Abstract
Musashi1 (Msi1) is an RNA-binding protein that is highly expressed in neural stem cells. We previously reported that Msi1 contributes to the maintenance of the immature state and self-renewal activity of neural stem cells through translational repression of m-Numb. However, its translation repression mechanism has remained unclear. Here, we identify poly(A) binding protein (PABP) as an Msi1-binding protein, and find Msi1 competes with eIF4G for PABP binding. This competition inhibits translation initiation of Msi1's target mRNA. Indeed, deletion of the PABP-interacting domain in Msi1 abolishes its function. We demonstrate that Msi1 inhibits the assembly of the 80S, but not the 48S, ribosome complex. Consistent with these conclusions, Msi1 colocalizes with PABP and is recruited into stress granules, which contain the stalled preinitiation complex. However, Msi1 with mutations in two RNA recognition motifs fails to accumulate into stress granules. These results provide insight into the mechanism by which sequence-specific translational repression occurs in stem cells through the control of translation initiation.
Introduction
Posttranscriptional regulation is a key aspect of gene expression and includes RNA surveillance, RNA splicing, mRNA stability, and mRNA translation (Moore, 2005). In particular, translational regulation contributes to the spatio-temporal pattern of gene expression in animal development. For example, the expression of Oskar, which is required for pole plasm formation (Lehmann and Nusslein-Volhard, 1986), is regulated by translational repression via an RNA-binding protein, Bruno (Nakamura et al., 2004). RNA-binding proteins are important in translational regulation, with critical roles in stem cell maintenance in planarians (e.g., bruno-like protein [Guo et al., 2006]) and mammalian neural stem cells (NSCs) (e.g., Musashi1 protein [Imai et al., 2001; Sakakibara et al., 2002]).
The Musashi family is an evolutionarily conserved group of neural RNA-binding proteins that contain two RNA recognition motifs (RRMs) and has representatives in vertebrates and invertebrates (for review see Okano et al., 2002). We previously identified Musashi-binding sequences in mammals (Imai et al., 2001) and Drosophila (Okabe et al., 2001). In the mammalian nervous system, Musashi1 (Msi1) is expressed in neural precursor cells, including NSCs (Sakakibara et al., 1996). Our previous studies revealed that Msi1 contributes to NSC maintenance by binding to the 3′-untranslated region (UTR) of one of its target mRNAs, m-Numb, and repressing its translation (Imai et al., 2001). m-Numb encodes a membrane-associated protein that inhibits Notch signaling (Spana and Doe, 1996). Thus, Msi1 probably contributes to maintenance of the stem cell state by repressing the translation of its downstream target genes. In addition, Msi1 acts cooperatively with Musashi2 (a second mammalian Msi protein) in the proliferation and maintenance of NSCs (Sakakibara et al., 2002). However, the detailed molecular mechanism underlying the Msi1-mediated translational repression has not been clarified.
Translational regulation usually occurs at the translation initiation step, in which recruitment of the 40S ribosome to the mRNA is rate limiting (Muckenthaler et al., 1998; Chekulaeva et al., 2006). Numerous eukaryotic initiation factors (eIFs) contribute to translation initiation. One of these, eIF4G, is an essential and multifunctional scaffold protein (Gingras et al., 1999). It is a subunit of the heterotrimeric eIF4F complex, which associates with the mRNA m7G cap and facilitates ribosome joining to the mRNA (Kahvejian et al., 2005). Two other components are the cap-binding protein eIF4E and the ATP-dependent RNA helicase eIF4A. eIF4G interacts with eIF4E, eIF4A, and the poly(A) binding protein (PABP), which stimulates initiation factor recruitment to mRNA and leads to mRNA circularization (Imataka et al., 1998; Kahvejian et al., 2005). The PABP–eIF4G interaction seems to be vital for efficient translation, and it stimulates the formation on mRNA of both the 48S and 80S ribosome complexes (Kahvejian et al., 2005).
Recent studies show that, in response to stress conditions, aggregates of stalled translation initiation complex localize to cytoplasmic foci called stress granules (SGs) (Kedersha et al., 2002; Anderson and Kedersha, 2006). SGs contain most components of the 48S preinitiation complex, which contains proteins involved in translational regulation and mRNA. SGs containing microRNA are thought to be involved in mRNA recycling, and its shift to polysomes for translation or to docked processing bodies (PBs) for degradation in the cytoplasm (Anderson and Kedersha, 2006; Tanaka et al., 2006; Parker and Sheth, 2007). PBs contain RNA (untranslated mRNA and noncoding RNA) and proteins involved in mRNA decapping, nonsense-mediated decay, and translational repression. Thus, although SGs and PBs are distinct RNA-containing granules, they share some components and seem to physically and functionally interact with each other.
Here, we found that PABP is a direct binding partner of Msi1. A time-course reporter assay with the Msi1-D2 mutant, which lacks the PABP-interacting domain, failed to bind PABP, and revealed a correlation between failure to bind PABP and failure to repress translation. We also found that Msi1 localizes to the cytoplasm and accumulates in SGs under stress, where it colocalizes with PABP. Our results indicate that Msi1 inhibits the cap-dependent translation of its target mRNAs by competing with eIF4G to bind PABP, and inhibiting formation of the 80S ribosome complex. Thus, we present a mechanism for selective posttranslational regulation by the neural RNA-binding protein Msi1.
Results
Identification of PABP as an Msi1-specific binding protein
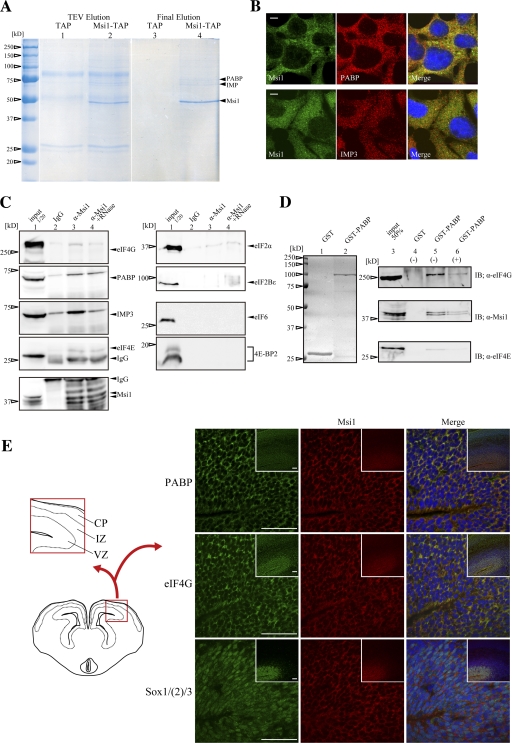
To clarify the role of Msi1 in translational repression, we sought to identify Msi1-binding partners using the TAP (tandem affinity puification) method (Rigaut et al., 1999). An Msi1-TAP fusion protein was expressed in 293T cells, and its associated molecules were recovered from purified final extracts. Several bands, representing proteins in Msi1-containing protein complexes, were detected by Coomassie brilliant blue (CBB) staining (Fig. 1 A, lane 4). Two of these, migrating at 63 and 70 kD, were identified as insulin-like growth factor 2 mRNA binding protein (IMP) and poly(A) binding protein (PABP), respectively, by MALDI-TOF mass spectrometry. By immunostaining, we showed that Msi1 was expressed diffusely in the cytoplasm, where it colocalized with PABP and IMP3 in P19 cells (Fig. 1 B).
Figure 1.
Identification of PABP as an Msi1-specific binding protein by the TAP method. (A) Msi1-bound proteins that were extracted from 293T cells expressing Flag-Msi1-TAP were resolved by SDS–PAGE, visualized by CBB staining (lanes 2 and 4), and compared with those of control Flag-TAP-expressing cells (lanes 1 and 3). The bound proteins in the TEV-digested extracts are shown in lanes 1 and 2; similarly, those of in the final extracts are shown in lanes 3 and 4. CBB-stained PABP, IMP, and Msi1 are indicated with arrowheads. (B) Msi1 colocalized with PABP and IMP3 in the cytoplasm. P19 cells were stained with anti-Msi1 (green) antibody, and anti-PABP (red, top) or anti-IMP3 (red, bottom) antibodies. Nuclei were stained with Hoechst (blue in P19 cells) in the merged image. (C) Immunoblottings after immunoprecipitation with ant-Msi1 antibody using E14 mouse brain extracts were performed with each antibody, respectively. (D) Protein extracts prepared from mouse brain at E16 were mixed with bacterially expressed and purified GST or GST-PABP fusion proteins. The GST fusion proteins were stained with CBB (lanes 1 and 2). Elutes were analyzed by immunoblotting using anti-eIF4G, anti-Msi1 14H1, or anti-eIF4E antibodies (lanes 3–6). (E) Double-immunohistochemistry of Msi1 (red) and PABP (green), eIF4G (green), or Sox1/(2)/3 (green) in coronal sections of the E14 forebrain. Sox1/(2)/3 is a marker for neural precursor cells. Inset in E shows a low magnification view of the main Msi1-expressing regions. CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone. Bars: 5 μm (B), 50 μm (E).
To verify whether Msi1 binds to PABP and IMP3, we performed immunoprecipitation experiments with an anti-Msi1 antibody and GST pull-down assays using tissue extracts prepared from mouse brain at embryonic day (E) 14 and 16, respectively. As shown in Fig. 1 C, the interaction between PABP and Msi1 was less sensitive to RNase A than that between IMP3 and Msi1 (lanes 3 to 4). These results suggest that Msi1 interacts directly with PABP, but indirectly with IMP3. This idea was supported by in vivo and in vitro GST pull-down assays using GST-PABP (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708004/DC1). In addition, the interaction between eIF4E and Msi1 was only weakly detectable in this assay, and that between eIF4G and Msi1 was even less clear. We therefore focused on PABP, as a protein that directly binds Msi1.
We next used GST pull-down assays to examine the interaction between endogenous Msi1 and GST-PABP. Both Msi1 and the PABP-binding protein eIF4G bound to PABP similarly in the presence and absence of RNase A. However, the coprecipitation of PABP and eIF4E was sensitive to RNase A (Fig. 1 D). Collectively, these immunostaining and binding experiments show that PABP is included in a complex in which it interacts directly with Msi1 and eIF4G, and indirectly with eIF4E.
To examine the colocalization of Msi1 and PABP in neural stem/precursor cells (NSPCs) of the developing embryonic neural tube, we conducted immunohistochemical analyses with an anti-Msi1 antibody and anti-PABP, eIF4G, or Sox1/(2)/3 antibodies, using E14 mouse brain sections. Msi1 colocalized with PABP and eIF4G in the cytoplasm of putative NSPCs in the ventricular zone (VZ) (Fig. 1 E). These results indicate that Msi1 colocalizes with both PABP and eIF4G in NSPCs.
The C-terminal domain of Msi1 is necessary for its interaction with PABP and translation repression activity
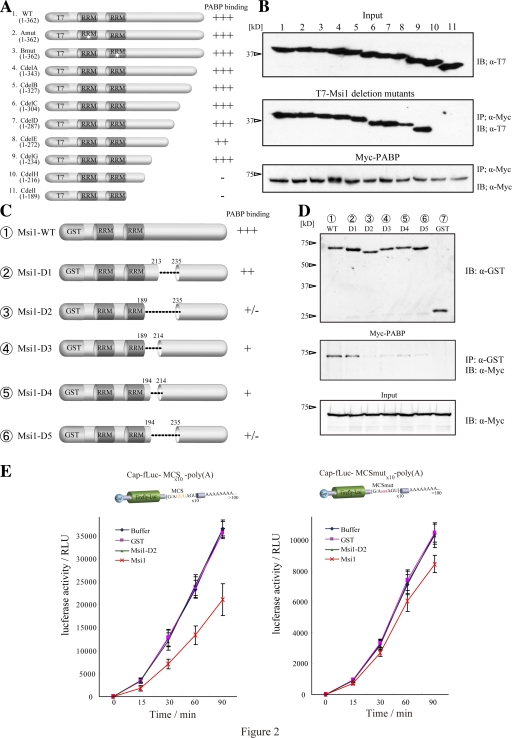
To identify the PABP-binding site in Msi1, a series of C-terminal deletion mutants of T7-Msi1 (Kaneko et al., 2000) was coexpressed with Myc-PABP in 293T cells, followed by coimmunoprecipitation. Immunoblot analysis revealed that two truncated Msi1 proteins lacking almost half the C terminus, 1–216 (Fig. 2, A and B, lane 10) and 1–189 (lane 11), failed to interact with Myc-PABP. A GST pull-down assay (Fig. 2, C and D) showed that GST-Msi1-D2 and -D5, in which the region proximal to RRM was deleted, corresponding to amino acids 190–234 (Msi1-D2, lane 3) and 195–234 (Msi1-D5, lane 6), respectively, showed almost no interaction with Myc-PABP, defining the PABP-binding region.
Figure 2.
The C-terminal region of Msi1 that bound PABP is necessary for its function. (A) Illustration of proteins containing the T7-Msi1 variants: Msi1Amut (mutation in RRM1, and fails to bind mRNA: lane 2), Msi1Bmut (mutation in RRM2: lane 3), and a series of Msi1C-terminal deletions (lanes 4–11). (B) Immunoprecipitation using the T7-Msi1 variants was performed and various T7-Msi1 mutants bound to Myc-PABP (middle). The intensities of binding with PABP are illustrated to the right of panel A. (C) Illustration of the GST-Msi1 variants. (D) GST-Msi1 variants or GST as a control were coimmunoprecipitated with Myc-PABP in 293T cells using glutathione-Sepharose 4B (middle). PABP bound to Msi1 variants was immunoblotted using an anti-Myc antibody and is indicated (middle). (E) The in vitro–transcribed reporter mRNAs are illustrated at top (left, mRNA containing MCS; right, mRNA containing MCSmut), were translated in RRL with equimolar amounts of purified various GST proteins, and the luciferase activity was measured at each time point (0–90 min). The values represent mean ± SD; n = 5.
The N-terminal half of Msi1 protein contains two RRMs, which are essential for RNA binding, but the function of the C-terminal region has not been elucidated. To examine whether the C-terminal PABP-binding region is required for Msi1's function as a translational repressor, we performed assays using a chimeric reporter mRNA consisting of a 5′-cap, the luciferase gene, 10 repeats of Msi1-binding consensus sequence (MCS; (G/AUUUAGU) derived from SELEX [Imai et al., 2001]) or of a mutated Msi1-binding sequence (MCSmut; (G/AaaaAGU)), and a poly(A)-tail. The cap-fLuc-MCS-poly(A) reporter mRNA (Fig. 2 E, left) was translated in rabbit reticulocyte lysate (RRL) in the presence of buffered saline, GST, GST-Msi1, or GST-Msi1-D2. GST-Msi1 decreased the luciferase activity by ∼50.7% (n = 5) after 15 min (Fig. 2 E, left). The recombinant GST-Msi1-D2, which lacked the PABP-binding region, did not inhibit translation (Fig. 2 E, left). In contrast, the cap-fLuc-MCSmut-poly(A) mRNA was translated in RRL equally well with GST proteins (Fig. 2 E, right). Thus, intact Msi1, but not Msi1-D2, caused sequence-specific translational repression (Fig. 2 E). These results indicate that Msi1's interaction with PABP is essential for Msi1's function as a translational repressor.
Msi1 can compete with eIF4G for PABP binding
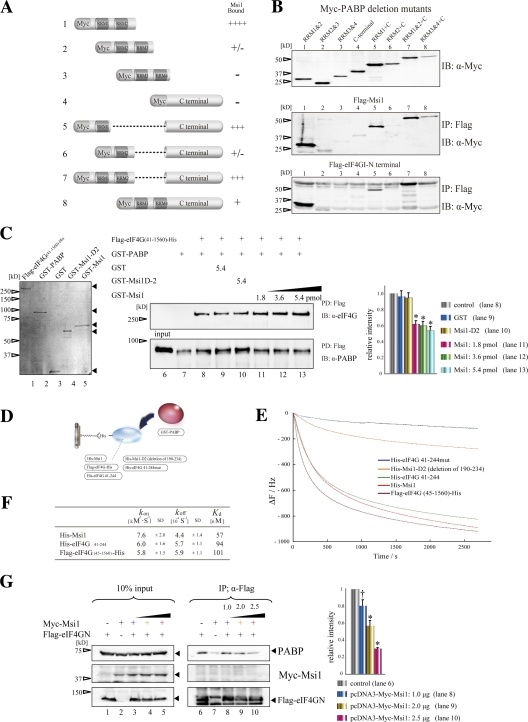
PABP includes four RRMs and a C-terminal domain (PABC), which contains a 60-amino acid sequence that is highly conserved among species (Deo et al., 2001). Each domain recruits several binding proteins and elicits diverse cellular functions: eIF4G binds to RRM1 and RRM2; Paip1 interacts with RRM1, RRM2, and the PABC domain; Paip2 interacts with RRM2, RRM3, and PABC; and GSPT/eRF3 binds to PABC (Preiss and Hentze, 2003). To locate the Msi1-binding domain within PABP, a Myc-tagged series of deletion mutants in which the PABP functional domain was deleted (Myc-PABP and Myc-PABP variants) was coexpressed in 293T cells with Flag-Msi1, coimmunoprecipitated with Flag-Msi1 (Fig. 3 A), and detected by immunoblotting with an anti-Myc tag antibody. Msi1 interacted strongly with the RRM1 and RRM2 of PABP, although the presence of PABC diminished the Msi1-binding activity of RRM1 or RRM2 (Fig. 3 B, middle, lanes 1, 5, and 7). Notably, these results were similar to the binding pattern of the Flag-tagged N-terminally truncated protein (1–585 aa) of mouse eIF4G1 (Flag-eIF4GN) (Fig. 3 B, bottom, lanes 1, 5, and 7), suggesting that Msi1 shares the binding region within PABP with eIF4G. Thus, Msi1 may compete with eIF4G for binding to the same domain of PABP.
Figure 3.
Msi1 competed with binding of eIF4G to PABP. (A) Illustration of the PABP variants. (B) Flag-Msi1 or Flag-eIF4GN-(1–582) was coimmunoprecipitated with Myc-PABP variants in 293T cells using anti-FLAG resin. Notably, Msi1 and eIF4G bound to a common domain within PABP (middle, bottom). (C) In vitro competition assay between purified GST-Msi1 and purified Flag-eIF4G (45–1560)-His immobilized FLAG resin. The CBB-stained, purified fusion proteins Flag-eIF4G (41–1560)-His, GST-PABP, GST, GST-Msi1-D2, and GST-Msi1 are shown (left panel, lanes 1–5). (D–F) Analysis of the kinetics of PABP's interaction with Msi1 or eIF4G by the QCM. (D) Illustration of the His-tagged proteins immobilized on the QCM plate and GST-PABP. The His-tag proteins were anchored to the QCM plate by an anti-His antibody. (E) Curves showing the time course of the changes in frequency for the proteins coated on the QCM plate, His-eIF4G 41-244mut, His-Msi1-D2, His-eIF4G 41–244, His-Msi1, and Flag-eIF4G (41–1560)-His, in response to the addition of 100 nM GST-PABP. (F) Summary of the kinetics parameters for the binding of PABP to Msi1 or eIF4G on the QCM; for a more detailed description see Materials and methods. (G) In vivo competition assay using 293T cells expressing Flag-eIF4GN and Myc-Msi1. The quantitative analysis was performed with Multigauge software (Fujifilm) in C and G (n = 5, mean ± SEM; *, P < 0.01 vs. control; †, P < 0.05 vs. control).
To examine this possibility, the interaction between PABP and eIF4G was tested in the presence of Msi1 by an in vitro pull-down assay, modified as described previously (Khaleghpour et al., 2001). Mixtures of GST-PABP and various proteins (GST, GST-Msi1, or GST-Msi1-D2) were added to Flag-eIF4G (41–1560) immobilized on FLAG resin. After stringent washing, the bound proteins were subjected to immunoblotting with an anti-PABP antibody. Almost no PABP coprecipitated with FLAG resin alone (Fig. 3 C, right, lane 7). Preincubation with 5.4 pmol GST or GST-Msi1-D2 did not interfere with PABP's association with Flag-eIF4G (compare lane 9 with 10), whereas GST-Msi1 (1.8, 3.6, or 5.4 pmol) decreased the amount of GST-PABP precipitated. These data suggest that Msi1 and eIF4G directly compete for binding to PABP in vitro.
To quantify the strength and kinetics of the Msi1–PABP interaction, we measured kinetics parameters using a 27-MHz quartz crystal microbalance (QCM) resonator, the AffinixQ (Initium), whose oscillatory resonance frequency decreases linearly with the mass on the QCM electrode (Okahata et al., 1998). In this system, the action of an object binding to its ligand is detected as a decrease in resonance frequency (mass increase). Purified Msi1 or eIF4G was immobilized on the Au-surface-coated QCM plate by binding the His-tag of His-Msi1 or His-eIF4G to orient the molecules, and purified GST-PABP in an aqueous solution was injected as the binding counterpart (Fig. 3 D). The addition of GST-PABP (100 nM) to the reaction solution with immobilized Msi1, eIF4G, or eIF4G 41–244 resulted in typical frequency decreases, representing the frequency of immobilized PABP (Fig. 3 E). However, the change in individual resonance frequency when PABP was added to immobilized molecules that were not expected to bind it (eIF4G 41-244mut and Msi1-D2) was barely detectable (Fig. 3 E), indicating that the specific interactions of Msi1–PABP and eIF4G–PABP were detectable by the resonance frequency curve.
To calculate the association rate constant (kon) and dissociation rate constant (koff), four concentrations of GST-PABP (25 to 100 nM) were individually injected into the reaction solution with immobilized Msi1 or eIF4G, and the time dependence of the frequency decrease was observed (Fig. 3 E). We analyzed these data by curve fitting, according to the manufacturer's instructions (see Materials and methods for details). The Kd value was estimated from the ratio of koff to kon. The results are summarized in Fig. 3 F. The kon and koff were almost the same for the eIF4G (45–1560)/PABP and eIF4G (41–244)/PABP interactions. Although the constants for the Msi1-PABP interaction were not very different from those for eIF4G/PABP, the Kd value calculated for Msi1–PABP was approximately half that for eIF4G/PABP (Fig. 3 F). Collectively, our data support the view that Msi1 can compete with eIF4G to associate with PABP in vitro.
To investigate the in vivo significance of Msi1–PABP interaction, in vivo competition assays were performed as described previously (Khaleghpour et al., 2001). In this assay, the N-terminal portion of eIF4G (Flag-eIF4GN) was used for immunoprecipitation bait because its PABP-binding activity was stronger than that of full-length eIF4G in the QCM assay. Flag-eIF4GN expressed in 293T cells coimmunoprecipitated with endogenous PABP (Fig. 3 G, lanes 6–10, top panel). The amount of coprecipitating PABP decreased dose-dependently when Myc-Msi1 was coexpressed (compare lane 6 to lanes 8–10, top panel), indicating that Msi1 and eIF4G directly compete for binding to PABP in vivo. Together these data indicate that Msi1 competitively inhibits the interaction between PABP and eIF4G through its PABP-binding domain.
Msi1 localizes to the cytoplasm and accumulates in SGs, where it colocalizes with PABP
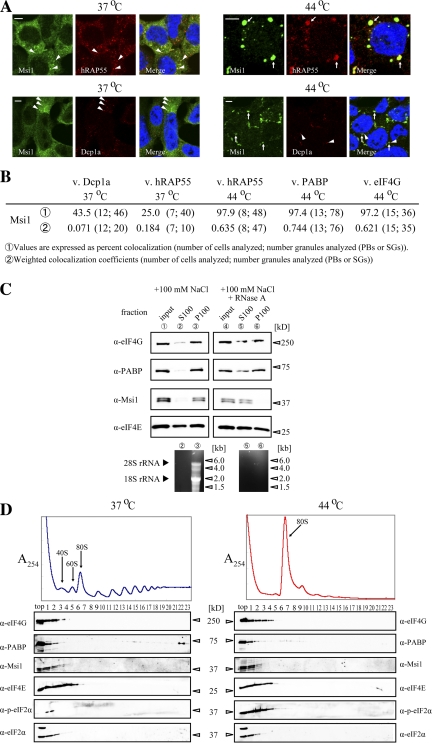
To examine whether Msi1 and PABP colocalize subcellularly, P19 cells were immunostained with anti-Msi1 (Kaneko et al., 2000) and anti-PABP antibodies. Both Msi1 and PABP predominantly showed diffuse cytoplasmic staining; Msi1 also accumulated in discrete cytoplasmic foci (Fig. 1 B). We then investigated the colocalization of endogenous Msi1 with markers for PBs, Dcp1a and hRAP55 (Tanaka et al., 2006; Parker and Sheth, 2007), and found that Msi1 colocalized with these markers in cytoplasmic foci in P19 cells at 37°C (Fig. 4 A), suggesting that Msi1 also localized to PBs. Because PABP is a component of SGs (Kedersha et al., 1999, 2002), we tested whether Msi1 and PABP colocalized in SGs under stress conditions. After P19 cells were heat stressed at 44°C, we found that Msi1 colocalized with PABP (see Fig. 5 C) and with hRAP55 in the SGs (Fig. 4 A). We observed the colocalization of Msi1 and PABP at a frequency of ∼97% under stress, but Msi1 did not always colocalize with PB markers under normal conditions (Fig. 4 B). These results indicate that Msi1 is recruited into SGs and is probably involved in SG function, in association with PABP.
Figure 4.
Cellular localization of Msi1. (A) Msi1 localizes to cytoplasmic foci. P19 cells treated with (right columns; 44°C for 30 min) or without (left columns) heat stress were stained with anti-Msi1 (green), and anti-hRAP55 (red, top) or anti-Dcp1a (red, bottom) antibodies, respectively. Nuclei were stained with TO-PRO-3 (blue) in the merged images. The white arrowheads and white arrow indicate PBs and SGs, respectively. Bars, 5 μm. (B) Msi1-positive granules were analyzed by two methods assessing the percent colocalization (1) or weighted colocalization coefficient (2) of their ratio to Dcp1a-, hRAP55-, PABP-, and eIF4G-containing granules. Msi1 mostly localized to SGs, but some was localized to PBs. (C) Association of Msi1 with heavy-sedimenting particles in an RNA-dependent manner. Each subcellular fraction with (lanes 4–6) or without (lanes 1–3) RNase A treatment after ultracentrifugation (top panels). S100, supernatants after ultracentrifugation; P100, pellets after ultracentrifugation. Total RNAs purified from S100 (lanes 2 and 5) and P100 (lanes 3 and 6) are shown (bottom panel). (D) P19 cells treated with (left) or without (right) heat stress were separated by 15–40% sucrose density gradients. Immunoblots of the gradient fractions were probed using antibodies against the indicated proteins (bottom panels).
Figure 5.
Msi1 inhibited the 80S ribosome initiation complex formation on mRNA. (A) Illustration of the in vitro–transcribed reporter mRNAs (top). The reporter mRNA and purified GST-tagged proteins were incubated with RRL at 30°C for 90 min. Msi1 repressed the cap-dependent and IRES-dependent translation. The relative luciferase activity value represents the mean ± SD: (n = 4; *, P < 0.01 vs. buffer). (B) Cap column assay was performed in HeLa cells expressing Flag-GST, Flag-Msi1, or Flag-eIF4G-MD, which contains the eIF4E-binding domain. (C and D) Msi1 colocalized with translation initiation factors in P19 cells (C) and in cultured hippocampal neurons (D). Treatment with heat stress (44°C for 30 min) is indicated at the right (C) and bottom (D) of columns. Cells were stained with anti-Msi1 (green), anti-eIF4E, anti-eIF4G, and anti-PABP antibodies (red). Nuclei were stained with Hoechst (blue) in the merged images. Msi1 accumulates in SGs under heat stress. Bars, 5 μm. (E and F) 80S or 40S ribosome binding assay using in vitro–transcribed reporter mRNAs containing MCS-poly(A). Curves show the relative radioactivity of each fraction from reaction mixtures supplemented with equimolar amounts of GST (purple line), GST-Msi1 (green line), or GST-Msi1-D2 (red line), or buffer as a control (blue line). The percentage of the total recovered count was plotted against the fraction number (top panels). The RNAs purified from each fraction are shown (bottom panels). These results were reproduced in three independent experiments. (E) Peaks (fraction 19 or 20) corresponding labeled reporter mRNA in a complex with 80S ribosomes are indicated with arrows (top). The peak of 28S and 18S rRNA was found in fraction 19 or 20 (bottom). With GST-Msi1 addition, the 80S ribosome complex formation decreased to 52.7 ± 3.0% (n = 4; mean ± SD; P < 0.001) of the buffer control level. (F) Peaks (fraction 20 or 21) corresponding to labeled reporter mRNA in a complex with 48S ribosomes are indicated with arrows (top). The peak of 18S rRNA was found in fraction 20 or 21 (bottom).
To verify the presence of Msi1 and PAPB in the mRNPs (i.e., RNA granules), we examined whether Msi1 and PABP were associated with heavy-sedimenting particles, using P19 cells. Lysates were subjected to subcellular fractionation as described previously (Aoki et al., 2002). Msi1 and PABP were detected mainly in the heavy-sedimenting particles in P100 fraction (Fig. 4 C, lane 3), with most of the ribosomes (Fig. 4 C, bottom, lane 3). After RNase treatment, both Msi1 and PABP were also detected in the soluble S100 fraction (Fig. 4 C, lane 5), suggesting that Msi1 coexists with PABP in heavy-sedimenting RNP particles, which are likely to represent mRNPs. Furthermore, we investigated the polysome profiles in 15–40% sucrose gradients using lysates of stressed or unstressed P19 cells. After heat stress, although some of the PABP was partly detected in the heavy (polysome) fractions, most of it was in the light fractions (Fig. 4 D). Msi1 and eIFs remained mostly in the light fractions after both conditions, and the fractionation pattern of eIFs coincided with that of a previous study (Kedersha et al., 2002). These data indicate that Msi1 comigrates with eIFs and PABP, and is likely to be involved in the regulation of translation initiation.
Msi1 is involved in translation initiation regulation
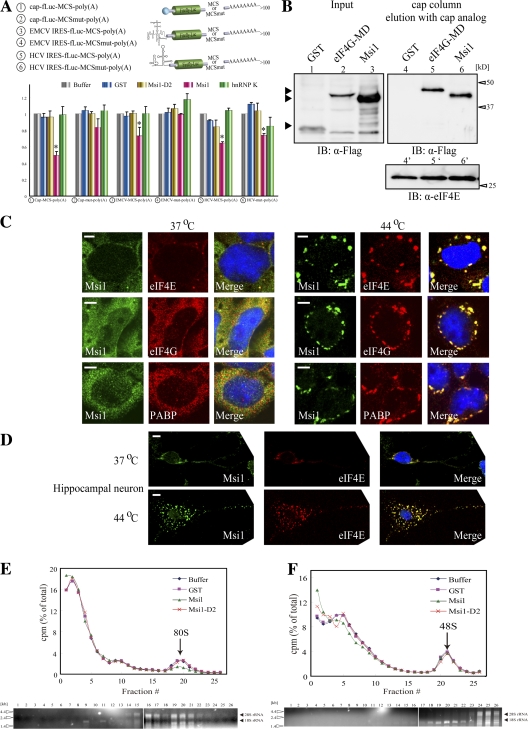
Because SGs contain the stalled translation initiation complex (Kedersha et al., 2002), Msi1's localization to SGs, its comigration with eIFs (Fig. 4), and its binding to PABP in competition with eIF4G (Fig. 3), suggested that Msi1 could be involved in the regulation of translation initiation. To examine whether the translational repression by Msi1 is cap dependent, we added GST-Msi1 to RRL, using either cap, EMCV IRES, or HCV IRES to drive the translation of fLuc reporter mRNA containing MCS or MCSmut in its 3′ UTR before poly(A) (Fig. 5 A), and measured the amount of reporter produced. HCV IRES-, but not EMCV IRES-directed translation initiation is also independent of eIF4G (Pestova et al., 1998). In RRL, endogenous Msi1 is not detectable (unpublished data), but eukaryotic translation initiation factors and PABP are present (Imataka et al., 1998; Kahvejian et al., 2005). We found ∼50.6 ± 3.0% (n = 4) inhibition of the cap-dependent translation by GST-Msi1, compared with ∼27.0 ± 6.2% and ∼35.9 ± 1.2% decreases in the EMCV IRES- and HCV IRES-directed translation of the MCS-containing reporter mRNA, respectively, whereas GST-Msi1-D2 had little effect on reporter mRNA containing MCS or MCSmut (Fig. 5 A). We also conducted Northern blot analyses to evaluate the amount of reporter RNA (cap-rfLuc-MCS-poly(A) RNA) in each assay, and found that it was barely altered by the addition of the tested proteins (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200708004/DC1). These observations suggest that Msi1 represses cap-dependent translation and exerts selective translational repression through Msi1-binding sequences in the 3′ UTR of its target mRNAs.
To investigate whether Msi1 is contained in the cap-binding complex, we performed a cap pull-down assay (Stebbins-Boaz et al., 1999). Several Flag-tagged proteins, Msi1, eIF4G MD (middle domain) including the eIF4E-binding domain, and GST as a control, were expressed in HeLa cells, and tested for their ability to bind a cap analogue column. Immunoblotting with an anti-Flag tag antibody showed that Msi1 was included in the cap-binding complex (Fig. 5 B, lane 6).
Next, to ascertain whether Msi1 was included in a translational initiation complex, we further examined the intracellular localization of translation initiation factors, by immunostaining. Msi1 and translation initiation factors PABP, eIF4G, and eIF4E predominantly showed diffuse cytoplasmic staining, and Msi1 and eIF4E were also detected in the PBs, and in SGs under heat stress in P19 cells (Fig. 5 C). Msi1 also colocalized with eIF4E in cultured hippocampal neurons at 37 and 44°C (Fig. 5 D).
Several RNA-binding proteins involved in the regulation of translational repression are known to inhibit ribosome complex formation (Muckenthaler et al., 1998; Stebbins-Boaz et al., 1999; Ostareck et al., 2001; Chekulaeva et al., 2006). In addition, the 48S preinitiation complex is selectively recruited to SGs (Kedersha et al., 2002). To examine whether Msi1 is involved in ribosome complex formation, 80S ribosome binding assays were performed with cap-fLuc-MCS-poly(A) reporter mRNA as described previously (Kahvejian et al., 2005). The 3′-end 32P-labeled reporter mRNA and various GST proteins were incubated with RRL in the presence of cycloheximide, and the mixtures were applied to a 15–30% sucrose density gradient (Kahvejian et al., 2005), subjected to ultracentrifugation, fractioned, and the radioactivity of each fraction was counted. The addition of GST-Msi1 strikingly decreased the 80S ribosome complex formation, to 52.7% (n = 4) of the control level, represented as a peak in the 19th or 20th fraction, whereas the addition of equimolar amounts of GST-Msi1-D2 had no effect on the 80S ribosome complex formation (Fig. 5 E), suggesting the Msi1–PABP interaction is required for this inhibitory mechanism. Therefore, Msi1 inhibits the recruitment of the large ribosomal subunit onto mRNA, and the Msi1–PABP interaction is probably required to inhibit the assembly of the 80S ribosome initiation complex.
To examine whether Msi1 also influences the 48S preinitiation complex formation on reporter mRNA, we performed a 40S ribosome binding assay by adding GMP-PNP, an unhydrolyzable analogue of GTP, which blocks 60S ribosomal subunit recruitment, followed by sucrose density gradient analysis as described previously (Kahvejian et al., 2005). We found that the 48S complex formation, which was represented as a peak in the 20th or 21st fraction, was unaffected by equimolar amounts of GST-Msi1 compared with controls (Fig. 5 F). Therefore, the Msi1–PABP interaction did not inhibit the 48S preinitiation complex formation. Nevertheless, Msi1 is involved, through its inhibition of 80S formation, in repressing the formation of a functional translational initiation complex.
The intracellular localization of Msi1 depends on two RRMs
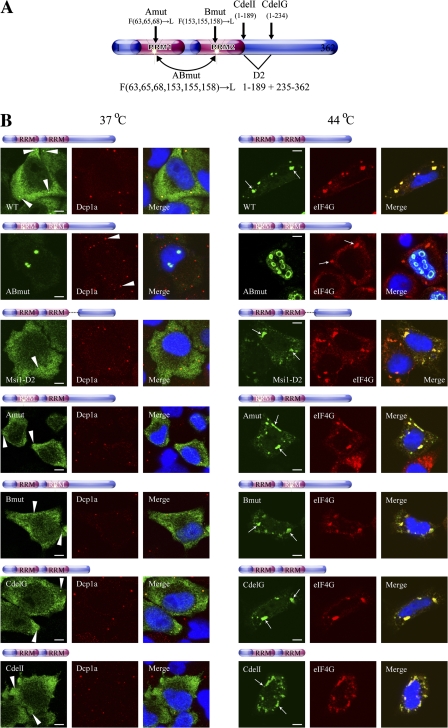
Msi1's function as an RNA sequence-dependent translation inhibitor and its requirement for PABP to carry out this function may correlate with Msi1's accumulation in SGs under stress. To examine this possibility, HeLa cells under stress were transiently transfected with the deletion and point mutants of Msi1 described in Fig. 2 A and p3xFlag-Msi1-D2. All the deletion mutants—CdelG, CdelI, and Msi1-D2 (which lack the PABP-interacting domain)—colocalized to eIF4G as clear marker of SGs (Fig. 6 B). In contrast, the point mutants of each single RRM—Msi1Amut and Msi1Bmu—colocalized with eIF4G, while that of double RRM—Msi1ABmut (which cannot bind RNA)—instead accumulated predominantly in discrete aggregates in the nucleus (Fig. 6 B). Meanwhile, under unstressed conditions, none of the Msi1 mutants in the PBs, except for Msi1ABmut, yielded intense staining patterns, compared with the wild type (Fig. 6 B). The intracellular distribution pattern of Msi1ABmut was similar to that under stressed conditions. These data suggest that the localizations of Msi1 mutants in PBs are sensitive to the natures of the mutant proteins themselves, and data from all the deletion mutants (CdelG, CdelI, and Msi1-D2) indicate that the C-terminal region of Msi1 plays an important role in its recruitment to PBs, probably via molecules that interact with this region. Also, all the mutants appeared to have little direct effect on the distribution of Dcp1-positive PBs in an expression dose-dependent manner (unpublished data). Collectively, these data suggest that an interaction with RNA is required for the appropriate cellular localization of Msi1.
Figure 6.
Two RRMs of Msi1 as a regulated modifier domain of its cytoplasmic localization. Illustration of Msi1 variants that were modifications of the constructs described in Fig. 2 A. (B) HeLa cells were transfected with constructs expressing Flag-Msi1, Flag-Msi1ABmut, 3xFlag-Msi1-D2 (1–189 and 235–362), T7-Msi1, T7-Msi1Amut, T7-Msi1Bmut, T7-Msi1CdelG (1–234), and T7-Msi1CdelI (1–189). HeLa cells treated with (44°C for 30 min; left panels) or without (right panels) heat stress were stained with anti-Flag (green), anti-T7 (green), anti-eIF4G (red), and anti-Dcp1a (red, C) antibodies, respectively. Nuclei were stained with Hoechst (blue) in the merged images. The white arrowheads and white arrow indicate PBs and SGs, respectively. Bars, 5 μm.
Discussion
Molecular mechanism of translational repression and intracellular localization by Msi1
Here, we identified PABP as an Msi1-binding protein, and found that Msi1 competes with another PABP binding partner, eIF4G, to bind PABP; the eIF4G–PABP interaction is required for the formation of the translational initiation complex in mammalian cells (Kahvejian et al., 2005) and for promoting the circularization of mRNA (Wells et al., 1998). Therefore, we show here that Msi1's function as a translational repressor of downstream target mRNAs is exerted by its competition with eIF4G for PABP.
Recently, Paip2 was reported to act as a translational repressor via competition with eIF4G for PABP binding (Karim et al., 2006). However, the mechanisms used by Paip2 and Msi1 to repress translation are different. Msi1 recognizes a specific RNA sequence (G/A)UnAGU (n = 1∼3) with a relatively high affinity (Kd value 4 nM) (Imai et al., 2001) and represses the translation of mRNA in a sequence-dependent manner, as shown in Figs. 2 E and 5 A. Paip2, however, inhibits the translation of mRNA in a sequence-independent manner. Thus, we propose that the Msi1-mediated inhibition of the eIF4G–PABP interaction is a novel mechanism for the translational repression of mRNAs that are specifically bound by Msi1.
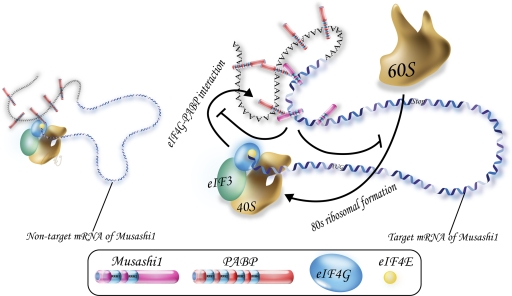
Given that Msi1 physically and functionally interacts with PABP and that it colocalizes with PABP and eIF4G in SGs under stress, we propose that the PABP-mediated translational repression by Msi1 is, at least in part, the mechanism of translational repression that occurs in SGs. SGs contain aggregates of stalled initiation complex, and are thought to be involved in “the mRNA cycle,” which maintains an appropriate ratio of translation machinery to the amount of mRNAs being translated (Parker and Sheth, 2007). Recent findings revealed that the inhibition of ribosome recruitment caused by perturbing the eIF4F (eIF4A and eIF4G) function induces SG formation without the phosphorylation of eIF2α (Mazroui et al., 2006). In addition, although SGs contain most of the components of the translational 48S preinitiation complex, such as eIF4E, eIF4G, PABP, and the small ribosomal subunit, they do not include the large ribosomal subunit (Kedersha et al., 2002; Anderson and Kedersha, 2006). Our present findings suggest that Msi1 represses translation initiation, possibly by perturbing the function of eIF4G, and these events may take place in SGs under stress. That SGs lack the large ribosomal subunit is consistent with our model that Msi1 inhibits the formation of the 80S ribosomal complex (Fig. 7). In addition, because two RRMs are required for the accurate intracellular distribution of Msi1, the localization of Msi1 may occur via capture of its target mRNA.
Figure 7.
A working model for targeted translational repression by Msi1. Msi1 interacts with the 3′ UTR of its target mRNA and PABP, and subsequently inhibits translation initiation by competing with eIF4G for PABP. These sequential events inhibit formation of the 80S ribosome complex.
PBs participate not only in mRNA decay but also in microRNA-mediated translational repression in response to stress. For example, this repression is reversed by HuR, which interacts with the 3′ UTR of its target mRNA and depresses the repression (Bhattacharyya et al., 2006). As shown in Figs. 4–6, Msi1 also localizes to PBs, and it does not participate in its target mRNA decay (Fig. S3; Imai et al., 2001). It is possible that Msi1 is involved, via microRNA, in translational repression in PBs as well as in SGs.
Msi1 inhibits translation initiation in a sequence-specific manner
The results of Fig. 5 (A and B) suggest that Msi1 repressed cap-dependent translation but also modestly inhibited EMCV-IRES- and HCV-IRES-dependent translation. Notably, Hentze's group showed that hnRNP K, which inhibits cap-dependent and IRES-dependent translation, is involved in the inhibition of the recruitment of the 60S ribosomal subunit but not of the 40S subunit (Ostareck et al., 1997, 2001). Thus, these functions of hnRNP K are likely to be similar to those of Msi1. Furthermore, a recent study demonstrated that domain II of HCV IRES is required for the 80S ribosome assembly process after 48S complex formation (Locker et al., 2007), which could be relevant to the repressive actions of Msi1 upon the recruitment of the 60S ribosomal subunit and HCV-IRES-dependent translation.
Our ribosome-binding assays (Fig. 5, E and F) suggest that Msi1 inhibits the translation of the targets at steps between the formation of the 48S preinitiation complex and the formation of the 80S complex, which requires the PAPB-binding domain (D2). Relevantly, Sonenberg's group suggested that both the 40S and the 60S ribosomal subunit recruitment steps are separate targets of PABP, although their underlying molecular mechanisms remain to be elucidated (Kahvejian et al., 2005). Thus, Msi1 could exert its regulatory function at some distinct steps among the multiple stages of translational initiation by binding to PABP. According to these previous reports and based on our present results (Fig. 5, F and G), we consider that Msi1 is involved in inhibiting the formation of the 80S ribosomal complex through an interaction with PABP, without affecting the formation of the 48S complex.
Thus, these actions of Msi1 on translational initiation differ from those of Bruno-Cup's and CPEB-Maskin, which inhibit the eIF4E–eIF4G interaction and the recruitment of the 40S ribosomal subunit in a sequence-specific manner (Stebbins-Boaz et al., 1999; Nakamura et al., 2004; Chekulaeva et al., 2006), but can be explained by one of the following four possibilities. First, Msi1 might be post-translationally modified and inactivated by factors in the RRL, rendering it incapable of binding PABP; an example of this is the Maskin–CPEB interaction, which is regulated by phosphorylation (Groisman et al., 2002). Second, Msi1 binding may be hindered by a unilateral PABP-binding protein like Paip1 (Roy et al., 2002) or an unknown factor. Third, to compete with the eIF4G-PABP, Msi1 may need to recognize an accessible conformation of the 3′ UTR in its target mRNA. Most 3′ UTRs containing poly(A) tails, which bind to multiple PABPs, are probably too flexible. Such structural flexibility leads to a situation in which Msi1 incompletely inhibits the interaction between PABP and eIF4G, even if the number of Msi1 molecules on the 3′ UTR is greater than that of PABP. Indeed, Msi1 incompletely represses the translation of its target mRNA (Figs. 2 E and 5 A; Imai et al., 2001). Thus, because the competition with the eIF4G–PABP interaction by Msi1 may be necessary for the flexibility and energy-requiring dynamic conformational changes of the 3′ UTR, no inhibition of 48S formation and incomplete inhibition of 80S formation may occur. Fourth, Msi1 may indirectly regulate molecules that are influenced by PABP and promote the 80S ribosomal complex. For example, in yeast, the poly(A)/PABP interaction inhibits Slh1p and Ski2p, which in turn inhibits eIF5 and eIF5B, which promote 80S ribosomal formation (Searfoss et al., 2001). To elucidate these events, further work is needed to clarify the relationship between Msi1's function and the detailed molecular mechanisms of ribosome formation.
A recent study showed that Xenopus-Msi regulates the polyadenylation of multiple mRNAs during early Xenopus oocyte maturation and activates translation (Charlesworth et al., 2006). The opposite effects of Msi1 on translational regulation in Xenopus may depend on whether xMsi is involved in cytoplasmic polyadenylation or not. The poly(A) tails of mRNAs expressed in the oocyte are often relatively short. Our model postulates that Msi1 participates in translational regulation by binding to PABP that is coupled with an elongated poly(A) tail. Msi1 may promote PABP stability by binding to PABP, and the stabilized PABP may maintain an extended poly(A) tail in the oocyte. Collectively, Msi could act as a bi-directional regulator of translation in a context-dependent fashion.
Translational control of stem cell characteristics
The biological activity of stem cells in many tissues is regulated by translational and transcriptional controls. In particular, Msi1 helps establish stem cell identity and/or the maintenance of stem cell status, given that Msi1 is strongly expressed in various types of stem cells, including NSCs (Okano et al., 2005) and intestinal stem cells (Potten et al., 2003), and its target mRNAs are involved in stem cell regulation. A recent report indicates that translational repression by Musashi is required intrinsically to maintain Drosophila germline stem cell identity (Siddall et al., 2006). Another group reported that the Msi1-mediated translational repression of p21waf1 mRNA is needed for cell cycle progression (Battelli et al., 2006). These Msi1 functions depend on the translational repression mechanism revealed in this paper. For example, in response to environmental stress (e.g., hypoxia), cells, probably including neural stem/precursor cells (NSPCs), reprogram their translational machinery and sort mRNAs that are released from polysomes to SGs (Kedersha et al., 1999; Stohr et al., 2006). Hypoxia promotes the survival and proliferation of several NSPCs (Studer et al., 2000), indicating that NSPCs may elicit SGs and respond to stress via translational repression. In the present study, we show that Msi1 represses translation initiation under ordinary conditions, and sometimes repression events also take place in SGs under stress. Our previous studies revealed that Msi1 functions in neural stem cell maintenance by binding to its target gene, m-Numb, and repressing its translation (Imai et al., 2001), and that it is involved in the self-renewal of neural stem cells (Sakakibara et al., 2002). Collectively, these results indicate that Msi1 is likely to play an important role in translation in the cytoplasm under ordinary conditions and in SGs under stress conditions, via its inhibition of translational initiation. In addition, the colocalization of Msi1 and eIF4G was high in the VZ (where neural precursor cells are dominant) but not in the cortical plate (where differentiated neuronal cells dominate), in good agreement with our model, and these findings lead to further research focused on the translational control of NSCs. However, the mechanism for the indirect and partial inhibition of eIF4G functions remains to be elucidated.
Materials and methods
Vectors, buffers, and antibodies
Details regarding the plasmid constructs expressing recombinant Msi1, PABP, and eIF4G in this study are available in the supplemental tables (http://www.jcb.org/cgi/content/full/jcb.200708004/DC1). Buffers and antibodies used in this study are also described in the supplemental tables.
Screening for Msi1-binding proteins
293T cells were transfected with a plasmid to express the Flag-Msi1-TAP tag or the Flag-TAP tag alone. After 2 d of culture, the cells were lysed in IPP150 buffer with Complete Protease Inhibitor Cocktail (Roche), and the TAP-fusion proteins in the lysate were purified using the Séraphin Laboratory TAP protocol (http://www.cgm.cnrs-gif.fr/epissage/). The Msi1-binding proteins, the specific bands of which are shown in Fig. 1 A, were identified by MALDI-TOF mass spectrometry (Apro-Science).
Cell culture and transfection
All cell lines (293T, P19, HeLa) were cultured as described previously (Imai et al., 2001), and transfections of Msi1 variants, PABP variants, and eIF4G variants in 293T or HeLa cells were performed using the Fugene 6 transfection reagent (Roche) or Lipofectamine 2000 (Invitrogen). Cultures of dissociated rat primary hippocampal neurons were prepared as described previously (Iijima et al., 2005), and then staining was examined in stage-5.
Protein purification and immunoblotting
GST- and His-tagged proteins were expressed in Escherichia coli strain BL21 and purified by glutathione-Sepharose 4B and ProBond resin as described by the manufacturer (GE Healthcare and Invitrogen) and the previous study (Imai et al., 2001). Immunoblotting was performed using methods described previously (Kaneko et al., 2000). To detect and quantify the probed proteins, ECL reagent (GE Healthcare) and the LAS 3000 mini PhosphorImager (Fujifilm) and its software were used.
In vitro translation assay
For the in vitro translation assays, luciferase reporter mRNAs containing the cap and poly(A)-tail were synthesized following the standard procedure for mMESSAGE mMACHINE T7 Ultra (Ambion) after pT7-rLuc-MCS and pT7-fLuc-MCSmut were linearized with XhoI digestion, respectively. In this kit (Ambion), these capped mRNAs were synthesized by using ARCA (anti-reverse cap analogue; Ambion). Similarly, luciferase reporter mRNAs containing the IRES and poly(A)-tail were synthesized without the cap analogue, according to standard procedures (Ambion), after pT7-HCV IRES rLuc-MCS, pT7-HCV IRES rLuc-MCSmut, pT7-EMCV IRES-fLuc-MCS, or pT7-EMCV IRES-fLuc-MCSmut was linearized with XhoI digestion, respectively. The in vitro translation reactions were performed as described below, according to the manufacturer's protocol (Promega). Each reaction mixture (total volume 12 μl) contained: 8.0 μl of nuclease-treated rabbit reticulocyte lysate (RRL), 0.50 μl of complete amino acid mix (1 mM stock; Promega), 0.25 μl of RNasin (40 U/μl l stock; Promega), 0.28 μl of 2 M KCl, 0.075 pmol of luciferase reporter mRNA, and 7.5 pmol of recombinant proteins (GST, GST-Msi1, GST-Msi1-D2). The reaction mixtures were incubated at 30°C for 0–90 min, and the luciferase activity was measured at time points throughout the incubation period. To assay luciferase activity, 1 μl of the translation reaction was added to 25 μl luciferase assay reagent (Picka-Gene Dual; Toyo B-net Co., ltd) and immediately measured in a 0.1-s reading using a Luminometer (Lumat LB 960).
Immunohistochemistry and immunofluorescence
The immunohistochemical staining of E14 mouse brain coronal sections with anti-Msi1 (Mab 14H1), PABP, eIF4G, or Sox1/(2)/3 antibodies were performed as described previously (Okada et al., 2004; Tokunaga et al., 2004). Immunocytochemistry was performed as described previously (Tanaka et al., 2006), and CSK buffer was used to wash before cells were fixed. The stainings were visualized by AlexaFluor 488-, 555-, or 568-conjugated secondary antibodies (Invitrogen). The digital images of cells were captured by a laser confocal microscope (LSM510; Carl Zeiss, Inc.), using in immunohistochemistry either a 20x/0.5 NA or 63x/1.2 NA water objective lens and in immunocytochemistry a 100x/1.45 NA oil or 63x/1.4 NA oil objective lens. Image acquisition was performed with LSM Image Browser software (Carl Zeiss, Inc.).
Subcellular fractionation and sucrose gradient analysis
Subcellular fractionation and sucrose gradient analysis were performed as described previously (Matsumoto et al., 2000; Aoki et al., 2002), using P19 cells treated with heat shock at 44°C or untreated. Ultracentrifugation was performed using either the MLS 50 rotor (Beckman Coulter) at 100,000 g for 1 h at 4°C (Fig. 4 C) or a Hitachi P50S2 rotor at 48,000 rpm for 0.8 h at 4°C (Fig. 4 D). The gradients in Fig. 4 D were then sequentially fractionated into 230-μl fractions by a piston gradient fractionator (Biocomp).
Immunoprecipitation and in vitro and in vitro competition assays
Immunoprecipitations were performed in TC buffer with Complete Protease Inhibitor Cocktail (Roche). E14 mouse brain was homogenized in TC buffer and spun at 20,400 g. The supernatants, treated with RNase A or untreated, were precleared with protein G–Sepharose (GE Healthcare) for 1 h at 4°C, followed by incubation with either an anti-Msi1 antibody (28 μg/ml) or equimolar amounts of control purified IgG (R&D Systems) and protein G–Sepharose, and the beads were then washed extensively. The precipitated proteins were detected by immunoblotting. In vitro competition assays were performed in TC buffer without 1 mM DTT as described previously (Khaleghpour et al., 2001). First, GST-PABP (5.4 pmol) was mixed with 1.8, 3.6, or 5.4 pmol of GST-Msi1 in 10 μl of reaction mixture, and incubated for 1 h at 4°C. Second, the mixture was incubated again with 10 μl of anti-FLAG M2 resin (Sigma-Aldrich) conjugated with Flag-eIF4G (41–1560)-His for 1 h at 4°C. Third, to remove unbound GST-PABP, the resin was washed three times with 1 ml of TC buffer. In vivo competition assays were also performed in TC buffer without 1 mM DTT, as described previously (Khaleghpour et al., 2001). 293T cells expressing both Flag-eIF4GN and Myc-Msi1 were homogenized and spun at 20,400 g. The supernatants, with RNase A treatment, were then incubated with anti-FLAG M2 resin for 1 h at 4°C. The resin was washed extensively in 1 ml of TC buffer. Proteins were eluted with 2× Laemmli sample buffer, and processed for immunoblotting.
Kinetics measurements by AffinixQ
Each of the His-tag proteins was immobilized onto the QCM Au electrode, as described in the manufacturer's protocol (AffinixQ; Initium Inc.) and the previous studies (Okahata et al., 1998; Sato et al., 2004). Measurements were performed under the following conditions: QCM Assay buffer-8, 750 rpm, at 25°C. The binding between PABP and QCM-immobilized Msi1 (or eIF4G) was determined by Equation 1.
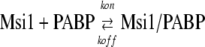 |
(1) |
The concentration of Msi1/PABP complex formed at time t after injection is given by Equations 2–4. The time dependence of the increase in mass is indicated by Δmt.
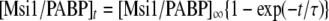 |
(2) |
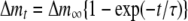 |
(3) |
 |
(4) |
To obtain kinetics constants from the linear reciprocal plots of the relaxation time τ against the concentration of GST-PABP according to Equation 4, the relaxation time τ was used in the time range from 0 to 45 min. Dissociation constants (Kd) were obtained with the equation [Kd = koff /kon]. We analyzed these data by curve fitting following the manufacturer's procedures.
Quantitation of colocalization in granules
The ratio of Msi1-containing granules to total marker-containing granules was determined in Fig. 4 B. The percent colocalization (Fig. 4 B; 1) was estimated as described previously (Barbee et al., 2006). The weighted colocalization coefficients (Fig. 4 B; 2) were calculated using LSM Image Examiner software (Carl Zeiss, Inc.).
Cap column assay
Cap chromatography was performed as described previously (Stebbins-Boaz et al., 1999). Transfected HeLa cell lysate (supplemented with 0.2 mM GTP) was incubated at 4°C for 1 h with 15 μl of m7GTP resin (GE Healthcare) in TC buffer, and the resin was then washed extensively in TC buffer. The cap-binding complex was eluted with m7GpppG (0.2 mM; Ambion), and proteins were resolved by SDS-PAGE.
Ribosome binding assay
The 80S ribosome binding assays were performed with the following steps: (1) RRL was preincubated at 30°C for 20 min; (2) the RRL was then incubated with radio-labeled (3′-end) reporter mRNA (0.36 pmol), equimolar amounts of Msi1 (3.6 pmol) or GST, cycloheximide (0.6 mM) (EMD), complete amino acid mix (0.05 mM) (Promega), RNasin (40 U) (Promega), and high salt buffer, in a total volume of 37.5 μl, at 30°C for 20 min. The following steps; (3) stop-reaction, (4) ultracentrifugation, (5) fractionation, were performed as described previously (Kahvejian et al., 2005). 40S ribosome binding assays were performed similarly to the 80S ribosome binding assay except for using low salt buffer (LSB) and adding GMP-PNP (2 mM) (Sigma-Aldrich) to prevent 60S subunit joining in incubation step (2).
Northern blot analysis
After in vitro translation (Fig. 5 A), each total RNA from each fraction was prepared from the RRL lysate as described previously (Matsumoto et al., 2000). Northern blot analyses were performed as described previously (Iijima et al., 2005). Hybridization signals were detected using BAS2500 (Fujifilm).
Online supplemental material
Fig. S1 shows the specific interaction between Msi1 and PABP by coimmunoprecipitation assay and GST pull-down assay using purified proteins. Fig. S2 shows the biophysical analysis using a QCM-resonator, the AffinixQ (Initium Inc.). Fig. S3 shows Northern blot analysis of the reporter mRNAs isolated from the RRL after in vitro translation in Fig. 5 A. The supplemental tables include lists of plasmids, buffers, and antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708004/DC1.
Supplementary Material
Acknowledgments
We greatly appreciate our fruitful discussions with Drs. K.J. Tanaka and S. Yamasaki. We are grateful to Drs. M. Yano and Y. Okada for technical advice; Drs. T. Iijima, K. Sawai, H. Tada, S. Shibata, T. Yoshizaki, and T. Nishikata for help and encouragement; Drs. S. Hoshino and T. Katada for the pUC18-T7-R-luc-HCV IRES-F-luc plasmid; Dr. B. Séraphin and Cellzome for the pBS1539 plasmid; Dr. N. Standart for the pGEX2T-PABP plasmid; Dr. Y. Okada for the plasmid containing EMCV-IRES; Drs. H.J. Okano and M. Yano for several plasmids containing PABP mutants and pGEX6P-hnRNPK; Dr. H. Kondoh for the anti-Sox1/(2)/3 antibody; and Dr. T. Shimogori and Mr. T. Sunabori for information on the anti-eIFs antibodies.
This work was supported by grants from the Japanese Ministry of Education, Sports and Culture of Japan (MEXT) to H. Okano and T. Imai, the 21st Century COE Program at Keio University, and Keio University Grant-in-Aid for Encouragement of Young Medical Scientists.
Abbreviations used in this paper: CBB, Coomassie brilliant blue; fLuc, firefly luciferase; IMP, insulin-like growth factor 2 mRNA binding protein; IRES, internal ribosomal entry site; MCS, Msi1-binding consensus sequence; MCSmut, mutated MCS; Msi, Musashi; NSC, neural stem cell; NSPC, neural stem/precursor cell; PABP, poly(A) binding protein; PB, processing body; QCM, quartz crystal microbalance; RRL, rabbit reticulocyte lysate; RRM, RNA recognition motif; SG, stress granule; TAP, tandem affinity purification; UTR, untranslated region; VZ, ventricular zone.
References
- Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Y. Ishii, K. Matsumoto, and M. Tsujimoto. 2002. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30:5182–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee, S.A., P.S. Estes, A.M. Cziko, J. Hillebrand, R.A. Luedeman, J.M. Coller, N. Johnson, I.C. Howlett, C. Geng, R. Ueda, et al. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 52:997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelli, C., G.N. Nikopoulos, J.G. Mitchell, and J.M. Verdi. 2006. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 31:85–96. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S.N., R. Habermacher, U. Martine, E.I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 125:1111–1124. [DOI] [PubMed] [Google Scholar]
- Charlesworth, A., A. Wilczynska, P. Thampi, L.L. Cox, and A.M. MacNicol. 2006. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 25:2792–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva, M., M.W. Hentze, and A. Ephrussi. 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 124:521–533. [DOI] [PubMed] [Google Scholar]
- Deo, R.C., N. Sonenberg, and S.K. Burley. 2001. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc. Natl. Acad. Sci. USA. 98:4414–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963. [DOI] [PubMed] [Google Scholar]
- Groisman, I., M.Y. Jung, M. Sarkissian, Q. Cao, and J.D. Richter. 2002. Translational control of the embryonic cell cycle. Cell. 109:473–483. [DOI] [PubMed] [Google Scholar]
- Guo, T., A.H. Peters, and P.A. Newmark. 2006. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 11:159–169. [DOI] [PubMed] [Google Scholar]
- Iijima, T., T. Imai, Y. Kimura, A. Bernstein, H.J. Okano, M. Yuzaki, and H. Okano. 2005. Hzf protein regulates dendritic localization and BDNF-induced translation of type 1 inositol 1,4,5-trisphosphate receptor mRNA. Proc. Natl. Acad. Sci. USA. 102:17190–17195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, T., A. Tokunaga, T. Yoshida, M. Hashimoto, K. Mikoshiba, G. Weinmaster, M. Nakafuku, and H. Okano. 2001. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21:3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Y.V. Svitkin, R. Sukarieh, M.N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, Y., S. Sakakibara, T. Imai, A. Suzuki, Y. Nakamura, K. Sawamoto, Y. Ogawa, Y. Toyama, T. Miyata, and H. Okano. 2000. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 22:139–153. [DOI] [PubMed] [Google Scholar]
- Karim, M.M., Y.V. Svitkin, A. Kahvejian, G. De Crescenzo, M. Costa-Mattioli, and N. Sonenberg. 2006. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. USA. 103:9494–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N.L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., S. Chen, N. Gilks, W. Li, I.J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., A. Kahvejian, G. De Crescenzo, G. Roy, Y.V. Svitkin, H. Imataka, M. O'Connor-McCourt, and N. Sonenberg. 2001. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21:5200–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, R., and C. Nusslein-Volhard. 1986. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 47:141–152. [DOI] [PubMed] [Google Scholar]
- Locker, N., L.E. Easton, and P.J. Lukavsky. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, K., K. Aoki, N. Dohmae, K. Takio, and M. Tsujimoto. 2000. CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes. Nucleic Acids Res. 28:4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui, R., R. Sukarieh, M.E. Bordeleau, R.J. Kaufman, P. Northcote, J. Tanaka, I. Gallouzi, and J. Pelletier. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 17:4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science. 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- Muckenthaler, M., N.K. Gray, and M.W. Hentze. 1998. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell. 2:383–388. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., K. Sato, and K. Hanyu-Nakamura. 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 6:69–78. [DOI] [PubMed] [Google Scholar]
- Okabe, M., T. Imai, M. Kurusu, Y. Hiromi, and H. Okano. 2001. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 411:94–98. [DOI] [PubMed] [Google Scholar]
- Okada, Y., T. Shimazaki, G. Sobue, and H. Okano. 2004. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 275:124–142. [DOI] [PubMed] [Google Scholar]
- Okahata, Y., K. Niikura, Y. Sugiura, M. Sawada, and T. Morii. 1998. Kinetic studies of sequence-specific binding of GCN4-bZIP peptides to DNA strands immobilized on a 27-MHz quartz-crystal microbalance. Biochemistry. 37:5666–5672. [DOI] [PubMed] [Google Scholar]
- Okano, H., T. Imai, and M. Okabe. 2002. Musashi: a translational regulator of cell fate. J. Cell Sci. 115:1355–1359. [DOI] [PubMed] [Google Scholar]
- Okano, H., H. Kawahara, M. Toriya, K. Nakao, S. Shibata, and T. Imai. 2005. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306:349–356. [DOI] [PubMed] [Google Scholar]
- Ostareck, D.H., A. Ostareck-Lederer, M. Wilm, B.J. Thiele, M. Mann, and M.W. Hentze. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 89:597–606. [DOI] [PubMed] [Google Scholar]
- Ostareck, D.H., A. Ostareck-Lederer, I.N. Shatsky, and M.W. Hentze. 2001. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 104:281–290. [DOI] [PubMed] [Google Scholar]
- Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., I.N. Shatsky, S.P. Fletcher, R.J. Jackson, and C.U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten, C.S., C. Booth, G.L. Tudor, D. Booth, G. Brady, P. Hurley, G. Ashton, R. Clarke, S. Sakakibara, and H. Okano. 2003. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 71:28–41. [DOI] [PubMed] [Google Scholar]
- Preiss, T., and W.M. Hentze. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 25:1201–1211. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Roy, G., G. De Crescenzo, K. Khaleghpour, A. Kahvejian, M. O'Connor-McCourt, and N. Sonenberg. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 22:3769–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, S., T. Imai, K. Hamaguchi, M. Okabe, J. Aruga, K. Nakajima, D. Yasutomi, T. Nagata, Y. Kurihara, S. Uesugi, et al. 1996. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 176:230–242. [DOI] [PubMed] [Google Scholar]
- Sakakibara, S., Y. Nakamura, T. Yoshida, S. Shibata, M. Koike, H. Takano, S. Ueda, Y. Uchiyama, T. Noda, and H. Okano. 2002. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. USA. 99:15194–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., I. Sagami, and T. Shimizu. 2004. Identification of caveolin-1-interacting sites in neuronal nitric-oxide synthase. Molecular mechanism for inhibition of NO formation. J. Biol. Chem. 279:8827–8836. [DOI] [PubMed] [Google Scholar]
- Searfoss, A., T.E. Dever, and R. Wickner. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21:4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddall, N.A., E.A. McLaughlin, N.L. Marriner, and G.R. Hime. 2006. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl. Acad. Sci. USA. 103:8402–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana, E.P., and C.Q. Doe. 1996. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 17:21–26. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz, B., Q. Cao, C.H. de Moor, R. Mendez, and J.D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 4:1017–1027. [DOI] [PubMed] [Google Scholar]
- Stohr, N., M. Lederer, C. Reinke, S. Meyer, M. Hatzfeld, R.H. Singer, and S. Huttelmaier. 2006. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer, L., M. Csete, S.H. Lee, N. Kabbani, J. Walikonis, B. Wold, and R. McKay. 2000. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J. Neurosci. 20:7377–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K.J., K. Ogawa, M. Takagi, N. Imamoto, K. Matsumoto, and M. Tsujimoto. 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 281:40096–40106. [DOI] [PubMed] [Google Scholar]
- Tokunaga, A., J. Kohyama, T. Yoshida, K. Nakao, K. Sawamoto, and H. Okano. 2004. Mapping spatio-temporal activation of Notch signaling during neurogenesis and gliogenesis in the developing mouse brain. J. Neurochem. 90:142–154. [DOI] [PubMed] [Google Scholar]
- Wells, S.E., P.E. Hillner, R.D. Vale, and A.B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 2:135–140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.