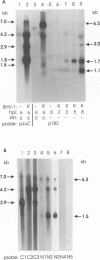
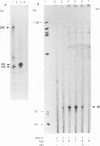
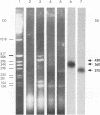
Abstract
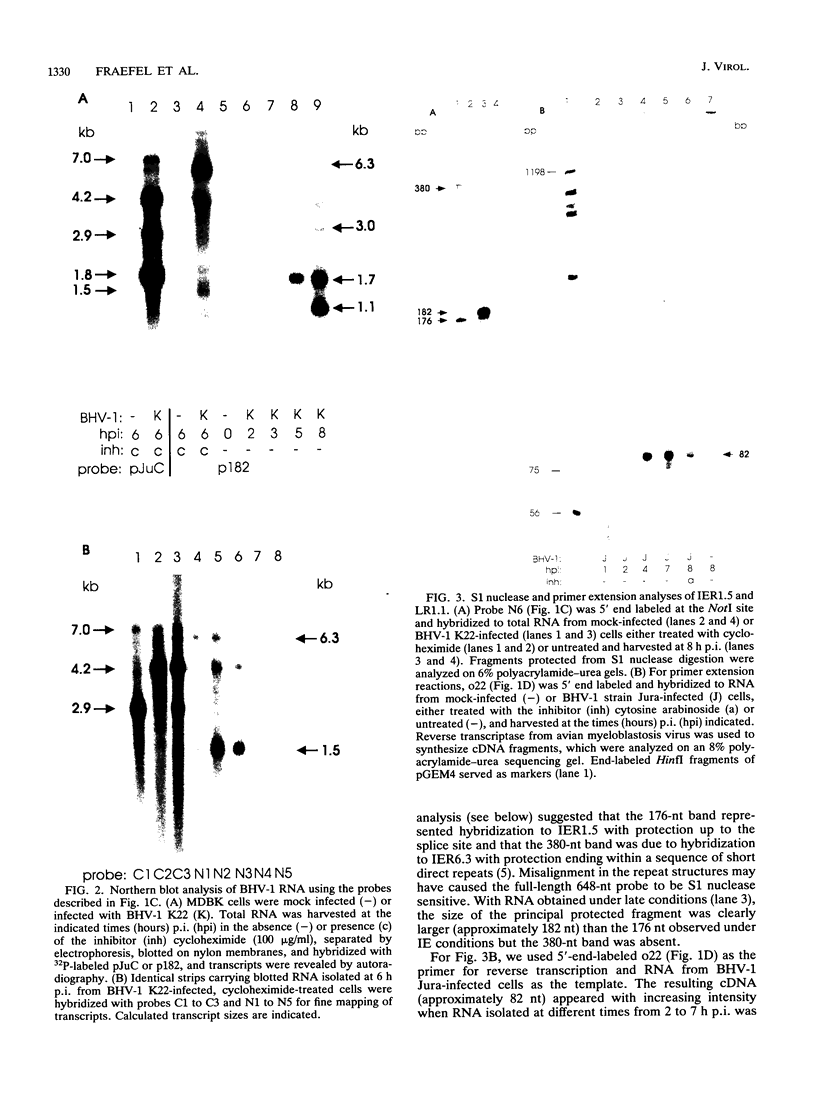
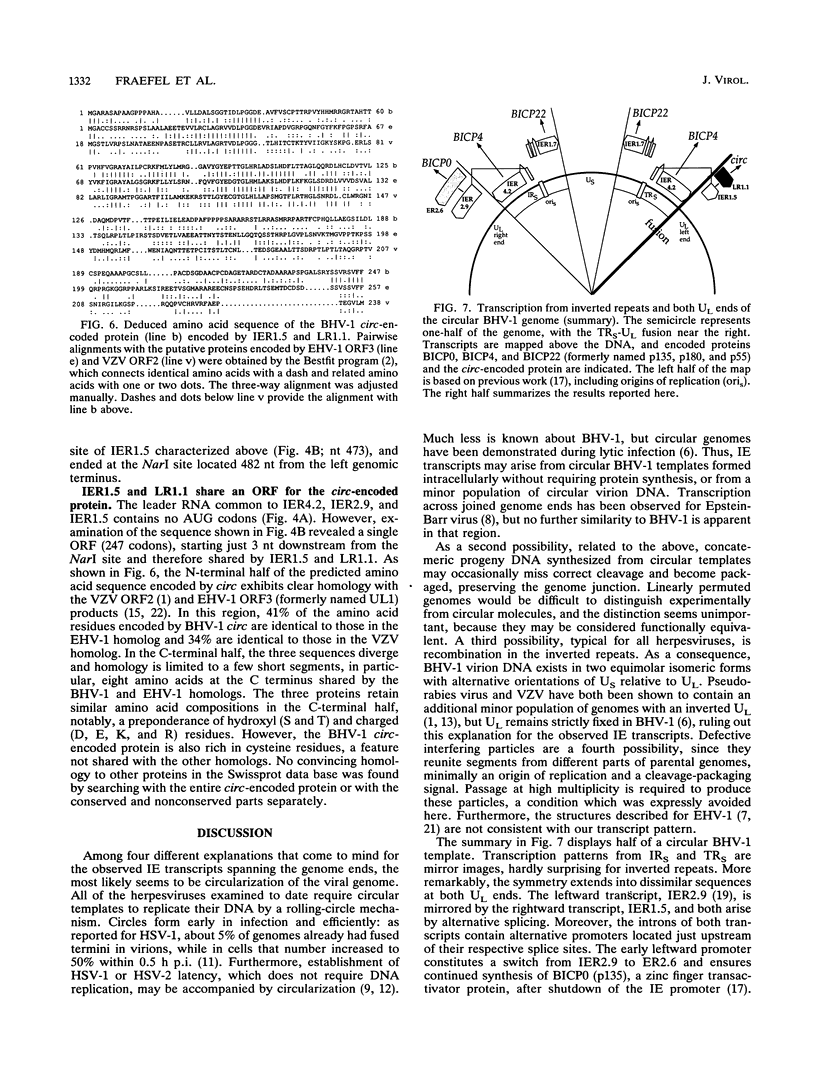
Herpesvirus genomes are linear molecules in virions. Prior to replication in host cells, they form circular templates by unknown mechanisms. Examining lytic infection with bovine herpesvirus 1, we observed immediate-early transcription over joined genome ends, which suggested that circles are present at the initial stage of infection. Among the transcripts was a spliced immediate-early RNA (1.5 kb) sharing exon 1 with previously described major immediate-early transcripts from the right genome end and exon 2 with a late transcript located near the left genome end. Exon 2 encodes a putative circ-encoded protein with homology to the varicella-zoster virus open reading frame 2 and equine herpesvirus 1 open reading frame 3 products. The novel features reported here for bovine herpesvirus 1 may constitute a more general property of herpesviruses.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels M., Giuliani C., Wild P., Beck T. M., Loepfe E., Wyler R. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 1986 Oct;6(1):57–73. doi: 10.1016/0168-1702(86)90057-2. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Ludwig H., Buhk H. J. Short repeats cause heterogeneity at genomic terminus of bovine herpesvirus 1. J Virol. 1986 Apr;58(1):43–49. doi: 10.1128/jvi.58.1.43-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W., Ludwig H., Buhk H. J. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J Virol. 1988 Apr;62(4):1355–1363. doi: 10.1128/jvi.62.4.1355-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty R. N., Yalamanchili R. R., O'Callaghan D. J. Transcriptional analysis of the UL1 gene of equine herpesvirus 1: a gene conserved in the genome of defective interfering particles. Virology. 1991 Aug;183(2):830–833. doi: 10.1016/0042-6822(91)91020-h. [DOI] [PubMed] [Google Scholar]
- Laux G., Economou A., Farrell P. J. The terminal protein gene 2 of Epstein-Barr virus is transcribed from a bidirectional latent promoter region. J Gen Virol. 1989 Nov;70(Pt 11):3079–3084. doi: 10.1099/0022-1317-70-11-3079. [DOI] [PubMed] [Google Scholar]
- Mellerick D. M., Fraser N. W. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology. 1987 Jun;158(2):265–275. doi: 10.1016/0042-6822(87)90198-x. [DOI] [PubMed] [Google Scholar]
- Misra V., Blumenthal R. M., Babiuk L. A. Proteins Specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus). J Virol. 1981 Nov;40(2):367–378. doi: 10.1128/jvi.40.2.367-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M., Russell J. Retention of nonlinear viral DNA during herpes simplex virus latency in vitro. Intervirology. 1991;32(2):69–75. doi: 10.1159/000150187. [DOI] [PubMed] [Google Scholar]
- Rall G. F., Lu Z. Q., Sugg N., Veach R. A., Ben-Porat T. Acquisition of an additional internal cleavage site differentially affects the ability of pseudorabies virus to multiply in different host cells. J Virol. 1991 Dec;65(12):6604–6611. doi: 10.1128/jvi.65.12.6604-6611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford E. A., Watson M. S., McBride K., Davison A. J. The DNA sequence of equine herpesvirus-1. Virology. 1992 Jul;189(1):304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- Wirth U. V., Fraefel C., Vogt B., Vlcek C., Paces V., Schwyzer M. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3' coterminal and encode a putative zinc finger transactivator protein. J Virol. 1992 May;66(5):2763–2772. doi: 10.1128/jvi.66.5.2763-2772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth U. V., Gunkel K., Engels M., Schwyzer M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J Virol. 1989 Nov;63(11):4882–4889. doi: 10.1128/jvi.63.11.4882-4889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth U. V., Vogt B., Schwyzer M. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J Virol. 1991 Jan;65(1):195–205. doi: 10.1128/jvi.65.1.195-205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili R. R., Raengsakulrach B., Baumann R. P., O'Callaghan D. J. Identification of the site of recombination in the generation of the genome of DI particles of equine herpesvirus type 1. Virology. 1990 Apr;175(2):448–455. doi: 10.1016/0042-6822(90)90429-u. [DOI] [PubMed] [Google Scholar]
- Yalamanchili R. R., Raengsakulrach B., O'Callaghan D. J. Equine herpesvirus 1 sequence near the left terminus codes for two open reading frames. Virus Res. 1991 Mar;18(2-3):109–116. doi: 10.1016/0168-1702(91)90012-k. [DOI] [PubMed] [Google Scholar]