Abstract
During polarized growth and tissue morphogenesis, cells must reorganize their cytoplasm and change shape in response to growth signals. Dynamic polymerization of actin filaments is one cellular component of polarized growth, and the actin-related protein 2/3 (ARP2/3) complex is an important actin filament nucleator in plants. ARP2/3 alone is inactive, and the Arabidopsis thaliana WAVE complex translates Rho-family small GTPase signals into an ARP2/3 activation response. The SCAR subunit of the WAVE complex is the primary activator of ARP2/3, and plant and vertebrate SCARs are encoded by a small gene family. However, it is unclear if SCAR isoforms function interchangeably or if they have unique properties that customize WAVE complex functions. We used the Arabidopsis distorted group mutants and an integrated analysis of SCAR gene and protein functions to address this question directly. Genetic results indicate that each of the four SCARs functions in the context of the WAVE-ARP2/3 pathway and together they define the lone mechanism for ARP2/3 activation. Genetic interactions among the scar mutants and transgene complementation studies show that the activators function interchangeably to meet the threshold for ARP2/3 activation in the cell. Interestingly, double, triple, and quadruple mutant analyses indicate that individual SCAR genes vary in their relative importance depending on the cell type, tissue, or organ that is analyzed. Differences among SCARs in mRNA levels and the biochemical efficiency of ARP2/3 activation may explain the functional contributions of individual genes.
INTRODUCTION
In plants, the shapes and mechanical properties of cells are intimately linked to their function. For example, in the leaf epidermis, the pavement cells, guard cells, trichomes, and their support cells form an interlocking network of highly specialized cells. The epidermis not only defines the interface of the plant with the environment, but this organ strongly affects the growth properties of underlying tissues and the overall architecture of the plant (Savaldi-Goldstein et al., 2007). Consequently, there is great interest in understanding, at a molecular level, how epidermal cells define the location and extent of growth. It is clear that the cytoskeleton is a critical regulatory target, and the localized polymerization and stabilization of microtubules and actin filaments control the spatial patterns of cell expansion (reviewed in Panteris and Galatis, 2005; Smith and Oppenheimer, 2005; Kotzer and Wasteneys, 2006). This article focuses on understanding the mechanisms by which cellular signals are converted to an actin filament nucleation response.
Actin filaments have numerous functions in the cell. Leaf epidermal cells, like most expanding vacuolated cells, contain a loose web of actin filaments and bundles that populate the cortical and transvacuolar cytoplasm. These bundles not only provide a substrate for the long-distance intracellular transport of a variety of organelles (Boevink et al., 1998; Nebenfuhr et al., 1999; Mathur et al., 2002), but they also function as a scaffold for stable organelle positioning. For example, in pavement cells, an anastomosing network of the endoplasmic reticulum partially mirrors the locations of actin bundles (Boevink et al., 1998). In trichomes, disruption of actin bundle organization in the core cytoplasm leads to defects in the positioning of the central vacuole and disrupts polarized growth (Le et al., 2003). Similar effects of actin filament disruption on vacuole positioning have been described in tip-growing pollen tubes (Kost et al., 1999; Kaothien et al., 2005). Actin filaments have been frequently hypothesized to mediate the localized delivery of post-Golgi vesicles that support polarized growth (reviewed in Hussey et al., 2006). However, in general, the functions of dynamic actin filaments and cortical arrays are not well understood in plants.
It is clear that plants employ conserved actin filament nucleators during growth and development. Recently, several proteins that link actin polymerization to cell growth have been identified (reviewed in Smith and Oppenheimer, 2005; Szymanski, 2005; Hussey et al., 2006; Staiger and Blanchoin, 2006). Numerous mutants in Arabidopsis thaliana and in the moss Physcomitrella patens provide clear genetic proof that the actin-related protein 2/3 (ARP2/3) complex is a key actin filament nucleator (Le et al., 2003; Li et al., 2003; Mathur et al., 2003a; Harries et al., 2005). ARP2/3 controls many aspects of plant morphogenesis (Li et al., 2004), and arp2/3 mutant cells undergoing both diffuse growth (Mathur et al., 1999; Szymanski et al., 1999) and tip growth (Finka et al., 2007) have clear defects in the organization of the actin cytoskeleton. There is a correlation between the localization of plant ARP2/3 and actin with broad cytoplasmic domains of polarized growth (Hable and Kropf, 2005; Fiserova et al., 2006; Perroud and Quatrano, 2006; Finka et al., 2007).
The ARP2/3 complex alone is inactive, and diverse classes of proteins termed nucleation promoting factors or activators assemble into heteromeric complexes that can increase the actin filament nucleation activity of ARP2/3 (reviewed in Welch and Mullins, 2002; Stradal and Scita, 2006). Multicellular organisms employ a heteromeric complex termed WAVE to convert Rac/ROP small GTPase signals into an ARP2/3 activation response (Eden et al., 2002; Basu et al., 2004; Steffen et al., 2004). The eight known Arabidopsis distorted group genes define a WAVE-ARP2/3 growth control pathway, and double mutant analyses prove that WAVE positively regulates ARP2/3 (reviewed in Szymanski, 2005). The WAVE complex protein termed SCAR (for suppressor of cAMP receptor) (Bear et al., 1998), also referred to as WASP family Verprolin-homologous protein (WAVE) in vertebrates (Miki et al., 1998), is the WAVE complex subunit that binds to and activates ARP2/3 (Machesky et al., 1999; Eden et al., 2002; Innocenti et al., 2004; Basu et al., 2005; Stovold et al., 2005). In this article, we refer to the ARP2/3 activator as SCAR and the entire heteromeric complex as WAVE according to its original name (Eden et al., 2002). One of the Arabidopsis distorted group genes known either as DISTORTED3 (DIS3) (Basu et al., 2005) or as IRREGULAR TRICHOME BRANCH1 (Zhang et al., 2005) corresponds to SCAR2. SCAR2 is a potent activator of vertebrate ARP2/3 (Basu et al., 2005), and the gene clearly functions in the WAVE-ARP2/3 pathway (Basu et al., 2005; Zhang et al., 2005; Le et al., 2006).
Plant SCAR genes encode a conserved N-terminal SCAR homology domain (SHD) and a C-terminal WCA/VCA (WA) domain that is separated by a large and highly diverged internal segment of no known function. The SHD appears to regulate protein stability via an interaction with the WAVE complex protein BRICK1 (Djakovic et al., 2006; Le et al., 2006) and the assembly of SCAR into the WAVE complex (Basu et al., 2005). The acidic WA domain is highly conserved in all WASP/SCAR family members, and WA is sufficient for ARP2/3 activation activity (Panchal et al., 2003). However, plant SCAR proteins may differ in the efficiency with which they activate ARP2/3. For example, in one comparative study, Zea mays SCAR1 potently activated ARP2/3, whereas Arabidopsis SCAR4 had extremely limited activity (Frank et al., 2004).
It is possible that SCAR genes have evolved unique morphogenetic functions. Vertebrate genomes encode at least three SCAR isoforms with distinct expression patterns (Sossey-Alaoui et al., 2003). Mutation of SCAR2/WAVE2 causes severe embryonic defects (Yan et al., 2003). SCAR1/WAVE1 may have unique signaling functions that are independent of ARP2/3 (Westphal et al., 2000; Soderling et al., 2007). Multiple SCARs may have independent functions within an individual cell. In one study of cultured fibroblasts, SCAR1/WAVE1 and SCAR2/WAVE2 are reported to independently regulate the formation of dorsal and peripheral ruffles, respectively (Suetsugu et al., 2003). However, in a second similar study employing an identical cell type, the separate functions of these SCAR isoforms were not confirmed (Legg et al., 2007). Furthermore, multiple SCARs are not required in animals because Drosophila melanogaster employs a single SCAR during development (Zallen et al., 2002). In Arabidopsis, the importance of the SCAR family is not well characterized. Arabidopsis encodes four SCARs, each with a similar domain organization (Frank et al., 2004).The overlapping expression patterns of the four genes and the relatively weak phenotypes of scar2 plants compared with other distorted mutants led to the suggestion that that there is functional redundancy within the gene family (Basu et al., 2005; Le et al., 2006). Consistent with this hypothesis, SCAR2 and SCAR4 function redundantly and are the lone SCAR genes that regulate trichome morphogenesis because only the scar2;scar4 double mutant (the semicolon indicates unlinked genes) had a phenotype that was indistinguishable from arp2/3 null strains (Uhrig et al., 2007). On the other hand, based on the distinct yeast two-hybrid and bimolecular fluorescence complementation interactions of individual SCARs with other WAVE complex and signaling proteins, it has also been proposed that highly specialized WAVE complexes assemble to perform distinct functions (Uhrig et al., 2007).
In this article, a reverse genetic analysis was undertaken to define more clearly the functional assignments and regulatory schemes of the Arabidopsis SCAR gene family. Based on the quantitative comparisons of double, triple, and quadruple scar mutants with arp2/3 null plants, we find that SCAR and the WAVE pathway appear to define the sole mechanism for ARP2/3 activation, and we find no evidence for SCAR functions that are independent of ARP2/3. Genetic interactions and gene dosage sensitivity among scar mutants reveal widespread redundancy and clear thresholds for SCAR function and ARP2/3 activation. Interestingly, the relative contribution of individual SCARs is clearly not equal, and the exact nature of this unequal genetic redundancy varies depending on the cell type or tissue examined. Our genetic analysis coupled with cell type–specific measurements of SCAR mRNA levels and comparisons of the biochemical efficiency of ARP2/3 activation point to a threshold model. In this regulatory scheme, cell types have a capacity to use different combinations of SCAR homologs in an interchangeable manner to meet defined cellular requirements for ARP2/3 activation.
RESULTS
Evolution and Reverse Genetic Analysis of Arabidopsis SCAR Genes
Arabidopsis encodes four SCAR homologs with conserved SHD and WA domains (Figure 1A), but the importance of individual family members during plant development is not well understood. Therefore, we initiated a broader functional analysis of the SCAR gene family by comparing evolutionary relationships among plant SCAR genes from species with an available genome sequence. In Arabidopsis, SCAR1 and SCAR3 are parts of two large unlinked, duplicated blocks of genes that are by-products of a relatively recent whole-genome duplication event (Blanc et al., 2003). SCAR2 and SCAR4 are proposed to reflect a more ancient duplication, and our phylogenetic analyses using plant SCAR proteins from highly diverged species were consistent with SCAR duplication events occurring prior to (SCAR2/4) and after (SCAR1/3) the speciation of Arabidopsis (Figure 2). It is interesting that both of the strongly supported SCAR1/3 and SCAR2/4 clades contained distinct SCAR homologs from rice (Oryza sativa) and poplar (Populus trichocarpa).
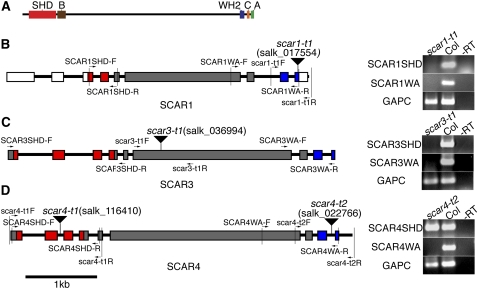
Figure 1.
SCAR Protein Domains and Physical Maps of the Arabidopsis scar T-DNA Insertion Alleles.
(A) Domain organization of a typical plant SCAR family protein, Arabidopsis SCAR4.
(B) to (D) Molecular characterization of SCAR T-DNA insertion alleles. The presence of SHD and WA encoding regions of scar transcripts was assayed using RT-PCR (right panels). Detection of the GAPC gene served as a positive control. Wild-type (Col-0) cDNA and a no-RT reaction served as the positive and negative controls, respectively. The position of the primers used for characterizing T-DNA insertions and the transcription of SHD and WA domains are marked on the physical map by arrows. Red boxes indicate SHD-encoding exons, blue boxes indicate exons encoding the WA domain, gray boxes indicate the regions that encode the nonconserved region in the SCAR proteins, and white boxes indicate the untranslated regions. The inverted triangles label the position of the T-DNA insertions.
(B) Analysis of scar1-t1 (salk_017554) transcripts.
(C) Analysis of scar3-t1 (salk_036994) transcripts.
(D) Analysis of scar4-t2 (salk_022766) transcripts.
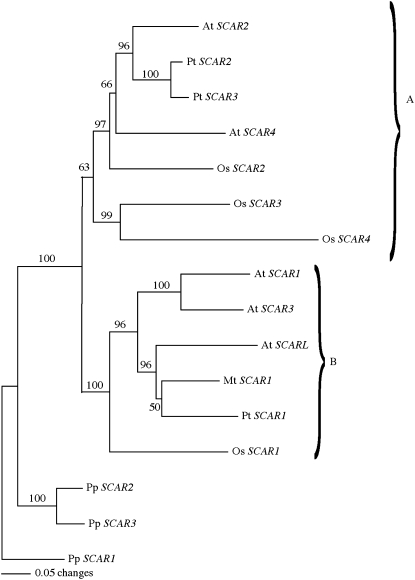
Figure 2.
Distance-Based Gene Tree of the SHD and WA Domains Manually Fused Together for Arabidopsis, M. truncatula, P. trichocarpa, O. sativa, and P. patens.
We designated SCARs within the Arabidopsis SCAR2/4 clade as the A clade and those within the SCAR1/3 clade as the B clade. The numbering scheme for the SCAR genes is not intended to reflect ancestral relationships. With the exception of Arabidopsis SCARL that contains only an SHD domain, SCAR-like proteins from other species that did not contain both a SHD and WA domain were not included in this tree. The resulting tree had a minimum evolution score of 567.3. Corresponding accession numbers for each gene can be found under accession numbers and the corresponding Nexus alignment as Supplemental Data Set 1 online.
The A family included SCARs most similar to the SCAR2/4 homologs, and the B family included SCARs that were more closely related to the SCAR1/3 paralogs. This evolutionary model is consistent with an ancient split in the SCAR gene family that predates the monocot and dicot divergence (Figure 2). Similar ancient divergences have been studied for other plant cytoskeletal proteins, including actin (Meagher et al., 1999). If Arabidopsis SCARs encode functional proteins that are subject to selective pressure over evolutionary time scales, one would expect to observe a low frequency of DNA sequence changes that alter the protein-coding information. Indeed, we detected an extremely low ratio of nonsynonymous (Ka) to synonymous (Ks) nucleotide sequence changes for each of the four SCARs (Table 1). Low Ka/Ks ratios are common for functional proteins, and values in this range are consistent with the hypothesis that each SCAR is functional and subject to purifying selection that maintains amino acid sequences (Fay and Wu, 2003).
Table 1.
Ratio of Nonsynonymous to Synonymous Distance between Arabidopsis SCAR Genes
SCAR genes were identified as described in Methods.
The ratio of nonsynonymous to synonymous distances between SCAR genes. Pairwise alignments using virtual translation between full-length SCAR genes were generated using DNAStar Lasergene. Synonymous and nonsynonymous distances were determined using PAML (Yang, 1997).
We therefore set out to test the hypothesis that the distinct SCAR clades reflect specialized functions within the gene family. Because removal of the SCAR WA-encoding domain of SCAR2 creates null alleles (Basu et al., 2005; Zhang et al., 2005), it was relatively straightforward to use the publicly available T-DNA insertion lines (Alonso et al., 2003) to isolate knockouts for the remaining SCARs (Figures 1B to 1D). We isolated RNA from each of the putative SCAR knockout lines and analyzed the transcriptional effects of the T-DNA insertions. The quality and purity of the RNA samples used to characterize scar alleles were confirmed by the efficient amplification of GAPC and by the lack of signal in RNA samples that were not treated with reverse transcriptase, respectively. We found that the T-DNA insertion lines salk_017554 (scar1-t1) and salk_036994 (scar3-t1) destabilized the transcripts encoded by SCAR1 and SCAR3, respectively (Figures 1B and 1C). The insertion salk_022766 (scar4-t2) causes premature transcription termination of the SCAR4 gene and removes the coding information for the WA (Figure 1D). Therefore, the scar1, -3, and -4 T-DNA alleles are likely to be null. We also generated a null allele of the SCAR-like (SCARL) gene AT4G18600 using salk_055413 (scarl-t1). SCARL lacks a WA domain and is therefore predicted to have no functional relevance in the WAVE-ARP2/3 pathway. We found that the scarl-t1 allele did not show any noticeable genetic interactions during morphogenesis with scar2 or any other scar mutants in a variety of triple, quadruple, and quintuple mutant combinations. Because these results were negative, we did not do additional genotyping to define precisely the individual genotypes of the combinations that showed no interactions.
SCAR Functions during Trichome Development: Threshold Models for Gene Function
Wild-type Arabidopsis trichomes have three or more highly elongated branches (Figure 3A). As shown previously (Basu et al., 2005), scar2 null alleles caused weak cell swelling and twisting as well as a significant reduction in branch length (Figure 3B, Table 2). The scar1, -3, and -4 single mutants did not have a trichome phenotype (Figure 3C, Table 2). We tested for functional redundancy within the gene family by constructing combinations of double, triple, and quadruple scar mutants. The double mutant combination of scar2;scar4 strongly enhanced the scar2 phenotype and caused trichome swelling, twisting, and a reduced branch length that were indistinguishable from the WAVE and ARP2/3 complex subunit mutants brk1/hspc300 and dis2/aprc2, respectively (Figure 3D, Table 2). The brk1 and dis2 phenotypes reflect wave and arp2/3 null phenotypes, respectively (El-Assal et al., 2004a; Djakovic et al., 2006; Le et al., 2006). The genetic interaction was not unique to the scar4-t2 and scar2 (dis3-1) alleles because a similar strong distorted phenotype was observed in scar2 (dis3-3);scar4-t2 and in scar2 (dis3-4);scar4-t2 plants. We also observed the strong phenotype when an independent scar4 allele, scar4-t1 (salk_116410), was in a double mutant combination with scar2 (dis3-1).
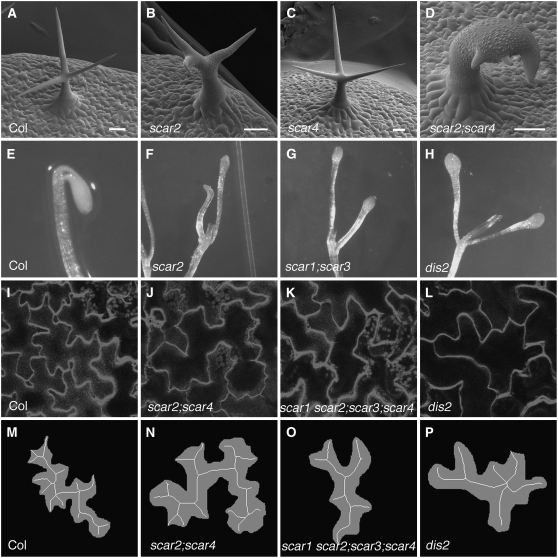
Figure 3.
SCAR Genes Function Redundantly during Morphogenesis and in Dark-Grown Seedlings.
(A) to (D) Scanning electron micrograph of mature trichomes. Bars = 50 μm.
(A) The mature wild-type (Col-0) trichome has three highly elongated branches.
(B) The mature scar2 (dis3-1) trichome with a typical weak distorted phenotype.
(C) The mature scar4 (scar4-t2) trichome has three highly elongated branches.
(D) The mature scar2;scar4 double mutant trichome has a very strong swollen and distorted phenotype.
(E) to (H) Shoots of 7-DAG dark-grown seedlings of wild-type (E), scar2 (F), scar1;scar3 (G), and arpc2 (dis2) (H).
(I) to (L) Wide-field fluorescence images of fields of cotyledon epidermal pavement cells of wild-type (I), scar2;scar4 (J), scar1 scar2;scar3;scar4 (K), and arpc2 (dis2) (L).
(M) to (P) Digitally processed images of individual pavement cells from (I) to (L) overlayed with their calculated skeletons.
Table 2.
Quantitative Analysis of the Trichome Branch Length Phenotype of scar, brk1, and dis2 Mutants
| Genotype | Branch 1 (μm) | Branch 2 (μm) | Branch 3 (μm) |
|---|---|---|---|
| Col | 223.5 ± 12.3 (16) a | 199.3 ± 13.2 (16) a | 171.1 ± 11.8 (16) a |
| scar1 | 208.9 ± 13.1 (17) a | 176.7 ± 10.5 (17) a | 145.0 ± 12.0 (17) a |
| scar2 | 136.6 ± 10.4 (22) b | 93.1 ± 6.4 (22) b | 60.7 ± 5.4 (22) b |
| scar3 | 212.4 ± 13.1 (16) a | 176.1 ± 11.0 (16) a | 145.8 ± 9.1 (16) a |
| scar4 | 219.7 ± 11.5 (19) a | 197.9 ± 9.8 (19) a | 158.6 ± 8.1 (19) a |
| scar1 scar2 | 103.4 ± 7.4 (22) bcd | 60.1 ± 4.7 (22) bc | 46.5 ± 3.4 (22) bc |
| scar2;scar3 | 146.9 ± 14.4 (21) b | 91.9 ± 7.6 (21) b | 62.1 ± 6.0 (21) b |
| scar2;scar4 | 59.5 ± 7.4 (22) de | 28.2 ± 2.0 (22) cd | 18.3 ± 1.2 (22) cd |
| scar1 scar2;scar3 | 110.9 ± 8.3 (20) bc | 66.8 ± 6.0 (20) b | 44.2 ± 5.7 (20) bcd |
| scar1;scar3;scar4 | 222.9 ± 8.4 (26) a | 194.6 ± 8.8 (26) a | 166.4 ± 8.2 (26) a |
| scar1 scar2;scar3;scar4 | 46.3 ± 5.5 (22) e | 25.7 ± 1.6 (22) d | 15.3 ± 0.7 (22) d |
| brk1-2 | 52.8 ± 6.4 (17) e | 23.2 ± 1.9 (17) d | 15.7 ± 1.2 (17) cd |
| dis2-1 (arpc2) | 66.0 ± 8.5 (20) cde | 25.1 ± 1.3 (20) d | 17.8 ± 1.1 (20) cd |
| scar2 (dis3-3) | 123.1 ± 5.2 (34) | 99.0 ± 3.9 (34) | 77.9 ± 4.4 (34) |
| dis3-3 35S:SCAR3 | 206.6 ± 8.8 (61)** | 154.3 ± 6.9 (61)** | 116.1 ± 7.3 (61)* |
Data are presented as mean values ± se. The numbers in the parentheses indicate the sample size. The letters indicate ANOVA Tukey multiple comparison test groupings. Mean values with the same letter in the group are not significantly different (α = 0.05). Student's t test with dis3-3. *, P < 0.0001; **, P < 10E-11.
In contrast with the previously reported arp2/3 null phenotype for scar2;scar4/+ plants (Uhrig et al., 2007), we detected intermediate phenotypes that correlated with SCAR4 gene dosage. For example, the trichomes on scar2;scar4/+ leaves had a mean branch 3 length of 44.7 ± 3.3 se (n = 39) μm, which was significantly different from and between the branch 3 lengths of the weak scar2 and the strong scar2;scar4 mutants (analysis of variance [ANOVA], α = 0.01). Therefore, in the scar2 background, the level of SCAR4 activity is critical and defines the morphogenetic output of the ARP2/3 pathway in trichomes. The trichome branch length phenotypes of scar1 scar2 and scar2;scar3 did not differ significantly from scar2 alone according to our ANOVA analyses. However, SCAR1 may make a minor contribution because the branch 1 lengths of scar1 scar2 were slightly shorter than scar2 (Student's t test, P < 0.05), and the scar1 scar2;scar3 triple mutant branches were shorter than those of the scar2;scar3 double mutant (Student's t test, P < 0.05).
Taken together, these results indicate that several SCAR genes regulate trichome growth, but SCAR2 and SCAR4 are clearly the most important. In fact, SCAR2 alone is sufficient to direct polarized growth in hair cells because scar1;3;4 triple mutant plants had no phenotype (Table 2). Furthermore, a single dose of SCAR2 appears to be sufficient for normal trichome growth because scar2/+;scar4/scar4 plants had wild-type trichomes. These genetic data indicate that, in trichomes, a threshold of SCAR gene activity is needed and that a weighted balance of SCAR2 and SCAR4 activity fulfills the requirement.
The strong distorted phenotype of scar2;scar4 suggests that the cell shape defects are caused by a failure to activate ARP2/3 and generate the actin filament arrays that maintain cellular regions of polarized growth. Analyses of the actin phenotypes of distorted trichomes using the live cell actin probe ABD2:green fluorescent protein (GFP), antibodies, and the actin filament binding drug phalloidin consistently identify a defect in the generation or maintenance of aligned actin bundles in the core cytoplasm of developing branches (Szymanski et al., 1999; Le et al., 2003; Deeks et al., 2004; Basu et al., 2005; Djakovic et al., 2006). The core bundle organization defects in stage 4 (Szymanski et al., 1998) trichomes are likely to be the most relevant because the frequently observed reductions in the quantity of core actin filaments is observed only in stage 5 cells (Le et al., 2003; Li et al., 2004; Zhang et al., 2005), which is well after the onset of the mutant phenotype. These later stage cytoskeletal defects may reflect an indirect effect of aberrant vacuole positioning or morphology.
Importantly, the misalignment of core actin bundles in stage 4 trichomes correlates with the severity of cell shape phenotypes. For example, the weak distorted mutant scar2/dis3 has intermediate actin cytoskeleton and trichome shape defects compared with strong wave and arp2/3 mutants (Basu et al., 2005). As previously reported, we detected a distinct population of aligned core bundles in 12 of 12 branches from stage 4 wild-type trichomes (Figures 4A and 4B). In similarly staged dis2 trichomes, the branch core bundles were randomly oriented in 78% (n = 28) of the branches examined (Figures 4I and 4J). Consistent with previous results (Basu et al., 2005), we found that scar2 stage 4 trichomes had an intermediate phenotype, and only 39% (n = 24) of the young trichomes had clearly disorganized core actin bundles. The actin bundle defects of the strong WAVE complex mutant brk1 (n = 18; Figures 4G and 4H), scar2;scar4 (n = 22; Figures 4C and 4D), and the scar quadruple mutant (n = 25; Figures 4E and 4F) were indistinguishable from dis2. These actin localization data provide a clear functional linkage of scar trichome cell shape with defects in the organization of the actin cytoskeleton.
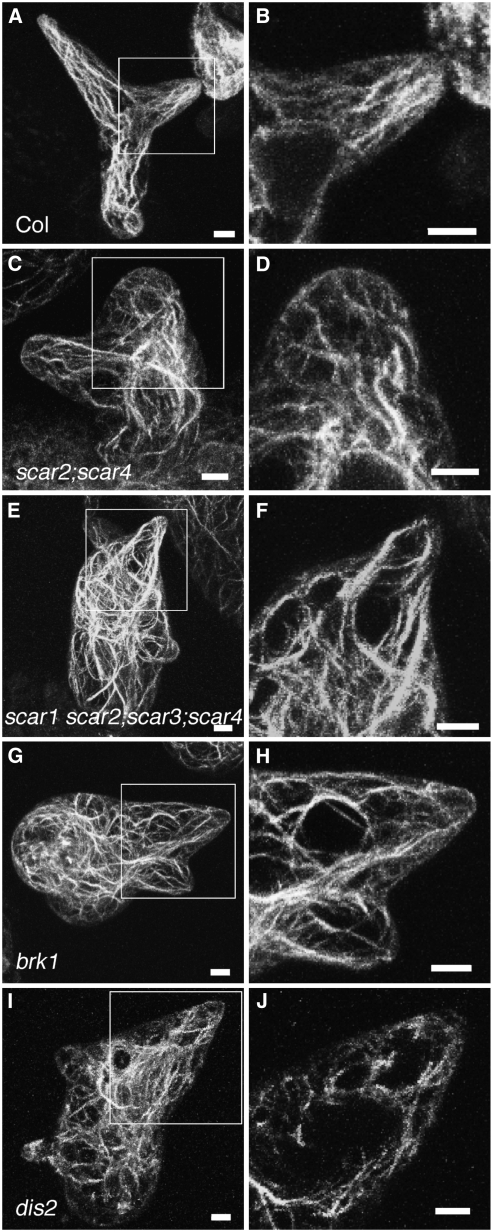
Figure 4.
The Trichome Actin Cytoskeleton Defects of the scar2;scar4 and the scar1 scar2;scar3;scar4 Mutants Are Indistinguishable from Strong wave and arp2/3 Mutants.
The left panels are the maximum projections of confocal image stacks of whole-mount trichomes probed with phalloidin. Boxes in the left panels show regions highlighted in adjacent panels to the right. The right panels are the projections of branch midplanes that contain core cytoplasmic bundles. Bars = 5 μm.
(A) and (B) F-actin organization in a wild-type stage 4 trichome showing actin bundles that are aligned with the long axis of trichome branch elongation.
(C) and (D) F-actin organization in a scar2;scar4 stage 4 trichome showing disorganized actin filaments or bundles.
(E) and (F) F-actin labeling of a similarly staged scar1 scar2;scar3;scar4 quadruple mutant.
(G) and (H) F-actin labeling of a brk1 stage 4 trichome.
(I) and (J) F-actin organization in an arpc2 (dis2) stage 4 trichome.
Unequal Redundancy and Interchangeable Functions during Shoot Development
Because ARP2/3 regulates multiple aspects of plant morphogenesis, we sought to determine the genetic interactions among scar alleles in other cell types and organs. The length of dark-grown hypocotyls can be measured accurately and is clearly reduced in arp2/3 (Mathur et al., 2003b) and wave mutants (El-Assal et al., 2004b; Le et al., 2006). Consistent with previous results (Basu et al., 2005), we found that scar2 expressed a hypocotyl phenotype during the mid to late stage of etiolated growth at 5.5 d after germination (DAG). The hypocotyl length of scar2 (17.4 ± 0.7 se mm; n = 16) at this stage was significantly reduced compared with the wild type (20.9 ± 0.4 se mm; n = 14, Student's t test, P < 0.001). Interestingly, an important contribution for SCAR4 was detected by comparing the hypocotyl growth of scar2 and the scar2;scar4 double after an additional 24 to 48 h in the dark. At these later time points, scar2;scar4 had a hypocotyl length of ∼16 mm that was indistinguishable from arp2/3 (dis2) and wave (brk1) seedlings (Table 3). Meanwhile, the slow-growing scar2 became indistinguishable from the wild type.
Table 3.
Morphogenetic and Developmental Phenotypes of Dark-Grown scar, brk1, and dis2 Seedlings
| Genotype | Hypocotyl Length (mm)a | Petiole Elongationb |
|---|---|---|
| Col | 21.5 ± 0.6 (20) ab | − |
| scar1 | 20.5 ± 0.4 (10) abc | − |
| scar2 | 19.8 ± 0.4 (22) abc | + |
| scar3 | 21.5 ± 0.3 (23) a | − |
| scar4 | 19.4 ± 0.3 (20) abc | − |
| scar1 scar2 | 20.1 ± 0.4 (21) abc | + |
| sar1;scar3 | ND | + |
| scar2;scar3 | 18.9 ± 0.4 (27) cd | + |
| scar2;scar4 | 16.0 ± 0.6 (21) f | + |
| scar1 scar2;scar3 | 18.3 ± 0.3 (22) cde | + |
| scar1;scar3;scar4 | 19.2 ± 0.9 (15) bc | + |
| scar1 scar2;scar3;scar4 | 16.8 ± 0.5 (22) def | + |
| brk1-2 | 16.4 ± 0.5 (14) ef | + |
| dis2-1(arpc2) | 16.8 ± 0.4 (20) def | + |
Mean values ± se. The numbers in the parentheses indicate the sample size. The letters indicate ANOVA Tukey multiple comparison test groupings. Mean values with the same letter in the group are not significantly different (α = 0.05). ND, not determined.
The plus sign indicates elongated petioles and premature shoot meristem activation in the dark.
These data indicate that SCAR4 has the ability to partially compensate for the absence of SCAR2. However, the functional power of SCAR4 was limited. SCAR4 is neither required, because the single mutant had no phenotype, nor is it sufficient, because scar1 scar2;scar3 hypocotyls were significantly shorter than the wild type (Student's t test, P < 0.0001; Table 3). The scar1,-2,-3 triple mutant also reveals minor and redundant functions for SCAR1 and SCAR3 because the triple mutant, but neither the scar1 scar2 nor the scar2;scar3 double mutant, had reduced hypocotyl growth compared with scar2 (Student's t test, P < 0.001). Therefore, dark-grown seedlings rely mostly on SCAR2 and SCAR4 to promote growth, and, as observed in trichomes, SCAR2 is the dominant regulator.
The wave and arp2/3 mutants have a weak but detectable effect on the polarized growth of epidermal pavement cells (Brembu et al., 2004; Djakovic et al., 2006; Le et al., 2006), but the role of specific SCAR genes is unknown (Basu et al., 2005; Le et al., 2006; Uhrig et al., 2007). Pavement cell shape and size vary greatly as they develop, and in developing leaves, cell division creates populations of asynchronously developing cells. Therefore, to reduce the variability in pavement cell measurements, we sampled fully expanded pavement cells within the apical third of 12-DAG cotyledons (see Methods; Le et al., 2006). In this region of the wild-type cotyledons, most of the cells are fully expanded and have a highly lobed morphology (Figures 3I and 3M).
Pavement cell shapes were quantitated using a dimensionless shape descriptor termed “circularity” and using a skeletonize algorithm to estimate lobe number (Russ, 2002). As pavement cell lobing increases, circularity becomes smaller and the number of skeleton end points, an estimate of lobe number, increases. SCAR single mutants had no detectable cotyledon pavement cell shape defect (Table 4). This result argues against independent functions for individual genes, but, based upon the phylogeny, it is possible that the SCAR1/3 and SCAR2/4 gene pairs function redundantly and mask independent functions. However, we found that the SCAR1/3 gene pair was not required because scar1;scar3;scar4 plants had no detectable pavement cell phenotype (Table 4). However, SCAR1 and SCAR3 functioned in pavement cells because we detected only a very modest reduction of cell shape complexity for the scar2;scar4 double mutant (Figures 3J and 3N, Table 4; Student's t test, P < 0. 05). Our double and triple mutant analyses provided additional evidence for redundant functions for SCAR1 and SCAR3 in pavement cells. The scar1,-2,-3 triple mutant had a more simple shape compared with both of the double mutants scar1,-2 and scar2,-3 (Table 4; Student's t tests, P < 0.001).
Table 4.
Quantitative Analysis of the Epidermal Pavement Cell Shape Phenotype of scar, brk1, and dis2 Mutants
| Genotype | Circularitya | Skeleton Endsb | Area (× 104 μm2) |
|---|---|---|---|
| Col | 0.20 ± 0.01 (30) ab | 12.1 ± 0.5 (n = 30) ab | 2.02 ± 0.07 (30) ab |
| scar1 | 0.21 ± 0.01 (30) ab | 12.0 ± 0.6 (n = 30) abc | 2.24 ± 0.11 (30) a |
| scar2 | 0.19 ± 0.01 (30) ab | 11.5 ± 0.5 (n = 30) bc | 1.59 ± 0.07 (30) cde |
| scar3 | 0.17 ± 0.01 (30) a | 14.0 ± 0.7 (n = 30) a | 1.84 ± 0.13 (30) abcde |
| scar4 | 0.18 ± 0.01 (30) ab | 12.3 ± 0.5 (n = 30) ab | 1.85 ± 0.08 (30) abcde |
| scar1 scar2 | 0.22 ± 0.01 (30) abc | 10.1 ± 0.5 (n = 30) bcd | 1.56 ± 0.07 (30) e |
| scar2;scar3 | 0.23 ± 0.01 (30) bc | 10.6 ± 0.4 (n = 30) bc | 1.52 ± 0.06 (30) ef |
| scar2;scar4 | 0.23 ± 0.01 (30) bc | 9.4 ± 0.4 (n = 30) cde | 2.00 ± 0.10 (30) abc |
| scar1 scar2;scar3 | 0.27 ± 0.01 (30) cd | 8.0 ± 0.4 (n = 30) de | 1.98 ± 0.08 (30) abcd |
| scar1;scar3;scar4 | 0.22 ± 0.01 (30) ab | 10.6 ± 0.4 (n = 30) bc | 1.99 ± 0.08 (30) abc |
| scar1 scar2;scar3;scar4 | 0.30 ± 0.01 (30) d | 7.8 ± 0.4 (n = 30) e | 1.57 ± 0.06 (30) de |
| brk1-2 | 0.29 ± 0.02 (30) d | 7.5 ± 0.4 (n = 30) e | 1.61 ± 0.10 (30) cde |
| dis2-1(arpc2) | 0.30 ± 0.01 (30) d | 7.5 ± 0.4 (n = 30) e | 1.65 ± 0.09 (30) bcde |
| brk1-2 er-105 | ND | ND | 1.14 ± 0.07 (20) f |
Circularity is a descriptor of shape complexity and is calculated as 4 π(area)/(perimeter)2. Data are mean values ± se. Numbers in the parentheses indicate the sample size. The letters indicate ANOVA Tukey multiple comparison test groupings. Mean values with the same letter in the group are not significantly different (α = 0.05). ND, not determined.
Number of skeleton end points calculated from the medial-axis transformation of thresholded individual pavement cell images.
Clear pavement cell shape differences between scar mutants and the wild type were observed only after removal of all four SCAR genes (Figures 3K and 3O). A similar cell shape phenotype was observed in the strong distorted mutant dis2 (Figures 3L and 3P). The mean pavement cell circularity values of ∼0.3 and the number of lobes for the scar quadruple mutant, brk1, and dis2 were not significantly different from each other (Table 4), but each of these mutants clearly had a more simple shape compared with the wild type (Student's t tests, P < 0.0001). These data indicate that each of the four SCAR genes function interchangeably during pavement cell growth. However, SCARs do not make equal contributions. For example, SCAR4 is not sufficient for pavement cell growth because scar1,-2,-3 cotyledon pavement cells had a shape that was more simple than the wild type (Table 4; Student's t test, P < 0.001). On the other hand, SCAR2 can support wild-type growth because scar1,-3,-4 triple mutants had no phenotype. Upon breaking linkage between brk1 and erecta (er-105), we discovered that the reduced size of brk1 er-105 pavement cells (Le et al., 2006) was due to an interaction between brk1 and the er-105 mutations (Table 4). Taken together, these results prove that in pavement cells, SCARs are broadly interchangeable, but the genes have unequal importance.
We wanted to test further the idea that SCAR genes could function interchangeably in other cell types. Trichome development provides a perfect system for this experiment because according to our mutant analysis SCAR3 does not regulate trichome morphogenesis. The lack of a detectable function for SCAR3 could merely reflect a low level of gene expression in this particular cell type, rather than an inability of the SCAR3 protein to satisfy ARP2/3 activation thresholds. If SCAR genes function interchangeably, then it should be possible to overexpress SCAR3 and test its ability to substitute for SCAR2. SCAR protein levels are under strict posttranslational control (Djakovic et al., 2006; Le et al., 2006). Expression driven by the 35S promoter can rescue scar2 phenotypes, but the viral promoter does not lead to detectable overexpression of GFP-tagged SCAR2 (Zhang et al., 2005; Uhrig et al., 2007). We also tested the ability of wild-type and GFP-tagged SCAR2 to rescue mutant phenotypes upon overexpression.
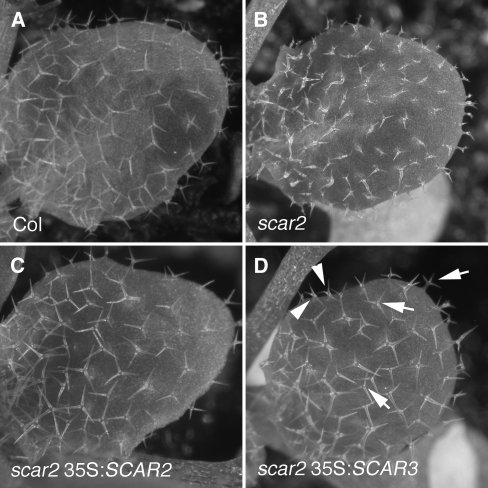
We found that 78% (n = 30) of primary scar2 35S:SCAR2 transformants had trichomes that were indistinguishable from the wild type (Figures 5A to 5C). GFP-tagged SCAR2 was also functional based on a similar efficiency of phenotypic rescue, but as reported previously, the tagged proteins were not detectable. We also found no evidence of morphological phenotypes that would be expected if SCAR2 was massively overexpressed. When SCAR3 was overexpressed in the scar2 background, trichome development in 10 of 19 primary transformants was nearly indistinguishable from the wild type (Figure 5D). For comparison, Figure 5B shows a population of scar2 trichomes. Although SCAR3 clearly had activity in trichomes, the scar2 35S:SCAR3 plants had a small fraction of trichomes, usually along the leaf perimeter, that had subtle branch position and/or elongation defects (Figure 5D). The spatial distribution of imperfectly rescued cells probably reflects variability in 35S-mediated expression in the leaf. An analysis of randomly selected trichomes from scar2 35S:SCAR3 plants indicates that, while all branch lengths were significantly longer compared with scar2, branch 2 and 3 length was reduced slightly compared with the wild type (Table 2). Nonetheless, these experiments clearly demonstrate that SCAR3 can substitute quite well for SCAR2 during trichome morphogenesis.
Figure 5.
SCAR3 Can Substitute for SCAR2 during Trichome Morphogenesis.
Full-length SCAR2 and SCAR3 cDNAs were transformed into the scar2 background. Trichome morphogeneis was assayed in expanding leaves.
(A) Wild-type leaf with highly elongated trichomes.
(B) The expanding leaf of a scar2 mutant with distorted trichomes.
(C) The expanding leaf of a scar2 mutant transformed with 35S:SCAR2.
(D) The expanding leaf of a scar2 mutant transformed with 35S:SCAR3. Both wild-type trichomes (arrows) and trichomes with a weak distorted phenotype (arrowheads) are present.
The wave and arp2/3 mutants have an additional dark-grown phenotype: enhanced cotyledon petiole elongation and a premature activation of the shoot apical meristem (Uhrig et al., 2007). Normally, petiole elongation and leaf initiation are inhibited in the dark (Figure 3E). In three independent experiments, we found that a vast majority of scar2 (Figure 3F), scar1;scar3 (Figure 3G), and dis2 (Figure 3H) plants display this strange dark-grown phenotype. Other distorted mutants, such as brk1 and the scar quadruple mutant, had an equally severe and incompletely penetrant elongated cotyledon petiole phenotype (>80% seedlings elongated, n > 36; Table 3). The wild-type plants expressed the phenotype at a low frequency (<10% seedlings elongated, n = 26; see also Roldan et al., 1999). The enhanced cotyledon petiole elongation in dark-grown wave and arp2/3 seedlings is opposite to the reduced cell elongation in the hypocotyl. Taken at face value, these genetic data indicate that ARP2/3 negatively regulates growth in the petiole (Uhrig et al., 2007). However, we found that this phenotype was conditional and might reflect a misregulated developmental process rather than an obvious effect on morphogenesis. For example, the petiole phenotype was sucrose dependent and occurred only when the shoot meristem was in physical contact with the sucrose-containing media. This reflects an enhancement of a known wild-type response (Roldan et al., 1999). Mutation of the putative transcription coactivator LEAFY COTYLEDON1 causes similar dark-grown phenotypes (Casson and Lindsey, 2006), further suggesting that this phenotype is developmental in nature.
Regardless of its functional basis, the genetics of the dark-grown cotyledon petiole phenotype is interesting. SCAR2 was required because the dark-grown scar2 phenotype was as severe as dis2, brk1, and scar quadruple mutants. SCAR1 and SCAR3 function with equal redundancy during normal petiole growth in the dark because the scar1;scar3 double mutant, but neither of the corresponding single mutants, had a strong phenotype. These data could be explained by a threshold model; however, in this case, an equally likely scenario is one in which parallel and necessary SCAR2 and SCAR1/3 pathways regulate shoot maturation in the dark: removal of either pathway would then cause strong loss-of-function phenotypes. Obligate dimerization of SCAR2 with SCAR1 or SCAR3 could also explain these genetic interactions.
Gene Expression Levels and Biochemical Efficiency Contribute to Unequal Redundancy
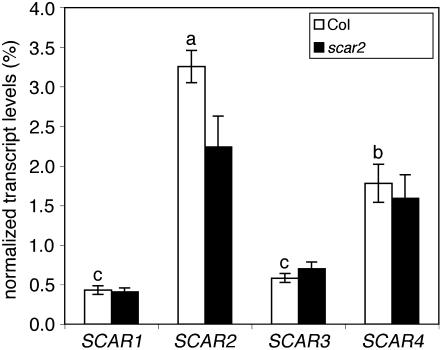
The genetic and morphometric analyses above indicate that multiple SCAR genes contribute toward meeting a threshold of ARP2/3 activation, but fail to explain why a particular homolog is more important than another. To determine if differences in steady state mRNA levels might contribute to unequal importance among the homologs, we used quantitative real-time RT-PCR (qRT-PCR) to measure the abundance of each of the SCAR transcripts in trichomes (see Methods). In RNA samples isolated from purified trichomes, SCAR2 message levels were higher by a factor of 6 compared with those of SCAR1 and SCAR3. SCAR2 mRNA levels were significantly different from and approximately twofold greater than SCAR4 (Figure 6). It was possible that the weak phenotype of scar2 was partially compensated for by increases in SCAR4 mRNA. We found no evidence for compensatory upregulation of any of the SCAR genes in the scar2 background (Figure 6). Therefore, the weak and variable phenotype of scar2 occurs because a normal level of SCAR4 expression is insufficient. However, gene expression levels may not be robust indicators of gene function in all cases, especially when measurements are taken from entire organs. For example, micorarray analysis of dark-grown hypocotyls (Schmid et al., 2005) shows that the expression levels of SCAR2 and SCAR4 are reduced, whereas SCAR1 is elevated in dark-grown compared with light-grown hypocotyls (see Supplemental Figure 1 online). These expression data conflict with the dominant functions of SCAR2 and SCAR4 during etiolated growth.
Figure 6.
Quantitative Analysis of SCAR Transcript Levels in Wild-Type and scar2 Trichomes.
Steady state mRNA levels of SCAR genes were quantified in trichomes purified from wild-type and scar2 plants (see Methods). mRNA levels were quantified by qRT-PCR, and SCAR expression was normalized to the GAPC (AT3G04120) gene. The normalized transcript values represent the means ± se of triplicate measurements as described in Methods. Groups with an identical superscripted letter were not significantly different according to an ANOVA Tukey multiple comparison test (P < 0.01). The scar2 (dis3-1) allele causes premature transcriptional termination after the SHD-encoding exons (Basu et al., 2005).
In Arabidopsis, the primary, if not sole, morphogenetic function of plant SCARs is to activate ARP2/3 because the pleiotropic and severe nature of scar double and quadruple mutant phenotypes matches exactly those of the strong arp2/3 (dis2) and wave (brk1) mutants (Tables 2 to 4). It is therefore possible that differences in the efficiency with which SCAR isoforms activate ARP2/3 could contribute to unequal redundancy. The WA domain of SCAR was sufficient for ARP2/3 activation (Machesky et al., 1999), and ARP2/3 activators are often assayed as glutathione S-transferase (GST)-tagged proteins (Machesky et al., 1999; Blanchoin et al., 2000; Duncan et al., 2001). A previous examination of GST-WA domains from plant SCAR proteins demonstrated that maize SCAR1 could efficiently activate ARP2/3 at nanomolar concentrations, whereas Arabidopsis SCAR3 and SCAR4 had very little activity, even at micromolar concentrations (Frank et al., 2004). To compare directly the relative activity of SCAR2, -3, and -4, we expressed similar WA domain–containing peptides from each protein and tested their ability to activate ARP2/3 in a pyrene-actin polymerization assay.
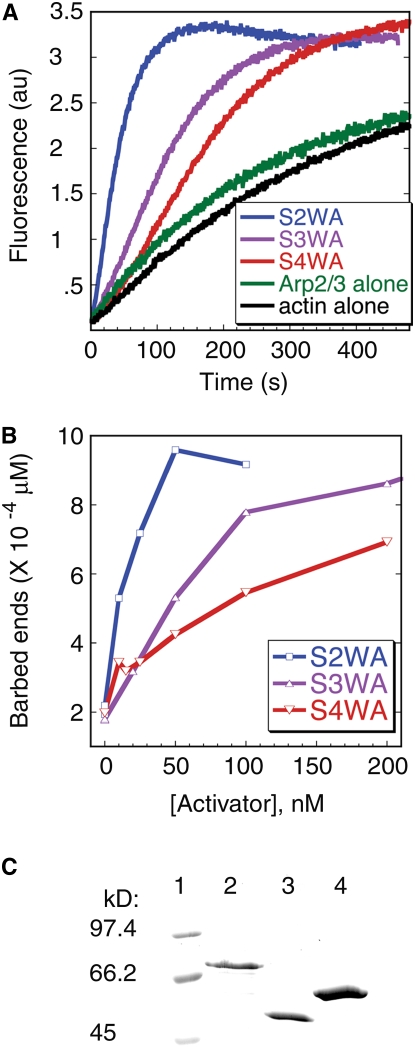
Each of the SCAR protein fragments, when tested at submicromolar concentrations, increased the actin filament nucleation activity of ARP2/3 (Figures 7A and 7B). However, at 10 nM or higher activator concentrations, SCAR2-WA was consistently the most active. SCAR2-WA decreased the time required to reach half-maximal actin polymerization to a greater extent when compared with SCAR3-WA and SCAR4-WA (Figure 7A). SCAR2-WA also consistently created the greatest number of actin filament barbed ends at several activator concentrations (Figure 7B). At the saturating concentration of 50 nM, SCAR2-WA produced ∼20 pM actin filament ends/nanomolar activator. Although active at nanomolar concentrations, SCAR3-WA and SCAR4-WA surprisingly require ∼1 μM concentrations to reach saturation (see Supplemental Figure 2 online), and they produced ∼2.0 and 0.5 pM actin filament ends/nanomolar activator, respectively. Similar results for SCAR2-, 3-, and 4-WA were observed with three separate batches of the recombinant proteins. The GST tags did not contribute to WA fragment activities because purified GST had no effect on ARP2/3 activity when tested at all activator concentrations that were used in this work, including the highest concentration of 3 μM.
Figure 7.
ARP2/3 Activation Efficiency of SCAR2, SCAR3, and SCAR4 WA Domains.
(A) Kinetic analysis of actin polymerization in the presence of ARP2/3 was monitored with the indicated GST-tagged activator protein, 3 μM actin (5% pyrene-labeled), and 10 nM bovine ARP2/3 complex.
(B) Comparison of the concentrations of barbed ends generated at half-maximal polymerization in the presence of varying amounts of GST-SCAR2-WA, GST-SCAR3-WA, and GST-SCAR4-WA. SCAR2-WA's end concentration plot typically saturates at 50 nM SCAR2-WA at 10 × 10−4 μM ends and then decreases at higher concentrations. SCAR3-WA's end concentration plot saturates at 1 μM SCAR3-WA at ∼20 × 10−4 μM ends and then decreases by ∼25% at 3 μM SCAR3WA. SCAR4-WA's end concentration plot saturates at 1.5 μM SCAR4-WA at ∼7 × 10−4 μM ends.
(C) Coomassie blue–stained SDS-PAGE gel showing molecular size standards (lane 1), GST-SCAR2-WA (lane 2), GST-SCAR3-WA (lane 3), and GST-SCAR4-WA (lane 4).
We are unsure if these high stoichiometries of activator:ARP2/3 complex are physiologically relevant, but clearly SCAR2-WA is a stronger activator when compared with the activating domains of SCAR3 and SCAR4. These biochemical results must be treated with some caution because these experiments, like prior analyses of plant SCAR proteins (Frank et al., 2004; Basu et al., 2005), employed vertebrate ARP2/3, and amino acid sequence divergence between plant and vertebrate ARP2/3 could contribute to some of the measured variability among plant SCARs. On the other hand, the source of ARP2/3 may be irrelevant. Plant, human, and yeast ARP3, ARPC2, and ARPC5 proteins function interchangeably in all cases examined (Le et al., 2003; Mathur et al., 2003b; El-Assal et al., 2004a). Furthermore, the WA domains of plant and animal SCARs are highly conserved and vertebrate ARP2/3 responds sharply to nanomolar concentrations of the plant activators (Figure 7B; Basu et al., 2005). Thus far, we and others have found that plant ARP2/3 is at very low concentrations and rather difficult to isolate; therefore, further substantial effort will be necessary to determine if plant ARP2/3 has unique properties. Nevertheless, our results demonstrate unambiguously that the Arabidopsis SCAR isoforms are not functionally equivalent when ARP2/3 activation is compared quantitatively.
DISCUSSION
WAVE is an evolutionarily conserved effector complex that converts activating Rho family small GTPase signals into a localized ARP2/3-dependent actin polymerization response (Eden et al., 2002; Basu et al., 2004; Innocenti et al., 2005). The evolutionarily conserved nature of WAVE and the experimental power afforded by the Arabidopsis distorted mutants makes this an ideal system to dissect the transmission of Rho-based signals to the cytoskeleton. Plant and animal SCAR proteins potently activate ARP2/3 (Machesky et al., 1999; Basu et al., 2005), and in this article, we demonstrate that multiple plant SCAR proteins function interchangeably as critical positive regulators of ARP2/3. Because the scar (Tables 2 to 4), wave (Brembu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004b; Djakovic et al., 2006; Le et al., 2006), and arp2/3 (Mathur et al., 2003a; El-Assal et al., 2004a) mutants have an identical array of phenotypes with indistinguishable severities, SCAR functions in the context of a WAVE complex and is the major, if not sole, pathway for ARP2/3 activation.
Our conclusion that Arabidopsis employs a single class of ARP2/3 activator is surprising, given that budding yeast, Dictyostelium, and vertebrate cells employ multiple ARP2/3 activators with nonoverlapping functions. For example, Saccharomyces cerevisiae recruits several different types of ARP2/3 activators sequentially to promote transport through the endocytic pathway (Kaksonen et al., 2003). Animal cells use the WASP and WAVE classes of ARP2/3 activators (reviewed in Stradal and Scita, 2006), and independent functions for WAVE1 and WAVE2 have been proposed. For example, loss of WAVE1 or WAVE2 causes distinct phenotypes in the formation of dorsal ruffles and lamellipodia, respectively (Suetsugu et al., 2003). However, the independent functions for vertebrate SCAR proteins are controversial, as a more recent article reports that dorsal ruffle formation is regulated by the ARP2/3 activator N-WASP (Legg et al., 2007).
The genetic interactions among Arabidopsis scar mutants depict a regulatory scenario in which SCARs function redundantly. In general, scar single mutants have weak or undetectable morphogenesis phenotypes, and growth defects become progressively worse as additional SCAR genes are removed (Tables 2 to 4). Interchangeable functions of SCAR isoforms are broadest during pavement cell morphogenesis. In this cell type, a wide array of SCAR genes can support wild-type growth, and a pavement cell shape equivalent to wave or arp2/3 subunit null mutants is obtained only after removal of all four genes (Table 4). SCARs can also function interchangeably in trichomes. Although our mutant analyses did not reveal a function for SCAR3 in trichomes, we found that the gene could substitute for SCAR2 when overexpressed. Most of the trichomes on scar2 35S:SCAR3 plants were indistinguishable from those of the wild type, indicating that SCAR3 can perform all SCAR2 functions in this cell type. However, a subset of scar2 35S:SCAR3 trichomes, usually those near the leaf margin, had subtle defects in branch position or elongation (Figure 5, Table 2). We suspect that these phenotypes reflect an uneven activity of the 35S promoter in the leaf.
Although there are clear examples in which isoforms of plant actin, profilin, and actin depolymerizing factor have unique functional properties (Kandasamy et al., 2002, 2007), this does not appear to be the case with SCAR. If cells assemble unique WAVE complexes containing specific SCAR isoforms that perform customized duties (Suetsugu et al., 2003; Uhrig et al., 2007), then single and/or double mutant combinations that disrupt the SCAR1/3 paralogs or the SCAR2/4 orthologs would have clear phenotypic effects. To the contrary, we fail to detect any morphogenetic phenotype that requires SCAR1, SCAR3, or SCAR4 (Tables 2 to 4). Only the developmental phenotype of enhanced petiole elongation (see below) is consistent with a model of independent and required functions for SCAR2 (A group) and the SCAR1/3 (B group) genes.
Interestingly, the genetic redundancy among SCARs is unequal. That is, SCAR genes function at a common point in the WAVE-ARP2/3 pathway, but the genes have unequal importance depending on the cell type or tissue. Unequal genetic redundancy is most apparent in trichomes and dark-grown hypocotyls (Tables 2 and 3). In these developmental contexts, growth is controlled by the SCAR2/4 pair because only the scar2;scar4 double mutant combination generates a phenotype that is equivalent to wave and arp2/3 null mutants. SCAR2 is clearly the major player because only the scar2 single mutant has a phenotype (Figure 2, Table 2), and in trichomes, a single dose of the gene is sufficient for wild-type growth. In the absence of SCAR2, cells appear to teeter on the edge of morphogenetic chaos. For example, our double mutant analyses and measurements of SCAR transcript levels indicate that the weak and variable phenotypes of scar2 trichomes reflect the inability of SCAR4 to positively regulate ARP2/3 with sufficient power. The fact that SCAR4 shows dosage sensitivity in the scar2 background suggests that SCAR4 approaches a minimal level of ARP2/3 activation, but growth is less stable due to small fluctuations in gene activity.
The genetic interactions between scar2 and scar4 in trichomes and dark-grown hypocotyls are explained most easily by a threshold of ARP2/3 activation model: a certain level of ARP2/3 activation is needed, and the weighted activities of SCARs provide it. A threshold of ARP2/3 activation could be met by the collective and shared biochemical properties of SCAR2 and 4. Cell type–specific differences in SCAR2 and SCAR4 mRNA levels (Figure 6) and differing potencies of ARP2/3 activation (Figure 7) correlate with the relative efficiencies of gene function.
Our experiments also raise a broader question. What determines ARP2/3 activation thresholds in vivo? Thresholds could be defined by either the relative importance of ARP2/3 functions for a particular cell type or the extent to which additional pathways can compensate for the loss of ARP2/3. For example, in the former scenario, the polarized growth of trichomes may include numerous and/or highly constrained ARP2/3-dependent functions and a high overall activation threshold. In this scenario, SCAR2 is critical because of its dominant contributions to ARP2/3 activation in this cell type. Pavement cells have more subtle wave and arp2/3 phenotypes. Perhaps in this cell type there are fewer or less important ARP2/3 functions, and a lower threshold can be met with relative ease by any one of several SCARs. Alternatively, the weak wave and arp2/3 pavement cell phenotype could reflect partial compensation by parallel, seemingly unrelated, growth control pathways (Fu et al., 2002, 2005; Qiu et al., 2002; Lavy et al., 2007). Broader knowledge about morphogenesis control and the cellular functions of actin filament nucleation will help to explain the variable responses of cells to the loss of ARP2/3.
Interestingly, although we detect in vivo functions and evidence of purifying selection for each of the SCARs, these genetic interactions do not provide clear insight into the biology that might have reinforced the divergence of separate SCAR1/3 and SCAR2/4 clades. SCAR duplication is not needed to buffer the plant against pathway loss because seven genes in the WAVE-ARP2/3 pathway are single copy and necessary (Szymanski, 2005). However, functional redundancy between duplicated genes can be stable over evolutionary time scales (Nowak et al., 1997), and we detect residual redundancy within the SCAR2/4 pair despite an ancient origin that predates the divergence of Arabidopsis and poplar. SCAR4, like the SHD domain–encoding gene AT4G18600 (SCARL), may be headed toward nonfunctionalization based on its minimal functional contribution, its low expression levels, and its weaker biochemical activity compared with SCAR2. The ancient divergence of SCAR clades is more easily understood in the context of independent functions for the SCAR1/3 and the SCAR2/4 gene pairs. However, only the developmental phenotypes of enhanced petiole elongation and premature shoot meristem activation in dark-grown seedlings are consistent with a model of independent and necessary SCAR functions. Perhaps the unknown functions of SCAR, WAVE, and ARP2/3 control aspects of sucrose signaling and development that have maintained the existence of A and B gene clades.
In conclusion, our quantitative analysis of SCAR and the heteromeric WAVE complex clearly defines a regulatory module in which four SCAR genes positively regulate ARP2/3. In the context of cell morphogenesis, SCAR function is best explained by a threshold model, one in which SCARs have an ability to substitute for one another to meet the cellular demands for ARP2/3 activation. However, SCARs clearly have unequal importance, and cell type–specific differences in steady state mRNA levels and the biochemical efficiency of ARP2/3 activation are two parameters that appear to contribute to unequal genetic redundancy. The above model can explain the observed strong purifying selection on SCAR genes and indicates that, at least in the context of morphogenesis, functional knowledge about SCAR2 can be extrapolated to the gene family as a whole.
METHODS
Plant Strains, Growth Conditions, and Allele Characterization
The SALK T-DNA insertion lines (Alonso et al., 2003) for SCAR1 (scar1-t1, salk_017554), SCAR3 (scar3-t1, salk_036994), and SCAR4 (scar4-t1, salk_116410 and scar4-t2, salk_022766), and SCARL/AT4G18600 (salk_055413) were obtained from the ABRC. The scar2/dis3-1 (Basu et al., 2005), brk-1 (Le et al., 2006), and dis2-1 (El-Assal et al., 2004b) alleles have been previously described. The soil-grown plants were planted in SunGro Redi-earth plug and seedling mix series growing medium (SunGro Horticulture) that was overlaid on an equal volume of coarse vermiculite under continuous illumination (110 μmol photons m−2 s−1) at 25°C. The trichome morphometry and the pavement cell shape data were collected using soil-grown plants. Aseptically grown plants were seeded on half-strength Murashige and Skoog plates with 1% sucrose under continuous illumination (110 μmol photons m−2 s−1) at 22°C. Dark-grown plants were grown aseptically and wrapped with three layers of aluminum foil. The T-DNA insertions in the SCAR genes were confirmed by PCR using primers that are named based on particular alleles. To analyze the expression of SCAR genes in mutant backgrounds, total RNA was extracted from the homozygous T-DNA insertion mutants by TRIzol reagent (Molecular Research Center), and 2 μg total RNA was reverse transcribed using 200 units of M-MLV reverse transcriptase (Promega) primed with 250 ng of Random Primer 6 oligo nucleotide (New England Biolabs). A volume (1/25) of the cDNA was used in each RT-PCR reaction using specific primers to the SHD domain and the WA domain of the corresponding SCAR genes. Primer sequences are included in Table 5.
Table 5.
The Sequence of the Oligonucleotide Primers Used in This Work
| Name | Sequence |
|---|---|
| Primers for the characterization of scar1-t1 | |
| SALK_017554-F | 5′-GGAATGAAATGGGAGAAATGA-3′ |
| SALK_017554-R | 5′-CGTGAAATTGTAAGAACTAAGAAGCA-3′ |
| Primers for the characterization of scar3-t1 | |
| SALK_036994-F | 5′-TCACTCCAGGAAGACATTTGC-3′ |
| SALK_036994-R | 5′-AAAACTGTTGGCCATAAGCC-3′ |
| Primers for the characterization of scar4-t1 and scar4-t2 | |
| SALK_116410-F | 5′-TCTTTCTCCTGACGCGTTTT-3′ |
| SALK_116410-R | 5′-TGAATCCTCAGGCGTCTCTT-3′ |
| SALK_022766-F | 5′-TCACTCCAGGAAGACATTTGC-3′ |
| SALK_022766-R | 5′-AAAACTGTTGGCCATAAGCC-3′ |
| Primers for the characterization of scarl-t1 | |
| SALK_055413-F | 5′-TGAACTAAAAGCTTGTCCCTTACCG-3′ |
| SALK_055413-R | 5′-CGTCAGTTAGGCGATCTCGCTG-3′ |
| T-LB | 5′-CCGTCTCACTGGTGAAAAGAA-3′ |
| RT-PCR primers | |
| SCAR1SHD-F | 5′-CACCAGAATTCACATGTATGACGAAT-3′ |
| SCAR1SHD-R | 5′-CTCGAGGTTTAGCCACAGAGGCAAGTCTTG-3′ |
| SCAR1WA-F | 5′-CACCAGAATTCTCCAAAATCCAGCTGAGCC-3′ |
| SCAR1WA-R | 5′-CGGCTCGAGTCATGTGTCGCTATCGC-3′ |
| SCAR3SHD-F | 5′-CACCATGGAATGCCACGGAATGTATACGG-3′ |
| SCAR3SHD-R | 5′- GAATTCGGCCAAACGTGACATATCTCGAC-3′ |
| SCAR3WA-F | 5′-CACCGAATTCCTCCAAAATCCTGCTGAGCCC-3′ |
| SCAR3WA-R | 5′-CTCGAGTTACGTTCACTCCATGTATCGC-3′ |
| SCAR4SHD-F | 5′-CACCGAATTCATGGCATTGACGAGATACCAG-3′ |
| SCAR4SHD-R | 5′-CTCGAGCGTCTCTTTAATAGTTGTGTGAG-3′ |
| SCAR4WA-F | 5′-CACCATGGTATCCCCTGAGTCCATAC-3′ |
| SCAR4WA-R | 5′-GCGGATCCTCACTCGCTCCAGCTATCTG-3′ |
| SCAR4C_F2 | 5′-CACCATGGTATCCCCTGAGTCCATAC-3′ |
| SCAR4C_R2: | 5′-GCGGATCCTCACTCGCTCCAGCTATCTG-3′ |
| qRT-PCR primers | |
| SCAR1QPCR-F | 5′-AAATGTCTCTTACCTCCAAAATCCA-3′ |
| SCAR1QPCR-R | 5′-TTCGAAACCATCCACTGTAACG-3′ |
| SCAR2QPCR-F | 5′-TGACCCTGAAGCTCTGCTTGA-3′ |
| SCAR2QPCR-R | 5′-CAGCGAGATCACCGAGTTGA-3′ |
| SCAR3QPCR-F | 5′-CTTCTTGCAACAAATCCGAACA-3′ |
| SCAR3QPCR-R | 5′-GCAGTAGCCGATGATGTCGTT-3′ |
| SCAR4QPCR-F | 5′-TGGTCTCGTCGGCGTTTT-3′ |
| SCAR4QPCR-R | 5′-CATGCAGGCAATGGAATACCT-3′ |
| GAPCQPCR-F | 5′-TCCCGTGTGGTCGACTTGA-3′ |
| GAPCQPCR-R | 5′-CCACTCCCTATCATTCGAGATCTG-3′ |
| GL2QPCR-F | 5′-TGTCAATGGCCGTCGACATGTCTT-3′ |
| GL2QPCR-R | 5′-TTGATCTGTGTGCGGCGGTTTT-3′ |
| Primers for the cloning of WA domains | |
| SCAR3WA-F | 5′-CACCGGATCCTCCTTGTCAAATTCTGAGCCGTGG-3′ |
| SCAR3WA-R | 5′-GTCGACTTACGTATCACTCCATGTATCGCTTTCAT-3′ |
| SCAR4WA-F | 5′-CACCGGATCCAACATCCAGAGGGATGAAAACACACA-3′ |
| SCAR4WA-R | 5′-GTCGACTCACTCGCTCCAGCTATCTG-3′ |
Construction of scar Mutant Combinations
The scar1 scar2 double mutant was made by crossing scar1-t1 with scar2/dis3-1. Rare recombination events between scar1 and scar2 were identified by screening visually for scar2 homozygotes that also harbored the kanamycin resistance marker from the scar1-t1 allele. Homozygous scar1 scar2 mutants were confirmed in F3 progeny by genotyping. All other double mutant combinations were identified using obvious combinations of phenotypic and PCR-based molecular markers. Quadruple and quintuple scar mutants were identified in F2 progeny derived from the cross scar1 scar2;scarl × scar3;scar4. The strong phenotype of scar2;scar4 and the linkage of scar1 and scar2 allowed us to easily identify a population of scar1 scar2;scar4 triple mutants from which quadruple and quintuple mutants were identified by PCR. The genotypes of all mutants were confirmed by progeny testing putative homozygous lines. Genotyping primers are the same as those that were used for scar allele characterization.
Phylogenetic Analyses
SCAR genes from the Oryza sativa, Populus trichocarpa, Medicago truncatula, and Physcomitrella patens genome sequences were identified by TBLASTX (Altschul et al., 1990). The conserved SHD and WA domains of Arabidopsis thaliana SCAR proteins were used in sequential SHD and WA searches against the available genomic sequence databases. Several SCARs were identified for each species. The identified genes were then used to rescreen the queried genome for additional SCAR-like genes. Annotations for O. sativa were from the National Center for Biotechnology Information, for P. trichocarpa from JGI v1.1 Jamboree Gene Models (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), P. patens from JGI v1.1 FilteredModels 3 annotations (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html), and M. truncatula from the IMGAG annotation of Medicago V1.0 genome release (http://www.medicago.org/genome/IMGAG). Only SCAR genes encoding both a SHD and a WA domain were used to generate a phylogenetic tree.
For all sequences, the predicted coding regions were translated into protein sequence and used for phylogenetic analysis. The SHD and WA domains from each protein were identified by ClustalW multiple sequence alignment to all of the Arabidopsis WA and SHD domains. Protein sequences were manually edited to remove dissimilar segments that probably represent splice prediction errors. For phylogenetic analyses, the protein encoding SHD and WA domains for three P. patens, four O. sativa, three P. trichocarpa, one M. truncatula, and four Arabidopsis SCAR genes were manually fused. These concatenated SHD-WA sequences were aligned using ClustalW (DNAStar Lasergene). That alignment was then imported into PAUP (Sinaur Associates), and a distance-based neighbor-joining tree was generated with manual rooting with P. patens as the outgroup, TBR branch swapping, and 1000 iterations to generate bootstrap values. A maximum likelihood tree for the fused nucleotide SHD-WA domains was also generated using PAUP with default settings of 10. The resulting tree follows the same topology as the distance tree with a -ln likelihood value of 3316.
Plant Morphometry and Statistical Analyses
Soil-grown, 12-DAG plants were used in trichome branch length and pavement cell shape quantification. To measure trichome branch lengths, images were acquired using a Nikon Coolpix 4500 digital camera mounted on a Leica SMZ 12.5 stereo microscope fitted with a ×1.6/0.2–numerical aperture objective. Image J software version 1.3 (http://rsb.info.nih.gov/ij) was used for the measurements. To measure the pavement cell shape, fully expanded cotyledons were dissected from plants and stained with 5 μM FM4-64 (Molecular Probes) for 2 h in the dark. Three images were taken from nonoverlapping regions in the apical third of the cotyledons using a MRC Bio-Rad 2100 laser scanning confocal microscope with a Nikon ×20/0.75–numerical aperture objective. The sampling of cells within a field was determined by selecting cells that made contact with a diagonal line. By analyzing three-dimensional stacks of images we could obtain measurements from nearly all of the cells that touched the diagonal. This sampling strategy included ∼60% of the cells whose boundaries were completely within the image field. In the pavement cell shape quantification, the cells ranked in the lowest quartile by cell area in each genotype were discarded because the small unlobed cells were usually associated with stomata. The cells that ranked in the top three quartiles by cell area were used for the comparison. The pavement cell circularity was calculated as previously described (Le et al., 2006). The hypocotyl elongation assay was performed as previously described (Basu et al., 2005) but at different time points. To test for differences among the mutants in trichome branch length, etiolated hypocotyl length, cell circularity, skeleton ends, and cell area in different mutants, the one-way ANOVA Tukey-Kramer multiple comparisons test was used at a significance level of α = 0.05. In addition to ANOVA, all possible pairwise Students t tests were conducted and a subset was used to resolve differences between key mutant combinations.
ARP2/3 Activation Assays, SCAR Cloning, and Production of Recombinant Proteins
Actin polymerization was performed as previously described (Basu et al., 2005), except that the actin concentration was 3 μM and ARP2/3 was 10 nM. GST-SCAR2WA was expressed and purified as described by Basu et al. (2005). The C-terminal regions of SCAR2, -3, and -4 were aligned, and peptides were chosen for expression so that each protein fragment contained a conserved Pro-rich segment that is near the WA domain. The gene regions representing SCAR3WA (amino acids 809 to 1020) and SCAR4WA (amino acids 922 to 1171) were PCR amplified with forward primers containing a BamHI site and reverse primers containing a SalI site. The template for SCAR3 PCR was pSK-29170FL (a gift from Laurie Smith). The template for SCAR4 was pCRIITopoScar4C, which was amplified from cDNA using SCAR4C_F2 and SCAR4C_R2 primers, cloned into pCRII-TOPO vector (Invitrogen), and sequenced. The PCR products were cloned into the Gateway vector pENTR/D/TOPO (Invitrogen), sequenced, and then subcloned into pGEX6P-1 to generate pGEX6P-SCAR3WA and pGEX6P-SCAR4WA (GE Healthcare) using BamHI and SalI. Full-length SCAR3 cDNA was subcloned into pSMB binary vector from pSK-29170FL using two SalI sites. Full-length SCAR2 cDNA was cloned to pENTR/D/TOPO, sequenced, and then subcloned into the pGWB2 binary vector by the LR reaction. GST-tagged proteins were purified using glutathione-agarose beads according to the manufacturer's specifications (Sigma-Aldrich). The protein concentration of purified SCAR-WA peptides was determined using the Bradford dye binding procedure using the Bio-Rad protein assay dye reagent. The activities of SCAR3 and SCAR4 peptides were significantly decreased by a freeze thaw cycle; therefore, these purified proteins were freshly prepared.
Gene Expression Analysis by qRT-PCR
Arabidopsis trichomes were isolated from the leaves of 21-DAG plants following a protocol adapted from Sinlapadech et al. (2007). To purify trichomes, the leaves were rapidly frozen in liquid nitrogen and were then filtered through two layers of 300-μm nylon mesh (Small Parts) after vortexing in liquid nitrogen for 10 s. To evaluate the quality of the trichome preparation, chlorophyll content was tested from each trichome sample (Porra, 2002). We also tested the expression level of the GLABRA2 (GL2) gene in the trichome sample by qRT-PCR. The chlorophyll content from the trichome samples are reduced by a factor of 16 to 20 compared with leaf samples, and the GL2 gene expression level is higher by a factor of 250 to 300 in trichomes compared with whole-leaf RNA samples. Total RNA was isolated from trichomes using the RNeasy plant mini kit (Qiagen). One microgram of total RNA was random primed and reverse transcribed to cDNA by M-MLV reverse transcriptase (Promega) and Random Primer 6 (New England Biolabs). Total cDNA (1/50) was used in each qRT-PCR reaction. Gene-specific primers for SCAR1, SCAR2, SCAR3, and SCAR4 were used in the experiment. GAPC (for glyceraldehyde-3-phosphate dehydrogenase C subunit) was used as the reference gene for normalization. Real-time PCR primers were designed using Primer Express Version 3.0 (Applied Biosystems). The qRT-PCR reactions were performed in the ABI Prism 7000 SDS system (Applied Biosystems). In each reaction, 10 μL SYBR Green PCR Master Mix (2×) (Applied Biosystems) was used in a 20 μL total volume. Triplicate reactions were performed for each cDNA preparation, and two biological replicates were performed for each sample. ΔCt (cycles to threshold) was calculated by substracting the Ct value of the GAPC gene from the Ct value of the SCAR gene [ΔCt = Ct (SCAR) − Ct (GAPC)]. The relative expression level of each SCAR was expressed as the percentage of the GAPC, which is calculated using the equation for normalized expression level = (2−ΔCt)*100%. The data shown are representative of two independent biological replicates. The normalized transcript levels for SCARs were tested for significant differences by one-way ANOVA with Tukey multiple comparisons. Primer names and sequences are listed in Table 5.
F-Actin Localization
F-actin localization and quantification methods are similar to Le et al. (2003). Young leaves from 14-DAG plants were dissected and fixed at room temperature in 2% formaldehyde in PEM buffer (100 mM PIPES-KOH, pH 6.9, 5 mM EGTA, and 4 mM MgCl2) for 30 min. After two washes (10 min for each wash) with PEM, the tissues were incubated in PEM containing 1% glycerol and 0.2μM Alexa Fluor 488 phalloidin (Invitrogen) overnight at 4°C. Samples were mounted in PEM containing 1% glycerol, and the images were collected using a MRC Bio-Rad 2100 confocal microscope with a Nikon ×60 Plan Apo/1.2-numerical aperture water immersion objective. Images were processed and analyzed using ImageJ and Adobe Photoshop software. Stage 4 trichomes with the length between 15 to 50 μm and with the branch long axis parallel to the XY plane were selected. Branches with aligned bundles were defined as those that contained more than three bundles that terminated within 3 μm of the branch apex and are aligned clearly with the long axis of the branch.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: Arabidopsis SCAR1 (AY743926), At2g34150; Arabidopsis SCAR2 (AY817016), At2g38440; Arabidopsis SCAR3 (AY743925), At1g29170; Arabidopsis SCAR4 (AY743927), At5g01730; Arabidopsis SCARL (AY743923), At4g18600; Arabidopsis GAPC, At3g04120; Arabidopsis GLABRA2 (GL2), At1g79840; Arabidopsis BRK1, At2g22640; Arabidopsis ARPC2, At1g30825. Sequence data for the Oryza sativa SCAR genes can be found in The Institute for Genomic Research Rice Genome Annotation database under the following locus ID numbers: O. sativa SCAR1 (Q5QNA6), Os1g11040; O. sativa SCAR2 (Q84TX2), Os3g60240; O. sativa SCAR3 (ABF95463), Os3g18710; O. sativa SCAR4 (BAC75554), Os07g49140. Proteins from P. trichocarpa are taken directly from the Populus trichocarpa v1.1 Jamboree Gene Models (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) and are as follows: P. trichocarpa SCAR1 (estExt_Genewise1_v1.C_LG_XI3306), P. trichocarpa SCAR2 (eugene3.00161237); P. trichocarpa SCAR3 (fgenesh4_pg.C_LG_VI000866). Proteins from P. patens are taken directly from the Physcomitrella patens v1.1 FilteredModels 3 annotations (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html) and are as follows: P. patens SCAR1 (fgenesh1_pg.scaffold_12000090); P. patens SCAR2 (estExt_fgenesh1_gp.C_220106; P. patens SCAR3 (fgenesh1_gp.scaffold_283000050). For Medicago, protein structures were taken from the IMGAG annotation of Medicago V1.0 genome release (http://www.medicago.org/genome/IMGAG): M. truncatula SCAR1 (2411.m00003).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SCAR Gene Expression in Light-Grown and Dark-Grown Hypocotyls.
Supplemental Figure 2. Arp2/3 Activation Efficiency of SCAR3 and SCAR4-WA Domains up to Saturating Generated Barbed End Concentrations.
Supplemental Data Set 1. Alignment of the SCAR Amino Acid Sequences That Was Used to Generate the Phylogeny Shown in Figure 2.
Supplementary Material
Acknowledgments
We thank Andew DeWoody and members of the Szymanski lab for helpful comments on the manuscript. The Purdue Cytoskeleton Group provided ongoing useful feedback for this project. Thanks to the ABRC for the distribution of SALK line strains. Thanks to Scott Jackson for help with the SCAR sequence comparisons. Laurie Smith kindly provided a cDNA clone for SCAR3. We thank Magdalena Benzanilla for providing the moss SCAR DNA sequences. Work in the lab of D.B.S. was supported by a grant from the Department of Energy–Energy Biosciences Division (DE-FG02-02ER15357) and a Purdue Agricultural Research Program fellowship. Work in the lab of C.J.S. was supported by a grant from the Department of Energy–Energy Biosciences Division (DE-FG02-04ER15526).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daniel B. Szymanski (dszyman@purdue.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Basu, D., El-Assal, S.E., Le, J., Mallery, E.L., and Szymanski, D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA-1 during leaf epidermal development. Development 131 4345–4355. [DOI] [PubMed] [Google Scholar]
- Basu, D., Le, J., El-Assal, S.E., Huang, S., Zhang, C., Mallery, E.L., Koliantz, G., Staiger, C.J., and Szymanski, D.B. (2005). DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17 502–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, J.E., Rawls, J.F., and Saxe, C.L.I. (1998). SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 142 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin, L., Amann, K.J., Higgs, H.N., Marchand, J.B., Kaiser, D.A., and Pollard, T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404 1007–1011. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant golgi apparatus traffics on an actin/ER network. Plant J. 15 441–447. [DOI] [PubMed] [Google Scholar]
- Brembu, T., Winge, P., Seem, M., and Bones, A.M. (2004). NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson, S.A., and Lindsey, K. (2006). The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of Auxin and sugars to promote embryonic cell identity. Plant Physiol. 142 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M.J., Kaloriti, D., Davies, B., Malho, R., and Hussey, P.J. (2004). Arabidopsis NAP1 is essential for ARP2/3-dependent trichome morphogenesis. Curr. Biol. 14 1410–1414. [DOI] [PubMed] [Google Scholar]
- Djakovic, S., Dyachok, J., Burke, M., Frank, M.J., and Smith, L.G. (2006). BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133 1091–1100. [DOI] [PubMed] [Google Scholar]
- Duncan, M.C., Cope, M.J.T.V., Goode, B.L., Wendland, B., and Drubin, D.G. (2001). Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3 687–690. [DOI] [PubMed] [Google Scholar]
- Eden, S., Rohatgi, R., Podtelejnikov, A.V., Mann, M., and Kirschner, M.W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418 790–793. [DOI] [PubMed] [Google Scholar]
- El-Assal, S.E., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004. a). DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 38 526–538. [DOI] [PubMed] [Google Scholar]
- El-Assal, S.E., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004. b). Arabidopsis GNARLED encodes a NAP125 homologue that positively regulates ARP2/3. Curr. Biol. 14 1405–1409. [DOI] [PubMed] [Google Scholar]
- Fay, J.C., and Wu, C.-I. (2003). Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genomics Hum. Genet. 4 213–235. [DOI] [PubMed] [Google Scholar]
- Finka, A., Schaefer, D.G., Saidi, Y., Goloubinoff, P., and Zryd, J.-P. (2007). In vivo visualization of F-actin structures during the development of the moss Physcomitrella patens. New Phytol. 174 63–76. [DOI] [PubMed] [Google Scholar]
- Fiserova, J., Schwarzerova, K., Petrasek, J., and Opatrny, Z. (2006). ARP2 and ARP3 are localized to sites of actin filament nucleation in tobacco BY-2 cells. Protoplasma 227 119–128. [DOI] [PubMed] [Google Scholar]
- Frank, M., Egile, C., Dyachok, J., Djakovic, S., Nolasco, M., Li, R., and Smith, L.G. (2004). Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc. Natl. Acad. Sci. USA 16 16379–16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G.O., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 11 687–700. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable, W.E., and Kropf, D.L. (2005). The Arp2/3 complex nucleates actin arrays during zygote polarity establishment and growth. Cell Motil. Cytoskeleton 61 9–20. [DOI] [PubMed] [Google Scholar]
- Harries, P.A., Pan, A., and Quatrano, R.S. (2005). Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17 2327–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, P.J., Ketelaar, T., and Deeks, M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57 109–125. [DOI] [PubMed] [Google Scholar]
- Innocenti, M., et al. (2005). Abi 1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7 969–981. [DOI] [PubMed] [Google Scholar]
- Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L.B., Steffen, A., Stradal, T.E.B., Di Fiore, P.P., Carlier, M.-F., and Scita, G. (2004). Abi1 is essential for the formation and activation of a WAVE2 signaling complex mediating Rac-dependent actin remodeling. Nat. Cell Biol. 6 319–327. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115 475–487. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.H., Burgos-Rivera, B., McKinney, E.C., Ruzicka, D.R., and Meagher, R.B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19 3111–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien, P., Ok, S.H., Shuai, B., Wengier, D., Cotter, R., Kelley, D., Kiriakopolos, S., Muschietti, J., and McCormick, S. (2005). Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J. 42 492–503. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer, A.M., and Wasteneys, G.O. (2006). Mechanisms behind the puzzle: Microtubule-micofilament cross-talk in pavement cell formation. Can. J. Bot. 84 594–603. [Google Scholar]
- Lavy, M., Bloch, D., Hazak, O., Gutman, I., Poraty, L., Sorek, N., Sternberg, H., and Yalovsky, S. (2007). A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17 947–952. [DOI] [PubMed] [Google Scholar]
- Le, J., El-Assal, S.E., Basu, D., Saad, M.E., and Szymanski, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13 1341–1347. [DOI] [PubMed] [Google Scholar]
- Le, J., Mallery, E.L., Zhang, C., Brankle, S., and Szymanski, D.B. (2006). Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr. Biol. 16 895–901. [DOI] [PubMed] [Google Scholar]
- Legg, J.A., Bompard, G., Dawson, J., Morris, H.L., Andrew, N., Cooper, L., Johnston, S.A., Tramountanis, G., and Machesky, L.M. (2007). N-WASP involvement in dorsal ruffle formation in mouse embyonic fibroblasts. Mol. Biol. Cell 18 678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Blanchoin, L., Yang, Z., and Lord, E.M. (2003). The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Sorefan, K., Hemmann, G., and Bevan, M.W. (2004). Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol. 136 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., Mullins, R.D., Higgs, H.N., Kaiser, D.A., Blanchoin, L., May, R.C., Hall, M.E., and Pollard, T.D. (1999). Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp 2/3 complex. Proc. Natl. Acad. Sci. USA 96 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., and Hulskamp, M. (2002). Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hulskamp, M. (2003. a). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B.P., and Hulskamp, M. (2003. b). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130 3137–3146. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126 5559–5568. [DOI] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasamyh, M.K. (1999). Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell 11 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Suetsugu, S., and Takenawa, T. (1998). WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17 6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, M.A., Boerlijst, M.C., Cooke, J., and Smith, J.M. (1997). Evolution of genetic redundancy. Nature 388 167–171. [DOI] [PubMed] [Google Scholar]
- Panchal, S.C., Kaiser, D.A., Torres, E., Pollard, T.D., and Rosen, M.K. (2003). A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10 591–598. [DOI] [PubMed] [Google Scholar]
- Panteris, E., and Galatis, B. (2005). The morphogenesis of lobed plant cells in the mesophyll and epidermis: organization and distinct roles of cortical microtubules and actin filaments. New Phytol. 167 721–732. [DOI] [PubMed] [Google Scholar]
- Perroud, P.-F., and Quatrano, R.S. (2006). The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil. Cytoskeleton 63 162–171. [DOI] [PubMed] [Google Scholar]
- Porra, R.J. (2002). The chequered history of the development and use of simltaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73 149–156. [DOI] [PubMed] [Google Scholar]
- Qiu, J.L., Jilk, R., Marks, M.D., and Szymanski, D.B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan, M., Gomez-Mena, C., Ruiz-Garcia, L., Salinas, J., and Martinez-Zapater, J.M. (1999). Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 20 581–590. [DOI] [PubMed] [Google Scholar]
- Russ, J.C. (2002). The Image Processing Handbook. (Boca Raton, FL: CRC Press).
- Savaldi-Goldstein, S., Peto, C., and Chory, J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446 199–202. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Sinlapadech, T., Stout, J., Ruegger, M.O., Deak, M., and Chapple, C. (2007). The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid:UDPG glucosyltransferase. Plant J. 49 655–668. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., and Oppenheimer, D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21 271–295. [DOI] [PubMed] [Google Scholar]
- Soderling, S.H., Guire, E.S., Kaech, S., White, J., Zhang, F., Schutz, K., Langeberg, L.K., Banker, G., Raber, J., and Scott, J.D. (2007). A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 27 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui, K., Head, K., Nowak, N., and Cowell, J.K. (2003). Genomic organization and expression profile of the human and mouse WAVE gene family. Mamm. Genome 14 314–322. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., and Blanchoin, L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9 554–562. [DOI] [PubMed] [Google Scholar]
- Steffen, A., Rottner, K., Ehinger, J., Innocenti, M., Scita, G., Wehland, J., and Stradal, T.E.B. (2004). Sra-1 and Nap1 link Rac to actin assembly driving Iamellipodia formation. EMBO J. 23 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovold, C.F., Millard, T.H., and Machesky, L.M. (2005). Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 6 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal, T.E., and Scita, G. (2006). Protein complexes regulating Arp2/3-mediated actin assembly. Curr. Opin. Cell Biol. 18 4–10. [DOI] [PubMed] [Google Scholar]
- Suetsugu, S., Yamazaki, D., Kurisu, S., and Takenawa, T. (2003). Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 5 595–609. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B. (2005). Breaking the WAVE complex: The point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8 103–112. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, M.D., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig, J.F., Mutondo, M., Zimmermann, I., Deeks, M.J., Machesky, L.M., Thomas, P., Uhrig, S., Rambke, C., Hussey, P.J., and Hulskamp, M. (2007). The role of Arabidopsis SCAR genes in ARP2–ARP3-dependent cell morphogenesis. Development 134 967–977. [DOI] [PubMed] [Google Scholar]
- Welch, M.D., and Mullins, R.D. (2002). Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18 247–288. [DOI] [PubMed] [Google Scholar]
- Westphal, R.S., Soderling, S.H., Alto, N.M., Langeberg, L.K., and Scott, J.D. (2000). Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 19 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C., et al. (2003). WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Zallen, J.A., Cohen, Y., Hudson, A.M., Cooley, L., Wieschaus, E., and Schejter, E.D. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Dyachok, J., Krishnakumar, S., Smith, L.G., and Oppenheimer, D.G. (2005). IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.