Abstract
The denatured state of a double mutant of the chemotactic protein CheY (F14N/V83T) has been analyzed in the presence of 5 M urea, using small angle X-ray scattering (SAXS) and heteronuclear magnetic resonance. SAXS studies show that the denatured protein follows a wormlike chain model. Its backbone can be described as a chain composed of rigid elements connected by flexible links. A comparison of the contour length obtained for the chain at 5 M urea with the one expected for a fully expanded chain suggests that ∼25% of the residues are involved in residual structures. Conformational shifts of the α-protons, heteronuclear 15N-{1H} NOEs and 15N relaxation properties have been used to identify some regions in the protein that deviate from a random coil behavior. According to these NMR data, the protein can be divided into two subdomains, which largely coincide with the two folding subunits identified in a previous kinetic study of the folding of the protein. The first of these subdomains, spanning residues 1–70, is shown here to exhibit a restricted mobility as compared to the rest of the protein. Two regions, one in each subdomain, were identified as deviating from the random coil chemical shifts. Peptides corresponding to these sequences were characterized by NMR and their backbone 1H chemical shifts were compared to those in the intact protein under identical denaturing conditions. For the region located in the first subdomain, this comparison shows that the observed deviation from random coil parameters is caused by interactions with the rest of the molecule. The restricted flexibility of the first subdomain and the transient collapse detected in that subunit are consistent with the conclusions obtained by applying the protein engineering method to the characterization of the folding reaction transition state.
Keywords: Denatured state, residual structures, folding initiation, heteronuclear NMR, backbone dynamics
The denatured state of a protein is formed by an ensemble of interconverting conformers. Interest in the role played by some preferential conformations of unfolded proteins in folding initiation has increased over the past 10 years, giving rise to several indepth studies of unfolded states. It is now recognized that denatured proteins may not only contain clues about early folding events, but also about stability or transport of protein across the membrane (Shortle 1996). Of particular interest is the presence of nascent structures. Indeed, a plausible first step in protein folding is considered to be the formation of local clusters in small regions of the polypeptide chain, even though this hypothesis often lacks direct experimental evidence (Wright et al. 1988).
During the last decade, improvements in various spectroscopic techniques and in particular in nuclear magnetic resonance (NMR) have allowed the observation of residual structure in the denatured state of a number of proteins (Evans et al. 1991; Neri et al. 1992; Logan et al. 1994; Lumb and Kim 1994; Arcus et al. 1995; Wong et al. 1996; Schwalbe et al. 1997; Blanco et al. 1998, Kortemme et al. 1999), and several reviews have stressed their possible role in the folding process (Miranker and Dobson 1996; Shortle 1996). Residual structures could correspond to the persistence of secondary structure in the denatured state (Bond et al. 1997), even though this conformation is populated only transiently (Smith et al. 1996b). These residual structures could also direct the search for the native structure toward "productive" regions of the conformational space (Fong et al. 1998). Recent studies of the spectrin SH3 domain (Blanco et al. 1998; Kortemme et al. 1999), the drk-SH3 domain (Zhang and Forman-Kay 1997; Mok et al. 1999), Staphylococcus nuclease (Sinclair and Shortle 1999) and a Protein G mutant (Sari et al. 2000) have offered indications that in the denatured state under native conditions, some of the molecules could possess some native-like topology. Finally, the existence of residual structures in certain regions of denatured barnase showed a good correlation with the regions found to fold early by protein engineering methods (Freund et al. 1996), which suggests that they can play an important role in the folding initiation. These results are also in good agreement with theoretical simulations that underline the importance of these residual structures in nucleating the folding process (Onuchic et al. 1995; Fersht 1997; Shakhnovich 1998; Thirumalai and Klimov 1998; Radford and Dobson 1999). The good agreement between theoretical results (lattice models, molecular simulations) and experimental data (folding studies at the residue level by protein engineering and NMR) has made possible a reliable representation of the first steps in the folding process and outline the role of sequence regions with restricted flexibility in the initiation process.
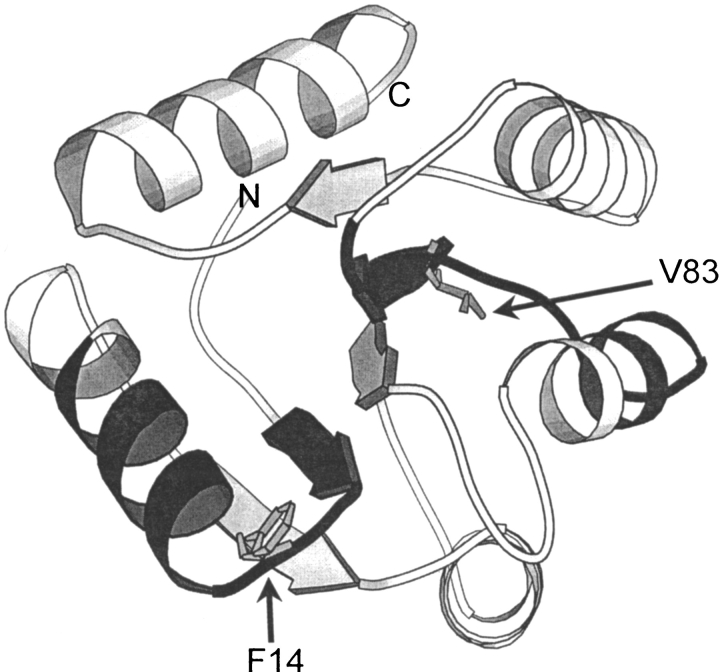
We present a structural description of the denatured state of a signal transduction chemotactic protein from Escherichia coli, CheY, and its relation with the initiation of the folding process. Three-dimensional X-ray crystal (Stock et al. 1993) and NMR solution (Moy et al. 1994; Santoro et al. 1995) structures of CheY, a 129-residue, α/β parallel protein, are available (Fig. 1 ▶). Its folding transition has been characterized thermodynamically (Filimonov et al. 1993) and its internal dynamics analyzed by H/D exchange combined with NMR (Lacroix et al. 1997). CheY belongs to the extremely restricted family of proteins for which the transition states have been structurally characterized by protein engineering methods (López-Hernández and Serrano 1996) and lattice model simulations (Mirny et al. 1998). In the protein engineering study, a partially folded structure of CheY folding transition state was deduced from the φ-value analysis of the folding/unfolding reaction. According to these data, the first half of the polypeptide chain would fold first, when the second half is still completely disorganized. In the transition state, a folding subdomain 1, up to β-strand 3, would be partially structured, on which the second half of the protein architecture would then condense. Two "folding subdomains" were then defined according to the folding pathway even when the native protein corresponds to a single, globular, domain (Lacroix et al. 1997). A nucleation/condensation mechanism, in which the folding nucleus corresponds to the packing of the first α-helix against β-strands 1 and 2, was proposed for the folding process. In a recent theoretical work (Mirny et al. 1998), lattice model simulation was used to investigate fast-folding dependence within the superfamily that includes CheY (Classical Monophosphate Binding Fold). The main conclusion of this study was that a small number of residue positions are crucial for a fast folding. Interestingly, there is a striking correlation between the positions detected theoretically and the residues participating in the folding nucleus observed experimentally.
Fig. 1.
Ribbon diagram of native CheY three-dimensional structure. Residues replaced in the mutant studied in the present work (F14N/V83T) are illustrated by their side chains. The two regions that show nonrandom coil behavior in the denatured protein (from residue 8 to 22 and 70 to 88) are represented in black. The diagram was produced using the program MOLSCRIPT (Kraulis 1991).
A good way to investigate the very first steps of a protein folding is to establish a correlation between the conformation of the protein at the starting point of this process, i.e., the denatured state, and the first elements of structure detected experimentally during folding. Because the CheY folding process has been extensively investigated, it should be very interesting to know if the denatured state contains clues about the earliest folding events and further compare them with the characterized transition state. This approach is somewhat different from other studies performed in milder denaturing conditions that favor a weak folding and formation of small structures that represent putative early folding steps (Zhang and Forman-Kay 1997; Blanco et al. 1998, Mok et al. 1999; Sari et al. 2000).
Previous studies of the folding transition of CheY mutants showed that the double mutant Phe14→Asn/Val83→Thr (here after F14N/V83T) has a particularly low equilibrium m-value when compared to the wild-type protein. m-values are approximately proportional to the change in buried surface that occurs between the unfolded and native states (Greene and Pace 1974). Once checked by circular dichroism (CD) and NMR that the native structure of the protein is conserved, the small m-value observed suggests that this mutant is likely to present a more compact unfolded state than the wild-type CheY. In this work, mutant F14N/V83T has been characterized by small angle X-ray scattering (SAXS) and NMR in the presence of 5 M urea, a concentration for which spectroscopic techniques such as CD or fluorescence show no evidence for structure. It is important to emphasize here that such studies in strong denaturing conditions do not determine stable and well defined structures but rather tendencies to "escape" the random coil conformation. The interesting aspect of these results is then the correlation of these indications with the conformation of early folding species such as the transition state, already characterized in the case of CheY. The evidence provided here on the existence of two subdomains can be easily correlated with the two subunits observed in folding of CheY. Moreover, it appears that residues 10–20, located in the first subdomain, show a nonrandom conformation that could be involved in the folding nucleus detected by protein engineering studies.
Results
Selection of a suitable CheY mutant
Equilibrium m-values are related to the difference in solvent accessibility between the folded and denatured states (Greene and Pace 1974). In the study of the folding pathway of CheY (López-Hernández and Serrano 1996) similar m-values were found for the majority of the mutants (within the experimental error). However, two mutants, F14N/V10T and F14N/V83T, showed significantly lower values (1.10 ± 0.08 kcal.mol−1.M−1 and 1.57 ± 0.04 kcal.mol−1.M−1, respectively, as compared to 1.80 ± 0.07 kcal.mol−1.M−1 for the wild-type protein). These results suggest that the denatured state of these two mutants could be more compact than that of the wild-type protein, once checked that the native state is unmodified (see below). Therefore, these two mutants were good candidates for the characterization of residual structures that could be involved in a nucleation process.
The very low stability of mutant F14N/V10T did not allow a complete analysis by NMR spectroscopy. A preliminary study of the native state by two-dimensional (2D) homonuclear experiments showed that, under native conditions, this mutant actually corresponds to a mixture of denatured and native populations (data not shown). We then focused our study on the more stable F14N/V83T mutant. This mutant is folded in aqueous solution and unfolded at 5M urea as indicated by CD and fluorescence.
NMR analysis of native mutant F14N/V83T
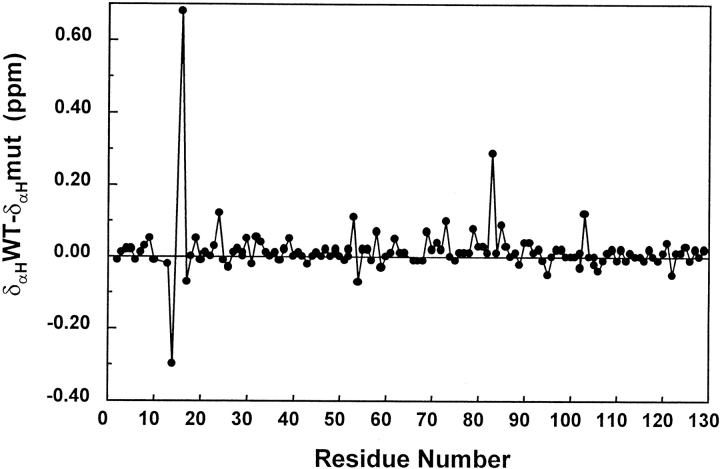
The NMR spectrum of F14N/V83T mutant shows that there are no important changes in the native structure as compared to the wild type. Indeed, no significant differences were observed between the chemical shifts of the wild-type protein and the mutant (Fig. 2 ▶), apart from the mutated residues and their sequential or spatial neighbors. The nuclear Overhauser enhancement (NOE) patterns defining the overall structure were carefully inspected and no significant change induced by the mutation was found. Also, the side-chain packing was checked and confirmed to be identical by the analysis of long-range NOE connectivities (data not shown). The three-dimensional fold of the single mutant F14N, as determined by NMR (Lacroix et al. 1997) and X-ray crystallography (Wilcock et al. 1998), is closely similar to the wild-type CheY solution structure. All this information strengthens the hypothesis that the lower m-value of this mutant is attributable to a compactation of the denatured state, or a folding intermediate accumulating in the midpoint of the transition and not to a substantial change in the three-dimensional (3D) structure.
Fig. 2.
Differences of Hα chemical shifts (ppm) between F14N/V83T mutant and wild-type CheY. Data are obtained under identical conditions (pH 7, 25°C).
Structural features of the denatured state: SAXS studies
The fact that small angle X-ray scattering technique provides useful low resolution information on the global structural features of denatured proteins has made it an important tool for folding studies (Sosnick and Trewella 1992; Chen et al. 1996; Smith et al. 1996a; Konno et al. 1997; Segel et al. 1998; Kamatari et al. 1999; Pollack et al. 1999; for review, see Lattman 1994; Kataoka and Goto 1996). Such information allows the determination of the overall dimensions, radius of gyration and shape of the polypeptide chain.
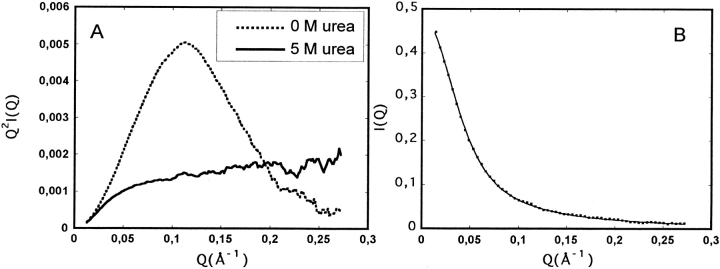
In its native form, mutant F14N/V83T has a radius of gyration (Rg) of 14.8 ± 0.2 Å, whereas in 5M urea a drastic increase of Rg up to 38.0 ± 1.0 Å is observed. Above 5M urea, Rg does not change within experimental error. Moreover, the scattering profiles at 5 and 7 M urea are nearly identical, which indicates that the protein does not undergo any major conformational change above 5M urea.
Additional structural information can be gained from SAXS data by inspecting the entire scattering patterns. In this respect, the Kratky representation (I(Q)Q2 vs. Q) gives rise to three different features, according to the nature of the protein conformation: First, when plotting I(Q)Q2 vs. Q, the scattering profile of a globular protein exhibits a characteristic bell-shape at high Q-values, because the scattering function satisfies Porod's law in a certain range of Q (i.e., I(Q) is proportional to 1/Q4). This is the case of CheY in native conditions represented in Figure 3A ▶ by the dashed line. On the contrary, the scattering profile of a Gaussian chain (equivalent to a random coil in the case of a infinitely thin chain) would display a plateau at high Q-values in this type of representation because I(Q) is proportional to Q2. A third instance corresponds to a persistence length chain model, where short-range interactions between adjacent segments produce stiffness of the chain. In this case, the Kratky representation gives an upward deviation from the profile of a Gaussian chain which depends on values of the apparent radius of gyration of cross-section, Rc. Such behavior is observed for CheY mutant in 5 M urea as shown in Figure 3A ▶ by the solid line.
Fig. 3.
(A) Kratky plots, I(Q)Q2 vs. Q, of mutant F14N/V83T in native conditions (dashed line) and of mutant F14N/V83T in 5 M urea (solid line). Each curve was normalized by the I(0) value obtained from the Guinier or Debye analysis. Both lines are the results of smooth fitting of the experimental data. (B) Experimental SAXS data (I(Q) vs. Q) for CheY in 5M urea. The solid line is the result of the fitting with equation (2).
A simple model to describe the SAXS spectrum of a persistence length chain of finite cross-section is the wormlike chain model of Porod and Kratky (Kratky and Porod 1949) with a finite apparent radius of gyration of cross-section Rc. The chain is regarded as rigid over a length b, which is called the statistical chain element. b is twice the persistence length. The chain can be considered as a sequence of N freely jointed rigid rods of length b. Its contour length is L = Nb. It may be noted that the contour length, L, is by no means the end-to-end distance between the amino and carboxyl termini of the polypeptide chain. It rather represents the sum of all the rigid segments lengths, b. According to Brûlet et al. (1996), the reduced structure factor I(Q)/I(0) of such a system can be approximately written as:
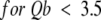 |
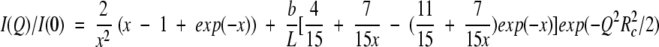 |
1 |
with x = Q2Lb/6
and for Qb > 3.5
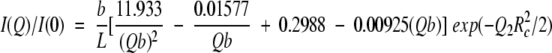 |
2 |
These expressions are valid for persistence chains without excluded volume effects, i.e., without long-range interactions between nonadjacent segments along the chain. However, they still can be used for chains with excluded volume interactions provided that N = L/b is rather small. In this case, the value obtained for the statistical element b is then slightly overestimated because the excluded volume interactions lead to an apparent increase of the stiffness.
Fitting the SAXS I(Q) vs. Q curve obtained for mutant F14N/V83T in 5 M urea to equation (2) (Fig 3 ▶) provides the following values for the L, b and Rc parameters: L = 340 ± 50 Å, b = 28 ± 3 Å and Rc = 3.3 ± 0.5 Å. A more "biological" interpretation of these parameters is that the denatured protein would be represented by 12 rigid segments of 28 Å connected to each other by flexible links and the sum of all these segment lengths is 340 Å.
It is interesting to compare these values with the parameters calculated for a fully expanded polypeptide chain of n amino acids without residual structure elements. In this case, the contour length, L, gives important information. For a random coil, L corresponds to the sum of all amino acid length: L = fnl. The factor f depends on bond angles in the real chain because, when fully extended, the chain is not a straight line but a zigzag line. For a polypeptide chain, f = 0.95 (Cantor and Schimmel 1980). Consequently, the contour length calculated for a fully expanded polypeptide chain of 129 amino acids is: L = 0.95 × 3.8 × 129 = 466 Å. Interestingly, the experimental value obtained for CheY mutant F14N/V83T in 5 M urea, L = 340 ± 50 Å, is significantly smaller than the value expected for a fully expanded polypeptide chain of 129 amino acids. This indicates that the chain is locally more compact than an extended random coil. The comparison between these two L values suggests that about 25% of the residues may be involved in residual structures.
Finally, SAXS studies of CheY mutant F14N/V83T in 5 M urea were useful for another purpose, as this technique is extremely sensitive to aggregation. Because protein concentrations used for data acquisition are of the same order as the samples used for NMR experiments, it was checked that the protein in these conditions did not suffer any aggregation. This process gives rise to an anomalous increase of I(Q) at small Q values on SAXS data (I(Q) vs. Q). As shown by the fitting presented in Fig. 3B ▶, equation 2 (which does not include any parameter concerning aggregation) describes accurately the experimental data, even for small Q values. As a consequence, it appears that the CheY Y mutant samples under strongly denaturing conditions (above 5 M urea) did not show any process of aggregation.
Assignment of F14N/V83T mutant in 5 M urea
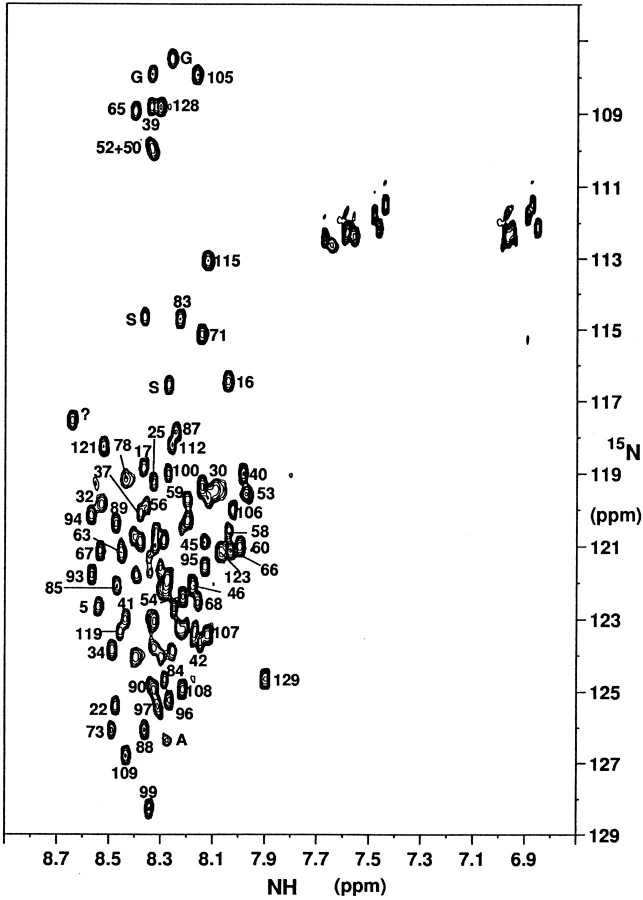
Assignment of the 1H and 15N resonances of CheY mutant F14N/V83T in 5 M urea has been achieved by the combined use of 3D TOCSY-HSQC and NOESY-HSQC experiments, through the classical sequence-specific assignment methods (Wüthrich 1986). As expected, resonances of all amide protons in the HSQC spectrum are clustered in a narrow 0.7 ppm range, between 7.9 and 8.6 ppm (Fig. 4 ▶). The 15N chemical shift dispersion is larger: ∼10 ppm for all amide protons of residues other than glycines. In the TOCSY-HSQC spectrum, almost all side chain protons are correlated to their intra-residue amide protons and resonate at the chemical shifts expected for unstructured peptides (Wishart et al. 1995). This fact, together with the detection of most sequential dαN cross-correlations in the NOESY-HSQC spectrum, allowed for the assignment of 114 amide 1H and 15N cross peaks and the corresponding proton spin systems, out of a total of 125 (after discounting the amino terminus and the three prolines).
Fig. 4.
15N-1H HSQC spectrum of mutant F14N/V83T denatured in 5 M Urea, at pH 7 and 25°C. Peaks are labeled with the residue number. Nonassigned peaks are labeled with a question mark. Signals associated with an identified amino-acid type but not sequentially assigned are labeled with the letter corresponding to the amino-acid type.
Deviation of chemical shifts from random coil values
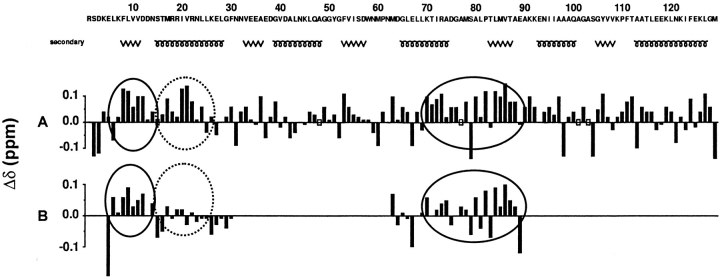
Conformational shifts are defined as the difference between the observed chemical shifts of nuclei and the ones measured in short peptides with a random coil distribution (Wishart et al. 1995). They are useful parameters to define regular secondary structure in folded proteins and to provide effective information on preferential conformations in unfolded proteins (Shortle 1996). The effect of 6 M urea on Hα chemical shifts of random Gly-Gly-X-Ala peptides has been shown to be negligible, just causing an average increase of 0.01 ppm for the central X residue (Jiménez et al. 1994). A more extensive study of small peptide chemical shifts in 8 M urea has been published recently (Schwarzinger et al. 2000). However, the highly acidic conditions of this study do not make it appropriate to use it as a good reference in our study, particularly for the residues more sensitive to important pH variations. The Hα conformational shifts of CheY mutant F14N/V83T in 5 M urea are illustrated in Figure 5A ▶. Most of the backbone chemical shifts show no significant deviation from random coil values. A few isolated residues show marked downfield or upfield deviations (A36, F53, M63, A98, S104, Y106, A113, and L127), but the fact that these residues are isolated makes it unsafe to draw any conclusion about the existence of residual structures. However, two regions, centered at S15 and M78, show an overall trend indicative of a nonrandom conformation. The first region, from residue 8 to 22, contains six residues with Hα conformational shifts equal to or higher than 0.1 ppm. The second region, 70 to 88, contains eight residues showing values equal to or higher than 0.1 ppm. Even though a 0.1 ppm chemical shift deviation is not usually indicative of structure in native protein studies, that value must be taken into consideration for disordered proteins (Schwalbe et al. 1997; Sari et al. 2000) especially when residues showing these shifts are grouped in the peptide sequence. In this respect it should be noted that the two regions mentioned above, which represent 25% of the sequence length, contain more than 50% of residues with conformational shifts ≥ 0.1 ppm.
Fig. 5.
Hα chemical shifts deviation from random coil values (Δδ = δexperim. − δrandom-coil) as a function of protein sequence. (A) Entire mutant F14N/V83T; (B) isolated peptides spanning regions 5–30 and 62–89 of the mutant. Experimental values are obtained for mutant F14N/V83T and peptides under 5 M urea. Random coil values are from Wishart et al. 1995. Regions showing an overall trend to be in a nonrandom conformation in the mutant and conserving this tendency in the peptides are displayed with solid circles. Regions loosing this trend in isolated peptides are displayed with dashed circles. The secondary structure in the native protein is also shown.
Peptides spanning regions 5–30 and 62–89
To check whether the clusters detected by the conformational shift analysis are stabilized by local or nonlocal interactions, two peptides spanning these regions were analyzed under identical denaturing conditions, 5 M urea. These peptides were designed to span the two clusters and a few additional residues. Therefore, hypothetical local interactions between residues involved in these regions and residues at their edges are taken into consideration, as well as minimizing any end-effects. The analyzed peptides correspond to residues 5 to 30 and 62 to 89 in the mutant protein. The TOCSY and NOESY spectra of the two peptides in 5 M urea also showed a strong overlap but, due to the small size of the molecule, the complete assignment could be achieved by homonuclear 2D NMR.
The conformational shifts of α-protons in the intact protein and in the isolated peptides are compared in Figure 5B ▶. The Hα conformational shift profile of the second peptide (63–89) is closely similar to the one observed in the intact protein, also showing upfield values around residues 70–88. This suggests that the putative residual structure observed in the full mutant is maintained in the peptide and it is mainly caused by local interactions.
The case of the 5–30 peptide is more straightforward. Only residues 8 to 12 show significant positive Hα conformational shifts in the isolated peptide. Residues 13 to 23 display values close to zero, indicating that their conformations are distributed randomly across the Ramachandran map. Interestingly, the major changes are observed for residues 20–21. The conformational shifts very close to zero determined for the isolated peptide reinforce the significance of the deviation observed in the entire polypeptide.
In conclusion, the comparison of the Hα conformational shifts in the isolated peptides and in the entire protein shows that residual structures around Ile20-Val21 in the intact CheY mutant would be stabilized by interactions between regions distant in the polypeptide chain. Conversely, local interactions would stabilize the cluster observed around residues 70–88. This conclusion is consistent with the different dynamics observed for the two subdomains in the unfolded protein (see below).
Heteronuclear relaxation data
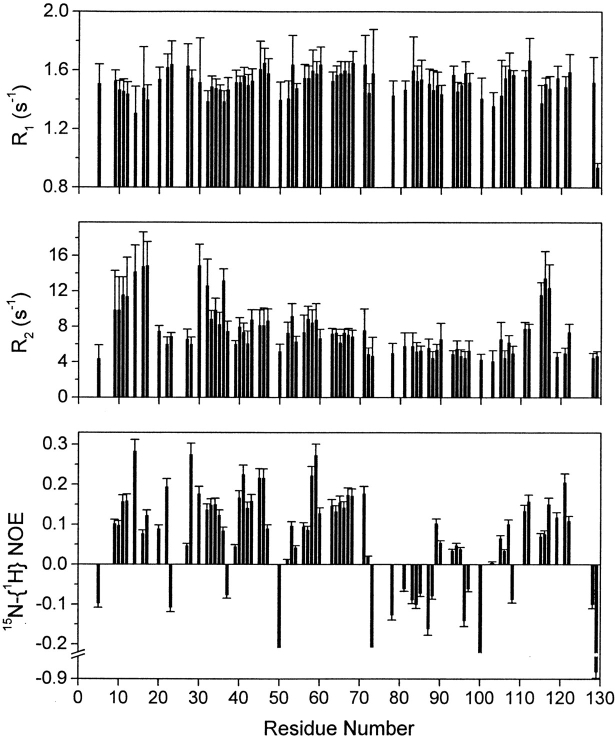
15N relaxation is a powerful tool for the study of protein dynamics. Backbone flexibility on the ns to ps time scale can be qualitatively described on the basis of 15N-{1H} NOE measurements. These data, presented in Figure 6 ▶, provide the clearest evidence for the existence of two different subdomains in the unfolded protein, even in a 5 M urea solution.
Fig. 6.
Relaxation data of backbone amide 15N in mutant F14N/V83T denatured in 5 M urea plotted as a function of protein sequence. R1: Longitudinal relaxation rates; R2: Transverse relaxation rates. Heteronuclear 15N-1H NOEs of backbone amide groups.
Highly rigid conformations are expected to give rise to 15N-{1H} NOE values up to 0.82 at a 15N resonance of 60 MHz (Kay et al. 1989) and the lower the value, the more dynamic the backbone. Negative values indicate a highly mobile chain. The distribution of heteronuclear NOEs along the sequence clearly shows that the denatured protein can be divided in two regions. In the first half of the protein (up to residue 70) heteronuclear NOEs are mainly positive. In the second half of the protein, heteronuclear NOEs are negative or close to zero, with the exception of residues between Phe111 and Lys122, which show slightly positive values. This suggests that the first half of the sequence behaves as a better structured entity with a slower effective tumbling than the second half.
A more quantitative description of the backbone dynamics is generally obtained for native proteins using the model free approach (Lipari and Szabo 1982). Although this method is very powerful for extracting useful dynamical parameters from relaxation data of folded proteins, its applicability to an unfolded chain, which is unlikely to tumble isotropically in solution, does not seem appropriate (Alexandrescu and Shortle 1994; Farrow et al. 1995; Wong et al. 2000). An alternative way to quantitatively interpret 15N relaxation data is the analysis of the value of the spectral density functions themselves by using the reduced spectral density mapping technique (Peng and Wagner 1992).
J(0), J(ωN) and J(ωH) (Fig. 7 ▶) have been determined from longitudinal (R1 = 1/T1), transverse (R2 = 1/T2) relaxation rates and heteronuclear 15N-{1H} NOEs measured for 77 out of the 125 observable backbone amide 15N nuclei. Even though 114 amide cross-peaks have been identified on the 3D spectra, the poor dispersion observed in the central region of the HSQC spectra (Fig. 4 ▶) severely hindered the determination of relaxation rates for 37 amide 15N-1H cross-peaks. However, the information on the backbone dynamics is well distributed all along the sequence and is representative of all the regions in the denatured protein.
Fig. 7.
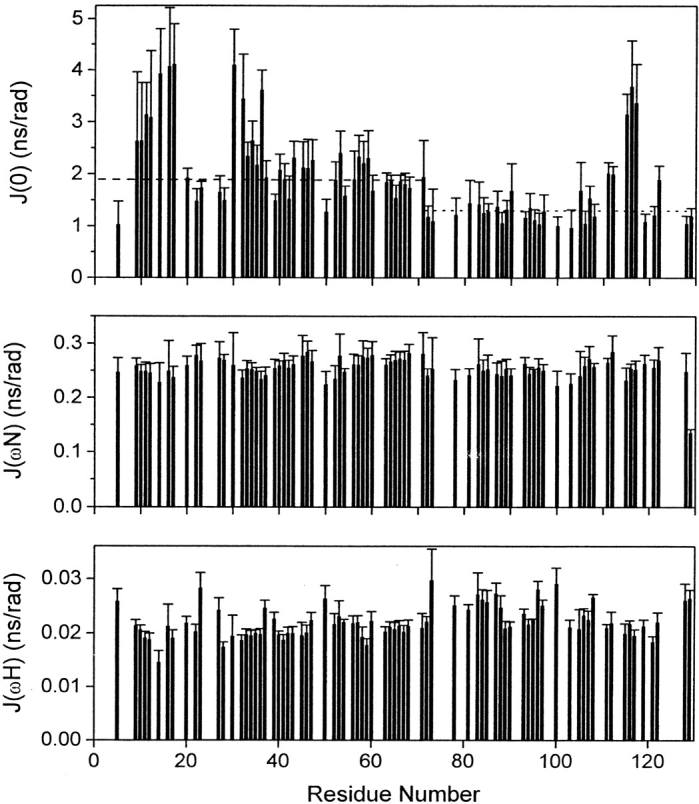
Calculated values of the reduced spectral density functions of residues as a function of protein sequence for CheY mutant F14N/V83T denatured in 5 M urea. J(0), J(ωN) and J(ωH) are the spectral densities at frequencies 0, 60 and 540 MHz, respectively. Dashed line represents the mean J(0) value calculated for residues 1–70, < J(0)1–70 > (excluding residues that show highest J(0) values: 9–17 and 30–36). Dotted line represents the mean J(0) value calculated for residues 71–129, < J(0)71–129 > (excluding residues that show highest J(0) values: 115–117).
J(0) has been demonstrated to be the most sensitive probe detecting dynamic heterogeneity along the protein backbone and has also been applied to the characterization of the dynamic properties of unfolded systems (Farrow et al. 1995; Meekhof and Freund 1999; Wong et al. 2000). Inspection of J(0) versus sequence supplies a rapid overview of the relative mobility along the protein backbone. In general, anomalously high J(0) values indicate significant chemical exchange contribution (Rex), whereas anomalously low J(0) values indicate the presence of fast internal motions. In the case of CheY, large variations of J(0) values are observed along the sequence. Nevertheless, the same trend is not followed by J(ωN) and J(ωH). NH vectors experiencing J(0) values anomalously high (J(0)> 2.77 ns/rad) with respect to the mean (J(0) = 1.95 ± 0.09 ns/rad) are located in three regions: from residues 9 to 17, from residues 30 to 36 and from residues 115 to 117. Because the same regions show high R2 values, the higher J(0) values are likely to be the result of a chemical exchange process (Wong et al. 2000). The rest of the sequence shows less variation in the J(0) values. However, it is clear from the data represented in Figure 7 ▶, that the mean value of residues up to 70 (excluding those with significant Rex contribution) is slightly higher than the corresponding of the second half (< J(0)1–70 >= 1.90 ± 0.06 and < J(0)71–129 >= 1.32 ± 0.05 ns/rad, respectively).
In conclusion, 15N relaxation studies show that three regions are associated with a slower motion on the millisecond time scale, probably arising from a chemical exchange process, and that the molecular motion of the first part is restricted with respect to the second part, on a nano- to picosecond time scale.
Indeed, it is not surprising that the collapse stabilized by nonlocal interaction, suggested on the basis of the chemical shifts, is located in the first subdomain of the unfolded protein, which appears to have a more restricted motion.
Discussion
A structural characterization of the denatured state of CheY mutant F14N/V83T has been undertaken here to investigate the folding initiation process. As stated above, mutant F14N/V83T was selected because of its larger probability of forming a more compact denatured state. The folding properties of this mutant were studied by SAXS and NMR in 5 M urea, a denaturant concentration for which neither secondary nor tertiary structure were detected by CD or fluorescence measurements. The first mutation (F14N) allows for the comparison with the kinetic folding pathway described previously (López-Hernández and Serrano 1996), as that study was based on the "pseudo wild-type" protein, already containing that mutation. The relevance of the second mutation (V83T) for the initiation of CheY folding will be discussed later. The location of these two mutations in the native structure is presented in Figure 1 ▶.
SAXS studies of the denatured CheY mutant showed that the protein in 5 M urea is well described as a wormlike chain composed by rigid elements connected to each other by flexible links. In addition, the contour length of the chain obtained by the scattering function analysis indicates that 25% of the residues could be involved in residual structures. This study also showed that aggregation is likely to be negligible, even in samples at protein concentration corresponding to NMR experiments.
The NMR spectrum of CheY mutant F14N/V83T under 5 M urea shows a very low dispersion of HN chemical shifts, suggesting a disordered conformation. As stated above, studies of proteins in strong denaturing conditions rarely provide information on well-defined structures. Instead, NMR parameters can be interpreted as indications for a region of the sequence to be in a nonrandom conformation or to present a restricted mobility. For that purpose, an analysis of backbone dynamics, using the reduced spectral density function approach and heteronuclear NOEs, is very helpful.
Clear evidence is provided here by the heteronuclear NOEs measurements, that the unfolded state of CheY mutant F14N/V83T shows two distinct subdomains, in terms of motion. The NOE behavior of NH vectors of the first half of the polypeptide chain, up to residue 70, suggests a lower mobility, when compared to the second half. In addition, two regions of interest appear in the NMR chemical shift profiles. The first cluster corresponds to residues 8–22 and is therefore located in the first part of the polypeptide chain. This region, which corresponds to the first β-strand and half of the first α-helix in the native protein, shows upfield deviations of the α-protons chemical shifts and larger J(0) values. The second region that can be traced out from Hα conformational shift measurements, 70–88, is located in the second, more dynamic, subdomain.
Backbone dynamics studies, based on the spectral density function method show three regions that may be in chemical exchange with more rigid conformations, giving rise to a slower motion on the micro- to millisecond time scale. The observation of the J(0) variation along the sequence also shows that this parameter is on average higher for the first half of the residues. Here again, the first subdomain in the denatured state shows itself to be less dynamic than the second one.
The general agreement observed in all NMR data for the first cluster indicates that it does not correspond to a random coil conformation. To determine the local or nonlocal nature of the interactions that stabilize that cluster, a comparison was made with the results obtained for an isolated peptide corresponding to that region. The conformational shifts profile then obtained clearly show that residues 13–23 are likely to be in a totally random coil conformation in the isolated peptide. This suggests that the cluster around residues 20–21 is partially stabilized by nonlocal interactions in the unfolded state of the intact protein.
Contrary to what is common in folded proteins, evidence for specific long-range interactions are not easy to obtain for proteins under denaturing conditions. 3D spectroscopy is necessary to overcome the strong overlap of cross peaks in 2D spectra. However, 15N single labeling is not very helpful in this concern, as 15N-edited NMR experiments do not allow for measurements of NOE cross-correlations between aliphatic and/or aromatic protons in side chains. These cross-correlations often respond to hydrophobic interactions that have a predominant role in the stabilization process of residual structures. Observation of NOEs between methyl and aromatic protons, for example, are invaluable for structure determination (Neri et al. 1992). It is then very unfortunate that NOE cross-correlations between the methyl groups of valines, leucines, and isoleucines and aromatic protons cannot be observed in NOESY-HSQC spectra. Thus, possible long-range interactions between residues belonging to the cluster around residues 20–21 and residues outside this sequence are not observed, even though they may actually exist. Furthermore, averaging of NOEs by a population of highly disordered conformers significantly lowers the intensity of the signals that could be observed otherwise (Wüthrich 1994). The NMR study of denatured CheY mutant F14N/V83T illustrates well this feature. Nonsequential NOEs between amide and methyl protons are occasionally observed in the NOESY-HSQC spectrum. However, the extremely poor dispersion of methyl proton chemical shifts of valines, leucines, and isoleucines (all of them in the range δ = 0.88–0.92 ppm) prevents the assignment of these correlations to individual residues, with the consequent loss of structural information.
In conclusion, all the available data suggest that the first half of the protein is less mobile than the second half, defining therefore two different subdomains in the denatured state. Within the first subdomain, a region with certain tendency to form transient structures stabilized by nonlocal interactions can also be pointed out. A second region, located in the more mobile subdomain, also shows signs of adopting partially nonrandom conformation, although this would be mainly stabilized by local interactions.
Restricted mobility and folding initiation
The data presented here are in good agreement with the results obtained from kinetic (López-Hernández and Serrano 1996) and H/D exchange studies (Lacroix et al. 1997) on the CheY wild-type and F14N mutant, which also indicate the existence of two folding subdomains in CheY. The correlation of the data obtained in the present study with the early folding species characterized previously gives important information about the origins of the very first folding events.
Overall, there is a good correlation between the differences of the dynamic behavior observed in the first and second part of unfolded protein and the characteristics of the transition state described previously (López-Hernández and Serrano 1996). The lower mobility of the first half of the unfolded protein suggests that the compact first subdomain present in the transition state may find its origin in the denatured state. Moreover, the cluster detected in this subdomain may also play a role in the formation of the folding nucleus reported previously. Given the absence of well-defined structures in the unfolded state, the early event in the folding pathway of this protein may well be a collapse favoring the formation of longer range interactions by keeping key residues closer together.
The role of the cluster involving residues 70–88 is less straightforward. Residues 86, 88, and 89 were observed to participate in a folding nucleus in a simulation of CheY folding (Mirny et al. 1998). However, experimental studies show no evidence for these residues to be involved in the nucleation process (López-Hernández and Serrano 1996). This region may thus be slightly more compact in the unfolded state, but it would not necessarily act as an initiation site. This might be caused by the higher dynamics of this region (when compared to the "productive" region 10–20) and the absence of longer range interactions, which are necessary to convert embryonic initiation sites into nucleation centers, as proposed by the nucleation-condensation mechanism (Fersht 1997). Also, the higher mobility of the second subdomain might hinder the formation of a folded subdomain. A similar fact has been reported previously in a study of initiation sites for the folding of barnase (Freund et al. 1996), in which the residual structure found in the second helix region did not turn out to be productive. It is also interesting to note that this region corresponds to the second mutation introduced in this protein (V83T). This, in part, hinders the comparison with respect to the pseudo wild-type protein (already containing the mutation F14N) and would explain why the cluster 70–88, probably arising from the mutation, does not correspond to any early event in the folding pathway of the wild-type protein.
The above structural description is in agreement with the nucleation-condensation mechanism for protein folding (Fersht 1997). Transient local collapses in the denatured state would facilitate the setting up of longer-range interactions, funneling the reaction toward the transition state and narrowing the search for the native structure.
Materials and methods
Protein expression and purification
The mutant F14N/V83T was designed, cloned, and sequenced as described previously (López-Hernández and Serrano 1996). The plasmid containing the mutated gene was transformed into a BL21 E. coli strain. Cells were grown on minimal medium with 15NH4Cl as the only nitrogen source and 15N-labeled proteins were purified as described previously (Filimonov et al. 1993).
Peptide synthesis
Peptide #1 (26 residues) and peptide #2 (28 residues) correspond to residues 5 to 30 (inclusive) and 62 to 89 (inclusive) in the mutant protein, respectively. Their molecular weights are 3105 kD and 2960 kD, respectively. The amino terminus of each peptide is unmodified, whereas the carboxyl terminus is an amide group.
The peptides sequences are: #1: Glu5-Leu-Lys-Phe-Leu-Val-Val-Asp-Asp-Asn-Ser-Thr-Met-Arg-Arg-Ile-Val-Arg-Asn-Leu-Leu-Lys-Glu-Leu-Gly-Phe30-NH2;
#2: Asn62-Met-Asp-Gly-Leu-Glu-Leu-Leu-Lys-Thr-Ile-Arg-Ala-Asp-Gly-Ala-Met-Ser-Ala-Leu-Pro-Thr-Leu-Met-Val-Thr-Ala-Glu89-NH2. The peptides were synthesized in the laboratory of Dr. R. Frank at the Centre for Molecular Biology, Heidelberg University (ZmBH) using standard 9-fluorenyl methocarboxyl chemistry. Purification ≤95% was achieved by reverse high pressure liquid chromatography. The molecular mass of the peptide was confirmed by mass spectrometry.
SAXS experiments
Data acquisition
Data were collected on the small angle scattering instrument D24 at LURE (Laboratoire pour l'Utilisation du Rayonnement Electromagnétique, Orsay, France) using the synchrotron source. The wavelength λ of the X-rays was selected by a bent Ge(111) monochromator and adjusted to 1.488 Å, calibrated by the nickel absorption edge. X-ray patterns were recorded by a linear position-sensitive detector filled with a 90% Xe/10% CO2 gas mixture. The sample-to-detector distance was 1320 mm corresponding to the scattering vector range: 0.012 Å−1 < Q < 0.27 Å−1 (where Q = 4πsinθ/λ, 2θ being the scattering angle). The sample was placed in a quartz capillary of 1.2 mm diameter. The temperature was controlled (T = 20°C) via water circulation and the background was considerably reduced by evacuating the cell. To avoid radiation-induced damage, an automatic drive for the syringe connected to the cell was installed. The displacement of the sample was adjusted in such a way that protein molecules would be exposed to the X-ray beam for <2 min. Several successive frames (usually 16), of 200 s each, were recorded for both the sample and the corresponding buffer. Each frame was carefully inspected to check the absence of X-ray damage.
All SAXS experiments were performed in a 75 mM phosphate buffer containing 1 mM Dithiothreitol (DTT) at pH 8. Samples were prepared so that the urea concentration was 5 or 7 M. Urea concentration was checked after acquisition of data by refractometry, using the relation provided by Warren and Gordon (1970). The concentration of CheY F14N/V83T mutant was ∼5 mg/ml.
SAXS data treatment
Each scattering spectrum was normalized for the detector response. It was also normalized for the transmitted intensity by recording the scattering from a reference carbon-black sample and integrating its scattering over a given angular range. For each urea concentration the buffer solution was measured as background and its curve was subtracted from the corresponding protein sample curve.
SAXS data were collected to determine the overall dimensions (radii of gyration and shape) of CheY in native and denaturing conditions (5 and 7 M urea). For the native (0 M urea) globular protein, the radius of gyration Rg was calculated according to the Guinier approximation (Guinier and Fournet 1955):
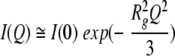 |
3 |
The fitting region spans from 0.013 to 0.090 Å−1 in Q. The scattering intensity at zero scattering angle, I(0), is proportional to the square of the excess electron density ρp-ρ0 between the protein molecule and the buffer background following the relation:
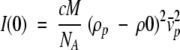 |
4 |
where c is the protein concentration, M the molecular weight, NA Avogadro's number and the partial specific volume of the protein. A careful examination of I(0) values allows us to check the absence of aggregation or association of molecules in the sample.
For an unfolded protein, such as a random coil, the Guinier approximation cannot be used because the previous approximation (3) is valid only for QRg ≤ 1. In this case, the scattering profile can be described at first approximation in the small Q region (QRg ≤ 3) by the Debye function (Calmettes et al. 1994):
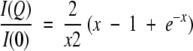 |
5 |
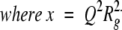 |
This equation was used to determine the Rg values for denatured CheY F14N/V83T in 5 and 7 M urea.
The concentration of the protein solutions used in this work was about 5 mg/ml in a 75 mM phosphate buffer containing 1 mM DTT. We expect that in these conditions the repulsive interactions between molecules are completely screened. Consequently, the SAXS scattering profiles are not significantly affected by interference effects. To confirm this point we calculated the scattering I(Q) curve at zero concentration for native CheY mutant using the CRYSOL program (Svergun et al. 1995) and the atomic coordinates of the F14N/V86T mutant (pdb 1ab6) (Wilcock et al. 1998) with a sequence very close to the mutant studied here. The good agreement between experimental and calculated curves corroborates the absence of significant interference effects in the native CheY mutant sample.
Denaturation experiments
All experiments were performed in a buffer containing 25 mM sodium phosphate at pH 7. All samples were prepared so that the urea concentration in the NMR tube was 5 M. Urea concentration was checked after acquisition of data by refractometry, using the relation provided by Warren and Gordon (1970). The final concentration of CheY F14N/V83T mutant was 1.6 mM for the 3D NMR experiments and 2.0 mM for 15N relaxation and heteronuclear NOE experiments. The temperature was set at 25°C. For the peptides, the concentration in the NMR tube was 2 mM. A set of TOCSY and NOESY spectra was recorded at 5°C for each peptide, so as to increase the NOE intensities. An additional TOCSY spectrum was recorded at 25°C to compare the data corresponding to the intact F14N/V83T mutant. All samples contained a minute amount of sodium 3-Trimethyl-silyl(2,2′,3,3′-2H4)-propionate (TSP) as the internal chemical shift reference.
NMR spectroscopy: Data acquisition and processing
All experiments were performed using a Bruker AMX-600 spectrometer with a 1H operating frequency of 600.13 MHz.
2D TOCSY (Bax and Davis 1985), NOESY (Kumar et al. 1980), and 15N-HSQC (Bodenhausen and Ruben 1980) experiments used for the native mutant F14N/V83T were recorded at 25°C using standard pulse sequences. Water suppression was achieved by selective presaturation of the water signal or by including the WATERGATE module (Piotto et al. 1992) in the original pulse-sequences. Mixing times were set to 80 ms and 50 ms in the TOCSY and NOESY experiments, respectively.
3D 15N-HSQC-TOCSY and 15N-HSQC-NOESY (Driscoll et al. 1990) spectra used for the unfolded mutant in 5 M urea were recorded with mixing times of 80 ms in the TOCSY and 200 ms in the NOESY experiments. The data set typically comprised 128, 64, and 1024 complex points in t1, t2, t3, respectively. Forward linear prediction was used to extend the data in the 15N dimension from 64 to 96 complex points and all 3D data sets were zero-filled to yield a final matrix size of 256, 128, and 2048 real data points.
All NMR data were processed and analyzed using standard Bruker software and NMRPipe/NMRDraw software (Delaglio et al. 1995). Various square sine bell functions with different shifts were applied along the data processing to obtain a good compromise between resolution and sensitivity.
Longitudinal (R1 = 1/T1) and transverse (R2 = 1/T2) relaxation rates as well as heteronuclear NOEs of 15N nuclei were measured using procedures described previously (Kay et al. 1989; Farrow et al. 1994). In the R1 and R2 relaxation measurements a series of eight experiments with relaxation times ranging from 40 to 1200 ms and 7 to 154 ms, respectively, was used. A relaxation delay of 2 sec was used for all relaxation experiments except for the NOE experiment in which the relaxation delay was 6 sec. The heteronuclear NOE effects were calculated as the ratio of peak heights in spectra recorded with and without 1H saturation. Relaxation rates were determined by fitting peak height data to single exponential decays. Errors in primary intensity data were taken as the standard deviation of spectral noise.
Reduced spectral density function analysis
The expressions given by Abragam (1961) relate the longitudinal and transverse relaxation rates and heteronuclear NOEs to the spectral density function at five distinct frequencies: 0, ωN, ωH − ωN, ωH, and ωH + ωN. The approach used in this study is close to the spectral density mapping method described by Peng and Wagner (1992), assuming that the spectral density function has a similar magnitude at high frequency. Thus, J(ωH)average = J(ωH − ωN) = J(ωH + ωN) = J(0.9.ωH). Values of the reduced spectral density function at 0 Hz (J(0)), 60 MHz (J(ωN)) and 540 MHz (J(ωH)) are calculated through measurements of 15N R1, R2 relaxation rates and 15N-{1H} NOE (Peng and Wagner 1992; Ishima and Nagayama 1995).
Acknowledgments
We thank Dr. Patrice Vachette (LURE) for his assistance during the SAXS experiments and for useful discussions about SAXS results, Dr. José Manuel Pérez-Cañadillas for performing the spectral density function analysis and Dr. Douglas Vinson Laurents for carefully reading the manuscript. This work was partially supported by an EU Human and Capital Mobility grant (contract N° ERBCHBGCI940530) and an EMBO short term fellowship (Ref. ASTF8611).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.52701.
References
- Abragam, A. 1961. In The principles of nuclear magnetism. Clarendon Press, Oxford, UK.
- Alexandrescu, A.T. and Shortle, D. 1994. Backbone dynamics of a highly disordered 131-residue fragment of staphylococcal nuclease. J. Mol. Biol. 242 527–546. [DOI] [PubMed] [Google Scholar]
- Arcus, V.L., Vuilleumier, S., Freund, S.M.V., Bycroft, M., and Fersht, A.R. 1995. A comparison of the pH, urea and temperature-denatured states of barnase by heteronuclear NMR: Implications for the initiation of protein folding. J. Mol. Biol. 254 305–321. [DOI] [PubMed] [Google Scholar]
- Bax, A. and Davis, D.G. 1985. MLEV-17-based two dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65 355–360. [Google Scholar]
- Blanco, F.J., Serrano, L., and Forman-Kay, J.D. 1998. High populations of non-native structures in the denatured state are compatible with the formation of the native folded state. J. Mol. Biol. 284 1153–1164. [DOI] [PubMed] [Google Scholar]
- Bodenhausen, G. and Ruben, D.J. 1980. Natural abundance 15N NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69 185–189. [Google Scholar]
- Bond, C.J., Wong, K.B., Clarke, J., Fersht, A.R., and Daggett, V. 1997. Characterization of residual structure in the thermally denatured state of barnase by simulation and experiment: Description of the folding pathway. Proc. Natl. Acad. Sci. 94 13409–13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brûlet, A., Boué, F., and Cotton, J.P. 1996. About the experimental determination of the persistence length of wormlike chains of polystyrene. J. Phys. II France 6 885–891. [Google Scholar]
- Calmettes, P., Durand, D., Desmadril, M., Minard, P., Receveur, V., and Smith, J.C. 1994. How random is a highly denatured protein? Biophys. Chem. 53 105–114. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry. Part III. The behavior of biological macromolecules. WH Freeman & Company, San Francisco.
- Chen, L., Hodgson, K.O., and Doniach, S. 1996. A lysozyme folding intermediate revealed by solution X-ray scattering. J. Mol. Biol. 261 658–671. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6 277–293. [DOI] [PubMed] [Google Scholar]
- Driscoll, P.C., Clore, G.M., Marion, D., Wingfield, P.T., and Gronenborn, A.M. 1990. Complete resonance assignment for the polypeptide backbone of interleukin 1 beta using three-dimensional heteronuclear NMR spectroscopy. Biochemistry 29 3542–3556. [DOI] [PubMed] [Google Scholar]
- Evans, P.A., Topping, K.D., Woolfson, D.N., and Dobson, C.M. 1991. Hydrophobic clustering in non native states of a protein: Interpretation of chemical shift in NMR spectra of denatured states of lysozyme. Proteins Struct. Funct. Genet. 9 248–266. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33 5984–6003. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Zhang, O., Forman-Kay, J.D., and Kay, L.E. 1995. Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry 34 868–878. [DOI] [PubMed] [Google Scholar]
- Fersht, A.R. 1997. Nucleation mechanism in protein folding. Curr. Opin. Struct. Biol. 7 3–9. [DOI] [PubMed] [Google Scholar]
- Filimonov, V.V., Prieto, J., Martínez, J.C., Bruix, M., Mateo, P.L., and Serrano, L. 1993. Thermodynamic analysis of the chemotactic protein from Escherichia coli, CheY. Biochemistry 32 12906–12921. [DOI] [PubMed] [Google Scholar]
- Fong, S., Bycroft, M., Clarke, J., and Freund, S.M. 1998. Characterisation of urea-denatured states of an immunoglobulin superfamily domain by heteronuclear NMR. J. Mol. Biol. 278 417–429. [DOI] [PubMed] [Google Scholar]
- Freund, S.M.V., Wong, K.B., and Fersht, A.R. 1996. Initiation sites of protein folding by NMR analysis. Proc. Natl. Acad. Sci. 93 10600–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, R.F., Jr. and Pace, N. 1974. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J. Biol. Chem. 249 5388–5393. [PubMed] [Google Scholar]
- Guinier, A. and Fournet, G. 1955. Small angle scattering of X-rays. Wiley, New York.
- Ishima, R. and Nagayama, K. 1995: Protein backbone dynamics revealed by quasi-spectral density function analysis of amide N-15 nuclei. Biochemistry 34 3162–3171. [DOI] [PubMed] [Google Scholar]
- Jiménez, M.A., Carreño, C., Andreu, D., Blanco, F.J., Herranz, J., Rico, M., and Nieto, J.L. 1994. Helix formation by the phospholipase A2 38–59 fragment: Influence of chain shortening and dimerization monitored by NMR chemichal shifts. Biopolymers 34 647–661. [DOI] [PubMed] [Google Scholar]
- Kamatari, Y.O., Ohji, S., Konno, T., Seki, Y., Soda, K., Kataoka, M., and Akasaka, K. 1999. The compact and expanded denatured conformations of apomyoglobin in the methanol-water solvent. Protein Sci. 8 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, M. and Goto, Y. 1996. X-ray solution scattering studies of protein folding. Folding Des. 1 R107–114. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Torchia, D.A., and Bax, A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28 8972–8979. [DOI] [PubMed] [Google Scholar]
- Konno, T., Kamatari, Y.O., Kataoka, M., and Akasaka, K. 1997. Urea-induced conformational changes in cold- and heat-denatured states of a protein, Streptomyces subtilisin inhibitor. Protein Sci. 6 2242–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme, T., Kelly, M.J., Kay, L.E., Forman-Kay, J., and Serrano, L. 1999. Similarities between the spectrin SH3 domain denatured state and its folding transition state. J. Mol. Biol. 297 1217–1229. [DOI] [PubMed] [Google Scholar]
- Kratky, O. and Porod, G. 1949. Röntgenuntersuchung Gelöster Fadenmoleküle. Recl. Trav. Chim. Pays-Bas Belg. 68 1106–1122. [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950. [Google Scholar]
- Kumar, A., Ernst, R.R., and Wüthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95 1–6. [DOI] [PubMed] [Google Scholar]
- Lacroix, E., Bruix, M., López-Hernández, E., Serrano, L., and Rico, M. 1997. Amide hydrogen exchange and internal dynamics in the chemotactic protein CheY from Escherichia coli. J. Mol. Biol. 271 472–487. [DOI] [PubMed] [Google Scholar]
- Lattman, E.E. 1994. Small angle scattering studies of protein folding. Curr. Opin. Struct. Biol. 4 87–92. [Google Scholar]
- Lipari, G. and Szabo, A. 1982. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104 4546–4559. [Google Scholar]
- Logan, T.M., Thériault, Y., and Fesik, S.W. 1994. Structural characterization of the FK506 binding protein unfolded in urea and guanidine hydrochloride. J. Mol. Biol. 236 637–648. [DOI] [PubMed] [Google Scholar]
- López-Hernández, E. and Serrano, L. 1996. Structure of the transition state for folding of the 129 aa protein CheY resembles that of a smaller protein, CI-2. Folding Des. 1 43–55. [PubMed] [Google Scholar]
- Lumb, K.J. and Kim, P.S. 1994. Formation of a hydrophobic cluster in denatured bovine pancreatic trypsin inhibitor. J. Mol. Biol. 236 412–420. [DOI] [PubMed] [Google Scholar]
- Meekhof, A.E. and Freund, S.M. 1999. Probing residual structure and backbone dynamics on the milli- to picosecond timescale in a urea-denatured fibronectin type III domain. J. Mol. Biol. 286 579–592. [DOI] [PubMed] [Google Scholar]
- Miranker, A.D. and Dobson, C.M. 1996. Collapse and cooperativity in protein folding. Curr. Opin. Struct. Biol. 6 31–42. [DOI] [PubMed] [Google Scholar]
- Mirny, L.A., Abkevich, V.I., and Shakhnovich, E.I. 1998. How evolution makes proteins fold quickly. Proc. Natl. Acad. Sci. 95 4976–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, Y.K., Kay, C.M., Kay, L.E., and Forman-Kay, J. 1999. NOE data demonstrating a compact unfolded state for an SH3 domain under non-denaturing conditions. J. Mol. Biol. 289 619–638 [DOI] [PubMed] [Google Scholar]
- Moy, F.J., Lowry, D.F., Matsumura, P., Dahlquist, F.W., Krywko, J.E., and Domaille, P.J. 1994. Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy. Biochemistry 33 10731–10742. [DOI] [PubMed] [Google Scholar]
- Neri, D., Billeter, M., Wider, G., and Wüthrich, K. 1992. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science 257 1559–1563. [DOI] [PubMed] [Google Scholar]
- Onuchic, J.N., Wolynes, P.G., Luthey-Schulten, Z., and Socci, N.D. 1995. Toward an outline of the topography of a realistic protein-folding funnel. Proc. Natl. Acad. Sci. 92 3626–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.W. and Wagner, G. 1992. Mapping of the spectral densities of N-H bond motions in eglin c using heteronuclear relaxation experiments. Biochemistry 31 8571–8586 [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Pollack, L., Tate, M.W., Darnton, N.C., Knight, J.B., Gruner, S.M., Eaton, W.A., and Austin, R.H. 1999. Compactness of the denatured state of a fast-folding protein measured by submillisecond small-angle x-ray scattering. Proc. Natl. Acad. Sci. 96 10115–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford, S.E. and Dobson, C.M. 1999. From computer simulations to human disease: Emerging themes in protein folding. Cell 97 291–298. [DOI] [PubMed] [Google Scholar]
- Santoro, J., Bruix, M., Pascual, J., López, E., Serrano; L. and Rico, M. 1995. Three-dimensional structure of chemotactic Che Y protein in aqueous solution by nuclear magnetic resonance methods. J. Mol. Biol. 247 717–725. [DOI] [PubMed] [Google Scholar]
- Sari, N., Alexander, P., Bryan, P.N., and Orban, J. 2000. Structure and dynamics of an acid-denatured protein G mutant. Biochemistry 39965–977. [DOI] [PubMed] [Google Scholar]
- Schwalbe, H., Fiebig, K.M., Buck, M., Jones, J.A., Grimshaw, S.B., Spencer, A., Glaser, S.J., Smith, L.J., and Dobson, C.M. 1997. Structural and dynamical properties of a denatured protein. Heteronuclear 3D NMR experiments and theoretical simulations of lysozyme in 8 M urea. Biochemistry 36 8977–8991. [DOI] [PubMed] [Google Scholar]
- Schwarzinger, S., Kroon, G.J.A., Foss, T.R., Wright, P.E., and Dyson, H.J. 2000. Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in MNRView. J. Biomol. NMR 18 43–48. [DOI] [PubMed] [Google Scholar]
- Segel, D.J., Fink, A.L., Hodgson, K.O., and Doniach, S. 1998. Protein denaturation: A small-angle X-ray scattering study of the ensemble of unfolded states of cytochrome c. Biochemistry 37 12443–12451. [DOI] [PubMed] [Google Scholar]
- Shakhnovich, E.I. 1998. Folding nucleus: Specific or multiple? Insights from lattice models and experiments. Folding Des. 3 R108–111. [DOI] [PubMed] [Google Scholar]
- Shortle, D. 1996. Structural analysis of non-native states of proteins by NMR methods. Curr. Opin. Struct. Biol. 6 24–30. [DOI] [PubMed] [Google Scholar]
- Sinclair, J.F. and Shortle, D. 1999. Analysis of long-range interactions in a model denatured state of staphylococcal nuclease based on correlated changes in backbone dynamics. Protein Sci. 5 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.K., Bu, Z., Anderson, K.S., Sturtevant, J.M., Engelman, D.M., and Regan, L. 1996a. Surface point mutations that significantly alter the structure and stability of a protein's denatured state. Protein Sci. 5 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.J., Fiebig, K.M., Schwalbe, H., and Dobson, C.M. 1996b. The concept of a random coil. Residual structure in peptides and denatured proteins. Folding Des. 1 R95–106. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R. and Trewella, J. 1992. Denatured state of ribonuclase A have compact dimension and residual secondary structures. Biochemistry 31 8329–8335. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Martinez-Hackert, E., Rasmussen, B.F., West A.H., Stock, J.B., Ringe, D., and Petsko, G.A. 1993. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32 13375–13380. [DOI] [PubMed] [Google Scholar]
- Svergun, D., Barberato, C., and Koch, M., 1995. CRYSOL — a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28 768–773. [Google Scholar]
- Thirumalai, D. and Klimov, D.K. 1998. Fishing for folding nuclei in lattice models and proteins. Folding Des. 3 R112–118. [DOI] [PubMed] [Google Scholar]
- Warren, J.R. and Gordon, J.A. 1970. Denaturation of globular proteins II. The interaction of urea with lysozyme. J. Biol. Chem. 245 4097–4104. [PubMed] [Google Scholar]
- Wilcock, D., Pisabarro, M.T., López-Hernández, E., Serrano, L., and Coll, M. 1998. Structure analysis of two CheY mutants: Importance of the hydrogen-bond contribution to protein stability. Acta Crystallogr. Sect. D Biol. Crystallogr. 54 378–385. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Bigam, C.G., Holm, A., Hodges, R.S., and Sykes, B.D. 1995. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5 67–81. [DOI] [PubMed] [Google Scholar]
- Wong, K.B., Freund, S.M.V., and Fersht, A.R. 1996. Cold denaturation of Barstar: 1H, 15N and 13C NMR assignment and characterisation of residual structures. J. Mol. Biol. 259 805–818. [DOI] [PubMed] [Google Scholar]
- Wong, K.B., Clarke, J., Bond, C.J., Neira, J.L., Freund, S.M., Fersht, A.R., and Daggett, V. 2000. Towards a complete description of the structural and dynamic properties of the denatured state of barnase and the role of residual structure in folding. J. Mol. Biol. 296 1257–1282. [DOI] [PubMed] [Google Scholar]
- Wright, P.E., Dyson, H.J., and Lerner, R.A. 1988. Conformation of peptide fragments of proteins in aqueous solution. Implications for initiation of protein folding. Biochemistry 27 7167–7175. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- ———. 1994. NMR assignments as a basis for structural characterization of denatured states of globular proteins. Curr. Opin. Struc. Biol. 4 93–99. [Google Scholar]
- Zhang, O. and Forman-Kay, J.D. 1997. NMR studies of unfolded states of an SH3 domain in aqueous solution and denaturing conditions. Biochemistry 36 3959–3970. [DOI] [PubMed] [Google Scholar]