Abstract
The c-Myb gene encodes a transcription factor required for proliferation and survival of normal myeloid progenitors and leukemic blast cells. Targeting of c-Myb by antisense oligodeoxynucleotides has suggested that myeloid leukemia blasts (including chronic myelogenous leukemia [CML]–blast crisis cells) rely on c-Myb expression more than normal progenitors, but a genetic approach to assess the requirement of c-Myb by p210BCR/ABL-transformed hematopoietic progenitors has not been taken. We show here that loss of a c-Myb allele had modest effects (20%-28% decrease) on colony formation of nontransduced progenitors, while the effect on p210BCR/ABL-expressing Lin− Sca-1+ and Lin− Sca-1+Kit+ cells was more pronounced (50%-80% decrease). Using a model of CML-blast crisis, mice (n = 14) injected with p210BCR/ABL-transduced p53−/−c-Mybw/w marrow cells developed leukemia rapidly and had a median survival of 26 days, while only 67% of mice (n = 12) injected with p210BCR/ABL-transduced p53−/−c-Mybw/d marrow cells died of leukemia with a median survival of 96 days. p210BCR/ABL-transduced c-Mybw/w and c-Mybw/d marrow progenitors expressed similar levels of the c-Myb–regulated genes c-Myc and cyclin B1, while those of Bcl-2 were reduced. However, ectopic Bcl-2 expression did not enhance colony formation of p210BCR/ABL-transduced c-Mybw/d Lin−Sca-1+Kit+ cells. Together, these studies support the requirement of c-Myb for p210BCR/ABL-dependent leukemogenesis.
Introduction
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder arising from neoplastic transformation of hematopoietic stem/progenitor cells1 by the p210BCR/ABL protein that is translated from bcr/abl transcripts generated by the bcr/abl chimeric gene of the Philadelphia chromosome.2–5 The oncogenic potential of p210BCR/ABL depends on its tyrosine kinase activity and rests on its ability to interact with and phosphorylate several substrates, which in turn activate cytoplasmic and nuclear effectors.6 Numerous studies on the mechanisms of p210BCR/ABL leukemogenesis have focused on the activation of cytoplasmic effectors such as RAS and PI-3K,7,8 but less is known about the nuclear effectors, the mechanisms of their activation or loss of function, and their contribution to the phenotype of transformed cells. The nuclear effectors of p210BCR/ABL include transcription factors whose activity is also modulated by the products of other leukemia-specific translocation or directly by mutation.6 One of the transcription factors whose expression in enhanced by p210BCR/ABL and may have an important role in BCR/ABL-dependent leukemogenesis is c-Myb.9
The importance of c-Myb in normal hematopoiesis is supported by studies showing that ablation of c-Myb expression leads to impaired liver hematopoiesis,10 reduced erythroid and myeloid colony formation,11 and defective B- and T-cell lymphopoiesis.12,13 The function of c-Myb may depend on its ability to modulate levels of genes (ie, CD34, c-Kit, Flt3) expressed by early progenitor cells and involved in the regulation of cell proliferation and survival.14–16 A target of c-Myb in hematopoietic cells is also Bcl-2.17
In CML cells, the c-Myb gene is intact18,19 but expression is often increased, in part, via enhanced protein stability regulated by the PI-3K/Akt pathway9
The c-Myb gene has been proposed as a therapeutic target in acute myelogenous leukemia (AML) and in CML based on the differential requirement of c-Myb by normal and leukemic cells.20,21 However, these studies were based on the use of antisense oligodeoxynucleotides in dose-response comparisons of normal and leukemic colony formation and on clonogenic assays of mixed normal and leukemic cells in which the phenotype of surviving cells was assessed by measuring bcr/abl transcripts. Since a genetic approach for assessing the role of c-Myb in BCR/ABL-dependent leukemogenesis has not been taken, we used hematopoietic progenitor cells from c-Myb knockout mice with 1 or 2 functional alleles to assess their reliance on c-Myb levels for p210BCR/ABL-dependent in vitro transformation and leukemogenesis. We show here that p210BCR/ABL-transduced Lin−Sca-1+ and Lin−Sca-1+ Kit+ marrow progenitors are more dependent than their normal counterpart on optimal c-Myb levels for primary and secondary colony formation. Moreover, in a mouse model of CML-blast crisis, p210BCR/ABL-expressing wild-type c-Myb marrow cells induced primary and secondary leukemia with a shorter latency of that induced by c-Myb+/− marrow cells.
Levels of Bcl-2, but not c-Myc, were decreased in p210BCR/ABL-expressing c-Myb+/− marrow cells; however, ectopic Bcl-2 expression did not enhance colony formation of p210BCR/ABL-transduced c-Myb+/− Lin−Sca-1+Kit+ cells, suggesting that lower expression of several c-Myb–regulated genes may be involved in the reduced leukemogenic potential of these cells.
Methods
Mice
c-Mybf/f and c-Mybf/d mice obtained from Dr Bender's laboratory were previously described.11 p53+/− C57BL6/J mice were obtained from Taconic (Hudson, NY). Mice were genotyped according to published procedures.12,22 All experiments on mice were performed with approval of and in accordance with Thomas Jefferson University‘s Animal Care and Use Committee.
Culture conditions, isolation, and retroviral transductions of primary bone marrow progenitor cells
Murine bone marrow cells were obtained from the femurs of c-Mybf/f, c-Mybf/d, or p53−/−c-Mybw/w or p53−/−c-Mybw/d mice after hypotonic lysis and enrichment for lineage negative (Lin−) hematopoietic precursors using the StemSep Mouse Progenitor Enrichment Kit protocol (StemCell Technologies, Vancouver, BC). For some experiments, Lin− cells were purified after lineage depletion of bone marrow cells using antibodies directed against CD3 (clone 145–2C11), CD45R/B220 (clone RA3–6B2), Ter-119, Ly6G and Ly-6C (Gr-1, clone RB6–8C5), and CD11b (Mac-1, clone M1/70) conjugated to a fluorochrome (BD Pharmingen, Franklin Lakes, NJ). Cells were then cultured for 24 hours in StemSpan SFEM medium (StemCell Technologies) supplemented with IL-3 (6 ng/mL), IL-6 (10 ng/mL), and Kit ligand (KL, 50 ng/mL). Lin−Sca-1+ and Lin−Sca-1+Kit+ cells were obtained by a MoFlo-MLS cell sorter (DAKO Cytomation, Carpinteria, CA) using PE-conjugated anti–Sca-1 (Ly-6A/E; clone E13–161.7) and APC-conjugated anti–c-Kit (CD117; clone 2B8) antibodies (BD Pharmingen). Cells were transduced with a retroviral stock of MigRI p210 BCR/ABL supernatant of transfected Phoenix cells in medium containing 50% (vol/vol) retroviral supernatant and 2 μg/mL polybrene. Virus and cells were cosedimented at 684g for 45 minutes in a Sorvall RT-6000 centrifuge (Newton, CT) at 32°C. Retroviral medium was changed after a 3- to 4-hour adsorption and a second round of transduction and cosedimentation, and replaced with normal medium after 2 to 3 hours. A third round of transduction and cosedimentation was performed 24 hours later, and the retroviral medium was replaced with normal medium after a 5-hour adsorption. After 24 hours, cells were sorted for GFP expression and plated for colony-forming assays.
For ectopic expression of c-Myb or Bcl-2 in p210BCR/ABL c-Mybf/d cells, transduced Lin−Sca-1+Kit+ cells p210BCR/ABL were sorted for GFP expression by a MoFlo-MLS cell sorter (DAKO Cytomation) and cultured in StemSpan SFEM medium (StemCell Technologies) supplemented with IL-3 (6 ng/mL), IL-6 (10 ng/mL), and KL (50 ng/mL) for 3 days. Then, cells were transduced with a retroviral stocks of murine stem cell virus (MSCV)/puro or MSCV/puro/c-Myb (19) or MSCV/puro/Bcl-2 (43) whose viral titer was determined by assessing puromycin-resistant colonies of transduced 32Dcl3 myeloid precursor cells. Since the viral titer of the MSCV/puro retroviral stock yielded 5% to 10% fewer colonies than the MSCV/puro/c-Myb or MSCV/puro/Bcl-2 stock, the amount of retroviral stock used was adjusted to give equivalent viral titers. After 24 hours, cells (8 × 104/plate) were plated in methylcellulose cells in the presence of 1 μg/mL puromycin. Colony formation was assessed 9 days later.
Colony formation assays
Clonogenic assays were carried out in Methocult H4230 (StemCell Technologies) in the presence of IL-3 (6 ng/mL), IL-6 (10 ng/mL), and Kit ligand (50 ng/mL) or 1/10th of the cytokines. Progenitor cells were plated in methylcellulose at different numbers (1 × 103 to 5 × 103) and colony formation was assessed after 7 days. Secondary colony formation was carried out using cells isolated from methylcellulose. After 3 washes with medium, cells were replated onto new plates and colonies counted after 7 days.
Bone marrow transplantation
p53+/−c-Mybw/d were generated by crossing the p53−/− mice with c-Mybf/d C57BL/6 mice. Intercrosses of p53+/−c-Mybw/d mice produced an offspring of p53−/−c-Mybw/w, p53−/−c-Mybw/d, p53+/−c-Mybw/w, and p53+/−c-Mybw/d mice with each genotype at the expected Mendelian ratio.
p53−/−c-Mybw/w and p53−/−c-Mybw/d mice were primed by intraperitoneal injection of 5-flurouracil (5-FU; 150 mg/kg) 4 days before harvest. Bone marrow cells were transduced with a retroviral stock of MigRIp-210BCR/ABL supernatant by serial spininfection as described above and then injected into lethally irradiated (11Gy in 2 split doses of 5.5 Gy each) syngenic C57BL/6J mice by lateral tail-vein injection (106 cells/mouse, 7% GFP+ cells). Mice showing signs of pain and suffering were killed and analyzed for the presence of leukemia. Diagnoses were established by pathologic and immunophenotypic analyses. Leukemic cells in bone marrow and spleen cells were subjected to immunophenotypic analyses for detection of the following markers: Gr-1, using a phycoerythrin (PE)–conjugated antimouse Ly-6G (Gr-1) antibody; Mac-1, using a phycoerythrin (PE)–conjugated antimouse CD11b (integrin αM chain, Mac-1 chain); CD19, using a phycoerythrin (PE)-conjugated antimouse CD19 antibody; or Sca-1, using a phycoerythrin (PE)–conjugated antimouse Ly-6A/E (Sca-1) antibody. The antibodies were obtained from BD Pharmingen.
For secondary transplantation assays, leukemic cells were sorted for GFP positivity by a MoFlo-MLS cell sorter (DAKO Cytomation), and 24 hours later injected in sublethally (5Gy) irradiated syngenic C57BL/6J mice. Mice were killed when terminally ill.
Western blotting
Cells (6 × 105/30 μL) were lysed in Laemmli buffer and heated to 100°C for 10 minutes, and lysates were used for Western blotting. Equal amounts of total cell lysate were loaded onto a 4% to 15% polyacrylamide gel (Ready Gel system; Bio-Rad Laboratories, Hercules, CA). Following electrophoresis, proteins were transferred to nitrocellulose (Whatman, Florham Park, NJ), using the mini Trans-Blot transfer apparatus for 1 hour at 100 V (Bio-Rad Laboratories). Nitrocellulose membranes were blocked by incubation in Tris (Tris-hydroxymethyl-aminomethane)–buffered saline (5 mM Tris, 135 mM NaCl, 5 mM KCl) containing 5% nonfat dry milk, 0.1% Tween 20 for 1 hour at room temperature followed by a 1-hour incubation with primary antibody diluted in blocking buffer. Proteins of interest were detected using anti–c-Myb monoclonal antibody (clone1.1; Upstate Biotechnology, Lake Placid, NY); anti-GRB2 monoclonal antibody (610112; BD Transduction Laboratories, Lexington, KY); PY20H anti-PTyr/HRP monoclonal antibody (P11265; BD Transduction Laboratories); anti–c-Myc polyclonal antibody (N-262; Santa Cruz Biotechnology, Santa Cruz, CA); anti–Bcl-2 monoclonal antibody (7/Bcl-2; BD Transduction Laboratories); anti–cyclin B1 monoclonal antibody (GNS1, Santa Cruz Biotechnology), antiactin polyclonal antibody (Sigma-Aldrich, St Louis, MO); anti–B-Myb (N-19; Santa Cruz Biotechnology); and anti–c-Abl (Ab-3; Calbiochem, San Diego, CA); monoclonal antibodies. Filters were then washed extensively in Tris-buffered saline containing 5% nonfat dry milk, 0.1% Tween-20 before incubation for 30 minutes with either a donkey antirabbit horseradish peroxidase–conjugated or a sheep antimouse horseradish peroxidase-conjugated antibody, each diluted 1:10 000 in blocking buffer (GE Healthcare Little Chalfont, United Kingdom). Filters were then washed as described above and developed using an enhanced chemiluminescence substrate (Pierce, Rockford, IL).
Results
Loss of a c-Myb allele has modest effects on myeloid colony formation of bone marrow progenitors
c-Myb is predominantly expressed by primitive hematopoietic progenitor cells and its levels decrease during differentiation.23 Moreover, c-Myb expression is elevated in many cases of acute myeloid and lymphoid leukemia,24 but the mechanisms underlying such an increase are unclear except in a cohort of pediatric T-cell acute lymphoblastic leukemia (T-ALL).25,26 To test the requirement of c-Myb in normal hematopoiesis and p210BCR/ABL leukemogenesis, we used heterozygous c-Myb knockout mice in which one allele has c-Myb exon 2 flanked by LoxP sites for eventual cleavage by the Cre recombinase, referred to here as the floxed Myb allele (Mybf), and the second has been deleted (Mybd).12
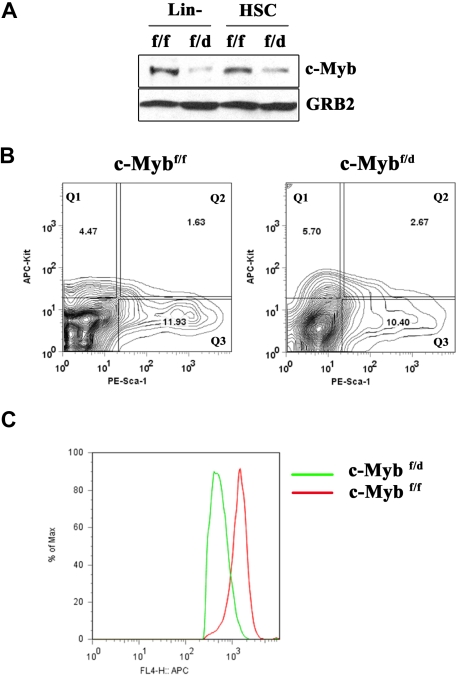
To determine whether loss of a c-Myb allele correlated with reduced protein levels, expression of c-Myb was examined by Western blotting in lysates of hematopoietic progenitors obtained from the marrow of c-Mybf/f and c-Mybf/d mice. Compared with c-Mybf/f marrow cells, we observed a 50% reduction of c-Myb levels in Lin−Sca-1+Kit+ cells from c-Mybf/d mice, while the decrease was more pronounced in “enriched Lin−” cells (Figure 1A), possibly reflecting enhanced differentiation of c-Myb+/−Lin− cells. To assess if loss of a c-Myb allele affects the hematopoietic stem cell (HSC) compartment, we monitored stem cell number and hematopoietic reconstitution ability of c-Mybf/f and c-Mybf/d bone marrow cells. By immunophenotype, the percentage of Sca-1+Kit+ cells in the Lin− fraction of bone marrow cells from c-Mybf/f and c-Mybf/d mice was essentially identical (2.23% ± 0.95% and 2.79% ± 0.82%, respectively) (Figure 1B). Instead, levels of c-Kit were lower in c-Mybf/d Lin−Sca-1+Kit+ cells (Figure 1C), consistent with the observation that c-Kit is a c-Myb–regulated gene.14
Figure 1.
Loss of a c-Myb allele does not affect the frequency of hematopoietic progenitors. (A) Western blot analysis of c-Myb expression in enriched Lin− and Lin−Sca-1+Kit+ cells from c-Mybf/f and c-Mybf/d mice. GRB2 expression served as a control. (B) Frequency of Lin−Kit+, Lin−Sca-1+, and Lin−Sca-1+Kit+ cells (Q1, Q3, and Q2) in enriched Lin− cells of c-Mybf/f and c-Mybf/d mice, assessed by use of PE-conjugated anti–Sca-1 and APC-conjugated anti–c-Kit antibodies on enriched Lin− marrow cells. (C) Flow cytometric analysis of c-Kit expression in Lin−Sca-1+Kit+ cells from c-Mybf/f and c-Mybf/d mice. Representative of 3 experiments.
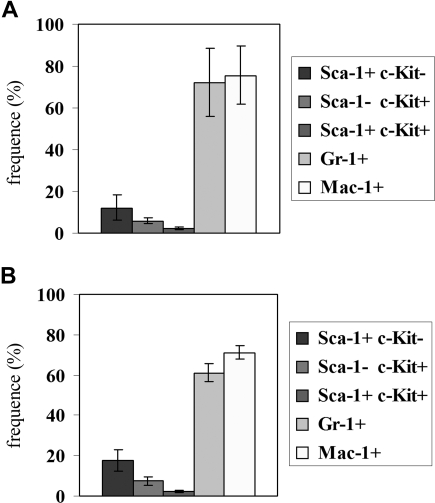
Hematopoietic reconstitution, tested 3 months after injecting 106 bone marrow cells from a c-Mybf/d or a c-Mybf/f mouse in 3 lethally irradiated syngenic mice, was also undistinguishable, based on peripheral blood and bone marrow cell counts (not shown) and the proportion of bone marrow cells expressing the Sca-1, c-Kit, Gr-1, and Mac-1 antigens (Figure 2).
Figure 2.
Hematopoietic reconstitution in mice injected with c-Mybf/d or c-Mybf/f bone marrow cells. Bone marrow cells (106) from a c-Mybf/f or a c-Mybf/d mouse were injected in 3 lethally irradiated syngenic mice (11 Gy, 2 split doses of 5.5 Gy rad/each). After 3 months, the proportion of bone marrow cells expressing the Sca-1, c-Kit, Gr-1, and Mac-1 antigens was tested by immunostaining. Histograms show frequency of Sca-1+c-Kit−, Sca-1−c-Kit+, Sca-1+c-Kit+, Gr-1+, and Mac-1+ cells in the bone marrow of mice injected with c-Mybf/f (A) or c-Mybf/d (B) bone marrow cells. None of the differences in the values of histograms in panels A,B is statistically significant. As control, lethally irradiated mice not injected with bone marrow cells died after 2 weeks; bone marrow cells purified from mice injected with c-Mybf/d cells were genotyped and showed the distinctive polymerase chain reaction (PCR) pattern of c-Mybf/d donor cells.
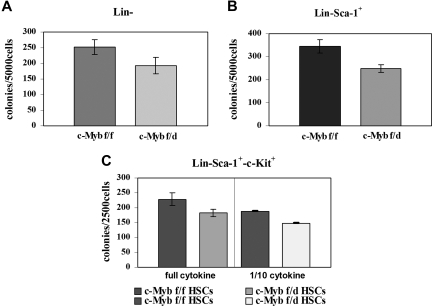
Since c-Myb is essential for myeloid bone marrow colony formation, we tested the clonogenic potential of the Lin−, Lin−Sca-1+, and the Lin−Sca-1+Kit+ progenitor subsets by methylcellulose colony formation assays. Enriched Lin− and Lin−Sca-1+ cells from c-Mybf/d mice formed approximately 25% fewer colonies than the counterpart from c-Mybf/f mice (Figure 3A,B). Colony formation and c-Myb expression were also assessed using “purified Lin−” cells isolated by lineage depletion after labeling with lineage-specific antibodies. By densitometry analysis, c-Myb levels were reduced 2.4-fold in Lin− cells from c-Mybf/d mice, while colony formation was reduced by 28% (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Lin−Sca-1+Kit+ marrow cells from c-Mybf/f or c-Mybf/d mice were plated in methylcellulose in the presence of optimal or suboptimal (1/10th) cytokine concentrations After 7 days, colonies were counted and there was an approximately 20% reduction in those arising from c-Mybf/d Lin−Sca-1+Kit+ cells compared with those from the c-Mybf/f counterpart (Figure 3C). Together, these data indicate that the reduced levels of c-Myb in marrow progenitors of c-Mybf/d knockout mice have modest effects on the colony formation capability of these cells, compared with the c-Mybf/f counterpart.
Figure 3.
Loss of a c-Myb allele has modest effects on colony formation of bone marrow progenitors. Colony formation assays were performed using (A) enriched Lin− marrow cells (5 × 103 cells/plate) plated in methylcellulose in the presence of GM-CSF (P = .015); (B) Lin−Sca-1+ marrow cells (5 × 103cells/plate) plated in methylcellulose in the presence of IL-3, IL-6, and KL (P = .008); and (C) Lin−Sca-1+Kit+ marrow cells (2.5 × 105/plate) plated in methylcellulose in the presence of optimal (IL-3, 6 ng/mL; IL-6, 10 ng/mL; KL, 50 ng/mL) or suboptimal (1/10th) cytokine concentrations; P = .011 and P ≤ .001, respectively. Representative of 3 different experiments performed in duplicate.
Low levels of c-Myb reduce colony formation of p210BCR/ABL-transduced marrow progenitors
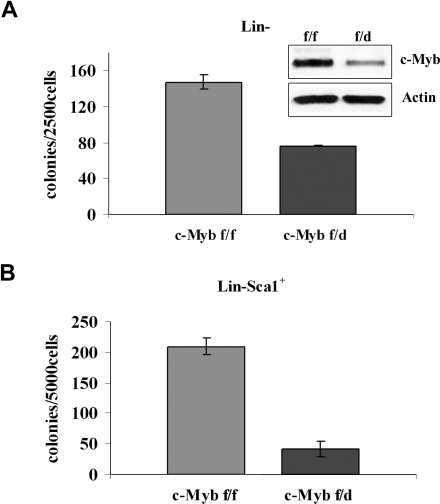
To assess the requirement of c-Myb in BCR/ABL-dependent transformation, different progenitor subsets were transduced with a p210BCR/ABL retrovirus and assessed for methylcellulose colony formation after GFP sorting. p210BCR/ABL-transduced purified Lin− cells (isolated by lineage depletion after labeling with lineage-specific antibodies) from c-Mybf/d mice formed approximately 50% fewer colonies than purified Lin− cells from c-Mybf/f mice (Figure 4A). The difference in colony formation was more striking for p210BCR/ABL-transduced Lin−Sca-1+ cells. In the presence of optimal cytokine concentrations, there was an 80% reduction of colony formation by the Lin−Sca-1+ progenitors of c-Mybf/d mice, compared with those from c-Mybf/f mice (Figure 4B).
Figure 4.
Low levels of c-Myb reduce colony formation of p210BCR/ABL-transduced marrow progenitors. (A) Methylcellulose colony formation of p210BCR/ABL-transduced purified Lin− cells (2.5 × 103/plate) in the presence of optimal cytokine concentrations from c-Mybf/f or c-Mybf/d mice. This assay was performed once in duplicate plates. Inset shows c-Myb levels detected by Western blotting. (B) Methylcellulose colony formation of p210BCR/ABL-transduced Lin−Sca-1+ cells (5 × 103cells/plate) isolated from c-Mybf/f or c-Mybf/d mice, in the presence of optimal cytokine concentrations. Values (P ≤ .001) are representative of 3 different experiments performed in duplicate.
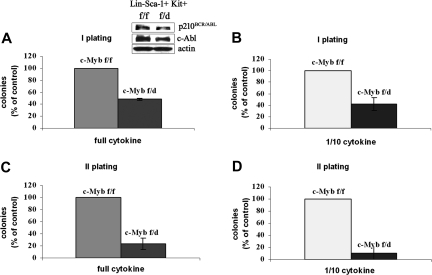
The clonogenic assays were also performed using the most primitive progenitors, the Lin−Sca-1+Kit+ HSCs. Thus, HSCs purified from c-Mybf/f or c-Mybf/d mice were retrovirally transduced with MigRI-p210BCR/ABL, GFP-sorted, assessed for p210BCR/ABL expression, and plated in methylcellulose in the presence of optimal or suboptimal cytokine concentration. Levels of p210BCR/ABL were essentially identical in transduced c-Mybf/f and c-Mybf/d HSCs (Figure 5 inset). Seven days after plating, HSCs from c-Mybf/d knockout mice formed 50% to 60% fewer colonies than the corresponding cells from c-Mybf/f mice, in the presence of an optimal cytokine concentration or with 1/10th of the cytokines, respectively (Figure 5).
Figure 5.
Colony formation of p210BCR/ABL-transduced c-Mybf/d HSCs. Lin−Sca-1+Kit+ cells from c-Mybf/f or c-Mybf/d mice were transduced with the MigRI-p210BCR/ABL retrovirus, GFP sorted, assessed for p210BCR/ABL levels, and plated in methylcellulose (2.5 × 103 cells/plate) in the presence of an optimal cytokine concentration (A) or 1/10th of the full cytokine doses (B). In panel A, inset shows p210BCR/ABL levels detected by anti–c-Abl Western blotting. Colonies were scored after 7 days. Values (P ≤ .001) are expressed as the percentage of the number of colonies derived from c-Mybf/f HSCs. Secondary colony formation assays were performed using cells isolated from methylcellulose. Cells were replated onto new plates in the presence of an optimal cytokine concentration (C) or 1/10th of the cytokines (D) and scored again after 7 days. Values (P ≤ .001) are expressed as the percentage of the number of colonies derived from c-Mybf/f HSCs. Representative of 3 different experiments.
Since secondary colony formation reflects the self-renewal potential of transformed cells, we performed secondary colony formation assays using cells isolated from methylcellulose. After primary plating of GFP+ HSCs, cells recovered from methylcellulose were replated in the presence of an optimal cytokine concentration or with 1/10th of the cytokines. p210BCR/ABL-transduced c-Mybf/f cells were 4- to 9-fold more clonogenic than c-Mybf/d cells in the presence of full dose of cytokines or 1/10th of the cytokines, respectively (Figure 5). Taken together, these data indicate that primary and secondary colony formation of p210BCR/ABL-transduced marrow progenitors is more efficient if both copies of the c-Myb gene are intact; moreover, by comparing colony assays of nontransduced and p210BCR/ABL-expressing hematopoietic progenitors, the latter appear to depend on c-Myb expression more than the nontransduced counterpart (Figure 3).
To further demonstrate that the reduced colony formation potential of p210BCR/ABL-transduced c-Mybf/d marrow progenitors is caused by lower c-Myb levels, we attempted to restore colony formation of c-Mybf/d p210BCR/ABL-transduced marrow progenitors by reconstituting c-Myb expression.
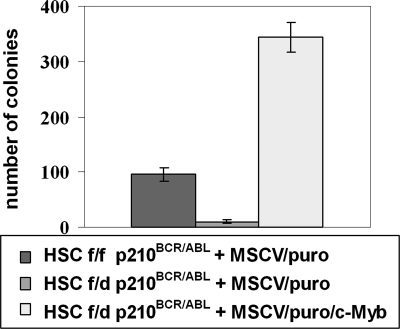
Thus, c-Mybf/d and c-Mybf/f Lin−Sca-1+Kit+ cells were transduced with the MigRI/p210BCR/ABL retrovirus and 24 hours after retroviral transduction purified by sorting for GFP expression. p210BCR/ABL-expressing c-Mybf/d Lin−Sca-1+Kit+ cells were subsequently transduced with a c-Myb/MSCV/puro retrovirus or the empty MSCV/puro vector as a negative control. p210BCR/ABL-expressing c-Mybf/f Lin−Sca-1+Kit+ cells were transduced with the empty vector MSCV/puro as a positive control. Transduced cells (80 000/each plate) were plated in methylcellulose in the presence of IL-3, IL-6, KL, and puromycin. Nine days after plating, c-Myb–transduced p210BCR/ABL c-Mybf/d HSCs formed 35-fold more colonies than p210BCR/ABL c-Mybf/d cells transduced with the empty vector (Figure 6). Moreover, the number of colonies formed by c-Myb–transduced cells was 4-fold higher than that of p210BCR/ABL c-Mybf/f HSCs transduced with the empty vector (Figure 6). Thus, the defect in colony formation by p210BCR/ABL c-Mybf/d marrow cells can be effectively rescued by restoring c-Myb expression.
Figure 6.
Restoration of c-Myb expression rescues colony formation of c-Mybf/d p210BCR/ABL-transduced marrow progenitors. c-Mybf/d and c-Mybf/f Lin−Sca-1+Kit+ cells were transduced with the MigRI/p210BCR/ABL retrovirus and 24 hours after retroviral transduction sorted for GFP positivity. p210BCR/ABL-expressing c-Mybf/d Lin−Sca-1+Kit+ cells were subsequently transduced with a c-Myb/MSCV/puro retrovirus or the empty MSCV/puro vector as a negative control. p210BCR/ABL-expressing c-Mybf/f Lin−Sca-1+Kit+ cells were transduced with empty vector MSCV/puro as a positive control. After 24 hours, cells (8 × 104/plate) were plated in methylcellulose in the presence of puromycin (1 μg/mL). Colony formation was assessed 9 days later. Representative of 3 different experiments. P ≤ .001.
Expression of c-Myb–regulated genes in c-Mybf/f or Mybf/d p210BCR/ABL-transduced progenitors
The reduced colony formation of transduced marrow progenitors may be caused by changes in the expression of c-Myb–regulated genes with a role in cell proliferation and survival.
One of the c-Myb targets implicated in these processes is c-myc.27 c-Myc is expressed in almost all proliferating normal cells, and down-regulation of c-Myc expression is required for terminal differentiation of many cell types, including myeloid cells.28–30 By assessing the expression of c-Myc in c-Mybf/f and c-Mybf/d enriched Lin− cells, we observed a decrease of its levels in c-Mybf/d Lin− cells (Figure 7A). However, the reduced levels of both c-Myb and c-Myc in c-Mybf/d Lin− cells did not appear to have a major effect on their colony formation potential as c-Mybf/d enriched Lin− cells plated in methylcellulose in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) formed only 25% fewer colonies than c-Mybf/f enriched Lin− cells (Figure 3).
Figure 7.
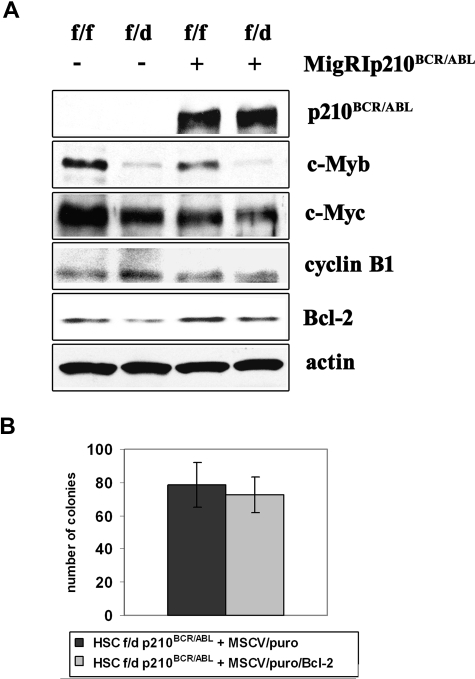
Ectopic Bcl-2 expression does not enhance colony formation of c-Mybf/d p210BCR/ABL-transduced HSCs. (A) Expression of c-Myb–regulated genes in p210BCR/ABL-transduced c-Mybf/f and c-Mybf/d progenitors. Western blot analysis of c-Myb, c-Myc, cyclin B1, and Bcl-2 expression in nontransduced and p210BCR/ABL-transduced enriched Lin− cells from c-Mybf/f and c-Mybf/d mice. Actin expression served as a control. (B) Colony formation of Bcl-2–transduced p210BCR/ABL-expressing c-Mybf/d progenitors. c-Mybf/d Lin−Sca-1+Kit+ cells were transduced with the MigRI/p210BCR/ABL retrovirus and 24 hours after retroviral transduction sorted for GFP expression. p210BCR/ABL-expressing c-Mybf/d Lin−Sca-1+Kit+ cells were subsequently transduced with a Bcl-2/MSCV/puro retrovirus or the empty MSCV/puro vector as a negative control. After 24 hours, cells (4 × 104/plate) were plated in methylcellulose in the presence of puromycin (1 μg/mL). Colony formation was assessed 9 days later. Representative of 3 different experiments.
Levels of c-Myc in p210BCR/ABL-transduced c-Mybf/d or c-Mybf/f were similar (Figure 7A), suggesting that the reduced colony formation of p210BCR/ABL-transduced c-Mybf/d progenitors is not caused by decreased c-Myc levels.
Levels of cyclin B1, a gene with an essential role in G2/M transition whose expression is regulated by c-Myb and B-Myb,31 were undistinguishable in uninfected and p210BCR/ABL-transduced c-Mybf/f or c-Mybf/d enriched Lin− cells (Figure 7A). Levels of B-Myb were similar in both cell subpopulations (not shown), possibly explaining why no changes in cyclin B1 levels were detected.
c-Myb has a role as a regulator of hematopoietic cell survival at least in part via the antiapoptotic Bcl-2 protein.17,32 c-Myb regulates Bcl-2 expression directly via a conserved Myb-binding site in the Bcl-2 promoter.32 Expression of Bcl-2 was lower in parental and p210BCR/ABL-transduced c-Mybf/d enriched Lin− cells, compared with that in c-Mybf/f cells (Figure 7A). However, ectopic Bcl-2 expression in p210BCR/ABL-transduced c-Mybf/d Lin−Sca-1+Kit+ cells did not enhance the colony formation of this progenitor subset (Figure 7B).
c-Myb is required for BCR/ABL-dependent leukemogenesis
Transplantation of lethally irradiated mice with p210BCR/ABL-retrovirally transduced bone marrow cells from 5-FU–treated mice to obtain a target population enriched in primitive hematopoietic cells induces a myeloproliferative syndrome resembling human CML. Since c-Mybf/d mice are more radiosensitive than normal mice33 and the recovery of HSCs and other progenitor subsets is lower from 5-FU–treated c-Mybf/d than c-Mybf/f mice (not shown), we assessed the role of c-Myb in p210BCR/ABL-dependent leukemogenesis using an in vivo model of CML-blast crisis.22 In this model, mice injected with p53-deficient, p210BCR/ABL-expressing marrow cells develop a more aggressive disease process than those injected with wild-type p53, BCR/ABL-expressing marrow cells.22
We obtained p53+/−Mybw/d by crossing the p53−/− mice with c-Mybf/d C57BL/6J mice. Intercrosses of p53+/−Mybw/d produced an offspring of mice with the p53−/−Mybw/w, p53−/−Mybw/d, p53+/−Mybw/w, and p53+/−Mybw/d genotypes at the expected Mendelian ratios with no apparent defect in growth, development, or fertility.
To test the role of c-Myb in leukemogenesis, p53−/−c-Mybww and p53−/−c-Mybw/d mice were treated with 5-FU (150 mg/kg for 4 days) to enhance the frequency of hematopoietic stem cells, and marrow cells were used for in vitro and in vivo studies. We tested first whether in vitro transformation by p53−/− marrow progenitors by p210BCR/ABL was more efficient when both copies of the c-Myb gene are intact.
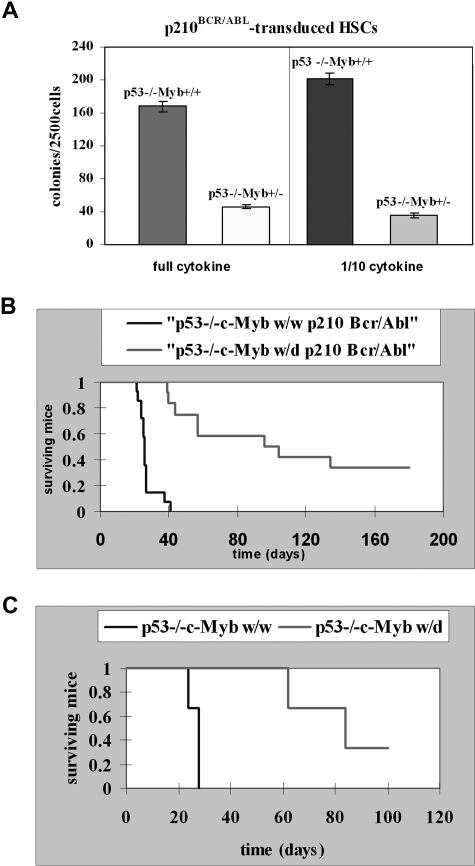
Lin−Sca-1+Kit+ HSCs isolated from 5-FU–treated p53−/−c-Mybww or p53−/−c-Mybw/d mice were retrovirally transduced with MigR1p210BCR/ABL and, after sorting for GFP expression, assessed in methylcellulose colony formation assays in the presence of optimal or suboptimal (1/10th) cytokine concentrations. Colonies were counted 7 days after plating transduced cells in methylcellulose. Lin−Sca-1+Kit+ cells from p53−/−c-Mybw/w mice produced 4- to 5-fold more colonies than Lin−Sca-1+Kit+ cells from p53−/−c-Mybw/d mice (Figure 8A). This result is consistent with data generated using p53-proficient progenitors from c-Mybf/f and c-Mybf/d mice, but the difference in colony formation is even more striking.
Figure 8.
c-Myb is required for p210BCR/ABL-dependent leukemogenesis. (A) Colony formation assays of p210BCR/ABL p53−/−c-Mybww or p53−/−c-Mybw/d marrow progenitors. Lin−Sca-1+Kit+ HSCs isolated from 5-FU–treated p53−/−c-Mybww or p53−/−c-Mybw/d mice were retrovirally transduced with MigR1p210BCR/ABL and, after GFP sorting, assessed for colony formation after plating in methylcellulose in the presence of a full dose or 1/10th of cytokines (IL-3, IL-6, and KL). Colonies were counted after 7 days. Representative of 3 different experiments performed in duplicate. P ≤ .001. (B) Survival of recipient mice injected with p210BCR/ABL-transduced marrow cells Kaplan-Meier plot shows survival of C57BL/6J recipient mice after transplantation with p210BCR/ABL p53−/−c-Mybw/w bone marrow cells (n = 14) or p210BCR/ABL p53−/−c-Mybw/d bone marrow cells (n = 12). Bone marrow cells from 5-FU–treated (150 mg/kg, 4 days), p53−/−c-Mybw/w or p53−/−c-Mybf/d mice were retrovirally transduced with the MigR1p210Bcr/Abl retrovirus and injected in lethally irradiated (1100 rad in 2 split doses of 550 each) syngenic C57BL/6J recipient mice. (C) Secondary leukemia in mice injected with GFP-positive cells isolated from mice with p210BCR/ABL-induced primary leukemia. Kaplan-Meier plot shows survival of C57BL/6J recipient mice (n = 3) injected with GFP-positive cells from a mouse with leukemia induced by p210BCR/ABL p53−/−c-Mybw/w cells or of recipient mice (n = 3) injected with GFP-positive cells from a mouse with leukemia induced by p210BCR/ABL p53−/−c-Mybw/d cells. Sublethally irradiated (500 rad) recipient mice each received with 1.0 × 105 GFP+ bone marrow cells.
To assess the role of c-Myb in BCR/ABL-dependent in vivo leukemogenesis, bone marrow cells from 5-FU–treated (150 mg/kg, 4 days), p53−/−c-Mybw/w, or p53−/−c-Mybf/d mice were transduced with the MigR1p210BCR/ABL retrovirus and injected into lethally irradiated (1100 rad in 2 split doses of 550 each) syngenic C57BL/6J recipient mice. Leukemia was assessed by analyzing the presence of GFP+ cells in bone marrow and spleen. Mice (n = 14) reconstituted with p210BCR/ABL-transduced p53−/−c-Mybw/w bone marrow cells died of leukemia 40 days after injection, and had a median survival of 26 days (Figure 8B). Bone marrow and spleen of 13 of 14 mice were immunophenotyped and GFP+ cells were myeloid (Gr-1 positive) in 5 mice, lymphoid (CD19+) in 5 mice, both myeloid and lymphoid in 2 mice, and neither myeloid nor lymphoid in 1 mouse.
Sixty-seven percent of mice (n = 12) reconstituted with p210BCR/ABL p53−/−c-Mybw/d marrow cells developed leukemia and died between 39 and 134 days after injection with a median survival of 96 days, more than 3-fold longer than that of p210BCR/ABL p53−/−c-Mybw/w–reconstituted mice (Figure 8B). By immunophenotype, GFP+ cells were lymphoid (CD19+) in 2 mice, and neither myeloid nor lymphoid in 1 mouse; the remaining mice had splenomegaly, but were not immunophenotyped because viable cells were not recovered from bone marrow or spleen.
The ability to serially transplant leukemic cells is a standard measure of their malignant potential. Sublethally irradiated (500 rad) recipient mice (n = 3) were injected with 1.0 × 105 GFP+ bone marrow cells from a leukemic p53−/−c-Mybw/w mouse, which had a mixed-lineage leukemia (GFP+Gr-1+, and GFP+CD19+ cells in bone marrow and spleen). The 3 injected mice developed a CD19+ lymphoblastic leukemia and died within 28 days (Figure 8C). Sublethally irradiated recipient mice (n = 3) were also injected with 1.0 × 105 GFP+ bone marrow cells from a leukemic p53−/−c-Mybw/d mouse. All GFP+ cells were Gr-1− and CD19−. Of the 3 injected mice, 2 developed leukemia with the GFP+ cells also expressing the IL-7R and died 62 and 84 days after transplantation, while 1 is still alive (Figure 8C). Thus, p210BCR/ABL-expressing p53−/−c-Mybw/d cells were less leukemogenic than p53−/−c-Mybw/w cells also in secondary recipient mice. Together, these data suggest that c-Myb is important for induction and maintenance of p210BCR/ABL-dependent leukemogenesis.
Discussion
Crucial roles for c-Myb during normal hematopoiesis have been established by a variety of strategies involving permanent and conditional inactivation of the c-Myb gene,10,12,13,34 expression of chimeric proteins with dominant-negative activity,35 and inhibition of gene expression by antisense oligodeoxynucleotides.20,21
Expression of c-Myb is often increased in acute leukemia blasts, but knowledge of the mechanisms underlying such an increase remains elusive. In a large cohort of CML-chronic phase and CML-blast crisis samples, we found no evidence of c-Myb mutations in the coding sequence or the exon/intron junctions,19 and amplification or truncation of c-Myb has previously been revealed only in 2 acute leukemia cell lines.36,37 Duplication or translocation of the c-Myb gene has been recently demonstrated in a cohort of pediatric T-ALL patients,25,26 essentially providing the first evidence that c-Myb can be activated by genetic mechanisms in human hematologic malignancies. Despite the lack of satisfactory mechanisms explaining the enhanced expression of c-Myb in leukemia blast cells, down-regulation of c-Myb expression in leukemic cells by use of antisense oligodeoxynucleotides was associated with reduced colony formation in vitro20,21 and suppression of leukemogenesis in mice.38 Of greater interest, leukemic blast cells appeared to rely on c-Myb expression more than normal progenitors, based on dose-response comparisons of normal and leukemic colony formation.20 However, the significance of these findings has remained somehow limited in the absence of genetic models in which to test the requirement of c-Myb expression for transformation by specific oncogenic proteins. In the present study, we assessed the requirement of c-Myb for p210BCR/ABL-dependent colony formation and leukemogenesis, using bone marrow cells from c-Myb knockout mice with only a single functional c-Myb allele.
Using different progenitor subsets, c-Mybf/d marrow cells yielded only 20% to 28% fewer colonies than c-Mybf/f marrow cells. These findings are in agreement with those showing that fetal liver colony formation was affected only if c-Myb levels were very low.39 Upon transduction with p210BCR/ABL, c-Mybf/d progenitors were markedly less clonogenic (50%-80% fewer colonies), both in primary and secondary colony formation assays and when transduced cells were plated in the presence of suboptimal cytokine concentrations.
The reduced colony formation potential of p210BCR/ABL-transduced c-Mybf/d HSCs was dependent on lower c-Myb levels because it increased dramatically after retroviral transduction with full-length c-Myb in experiments designed to restore c-Myb expression.
To test the requirement of c-Myb for p210BCR/ABL-dependent leukemogenesis, we could not rely on transplantation of lethally irradiated recipient mice with p210BCR/ABL-transduced bone marrow cells from 5-FU–treated mice: c-Myb heterozygous knockout are more radiosensitive than wild-type mice33 and, after 5-FU treatment, fewer marrow cells were recovered and number and distribution of progenitors subsets was also altered. Thus, we used marrow cells from p53−/−c-Mybw/w or p53−/−c-Mybw/d mice because the p53−/− background compensates the radiosensitivity of c-Mybw/d mice and the disease process induced by p210BCR/ABL-transduced p53−/− cells is rather aggressive, resembling CML-blast crisis.22
In agreement with the results of the colony formation assays, loss of c-Myb allele adversely affected p210BCR/ABL-dependent leukemogenesis as indicated by the longer survival of p53−/−c-Mybw/d mice.
Thus, using well-defined genetic models we have demonstrated that c-Myb is required for p210BCR/ABL-dependent leukemogenesis and that p210BCR/ABL-expressing progenitors rely on optimal levels of c-Myb more than the nontransduced counterpart.
The requirement of c-Myb in leukemogenesis induced by oncogenic proteins may not be limited to p210BCR/ABL. Indeed, a recent study has shown that c-Myb is an essential downstream target for in vitro transformation of hematopoietic cells by the MLL-ENL oncoprotein.40 In this study, c-Myb expression was activated through up-regulation of Hoxa9 and Meis1, which cooperate in leukemogenesis41 and are transcriptional targets of MLL-ENL.42 Since c-Myb is important for hematopoietic cell transformation by 2 oncogenic proteins, p210BCR/ABL and MLL-ENL, with distinct functions, activation of c-Myb might be an important step in many leukemogenic pathways and other mechanisms; besides gene duplication and rearrangement in pediatric T-ALL,25,26 transcription activation in leukemias with chimeric MLL-proteins42 and enhanced stability in some BCR/ABL-positive leukemias19 might be involved.
With regard to the molecular mechanisms responsible for the reduced colony formation and leukemogenesis of p210BCR/ABL-expressing c-Mybf/d progenitors, it is likely that attenuated expression of c-Myb targets plays an important role.
Of the c-Myb–regulated genes with a potential role in p210BCR/ABL leukemogenesis, expression of c-Myc was essentially identical in p210BCR/ABL-expressing c-Mybf/f and c-Mybf/d progenitors, but that of Bcl-2 was reduced in c-Mybf/d progenitors. The lower expression of Bcl-2 in these cells was potentially interesting because it has been shown that Bcl-2 cooperates with p210BCR/ABL in transformation of myeloid progenitors43,44; however, colony formation of p210BCR/ABL-expressing c-Mybf/d Lin− Sca-1+Kit+ cells was not enhanced by Bcl-2 transduction. Since expression of c-Kit is also reduced in c-Myb–deficient cells and c-Kit and Bcl-2 were previously shown to be required for survival and proliferation of hematopoietic stem cells,45 coexpression of Bcl-2 and c-Kit might rescue the lower colony formation and the reduced leukemogenesis of p210BCR/ABL-expressing c-Myb–deficient marrow cells.
Alternately, other c-Myb–regulated genes are required and a global approach might be needed to identify c-Myb–dependent changes in gene expression relevant for efficient p210BCR/ABL-transformation of marrow progenitors.
In summary, we have shown that optimal c-Myb levels are required for in vitro colony formation and in vivo leukemogenesis of p210BCR/ABL-expressing marrow progenitors. While these findings are consistent with previous studies based on the delivery of c-Myb antisense oligodeoxynucleotides to leukemic cells,20,21,38 the use of c-Myb–haplodeficient marrow cells will allow to assess mechanistically the role of c-Myb in p210BCR/ABL-dependent leukemogenesis.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grant CA95111 (B.C.). M.R.L. is supported by a fellowship of the American-Italian Cancer Foundation (AICF). F.C. is supported by a fellowship of the Associazione Italiana Ricerca sul Cancro (AIRC). T.W. is supported by a National Institutes of Health predoctoral fellowship (T32-CA09683-14).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.R.L. performed most of the studies and wrote the first draft of the paper; F.C. performed some of the in vitro studies; T.W. helped in the animal studies; T.P.B. provided c-Myb knockout mice and helped in writing the paper; B.C. designed the experiments and wrote the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruno Calabretta, Department of Cancer Biology, and Kimmel Cancer Center, Thomas Jefferson University Medical College, Philadelphia, PA, 19107; e-mail: B_Calabretta@mail.jci.tju.edu.
References
- 1.Kantarjian HM, Deisseroth A, Kurzrock R, Estrov Z, Talpaz M. Chronic myelogenous leukemia: a concise update. Blood. 1993;82:691–703. [PubMed] [Google Scholar]
- 2.Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497–1499. [Google Scholar]
- 3.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 4.Shtivelman E, Lifshitz B, Gale RB, Roe BA, Canaani E. Alternative splicing of RNAs transcribed from the human cell gene and from the bcr/abl fused gene. Cell. 1986;47:272–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific p210 protein is the product of the bcr-abl hybrid gene. Science. 1986;233:212–216. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 6.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 7.Goga A, McLaughlin J, Afar DE, Saffran DC, Witte ON. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 8.Skorski T, Bellacosa A, Nieborowska-Skorska M, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradini F, Cesi V, Bartella V, et al. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem. 2005;280:30254–30262. doi: 10.1074/jbc.M504703200. [DOI] [PubMed] [Google Scholar]
- 10.Mucenski ML, McLain K, Kier AB, et al. A functional c-myb gene is required for normal murine felt hepatic hemotopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 11.Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 12.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 13.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Melotti P, Ku DH, Calabretta B. Regulation of the expression of the hematopoietic stem cell antigen CD34: role of c-myb. J Exp Med. 1994;179:1023–1028. doi: 10.1084/jem.179.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratajczak MZ, Perrotti D, Melotti P, et al. Myb and ets proteins are candidate regulators of c-kit expression in human hematopoietic cells. Blood. 1998;91:1934–1946. [PubMed] [Google Scholar]
- 16.Melotti P, Ku DH, Calabretta B. Regulation of the expression of the hematopoietic stem cell antigen CD34: role of c-myb. J Exp Med. 1994;179:1023–1028. doi: 10.1084/jem.179.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frampton J, Ramquist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 18.Lutwyche JK, Keough RA, Hughes TP, Gonda TJ. Mutation screening of the c-MYB negative regulatory domain in acute and chronic myeloid leukaemia. Br J Haematol. 2001;114:632–634. doi: 10.1046/j.1365-2141.2001.02966.x. [DOI] [PubMed] [Google Scholar]
- 19.Bussolari R, Candini O, Colomer D, et al. Coding sequence and intron-exon junctions of the c-myb gene are intact in the chronic phase and blast crisis stages of chronic myeloid leukemia patients. Leuk Res. 2007;31:163–167. doi: 10.1016/j.leukres.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Calabretta B, Sims RR, Valtieri M, et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci U S A. 1991;88:2351–2355. doi: 10.1073/pnas.88.6.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratajczak MZ, Hijiya N, Catani L, et al. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood. 1992;79:1956–1961. [PubMed] [Google Scholar]
- 22.Skorski T, Nieborowska-Skorska M, Wlodarski P, et al. Blastic transformation of p53-deficient bone marrow cells by p210bcr/abl tyrosine kinase. Proc Natl Acad Sci U S A. 1996;93:13137–13142. doi: 10.1073/pnas.93.23.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood. 2003;101:4322–4332. doi: 10.1182/blood-2002-03-0835. [DOI] [PubMed] [Google Scholar]
- 24.Mavilio F, Sposi NM, Petrini M, et al. Expression of cellular oncogenes in primary cells from human acute leukemias. Proc Natl Acad Sci U S A. 1986;83:4394–4398. doi: 10.1073/pnas.83.12.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 26.Clappier E, Cuccuini W, Kalota A, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M, Nazarov V, Stevens L, Watson R, Wolff L. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol Cell Biol. 2000;20:1970–1981. doi: 10.1128/mcb.20.6.1970-1981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freytag SO. Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol. 1988;8:1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman-Liebermann B, Liebermann DA. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991;6:903–909. [PubMed] [Google Scholar]
- 30.Hoffman B, Liebermann DA, Selvakumaran M, Nguyen HQ. Role of c-myc in myeloid differentiation, growth arrest and apoptosis. Curr Top Microbiol Immunol. 1996;211:17–27. doi: 10.1007/978-3-642-85232-9_3. [DOI] [PubMed] [Google Scholar]
- 31.Nakata Y, Shetzline S, Sakashita C, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci U S A. 1997;94:3296–3301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsay RG, Micallef S, Lightowler S, Mucenski ML, Mantamadiotis T, Bertoncello I. c-myb Heterozygous mice are hypersensitive to 5-fluorouracil and ionizing radiation. Mol Cancer Res. 2004;2:354–361. [PubMed] [Google Scholar]
- 34.Allen RD, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 36.Pelicci PG, Lanfrancone L, Brathwaite MD, Wolman SR, Dalla-Favera R. Amplification of the c-myb oncogene in a case of human acute myelogenous leukemia. Science. 1984;224:1117–1121. doi: 10.1126/science.6585957. [DOI] [PubMed] [Google Scholar]
- 37.Tomita A, Watanabe T, Kosugi H, et al. Truncated c-Myb expression in the human leukemia cell line TK-6. Leukemia. 1998;12:1422–1429. doi: 10.1038/sj.leu.2401113. [DOI] [PubMed] [Google Scholar]
- 38.Ratajczak MZ, Kant JA, Luger SM, et al. In vivo treatment of human leukemia in a scid mouse model with c-myb antisense oligodeoxynucleotides. Proc Natl Acad Sci U S A. 1992;89:11823–11827. doi: 10.1073/pnas.89.24.11823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess JL, Bittner CB, Zeisig DT, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeisig BB, Milne T, García-Cuéllar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cirinnà M, Trotta R, Salomoni P, et al. Bcl-2 expression restores the leukemogenic potential of a BCR/ABL mutant defective in transformation. Blood. 2000;96:3915–3921. [PubMed] [Google Scholar]
- 44.Jaiswal S, Traver D, Miyamoto T, Akashi K, Lagasse E, Weissman IL. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100:10002–10007. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domen J, Weissman IL. Hematopoietic stem cells need two signals to prevent apoptosis; BCL-2 can provide one of these, Kitl/c-Kit signaling the other. J Exp Med. 2000;192:1707–1718. doi: 10.1084/jem.192.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.