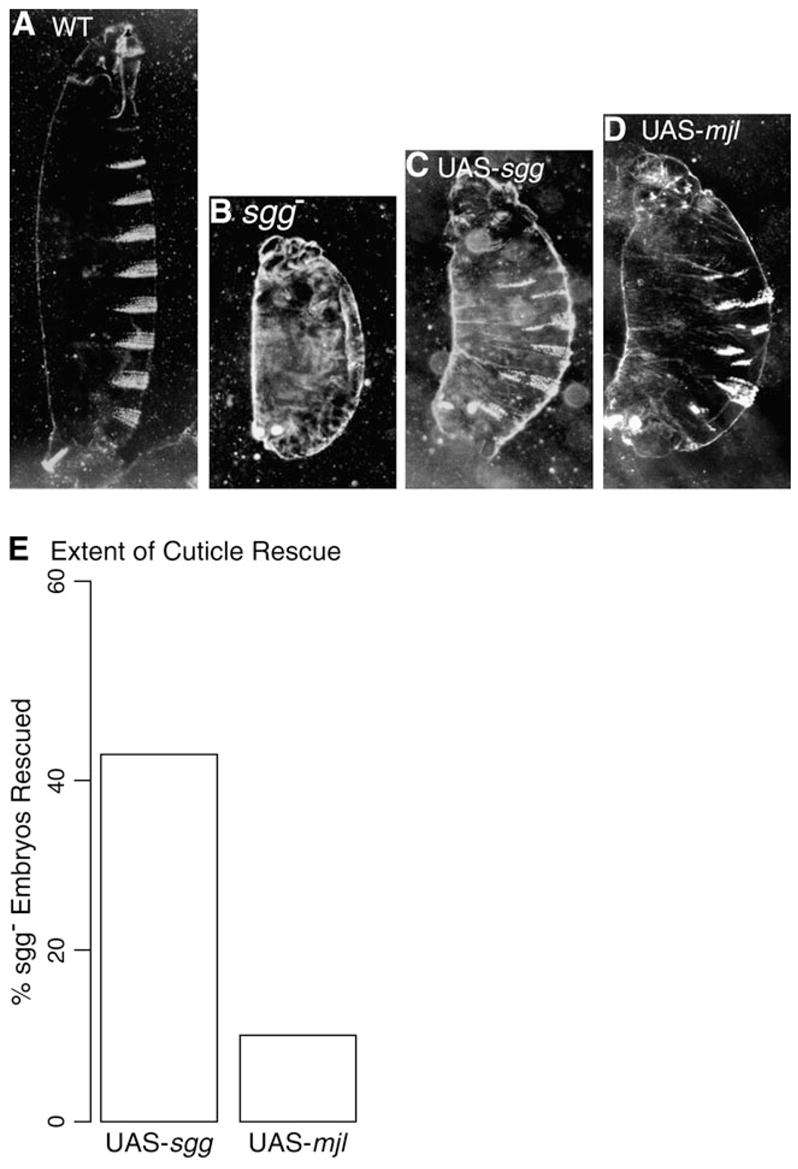
Mjl expression rescues the segmentation defect of sgg mutant embryos. Cuticle phenotypes of (A) wild-type embryo (B) sgg mutant embryo derived from a germ line clone, (C) sgg mutant embryo ectopically expressing a sgg rescue construct (sgg; arm-GAL4/UAS-sgg WDT7A), (D) sgg mutant embryo ectopically expressing a mjl-rescuing construct (sgg; arm-GAL4/UAS-mjl14B). (E) Comparison of efficiency of sgg mutant embryo rescue by the UAS-sgg transgene and UAS-mjl transgene.
Taken together the above data demonstrate that Mjl has at least partially retained its ability to substitute for Sgg and is indeed capable of genetically interacting with components of the canonical wg pathway. Functional divergence at the level of the proteins is subtle.
mjl Is Expressed in the Male Germ Line
Functional diversification often involves new spatial patterns of expression. Therefore, we explored the spatial expression pattern of mjl gene products by querying EST data, northern blotting, reverse transcriptase (RT)–PCR, western blotting, and antibody detection methods.
Multiple mjl ESTs have been identified in testis cDNA libraries, but not in ovary cDNA libraries (Andrews et al. 2000; Stapleton et al. 2002). One mjl EST from a head library exists (Stapleton et al. 2002). Although Mjl might also be expressed in other tissues, these EST data suggest that mjl expression is highly testes biased. To further explore mjl mRNA expression, we performed Northern blot analysis and RT-PCR experiments and confirmed that the expected 1.9-kb mjl transcript is expressed in the testis (data not shown). We have not detected mjl in females or in any other nongonadal tissues including head, either by RT-PCR or immunostaining techniques, thus confirming that mjl mRNA shows a highly testes-biased expression pattern.
To further characterize Mjl expression pattern in vivo, the divergent C-terminal portion of Mjl was used to produce an antibody that, predictably, does not cross-react with Sgg (data not shown). On western blots of protein extracts from adult males, male abdomens, and testes, a single band of the predicted 56-kDa mobility was detected with anti-Mjl antibody (fig. 5A). No protein was detected in extracts from females (fig. 5A).
Fig. 5.
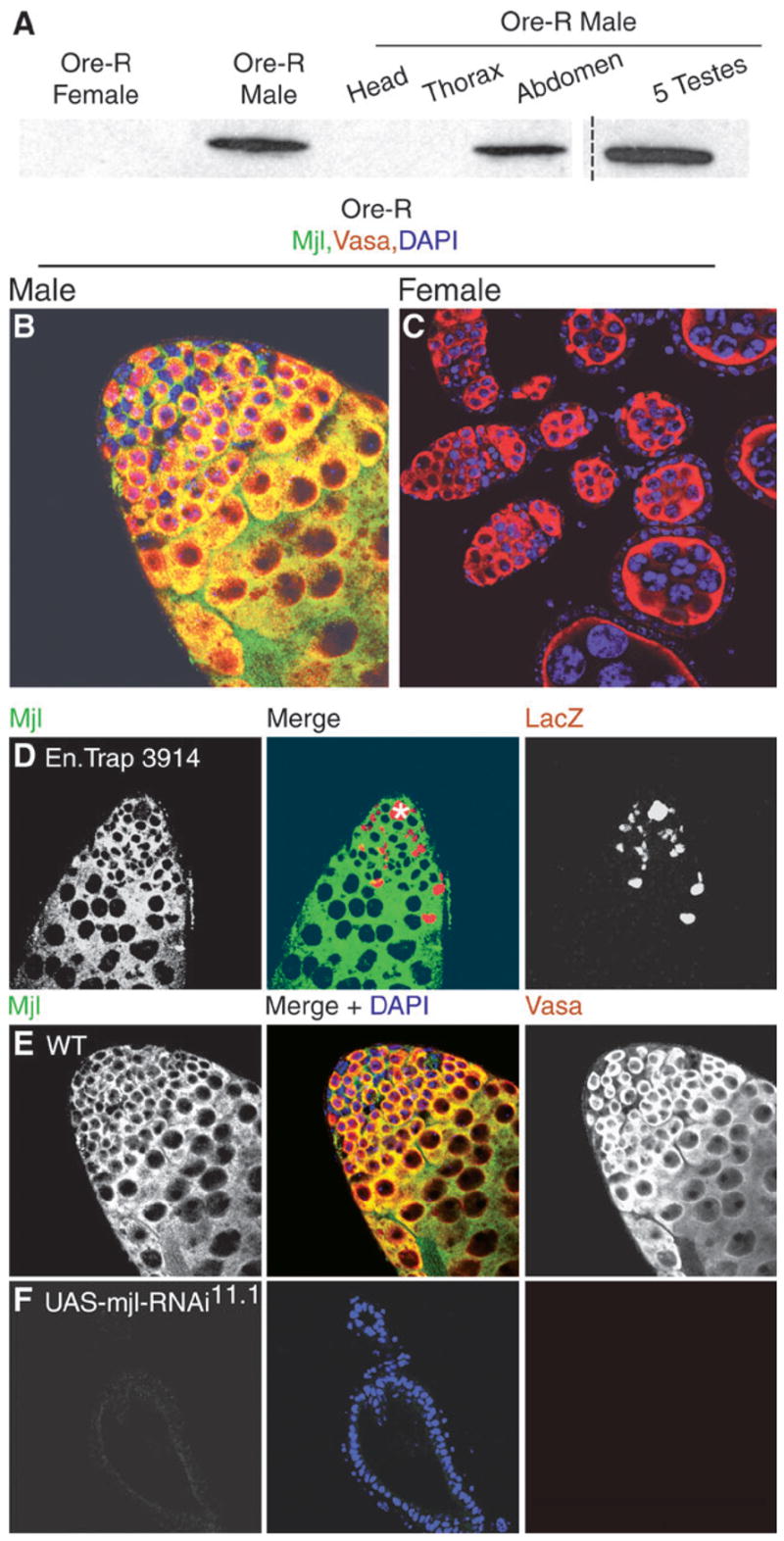
Mjl is enriched in testis and is required for male germ line viability. (A) Western blots containing samples as indicated. The broken line signifies that intervening lanes are not shown although all lanes correspond to a single blot. Wild-type male gonads (B) and female adult gonads (C) immunostained for anti-Mjl (green) and anti-Vasa (red). Nuclei are counterstained with DAPI (blue). Overlap of Vasa and Mjl can be seen (yellow). (D) Testis from line 3914 immunostained for anti-Mjl (green) and anti-β-galactosidase (red). The asterisk marks the hub. (E) Wild-type and (F) UAS-mjlRNAi11.1 testis immunostained for anti-Mjl (green) and anti-Vasa (red). Nuclei counterstained with DAPI (blue).
To determine what cell types express Mjl in the testis and where it is localized within cells, we performed immunostaining experiments. In D. melanogaster, the apical tip of the testis forms the germinal proliferation center (Fuller 1993). At the very tip of the testis a tight cluster of 12–16 nondividing somatic cells form a structure called the hub, which forms the niche (Kiger et al. 2001; Tulina and Matunis 2001). Closely attached to the hub is a layer of germ line stem cells (GSCs) interspersed with somatic stem cells.
Mjl was readily detected in adult testis, but not in ovary (fig. 5, compare panels B and C). The anti-Vasa antibody is a good marker of germ cells (Rongo et al. 1997), and anti-Mjl staining was detected in Vasa-positive cells of the testis (fig. 5B and E). Anti-Mjl staining revealed a cytoplasmic subcellular localization, as evidenced by the lack of overlap with DAPI nuclear counter staining (fig. 5E). This was expected because all GSK-3s are cytoplasmic (Woodgett 1990). Mjl is expressed in the GSCs, which immediately surround the hub, the gonialblasts, and the developing spermatogonia as evidenced by overlapping staining of anti-Mjl with anti-Vasa (fig. 5E). To examine if Mjl is expressed in somatic cells within the male gonad, we stained Enhancer trap line 3914 with anti-β-galactosidase and anti-Mjl antibodies. Enhancer trap line 3914 (Asaoka et al. 1998) marks the somatic cyst cells and hub cells in the gonad and Mjl does not colocalize with these cells (fig. 5D). We also immunostained wild-type testes with anti-Fas III antibody, which strongly outlines the somatic hub cells at the apical tip (Le Bras and Van Doren 2006), and Mjl is clearly excluded from those cells (data not shown). These data indicate that Mjl expression is restricted to the male germ line. This is highly significant in light of the fact that Sgg is expressed in multiple somatic tissues in both sexes (Bourouis et al. 1990; Siegfried et al. 1990). Mjl evolving a tissue-specific expression pattern is highly reminiscent of a model for evolution of new protein function following restriction of expression to specific tissue types (Hughes 1994).
mjl Is Required for Male Germ Line Survival
Although evolutionary conservation is a strong indicator of function, we were interested in directly testing for mjl genetic function. We utilized RNAi to disrupt mjl mRNA expression (Kennerdell and Carthew 2000). We generated mjl transgenic lines that express dsRNA under control of the GAL4/UAS system (see Methods). The UAS-mjlRNAi constructs strongly downregulate ectopically expressed mjl mRNA and Mjl protein and do not affect sgg function, mRNA expression, or Sgg protein levels (fig. 3F, additional data not shown).
Our studies on the endogenous expression pattern of Mjl establish that it is expressed in the male germ line. Hence, we used the nos-GAL4 driver to drive expression of UAS-mjlRNAi in the germ line. We also used a heat-shock GAL4 driver and obtained similar results (data not shown) indicating that the RNAi construct did not have overt off-target effects. The nos-GAL4 driver has been shown to activate expression in germ cells at the apical region of the testes, including stem cells, gonialblasts, spermatogonia, and early spermatocytes (Schulz et al. 2004). UAS-mjlRNAi/+nos-GAL4/+ males were mated to wild-type virgin females and the progeny were examined. The Mjl-depleted males were sterile. Female siblings as well as control males lacking either one of the UAS/GAL4 binary system components were fertile. These observations indicate that Mjl is crucial for maintaining male fertility in D. melanogaster.
To explore the cause of male sterility, testes of Mjl-depleted males were dissected and examined. Mjl-depleted males have grossly abnormal testes morphology when compared with wild-type males. The testes of Mjl-depleted males are stunted, as is seen in testes lacking a germ line (compare panels in fig. 5E and F). In a wild-type testis, Vasa-positive cells (germ line lineage) fill the testis (fig. 5E), whereas Mjl-depleted testes lack all germ cells (fig. 5F). The DAPI counterstain highlights the outer layer of sheath cells, which are the only cells left in the testes of Mjl-depleted adults. The Mjl-depleted testes often showed a complete loss of Vasa-positive cells, but a few isolated Vasa-positive cells were occasionally observed. These results showed that sterility was due to the loss of the entire germ line lineage in the adult testes of Mjl-depleted flies and that even when a few germ cells survive they are incapable of completing gametogenesis. This strongly suggests that Mjl has assumed an essential role for spermatogenesis in D. melanogaster.
Our analysis also indicates that Mjl has acquired a specific expression pattern and new function in the male germ line. Unlike sgg, which is required in a plethora of cell types and developmental stages, Mjl is only required in the male germ line. This can be inferred from the fact that global RNA interference of Mjl (using hs-GAL4) caused a very specific defect loss of the germ line in males. Taken together, our data argue for neofunctionalization of Mjl.
Discussion
Retroposition is a means by which organisms can expand their genomes (Brosius 1991). Here we demonstrate that a retroposed gene, mjl, has acquired an important function in Drosophila. The retroposition itself appears to have occurred near, or in, the last common ancestor of the Drosophila genus. The crucial function of mjl in maintaining male fertility is probably the reason it has been faithfully retained in every species of the genus examined.
Divergence of mjl and sgg
The most highly conserved domain between Mjl and Sgg is the kinase domain, which appears to be under strict evolutionary constraint. Structural conservation extends into functional relevancy, for when Mjl is placed in Sgg-centric contexts within D. melanogaster, it is capable of producing Sgg-like phenotypes. Thus, the core protein function is conserved in the retroposed copy following gene duplication. Does this highlight a lack of functional diversification or highlight the conserved core function of GSK-3 family members? The fact that Mjl, which was derived from just one of several isoforms of GSK-3 encoded by sgg over 50 MY A, rescues sgg mutants so well is striking, but not without precedent. Rat GSK-3 can also rescue sgg mutants (Siegfried et al. 1992). Clearly, the Sgg and Mjl proteins are under selection in the Drosophila genus. Therefore, we suggest that such cross-rescue is more a reflection of the large error in measuring phenotypes in a lab setting. Although we did see that altered rescue efficacy is consistent with Sgg being better adapted for sgg functions than Mjl, these transgenic experiments are inherently fraught with technical difficulties.
The best evidence for functional divergence is in the spatial expression (testes specific) as well as its sexually dimorphic expression (male-specific) pattern of Mjl and the largely uniform expression of Sgg. The GSK-3 kinase domain is used in multiple signaling pathways (Cohen and Frame 2001; Frame and Cohen 2001; Eldar-Finkelman 2002), but is not known to function in the male germ line. Mjl, on the other hand, is required only for maintaining male fertility. These data argue for mjl having undergone neofunctionalization following duplication (Hughes 1994; Lynch and Conery 2000; Zhang et al. 2004).
It is curious that the Ks values for Mjl and Sgg are not similar, as expected under nearly neutral models of DNA sequence evolution (Ohta 2002). It is possible that this is a reflection of selection that is independent of amino acid coding (Xing and Lee 2006). It is well documented that sgg “pre-mRNAs” undergo differential splicing giving rise to various functional isoforms, whereas the mjl pre-mRNAs are devoid of introns and therefore do not undergo posttranscriptional splice modifications (Bourouis et al. 1990). Higher Ks rates in Mjl might be the result of relaxed selection at sites that have a regulatory role in processing in sgg pre-mRNAs.
Selection or Chance
One of the more fascinating aspects of retroposition is the highly directional patterns of both distribution in the genome and the expression pattern of the daughter gene. In both Drosophila and mammals, the parental gene in a retroposed pair is usually located on the X chromosome and the daughter gene is autosomal (Betran et al. 2002; Emerson et al. 2004). As a general rule, retroposed genes in both mammals (Hisano et al. 2003; Rohozinski and Bishop 2004; Luo et al. 2006; Vinckenbosch et al. 2006) and Drosophila (Betran et al. 2002) tend to be expressed in testes. This preference is startling. In D. melanogaster, genes are preferential retroposed from the X chromosome onto autosomes (40% observed vs. 23.3% assuming random retroposition), where the majority (91%) of the retroposed genes evolved a testes-specific expression pattern (Betran et al. 2002). This pattern of expression along with growing evidence that the X chromosome is a disfavored location for genes with male-biased function (Parisi et al. 2003; Wu and Xu 2003) suggests that the 2 observations may be linked. Retroposition patterns may indicate that important male functions are being transferred to more favorable long-term locations in the genome (Betran et al. 2002, 2004).
In contrast to such functional movements, it is also possible that the testis is a permissive environment for gene expression (Vinckenbosch et al. 2006). Male-biased expression of retroposed genes is therefore a function of chance in this model. This is not well supported by global expression studies in D. melanogaster, which show a complex but highly specific pattern of testes gene expression (Andrews et al. 2000; Parisi et al. 2003, 2004). However, if genes retropose more frequently in the male germ line and then are preferential inserted due to the open chromatin structure of genes being highly expressed in the male germ line, then testis expression would also be expected (Khil et al. 2005).
If testes-biased expression of retroposed genes is neutral, then expression within the male germ line should not be evolutionarily stable. Indeed, a recent report shows that retroposed genes in primates initially tend to be transcribed in testes, but over time they tend to “come out” of the testes and be expressed more broadly (Vinckenbosch et al. 2006). The mjl gene does not fit this pattern, because it is an older retroposed gene, but its expression remains highly testes biased, and in fact, may be expressed specifically in the male germ line of D. melanogaster. Our data support the idea that retroposition may be one of the modes employed by genes with male-biased functions to emigrate from the disfavored X chromosome to an autosomal location.
Acknowledgments
We thank K. Howard, H. Lin, G. Thomas, K. Han, and A. Clark for reagents and flies. We thank M. Long, J. Dean, and members of the Oliver lab for critical reading of the manuscript and helpful discussions. We owe great thanks to providers of genomic sequence data: mjl and sgg sequences are from draft annotations (Iyer VN, Pollard DA, Eisen MB, personal communication) of assemblies generated by Agencourt (Smith D, personal communication) and Washington University (Wilson R, personal communication) genome sequencing centers. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health and the National Institutes of Diabetes & Digestive & Kidney Diseases.
Literature Cited
- Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. 195 co-authors. [DOI] [PubMed] [Google Scholar]
- Andrews J, Bouffard GG, Cheadle C, Lu J, Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka M, Sano H, Obara Y, Kobayashi S. Maternal Nanos regulates zygotic gene expression in germline progenitors of Drosophila melanogaster. Mech Dev. 1998;78:153–158. doi: 10.1016/s0925-4773(98)00164-6. [DOI] [PubMed] [Google Scholar]
- Betran E, Emerson JJ, Kaessmann H, Long M. Sex chromosomes and male functions: where do new genes go? Cell Cycle. 2004;3:873–875. [PubMed] [Google Scholar]
- Betran E, Long M. Expansion of genome coding regions by acquisition of new genes. Genetica. 2002;115:65–80. doi: 10.1023/a:1016024131097. [DOI] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourouis M, Moore P, Ruel L, Grau Y, Heitzler P, Simpson P. An early embryonic product of the gene shaggy encodes a serine/threonine protein kinase related to the CDC28/cdc2+ subfamily. EMBO J. 1990;9:2877–2884. doi: 10.1002/j.1460-2075.1990.tb07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant pheno-types. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brent R, Kingston RE, Seidman JG, Struhl K, Ansubel FM, Chanda VB, Moore DD. Current protocols in molecular biology. Hoboken (NJ): Wiley and Sons; 1987. [Google Scholar]
- Brosius J. Retroposons—seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Burnett C, Howard K. Fly and mammalian lipid phosphate phosphatase isoforms differ in activity both in vitro and in vivo. EMBO Rep. 2003;4:793–799. doi: 10.1038/sj.embor.embor900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. 32 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Eldar-Finkelman H. Glycogen synthase kinase 3: an emerging therapeutic target. Trends Mol Med. 2002;8:126–132. doi: 10.1016/s1471-4914(01)02266-3. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Fabrizio JJ, Boyle M, DiNardo S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol. 2003;258:117–128. doi: 10.1016/s0012-1606(03)00127-1. [DOI] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003;31:172–175. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1993. pp. 71–148. [Google Scholar]
- Hisano M, Yamada S, Tanaka H, Nishimune Y, Nozaki M. Genomic structure and promoter activity of the testis haploid germ cell-specific intronless genes, Tactl and Tact2. Mol Reprod Dev. 2003;65:148–156. doi: 10.1002/mrd.10276. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Khil PP, Oliver B, Camerini-Otero RD. X for intersection: retrotransposition both on and off the X chromosome is more frequent. Trends Genet. 2005;21:3–7. doi: 10.1016/j.tig.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEG A3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M. Development of the male germline stem cell niche in Drosophila. Dev Biol. 2006;294(1):92–103. doi: 10.1016/j.ydbio.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Long M, Deutsch M, Wang W, Betran E, Brunei FG, Zhang J. Origin of new genes: evidence from experimental and computational analyses. Genetica. 2003;118:171–182. [PubMed] [Google Scholar]
- Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87:348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford: Oxford University Press; 2000. [Google Scholar]
- Ohta T. Near-neutrality in evolution of genes and gene regulation. Proc Natl Acad Sci USA. 2002;99:16134–16137. doi: 10.1073/pnas.252626899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. 12 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohozinski J, Bishop CE. The mouse juvenile spermatogonial depletion (jsd) phenotype is due to a mutation in the X-derived retrogene, mUtp14b. Proc Natl Acad Sci USA. 2004;101:11695–11700. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Broihier HT, Moore L, Van Doren M, Forbes A, Lehmann R. Germ plasm assembly and germ cell migration in Drosophila. Cold Spring Harbor Symp Quant Biol. 1997;62:1–11. [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, Jones DL, Wood CG, Fuller MT. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 2004;167:707–723. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N. Wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Perkins LA, Capaci TM, Perrimon N. Putative protein kinase product of the Drosophila segment-polarity gene zeste-white3. Nature. 1990;345:825–829. doi: 10.1038/345825a0. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N. Components of wingless signalling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Clock mechanisms in Drosophila. Cell Tissue Res. 2002;309:11–26. doi: 10.1007/s00441-002-0569-0. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. 13 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz MC. An investigation of Zeste white-3 inactivation by wingless signalling. University Park (PA): Department of Biochemistry, Microbiology and Molecular Biology. The Pennsylvania State University; 2000. p. 173. [Google Scholar]
- Steitz MC, Wickenheisser JK, Siegfried E. Overexpression of zeste white 3 blocks wingless signaling in the Drosophila embryonic midgut. Dev Biol. 1998;197:218–233. doi: 10.1006/dbio.1998.8884. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat Struct Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nat Genet. 2004;36:523–527. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Xu EY. Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C. Can RNA selection pressure distort the measurement of Ka/Ks? Gene. 2006;370:1–5. doi: 10.1016/j.gene.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dean AM, Brunei F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci USA. 2004;101:16246–16250. doi: 10.1073/pnas.0407066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]