Abstract
Pseudomonas syringae pv. tomato strain DC3000 (PtoDC3000) is one of the most intensively studied bacterial plant pathogens today. Here we report a thorough investigation into PtoDC3000 and close relatives isolated from Antirrhinum majus (snapdragon), Apium graveolens (celery), and Solanaceae and Brassicaceae species. Multilocus sequence typing (MLST) was used to resolve the precise phylogenetic relationship between isolates and to determine the importance of recombination in their evolution. MLST data were correlated with an analysis of the locus coding for the type III secreted (T3S) effector AvrPto1 to investigate the role of recombination in the evolution of effector repertoires. Host range tests were performed to determine if closely related isolates from different plants have different host ranges. It was found that PtoDC3000 is located in the same phylogenetic cluster as isolates from several Brassicaceae and Solanaceae species and that these isolates have a relatively wide host range that includes tomato, Arabidopsis thaliana, and cauliflower. All other analyzed tomato isolates from three different continents form a distinct cluster and are pathogenic only on tomato. Therefore, PtoDC3000 is a very unusual tomato isolate. Several recombination breakpoints were detected within sequenced gene fragments, and population genetic tests indicate that recombination contributed more than mutation to the variation between isolates. Moreover, recombination may play an important role in the reassortment of T3S effectors between strains. The data are finally discussed from a taxonomic standpoint, and P. syringae pv. tomato is proposed to be divided into two pathovars.
Pseudomonas syringae pv. tomato DC3000 (PtoDC3000) is one of the most intensively studied plant pathogen isolates today. It was completely sequenced (6), and a large part of what is known about the plant immune system has been learned by studying the interaction of PtoDC3000 with its hosts Arabidopsis thaliana and tomato (Solanum lycopersicum), as can be seen from many recent high-profile publications (see references 39 and 47 for examples). However, much less is known about how PtoDC3000 relates to other P. syringae strains. Although PtoDC3000 is a rifampin-resistant derivative of the type strain of P. syringae pv. tomato (9; D. Cuppels, personal communication), its host range (which includes tomato, cauliflower [Brassica oleracea var. botrytis], and A. thaliana) was found to be more similar to that of pathovar maculicola isolates from Brassicaceae species than to the host range of other P. syringae pv. tomato strains (which are limited to tomato) (10, 58). Also, based on physiological (10) and molecular analyses (10, 63), PtoDC3000 was suggested to be more similar to pathovar maculicola strains than to other pathovar tomato strains. However, since strains of pathovars tomato, maculicola, antirrhini (isolated from ornamental snapdragon, Antirrhinum majus), and apii (isolated from celery, Apium graveolens) were all found to be closely related (18, 25), the precise relationship of PtoDC3000 with strains of these pathovars has been difficult to resolve. Therefore, PtoDC3000 is still generally considered to be a member of pathovar tomato.
Multilocus sequence typing (MLST) is a powerful approach to resolve the phylogenetic relationship at the inter- and intraspecies levels (34, 35). MLST is based on the sequencing of six or more fragments of housekeeping genes that are under purifying selection. Isolates with identical alleles at each locus are then grouped into sequence types (STs). Alternatively, sequences at all analyzed loci are concatenated, and phylogenetic trees can be constructed on the concatenated sequences. MLST allows for the determination of the contribution of homologous recombination (as a consequence of conjugation, transduction, or transformation [41] to variation between strains of a species). While homologous recombination has been found to play an important role in the evolution of several human pathogen species (54), MLST analysis of the plant pathogens P. syringae, Xylella fastidiosa, and Ralstonia solanacearum (see references 49, 50, and 7, respectively) indicated that these species are mostly clonal; i.e., the variation between strains of these species appears to be caused more by mutation than by recombination. However, closely related strains of the plant pathogen species Pseudomonas viridiflava were found to recombine frequently (19). Since only a relatively small number of closely related strains of P. syringae had been analyzed, it was suggested that recombination may also be found to occur at a high rate in P. syringae and other plant pathogen species if a larger number of closely related strains were to be analyzed (19).
What might be the role of homologous recombination in the evolution of plant-pathogenic bacteria? It is well known that horizontal gene transfer and the loss of plasmids and pathogenicity islands (PAIs) (22, 24) by conjugation and site-directed recombination play an important role in the evolution of virulence gene repertoires in plant-pathogenic bacteria (1, 43), in particular, in the acquisition and loss of genes coding for so-called effector proteins that are translocated from many plant-pathogenic bacteria into plant cells through the type III secretion (T3S) system and that have an important function in virulence (20). If recombination were found to be frequent between closely related P. syringae strains, homologous recombination may also play a similarly important role in reshuffling virulence genes between strains.
Here we report an MLST study and host range analysis of PtoDC3000 and a worldwide collection of closely related strains of pathovars tomato, maculicola, apii, and antirrhini that made it possible for us to precisely resolve their phylogenetic relationship. Moreover, recombination analysis suggests that homologous recombination significantly contributed to the variation between strains and to the evolution of T3S effector repertoires.
MATERIALS AND METHODS
Bacterial isolates.
The P. syringae isolates used in this study are given in Table 1. We are very grateful to our colleagues (Table 1) who generously shared their isolates with us.
TABLE 1.
P. syringae strains used in this study (listed in the same order as in Table 2)
| Pathovar | Strain name | Host of isolation (common name) | Host of isolation (scientific name) | Location | Collector of strain | Yr of collection | Source of strain | Reference |
|---|---|---|---|---|---|---|---|---|
| Antirrhini | 126 | Snapdragon | A. majus | M. Moffett | 1965 | D. Cuppels | ||
| Antirrhini | 152E | Snapdragon | A. majus | United Kingdom | J. Taylor | 1960 | D. Arnold | |
| Antirrhini | 4303 | Snapdragon | A. majus | United Kingdom | G. Jones | 1965 | D. Arnold | |
| Tomato | T1 | Tomato | S. lycopersicum | Canada | G. Bonn | T. Denny | 45 | |
| Tomato | Max1 | Tomato | S. lycopersicum | Italy | M. Zaccardelli | M. Zaccardelli | 62 | |
| Tomato | Max13 | Tomato | S. lycopersicum | France | M. Zaccardelli | 62 | ||
| Tomato | PST6 | Tomato | S. lycopersicum | Canada | D. Cuppels | T. Denny | 13 | |
| Tomato | PT13 | Tomato | S. lycopersicum | Gitaitis | J. Jones | |||
| Tomato | PT14 | Tomato | S. lycopersicum | G. Bonn | J. Jones | |||
| Tomato | PT18 | Tomato | S. lycopersicum | CA | C. Kado | T. Denny | 14 | |
| Tomato | PT2 | Tomato | S. lycopersicum | GA | S. McCarter | T. Denny | 14 | |
| Tomato | PT21 | Tomato | S. lycopersicum | FL | T. Howe | 1990 | J. Jones | |
| Tomato | PT26 | Tomato | S. lycopersicum | M. Ricker | 1990 | J. Jones | ||
| Tomato | PT32 | Tomato | S. lycopersicum | FL | J. Jones | 1993 | J. Jones | |
| Tomato | NCPPB1108 | Tomato | S. lycopersicum | Jersey, United Kingdom | R. A. Lelliott | 1961 | D. Arnold | |
| Tomato | B181 | Tomato | S. lycopersicum | GA | S. McCarter | 1981 | T. Denny | 14 |
| Tomato | 1318 | Tomato | S. lycopersicum | Switzerland | D. Cuppels | 10 | ||
| Tomato | 487 | Tomato | S. lycopersicum | Greece | D. Cuppels | 10 | ||
| Tomato | KS127 | Tomato | S. lycopersicum | Tanzania | K. C. Shenge | 2004 | M. Zaccardelli | 51 |
| Tomato | Max14 | Tomato | S. lycopersicum | Spain | M. Zaccardelli | 62 | ||
| Tomato | JL1065 | Tomato | S. lycopersicum | CA | J. Lindemann | R. Jackson | 57 | |
| Tomato | JL1031 | Tomato | S. lycopersicum | CA | J. Lindemann | 1983 | T. Denny | 14 |
| Tomato | PT28 | Tomato | S. lycopersicum | Mexico | J. Jones | 1992 | J. Jones | |
| Tomato | PT29 | Tomato | S. lycopersicum | Mexico | J. Jones | 1992 | J. Jones | |
| Tomato | PT30 | Tomato | S. lycopersicum | Mexico | J. Jones | 1992 | J. Jones | |
| Tomato | PST26L | Tomato | S. lycopersicum | South Africa | M. Hattingh | D. Cuppels | 10 | |
| Tomato | KS112 | Tomato | S. lycopersicum | Tanzania | K. C. Shenge | 2004 | M. Zaccardelli | 51 |
| Maculicola | F1 | Spinach mustard | Brassica rapa var. perviridis | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F7 | Spinach mustard | B. rapa var. perviridis | OK | 1995 | J. Damicone | 63 | |
| Apii | 1089 | Celery | A. graveolens | CA | D. A. Cooksey | D. Arnold | ||
| Maculicola | F15 | Turnip | B. rapa var. rapifera | OK | 1995 | J. Damicone | 63 | |
| Maculicola | M3 | Cauliflower | Brassica oleracea var. botrytis | United States | W. Burkholder | 1937 | J. Greenberg | 12 |
| Maculicola | M1 | Cauliflower | B. oleracea var. botrytis | United Kingdom | R. Lelliott | 1965 | J. Greenberg | 12 |
| Maculicola | M2 | Cauliflower | B. oleracea var. botrytis | New Zealand | D. Shackleton | 1965 | J. Greenberg | 12 |
| Maculicola | M6 | Cauliflower | B. oleracea var. botrytis | United Kingdom | G. E. Jones | 1965 | J. Greenberg | 12 |
| Maculicola | M8 | Kale | B. oleracea var. acephala | United Kingdom | J. Taylor | J. Greenberg | 12 | |
| Maculicola | 1766 | Cauliflower | B. oleracea var. botrytis | United Kingdom | G. E. Jones | 1965 | D. Cuppels | 10 |
| Tomato | ICMP3443 | Woolly nightshade | S. mauritianum | New Zealand | D. R. W. Watson | 1972 | J. Young | |
| Tomato | ICMP3449 | Woolly nightshade | S. mauritianum | New Zealand | D. R. W. Watson | 1972 | J. Young | |
| Tomato | DC3000a | Tomato | S. lycopersicum | Guernsey, United Kingdom | R. A. Lelliott | 1961 | J. Greenberg | 10 |
| Tomato | OH314 | Nettle | S. carolinenseb | OH | D. Coplin | 1978 | D. Cuppels | 10 |
| Maculicola | F6 | Kale | B. oleracea var. acephala | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F9 | Spinach mustard | B. rapa var. perviridis | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F10A | Turnip | B. rapa var. rapifera | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F18 | Kale | B. oleracea var. acephala | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F19 | Turnip | B. rapa var. rapifera | OK | 1996 | J. Damicone | 63 | |
| Maculicola | F16 | Turnip | B. rapa var. rapifera | OK | 1995 | J. Damicone | 63 | |
| Maculicola | F17 | Spinach mustard | B. rapa var. perviridis | OK | 1995 | J. Damicone | 63 | |
| Tomato | ICMP3435 | Woolly nightshade | S. mauritianum | New Zealand | D. R. W. Watson | 1972 | J. Young | 57 |
| Tomato | ICMP3455 | Woolly nightshade | S. mauritianum | New Zealand | D. R. W. Watson | 1972 | J. Young | 57 |
| Tomato | ICMP9305 | Woolly nightshade | S. mauritianum | New Zealand | D. R. W. Watson | 1987 | J. Young | |
| Maculicola | 84-59 | Cauliflower | B. oleracea var. botrytis | CA | W. Wiebe | D. Cuppels | 58 |
PtoDC3000 is a rifampin-resistant derivative of NCPPB1106 (ICMP2844, CFBP2212), which is the pathotype strain of P. syringae pv. tomato.
The botanical species was not determined at the time of collection (D. Coplin, personal communication). We believe it is probably S. carolinense (horse nettle), which is a close relative of tomato.
PCR and DNA sequencing of gene fragments.
Primers were designed on 24 P. syringae genes. Gene sequences of the three sequenced P. syringae genomes (6, 16, 28) were aligned in SeqMan (Lasergene; DNAStar, Madison, WI). Fifty- to 100-bp-long regions with high sequence identity between the three sequenced genomes at an approximate distance of 500 to 800 bp from each other were chosen as locations for forward and reverse primers. PtoDC3000 sequences from these regions were used for primer design in Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). While some primers annealed to all three genomes without the need for degeneration, other primers had to be degenerated to anneal to all three sequenced genomes. Primer sequences are given in Table S1 in the supplemental material.
Gene fragments were amplified from genomic DNA of P. syringae isolates extracted with the Puregene DNA purification system cell and tissue kit (Gentra Systems, Minneapolis, MN). The following DNA polymerases were used for PCRs: Eppendorf HotMaster Taq DNA polymerase (Brinkmann, Bestbury, NY) and Qiagen HotStarTaq and Qiagen Taq (Valencia, CA). Most primer pairs were used with a 58°C annealing temperature and 1 min extension time. For some primer pairs on some bacterial isolates, the annealing temperature was lowered or raised for optimal results. Instructions from polymerase manufacturers were followed for all other cycling conditions. All PCRs were performed on Eppendorf Mastercycler ep gradient thermocyclers (Brinkmann, Bestbury, NY). A total of 10 μl of PCR mixtures was cleaned for sequencing by using 1 unit shrimp alkaline phosphatase (USB Corp., Cleveland, OH) and 1 unit exonuclease I (USB Corp., Cleveland, OH).
DNA sequencing was carried out at the University of Chicago Cancer Research Center DNA Sequencing Facility. Chromatograms were reviewed and edited with SeqMan (Lasergene; DNAStar, Madison, WI).
Molecular evolutionary analysis.
Edited sequences were aligned in BioX 1.0b2 to 1.1b1 (E. Lagercrantz [http://www.lagercrantz.name/software/biox/]) using ClustalW 1.83 as the backend with default parameters. BioX is a graphical user interface for the eBiotools software package (http://www.ebioinformatics.org).
Bayesian trees were generated in MrBayes 3.1.2 (26, 46) using the Markov chain Monte Carlo method. The evolutionary model was set to GTR (general time reversible) with gamma-distributed rate variation across sites and a proportion of invariable sites. The program was run for 300,000 generations, which was long enough to ensure the standard deviation of split frequencies to be below 0.01. The sample frequency was 10. When summarizing the substitution model parameters and trees, 7,500 samples were used for the burn-in. Potential scale reduction factor values were all close to 1.0. The whole process was independently repeated three times to ensure convergence on the same tree. The Bayesian tree was rooted with P. syringae pv. syringae strain B728a (PsyB728a) as the outgroup in TreeView PPC 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
In order to determine the importance of recombination in the evolution of the analyzed isolates, genes with more than 10 informative sites were tested using the homoplasy test (37) in START 1.0.8 (29). DNA sequences were concatenated in frame for the homoplasy test. Phylogenetic networks were generated in SplitsTree 4.6 (27), using the NeighborNet (5) algorithm. The Web-based service GARD (genetic algorithm for recombination detection) (30) was employed to detect and locate recombination breakpoints. GARD's built-in tool was used to predict evolutionary models. The Shimodaira-Hasegawa (SH) test (52), implemented in PAUP* 4.0b10 (55), was performed on partitions flanking breakpoints predicted by GARD to determine their significance. The SH test determines the likelihood of a data set, given alternative trees.
Population recombination and mutation rates were estimated using the composite likelihood method in LDhat 2.0 (38). LDhat's built-in likelihood permutation test was used to test for the presence of recombination. The ratios of nonsynonymous to synonymous evolutionary changes for detecting positive selection were estimated by codonml of the PAML 3.15 package (61). Pair-wise sequence percent identities were calculated with MegAlign (Lasergene; DNAStar, Madison, WI).
BioX was used to convert sequence data file formats. Tree files were converted to SVG format with TreeView X 0.5.0 (http://darwin.zoology.gla.ac.uk/∼rpage/treeviewx/). Phylogenetic trees and network graphics were scaled and edited with Inkscape 0.45 (http://www.inkscape.org) and Illustrator CS3 (Adobe Systems Inc.) for publishing. ModelTest, MrBayes, LDhat, and PAML were compiled from the source in Mac OS X 10.4 to be run on Intel-based Mac computers.
Plant growth conditions and bacterial infections.
Plants were grown in a Percival Scientific CU-32L growth chamber (Perry, IA) in a 1:1 mixture of Pro-Mix BX and Pro-Mix PGX (Premier Horticulture Inc., Quakertown, PA). Plants were grown under 16-h days at 22°C and infected when 3 to 4 weeks old.
Isolates of P. syringae were streaked from glycerol stocks onto King's broth plates and grown at 30°C for 24 to 48 h. Bacteria were then restreaked onto new plates, covering the entire plate, and grown for another 24 h. Bacteria were scraped off the plates, resuspended, and diluted in sterile 10 mM MgSO4 for infections.
For the determination of disease symptoms, tomato cultivars Sunpride and Rio Grande, A. thaliana ecotype Columbia rps2, and cauliflower cultivar Early Snowball A were spray inoculated. Plants were placed into plastic bags and watered with 50 ml deionized water. Bags were sealed to maintain high humidity. Twenty-four hours later, the plants were sprayed with 20 ml of 10 mM MgSO4 containing 1.2 × 108 bacteria/ml. Twenty-four hours after inoculation, the plants were removed from the bags. Leaves were photographed 1 week after the date of infection. All pictures were taken with an Olympus Camedia C-765 digital camera. For the measurement of bacterial populations, plants were infected by dipping (tomato and cauliflower) or spraying (A. thaliana), including 0.02% of Silwet in the bacterial suspensions in order to obtain a uniform distribution of bacteria on the leaf surfaces.
Nucleotide sequence accession numbers.
All ST sequences were deposited in GenBank under the accession numbers EU296540 to EU296598.
RESULTS
The sequenced PtoDC3000 belongs to a group of closely related isolates from cultivated and wild plants around the world.
To confirm the close relationships between strains of pathovars tomato, maculicola, antirrhini, and apii reported in the literature, more than 100 isolates of these pathovars were assembled. A preliminary sequence analysis of the gyrB gene (data not shown) revealed that 83 isolates, including PtoDC3000, were closely related to each other, with DNA identities of more than 98.7%. In contrast, these isolates were only 92.3% identical, on average, to the other sequenced P. syringae isolates PsyB728a (16) and P. syringae pv. phaseolicola 1448A (Pph1448A) (28). Fifty-two representative isolates of the initial 83 isolates were chosen for further analysis to determine their phylogenetic relationship with PtoDC3000 and to test the hypothesis that closely related P. syringae isolates have high rates of recombination. Isolates were chosen based on preliminary sequence analysis of a subset of gene fragments, the geographic locations, the year of isolation, and the host of isolation, maximizing the diversity of the analyzed sample. Isolates are given in Table 1, including plant species, the year of collection, and the geographic location of isolation, when available.
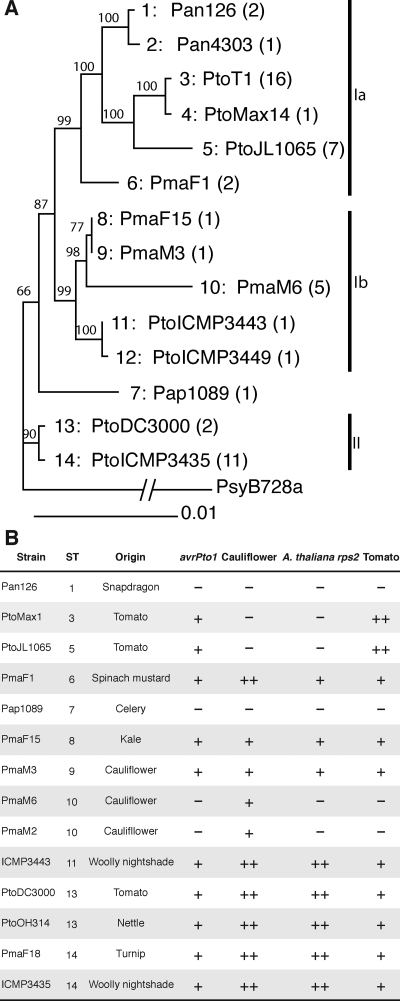
The gyrB gene was one of seven core genome genes (gyrB, rpoD, fruK, pgi, gap1, gltA, and acnB) used in a previous MLST study of P. syringae by Sarkar and Guttman (49). We used six of these genes and an additional three core genome genes located in proximity to avrPto1 to investigate the phylogeny of our collection in more detail; fruK was excluded from the analysis because of its high level of conservation. The positions of the nine analyzed genes in the PtoDC3000 genome are shown in Fig. 1. Table S1 in the supplemental material lists the primers used to amplify fragments of these genes. Fourteen different unique allele profiles were identified among the isolates (from now on called STs). Table 2 lists all isolates belonging to each ST. The sequences of the nine genes of each isolate were also concatenated in the order they are found in the PtoDC3000 genome. A Bayesian tree, a maximum likelihood (ML) tree, and a neighbor-joining tree were constructed with the concatenated sequences and rooted with the sequenced PsyB728a (and/or Pph1448A) isolate as the outgroup. The Bayesian consensus tree is shown in Fig. 2. The neighbor-joining tree (not shown) and the ML tree (not shown) have the same overall topology and similar branch lengths. Two main groups of isolates (I and II in Fig. 2) can be distinguished within all three trees. Group I contains the two subgroups Ia and Ib. Subgroup Ia contains all pathovar tomato isolates from tomato (with the exception of PtoDC3000), the P. syringae pv. antirrhini isolates, and two P. syringae pv. maculicola strains with identical STs (P. syringae pv. maculicola F1 [PmaF1] is shown in the tree). Several pathovar maculicola isolates and two Solanum mauritianum (wooly nightshade) isolates form group Ib. Group II contains two isolates with identical MLST profiles, PtoDC3000 from tomato and the PtoOH314 isolate from nettle (possibly horse nettle, Solanum carolinense, but unfortunately the species was not determined at the time of collection in 1978; D. Coplin, personal communication). Group II also contains 11 other almost-identical isolates (only one nucleotide difference in almost 6,000 bp compared to PtoDC3000), eight pathovar maculicola isolates from Brassicaceae and three S. mauritianum isolates. The P. syringae pv. apii 1089 isolate (Pap1089) is on a branch by itself but with low statistical support, making its placement outside or inside group I uncertain.
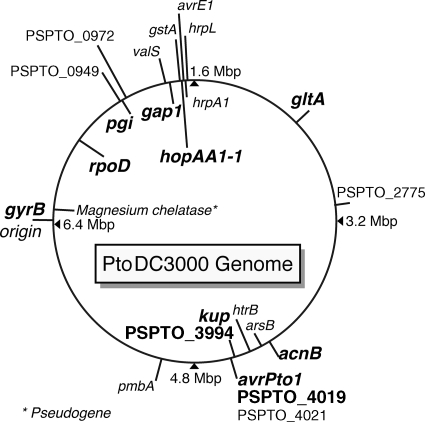
FIG. 1.
Positions of all analyzed genes in the genome of PtoDC3000. Genes that were sequenced for all isolates given in Table 1 are bold and in larger font. All other genes were sequenced only in five isolates used for the analysis shown in Fig. S3 in the supplemental material and in Table 6. For the exact gene location, primers used, and lengths of sequenced fragments, see Table S1 in the supplemental material.
TABLE 2.
Strains belonging to the STs in the Bayesian consensus tree (Fig. 2) and the NeighborNet network (Fig. 4)
| ST no. | Isolate(s)a |
|---|---|
| 1 | Pan126, Pan152E |
| 2 | Pan4303 |
| 3 | PtoT1, PtoMax1, PtoMax13, PtoPST6, PtoPT13, PtoPT14, PtoPT18, PtoPT2, PtoPT21, PtoPT26, PtoPT32, PtoNCPP1108, PtoB181, Pto1318, Pto487, PtoKS127 |
| 4 | PtoMax14 |
| 5 | PtoJL1065, PtoJL1031, PtoPT28, PtoPT29, PtoPT30, PtoPST26L, PtoKS112 |
| 6 | PmaF1, PmaF7 |
| 7 | Pap1089 |
| 8 | PmaF15 |
| 9 | PmaM3 |
| 10 | PmaM6, PmaM1, PmaM2, PmaM8, Pma1766 |
| 11 | PtoICMP3443 |
| 12 | PtoICMP3449 |
| 13 | PtoDC3000, PtoOH314 |
| 14 | PmaF9, PmaF6, PmaF10A, PmaF16, PmaF17, PmaF18, PmaF19, Pma84-59, PtoICMP3435, PtoICMP3455, PtoICMP9305 |
Representative strains shown in the tree in Fig. 2 are bold.
FIG. 2.
Bayesian tree of the concatenated gene fragments and host range of representative isolates with regard to A. thaliana, cauliflower, and tomato. (A) Each ST is identified by its number (preceding the colon) and by a representative isolate. The number of isolates belonging to each ST is indicated in parentheses. See Table 2 for a complete list of isolates belonging to each ST. The credibility values (×100) of clades, indicating the statistical significance of groupings, are given in front of each node. The sequenced isolate PsyB728a (16) was used as the outgroup. (B) The host of isolation (origin), the experimentally determined host range on selected plant species (−, no disease symptoms; +, mild disease symptoms; ++, severe disease symptoms), and the presence of the avrPto1 gene are indicated for representative isolates of most STs (some STs that differ from another ST by only a single nucleotide were not included in host range tests). A. thaliana rps2 is a mutant of A. thaliana with a nonfunctional RPS2 gene (40).
An ML tree built on only the six housekeeping genes acnB, gap1, gltA, gyrB, pgi, and rpoD had a topology that was very similar to the trees built on all nine genes, showing that the three genes kup, PSPTO_3994, and PSPTO_4019 close to the avrPto1 locus did not significantly alter tree topology (data not shown).
Isolates very closely related to PtoDC3000 have different host ranges.
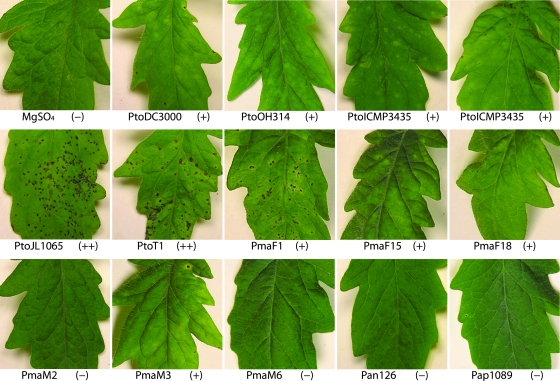
Since the analyzed isolates are all very closely related to each other—some of the pathovar maculicola isolates were reported to also cause disease on tomato (10, 58) and PtoDC3000 is well known to cause disease on A. thaliana, cauliflower, and tomato (6, 10)—representative isolates were inoculated under controlled conditions to determine whether host range differences existed between isolates. Inoculations were performed by spraying bacteria onto leaf surfaces without the addition of any surfactant to make them resemble natural infections as much as possible. In all cases, symptoms always appeared at least 3 days after infection, making it very unlikely that symptoms were caused by a hypersensitive defense response (21), which usually becomes macroscopically visible only when bacteria are directly infiltrated into leaves at high doses and appears within a day after infection. Figure 3 shows that the pathovar tomato isolates of ST3, -4 and -5 (see Table 2 for the list of isolates belonging to each ST) caused the most severe symptoms on tomato cultivar Sunpride. The isolates of ST6, -8, -9, and -11 to -15, including PtoDC3000, caused mild symptoms. Only when the concentration of the inoculum was increased 10-fold did we obtain disease symptoms on tomato, and these isolates were similar to those caused by the pathovar tomato isolates of ST3, -4, and -5 (data not shown). ST1, -2, -7, and -10 did not cause any disease symptoms on tomato. Inoculations of the tomato cultivar Rio Grande caused similar results, although differences in symptom severity between ST3, -4, and -5 on one hand and ST6, -8, -9, and -11 to -15 on the other hand were less pronounced (data not shown).
FIG. 3.
Virulence tests of representative P. syringae isolates on tomato. Entire tomato plants were sprayed with bacterial suspensions of P. syringae isolates. Photographs of representative leaves were taken 1 week after infection. ++, highly virulent P. syringae isolates causing severe disease symptoms (large numbers of necrotic pits with chlorotic haloes); +, isolates causing mild disease symptoms (few pits and diffused chlorosis); −, isolates causing no symptoms.
Cauliflower and A. thaliana infections (see Fig. S1A and S1C in the supplemental material) also revealed remarkable differences in symptoms caused by the different isolates, which are also summarized in Fig. 2. For example, the ability of PtoDC3000 to cause disease on A. thaliana and on cauliflower, reported in the literature, was confirmed. P. syringae pv. tomato isolates of ST3, -4, and -5 were found to be unable to cause any symptoms on these two plant species. Using an A. thaliana ‘Columbia’ accession mutated in the RPS2 resistance gene (40), it was shown that the inability of P. syringae pv. tomato isolates with ST3, -4, and -5 to cause disease on A. thaliana was not due to the long-known gene-for-gene interaction between the cognate A. thaliana resistance gene RPS2 (57) and the T3S effector gene avrRpt2 (which was confirmed by PCR to be present in all P. syringae pv. tomato isolates of ST3, -4, and -5, with the exception of isolate PtoMax13 [data not shown]). Moreover, these STs were even unable to cause any disease on the A. thaliana sgt1 (2), sid2 (59), and pad4 (64) (data not shown) defense mutants, suggesting multigenic nonhost resistance. A subset of isolates was tested on snapdragon and celery, which confirmed host range differences on these plant species as well (data not shown). For selected P. syringae pv. tomato and P. syringae pv. maculicola isolates, bacterial populations were measured on tomato, cauliflower, and A. thaliana 3 days after infection, revealing that symptom severity correlated well with bacterial population size (see Fig. S1B, S1D, and S1E in the supplemental material). For example, on tomato, the strains PtoT1 and PtoMax1 that caused the most severe symptoms grew to almost 100-fold-higher population densities than PtoDC3000 and PmaF18, which had caused only mild symptoms.
Recombination contributed to the evolution of the core genome of PtoDC3000 and closely related isolates.
Before using the nine sequenced gene fragments for recombination analyses, we determined that all genes are under purifying selection (Table 3) and are thus well suited for evaluating the importance of recombination in the evolution of the analyzed isolates.
TABLE 3.
Percent DNA identities between isolates used in this study and between the three sequenced P. syringae isolates and ratio of nonsynonymous to synonymous mutations for all analyzed genes
| Gene | % DNA identity
|
dN/dS ratiob | |||
|---|---|---|---|---|---|
| Avg | Minimum | Maximum | 3S Avga | ||
| acnB | 99.1 | 98.2 | 99.8 | 94.0 | 0 |
| gap1 | 99.7 | 99.3 | 99.8 | 92.4 | 0.0775 |
| gltA | 99.3 | 99.2 | 99.6 | 95.9 | 0 |
| gyrB | 99.3 | 98.7 | 99.9 | 92.3 | 0 |
| pgi | 99.4 | 98.9 | 99.8 | 90.9 | 0.0313 |
| kup | 98.7 | 97.2 | 99.9 | 91.1 | 0.0056 |
| PSPTO_3994 | 99.5 | 99.0 | 99.9 | 84.43 | NA |
| PSPTO_4019 | 98.8 | 98.3 | 99.4 | 92.6 | 0 |
| rpoD | 98.8 | 98.0 | 99.8 | 93.6 | 0 |
| All | 99.34 | 98.80 | 99.99 | NA | NA |
Percent DNA identities between the three completely sequenced P. syringae genomes (PtoDC3000, PsyB728a, and Pph1448A).
Ratios of nonsynonymous to synonymous evolutionary changes were calculated based on the sequences of all closely related isolates used in this study by using codonml of the PAML 3.15 package (61). NA, not applicable.
Since the homoplasy test (37) is well suited for very closely related isolates with a DNA identity of more than 98% (44), it was the first test applied to our data. Homoplasies are defined as mutations shared between different branches of a phylogenetic tree that have not been directly inherited from an ancestor. The homoplasy test calculates the ratio between homoplasies minus the expected homoplasies in the case of no recombination and calculates the expected homoplasies in the case of free recombination minus the expected homoplasies in the case of no recombination. The closer this ratio is to 1, the more recombination can be inferred. The homoplasy ratio for the genes rpoD, gyrB, and kup and for the concatenated sequence of all nine genes were found to be 0.408, 0.160, 0.623, and 0.413, respectively. The obtained ratios indicate the presence of recombination in two of three genes and in the concatenated sequence. It was not possible to calculate the homoplasy ratio for all other genes, since only rpoD, gyrB, and kup have the 10 or more informative sites required for this test in the START package (29).
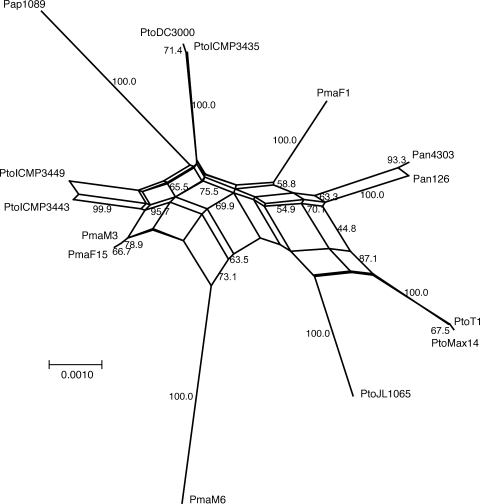
Because the homoplasy test indicated recombination, the phylogenetic tree shown in Fig. 2 may not be an accurate representation of the phylogeny of the analyzed isolates. Since alternative topologies cannot be represented in a tree, a tree cannot reflect recombination. A tree is always only a “compromise” of the different possible trees that can be built on an alignment when conflicting signals are present. Phylogenetic networks have been developed to overcome this inherent shortcoming of trees (17). In a network, alternative phylogenies are represented by splits. The more splits (or reticulations) there are in a network, the more conflicting signals—possibly due to recombination—exist in the data. We built a phylogenetic network on the concatenated sequences of the nine core genome genes using the NeighborNet algorithm (5), which is similar to the commonly used splits decomposition algorithm but is better suited for handling large data sets, and the generated networks are generally more resolved (5). Figure 4 shows that there are a large number of splits in the network built on the concatenated sequence, indicative of conflicting phylogenetic signals.
FIG. 4.
NeighborNet analysis of the nine concatenated housekeeping genes gyrB, rpoD, pgi, gap1, gltA, acnB, kup, PSPTO_3994, and PSPTO_4019. One representative isolate of each ST and bootstrap values higher than 40 are shown.
To look more carefully at the individual gene level, NeighborNet networks were built for each gene fragment (see Fig. S2 in the supplemental material). Six of the nine sequences show various degrees of reticulation. For the three gene fragments with the greatest percentage of DNA identity between isolates, no reticulation was identified, possibly because the number of nucleotide differences in these fragments was insufficient. To identify recombination breakpoints and subsequently determine if recombination could explain the reticulation in the other six genes, we used the GARD program (30), which is a likelihood-based model selection procedure. A breakpoint was predicted based on the improvement of the Akaike information criterion score for trees constructed on the partitions flanking the predicted breakpoint compared to a tree constructed on the entire sequence (30). Table 4 shows the GARD output for all genes. As suggested by the NeighborNet networks, GARD found breakpoints only in the most divergent genes. NeighborNet networks were then built on all predicted partitions of these genes. In many cases, NeighborNet did not find any reticulations in the gene partitions predicted by GARD, confirming the prediction of the recombination breakpoints (Table 4).
TABLE 4.
Recombination breakpoints predicted in analyzed gene fragments
| Gene | No. of breakpointsa | Length of fragments for each partition (nt) | NeighborNet results for partitionsc | No. of detectable breakpoints/total no. of breakpoints (P < 0.05)d |
|---|---|---|---|---|
| gyrB | 1 | 358, 343 | No reticulation | 1/1 |
| rpoD | 1 | 404, 241 | Fewer splits for both partitions | 1/1 |
| pgi | 0 | NAe | NA | NA |
| gap1 | 0 | NA | NA | NA |
| gltA | 0 | NA | NA | NA |
| acnB | 0 | NA | NA | NA |
| kup and PSPTO_3994b | 3 | 720, 273, 138, 812 | 2/4c | 3/3 |
| PSPTO_4019 | 3 | 233, 198, 189, 100 | 2/4c | 1/3 |
Number of breakpoints predicted by GARD (30).
kup and PSPTO_3994 is the sequence of part of the kup gene and part of the PSPTO_3994 gene, including the intergenic region between the two genes.
Number of partitions that show no reticulation out of the total number of partitions.
A breakpoint was considered detectable by the SH test if at least one flanking tree was significantly worse at fitting the partition on the other side of the breakpoint. Values represent detectable breakpoints over the number of breakpoints tested.
NA, not applicable.
The significance of the predicted breakpoints was further analyzed by performing the SH test (52) for all partition pairs separated by a breakpoint. The SH test is widely used to determine the significance of differences between tree topologies. If a tree built on one sequence alignment is statistically significantly different from the data derived from a second sequence alignment, recombination between the sequences can be inferred, as long as the sequences are under the same selection pressure. The SH test does not detect recombination events that change branch lengths without changing branching patterns (30). Nonetheless, the SH test showed that for six of eight breakpoints that were predicted by GARD (Table 4), trees built on flanking partitions were significantly different from each other, supporting the conclusion that recombination breakpoints exist in many analyzed genes.
The best way to compare recombination rates between species is to use a population genetics approach by expressing recombination rates in relation to mutation rates. The ratio between the population recombination rate (ρ) and the population mutation rate (θ) is often used for this purpose. ρ and θ can be calculated using a coalescent theory-based method developed by McVean and coworkers and implemented in the program LDhat (38). Applying the likelihood permutation test within LDhat, we found ρ to be significantly different from zero for several of the genes and ρ/θ values ranged from 0.259 for kup to 14.061 for PSPTO_3994 (Table 5). When the ρ/θ values obtained for the analyzed genes were simply averaged, the contribution of recombination was estimated to be more than five times greater than the contribution of mutation to variation between isolates. In comparison, by doing the same calculation using a mix of closely and distantly related isolates of P. syringae, the mean ρ/θ was found to be only 0.252 (49). The mean ρ/θ for P. viridiflava was 0.48 overall but 2.38 and 10.16, respectively, for closely related isolates in clades A and B (19). ρ/θ was found to be zero for Escherichia coli overall (42) but 2.139 for clade D of E. coli, which contains closely related, highly virulent isolates (60). Therefore, as for P. viridiflava and E. coli, recombination appears to greatly contribute to the variation between closely related isolates of P. syringae, while more distantly related isolates appear to mainly differ from each other because of mutation.
TABLE 5.
Estimates of ρ and θ
| Gene | ρa | Per site ρ | θ | Per site θ | ρ/θ | GARDb |
|---|---|---|---|---|---|---|
| gyrB | 14.0*** | 0.0200 | 4.949 | 0.00706 | 2.829 | Yes |
| rpoD | 100.0*** | 0.1550 | 7.328 | 0.01136 | 13.646 | Yes |
| pgi | 9.5 | 0.0167 | 4.380 | 0.00770 | 2.169 | No |
| gap1 | 13.5 | 0.0225 | 1.920 | 0.00320 | 7.031 | No |
| gltA | 5.0* | 0.0099 | 3.333 | 0.00657 | 1.500 | No |
| acnB | 7.0 | 0.00116 | 5.255 | 0.00867 | 1.332 | No |
| kup | 4.0*** | 0.0038 | 15.45 | 0.01450 | 0.259 | Yes |
| PSPTO_3994 | 70.5 | 0.0754 | 5.014 | 0.00536 | 14.061 | Yes |
| PSPTO_4019 | 80.0** | 0.1111 | 8.160 | 0.01133 | 9.804 | Yes |
| Mean | 33.722 | 0.046 | 6.198 | 0.008 | 5.847 |
Statistical significance for the presence of recombination determined by applying the likelihood permutation test. *, P < 0.10; **, P < 0.05; ***, P < 0.01; ρ, population recombination rate; θ, population mutation rate.
Yes, recombination breakpoints were found by the program GARD (30); No, recombination breakpoints were not detected by GARD.
Acquisition and loss of the avrPto1 PAI and its role in host range evolution.
AvrPto1 is a well-studied T3S effector protein. It contributes to virulence on susceptible tomato cultivars (8, 31, 33), although it induces plant defenses on tomato plants that carry the Pto resistance gene (45). Comparing the three sequenced P. syringae genomes by using the multiple genome alignment program MAUVE (11), the PtoDC3000 avrPto1 gene is located on a 23,532-bp-long, PtoDC3000-specific region between nucleotides 4506925 and 4530456. Besides avrPto1, the PtoDC3000-specific region contains several transposase genes and a defective prophage genome (6). A similar prophage genome is present adjacent to the avrPto1 gene in PsyB728a (16) but in a different genomic context. Together with the fact that avrPto1 is present in several P. syringae strains that are only distantly related to each other (48), this suggests that avrPto1 was acquired independently by several P. syringae strains, possibly as part of a bacteriophage. However, by correlating the phylogenetic tree with the distribution of avrPto1 in the PtoDC3000 relatives in Fig. 2, the absence of avrPto1 from the isolates of the P. syringae pv. antirrhini, Pap1089, and PmaM6 STs can be best explained through a one-time acquisition event of avrPto1 by an ancestor of all analyzed isolates and the later loss of avrPto1 from the ancestor of the P. syringae pv. antirrhini isolates, the ancestor of Pap1089, and the ancestor of the PmaM6 ST isolates (three separate events). Other evolutionary scenarios would require a greater number of events.
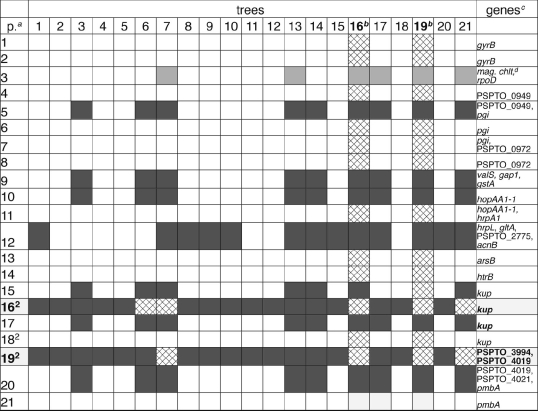
If avrPto1 were lost during evolution by the deletion of the avrPto1 gene alone or by the excision of the entire avrPto1 prophage region, similar to the excision of the PPHGI-1 PAI observed in P. syringae pv. phaseolicola 1301 (43), the regions flanking the excised region would not be affected by this event. However, the genes kup and PSPTO_3994 immediately upstream of the 23,532-bp-long, prophagelike PtoDC3000 avrPto1 region and the gene PSTPO_4019 immediately downstream of the same region have a very unusual nucleotide substitution pattern and contain several recombination breakpoints (Table 4). For most analyzed genes, the PmaM3 and PmaM6 alleles are either nearly identical to each other but different from the PtoDC3000 alleles or nearly identical to each other and nearly identical to the PtoDC3000 alleles. Only in the partitions flanking the avrPto1 region are PmaM3 and PtoDC3000 nearly identical to each other but different from PmaM6. We extended the sequence analysis for the five isolates PtoT1, PtoJl1065, PmaM3, PmaM6, and PtoDC3000 to 13 more genes in the genome and confirmed this observation (see Fig. 1 for the location of these genes in the PtoDC3000 genome and see Fig. S3 in the supplemental material for all nucleotide differences between alleles). We used GARD (30) to identify breakpoints in the concatenated sequence of all 23 genes. The SH test (52) shows that even when only the five analyzed isolates were used, the trees built on most gene partitions flanking the avrPto1 prophage region (in particular, partitions 16 and 19) are significantly different from almost all other analyzed genes in the genome (Table 6). Considering these results, a likely explanation for the loss of avrPto1 from an ancestor of the PmaM6 ST isolates is homologous recombination in core genome genes flanking the avrPto1 prophage region, during which this region was replaced with a genomic region of a donor strain that did not contain avrPto1. Note that this explanation assumes that avrPto1 was present in the same locus in the PmaM3-like ancestor of PmaM6 as it was in PtoDC3000, which is likely because of their close relationship but impossible to confirm based on our current data. Interestingly, PtoDC3000 and most of its relatives that cause weak or severe disease symptoms on tomato (ST3, -4, -5, -6, -8, -9, and -11 to -15) contain avrPto1 (as determined by PCR [data not shown]), while the isolates of ST1, -2, -7, and -10 (including PmaM6) that do not cause disease symptoms on tomato do not contain avrPto1 (Fig. 2), suggesting that the loss of avrPto1 from the PmaM6 ancestor may have been involved in host range evolution (see Discussion) below.
TABLE 6.
SH test of gene partitions flanking the avrPto1 prophage region compared to other regions in the P. syringae genome
p., partitions identified with the program GARD (30) in the concatenated sequence.
Partitions 16 and 19 are significantly different (dark grey, significantly different at a P of <0.01; light grey, significantly different at a P of <0.05) from most other partitions when data are compared with trees and trees are compared with data. Partition 19 is immediately flanking the avrPto1 prophage region on both sides. Other partitions (for example, partition 12) are significantly different from other genes only when data are compared with trees but not when trees are compared with data.
Genes of which the partitions are a part.
mag. chlt, magnesium chelatase.
DISCUSSION
Importance of recombination in the evolution of closely related P. syringae strains.
While all bacteria were assumed to be clonal, John Maynard Smith (53) revolutionized the field of microbial evolution by developing molecular evolutionary tests that showed that some bacterial species recombine frequently. However, only recently has it become clear that recombination is an important evolutionary mechanism in numerous bacterial species. A prominent example is E. coli, a species previously considered to be clonal. Extensive sequence analysis of many isolates has now confirmed earlier evidence (23) of frequent recombination in some E. coli clades (60). Recombination in many bacterial species was overlooked before the advent of MLST because recombination can efficiently occur only when DNA sequences of the donor and the recipient are very similar (36); however, it can be detected only when DNA sequences of the donor and the recipient are sufficiently different from each other for recombination events to be inferred from sequence data with statistical significance (44).
In the case of P. syringae, little recombination was detected when a mix of distantly and closely related isolates was analyzed, and mutation was found to contribute approximately four times more than recombination to variation (49). The reason for the relatively small contribution of recombination to variation between the analyzed isolates probably lies in the percentage of DNA sequence identity, which is as low as 91% between distantly related P. syringae isolates. Also, the adaptation of P. syringae pathovars to different plant species probably makes physical contact between distantly related isolates more difficult. The situation dramatically changed in the current study when closely related isolates with a DNA sequence identity of approximately 99% were analyzed; recombination was found to contribute 5.8 times more than mutation to variation between isolates (using the same population genetics test [38] that was used in the study by Sarkar and Guttman [49]). Moreover, many isolates that we studied have partially overlapping host ranges, which can be expected to give ample opportunity for recombination. Even in cases where a common host between isolates was not identified, isolates may have evolved only recently from ancestors with overlapping host ranges. We were also able to confirm high rates of recombination between closely related P. syringae isolates by identifying recombination breakpoints within several genes and by applying various recombination tests. However, for some gene fragments, the DNA percent identity between isolates was too high (more than 99.3%) to allow detection of recombination. The use of additional gene fragments with lower DNA percent identity (between 98% and 99%) will be advantageous in future studies.
Not only P. syringae (49) but also the plant pathogen species X. fastidiosa and R. solanacearum were found to evolve mainly by mutation rather than recombination when a mix of closely and distantly related isolates were analyzed (7, 50). However, when a large number of closely related isolates was analyzed, Goss and colleagues (19) found high recombination rates in P. viridiflava; R. solanacearum was found to be naturally competent during infection, potentially allowing a very high rate of recombination (4); and recombination rates were high enough in Xanthomonas to be detected in controlled coinfections (3). The combination of these results and our results showing that high recombination rates exist between closely related P. syringae isolates suggests that recombination probably plays an important role in the evolution of many bacterial plant pathogen species but that this is easily overlooked when an insufficient number of closely related isolates is sampled.
Role of recombination in determining the distribution of the effector avrPto1.
We found that avrPto1 was probably already present in an ancestor of all analyzed PtoDC3000 relatives and may have been lost later by the ancestor of PmaM6 through homologous recombination in genes flanking the avrPto1 prophage region. What could have led to the loss of avrPto1? The P. syringae ancestor containing avrPto1 can be assumed to have existed before the evolution of the plant resistance gene Pto, which elicits defenses upon recognition of avrPto1 (45), or to have been a pathogen of plants that did not carry the Pto resistance gene. Therefore, avrPto1 conferred a fitness advantage. However, after the evolution of the Pto resistance gene, it became a fitness advantage to lose avrPto1 when infecting a plant that expressed Pto. This is also supported by the fact that several P. syringae strains that contain avrPto but that are not tomato pathogens can cause disease only on tomato plants that do not carry the Pto resistance gene (32). Therefore, recombination leading to a replacement of the genomic region containing avrPto1 with a region from a P. syringae donor that did not contain avrPto1 (but that possibly contained a different virulence gene) became advantageous.
However, the loss of avrPto1 cannot explain why PmaM6 does not cause disease on tomato while other isolates which contain avrPto1 do. The presence/absence of avrPto1 is just one of several differences in the T3S effector repertoire between the analyzed isolates. Dot blot experiments revealed that PmaM6 has at least 16 differences in its effector repertoire compared to that of PtoDC3000 (data not shown). Moreover, PtoT1, a pathovar tomato isolate without avrPto1, was isolated from tomato and is still pathogenic on tomato (45). Thus, we believe that the loss of avrPto1 from an ancestor of PmaM6 was only one of the events in the adaptation of PmaM6 to plant species on which avrPto1 is detected by a Pto-like resistance gene.
Revealed identity of PtoDC3000.
In spite of the detected recombination between PtoDC3000 and its close relatives, it was possible to clearly resolve their phylogenetic relationship. Correlating the obtained phylogenetic data, the hosts of isolation, and the results from host range tests, it becomes clear that PtoDC3000 is not a typical P. syringae pv. tomato strain. Typical P. syringae pv. tomato strains form a distinct phylogenetic clade apart from PtoDC3000, have all been isolated from tomato, are more virulent on tomato than PtoDC3000, and do not cause disease on either A. thaliana or cauliflower. On the other hand, PtoDC3000 is part of a mixed group of almost identical P. syringae pv. maculicola and P. syringae pv. tomato isolates from cultivated Brassicacaeae and wild Solanaceae species that cause disease on tomato, cauliflower, and A. thaliana. Since, based on the definition of pathovar, strains are grouped into pathovars based on the host of isolation and the host range (15, 56), the typical P. syringae pv. tomato strains and the PtoDC3000-like strains should be separated into two distinct pathovars. In accordance with the rules on naming pathovars (15), the pathovar tomato name would need to remain associated with its deposited type strain, i.e., PtoDC3000/ PtoNCPPB1106. In fact, associating pathovar tomato with a new type strain would create ambiguity. Therefore, PtoDC3000-like strains would continue to be part of pathovar tomato, and a new pathovar with a new name could be introduced for the typical P. syringae pv. tomato strains from tomato that cause disease only on tomato.
Supplementary Material
Acknowledgments
This work was funded through Virginia Tech Start Up funds to Boris A. Vinatzer and a Virginia Tech ASPIRES grant to Boris A. Vinatzer and João C. Setubal.
We thank our generous colleagues (Table 1) for sharing the isolates and thank the undergraduate students Leiya Williams, Nina Long, Eric Hall, and Douglas Chandler for their help with PCR analysis, sequence analysis, and plant infections.
Footnotes
Published ahead of print on 31 March 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arnold, D. L., A. Pitman, and R. W. Jackson. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420. [DOI] [PubMed] [Google Scholar]
- 2.Austin, M. J., P. Muskett, K. Kahn, B. J. Feys, J. D. Jones, and J. E. Parker. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077-2080. [DOI] [PubMed] [Google Scholar]
- 3.Basim, H., R. E. Stall, G. V. Minsavage, and J. B. Jones. 1999. Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria. Phytopathology 89:1044-1049. [DOI] [PubMed] [Google Scholar]
- 4.Bertolla, F., A. Frostegard, B. Brito, X. Nesme, and P. Simonet. 1999. During infection of its host, the plant pathogen Ralstonia solanacearum naturally develops a state of competence and exchanges genetic material. Mol. Plant-Microbe Interact. 12:467-472. [Google Scholar]
- 5.Bryant, D., and V. Moulton. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 6.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed]
- 7.Castillo, J. A., and J. T. Greenberg. 2007. Evolutionary dynamics of Ralstonia solanacearum. Appl. Environ. Microbiol. 73:1225-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, J. H., J. P. Rathjen, A. J. Bernal, B. J. Staskawicz, and R. W. Michelmore. 2000. avrPto enhances growth and necrosis caused by Pseudomonas syringae pv. tomato in tomato lines lacking either Pto or Prf. Mol. Plant-Microbe Interact. 13:568-571. [DOI] [PubMed] [Google Scholar]
- 9.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuppels, D. A., and T. Ainsworth. 1995. Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl. Environ. Microbiol. 61:3530-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debener, T., H. Lehnackers, M. Arnold, and J. L. Dangl. 1991. Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1:289-302. [DOI] [PubMed] [Google Scholar]
- 13.Denny, T. P. 1988. Differentiation of Pseudomonas syringae pv. tomato from P. syringae with a DNA hybridization probe. Phytopathology 78:1186-1193. [Google Scholar]
- 14.Denny, T. P., M. N. Gilmour, and R. K. Selander. 1988. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J. Gen. Microbiol. 134:1949-1960. [DOI] [PubMed] [Google Scholar]
- 15.Dye, D. W., J. F. Bradbury, M. Goto, A. C. Hayward, R. A. Lelliott, and M. N. Schroth. 1980. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev. Plant Pathol. 142:153-158. [Google Scholar]
- 16.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitch, W. M. 1997. Networks and viral evolution. J. Mol. Evol. 44(Suppl. 1):S65-S75. [DOI] [PubMed] [Google Scholar]
- 18.Gardan, L., H. Shafik, S. Belouin, R. Broch, F. Grimont, and P. A. Grimont. 1999. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49:469-478. [DOI] [PubMed] [Google Scholar]
- 19.Goss, E. M., M. Kreitman, and J. Bergelson. 2005. Genetic diversity, recombination and cryptic clades in Pseudomonas viridiflava infecting natural populations of Arabidopsis thaliana. Genetics 169:21-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant, S. R., E. J. Fisher, J. H. Chang, B. M. Mole, and J. L. Dangl. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60:425-449. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg, J. T., and N. Yao. 2004. The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6:201-211. [DOI] [PubMed] [Google Scholar]
- 22.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 23.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 24.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 25.Hendson, M., D. C. Hildebrand, and M. N. Schroth. 1992. Relatedness of Pseudomonas syringae pv. tomato, Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. antirrhini. J. Appl. Bacteriol. 73:455-464. [Google Scholar]
- 26.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 27.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 28.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. DeBoy, A. S. Durkin, M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 30.Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk, and S. D. Frost. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891-1901. [DOI] [PubMed] [Google Scholar]
- 31.Lin, N. C., and G. B. Martin. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant-Microbe Interact. 18:43-51. [DOI] [PubMed] [Google Scholar]
- 32.Lin, N. C., and G. B. Martin. 2007. Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse Pseudomonas syringae pathovars to infect tomato. Mol. Plant-Microbe Interact. 20:806-815. [DOI] [PubMed] [Google Scholar]
- 33.Macho, A. P., A. Zumaquero, I. Ortiz-Martin, and C. R. Beuzon. 2007. Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol. Plant Pathol. 8:437-450. [DOI] [PubMed] [Google Scholar]
- 34.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561-588. [DOI] [PubMed] [Google Scholar]
- 35.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majewski, J., and F. M. Cohan. 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maynard Smith, J., and N. H. Smith. 1998. Detecting recombination from gene trees. Mol. Biol. Evol. 15:590-599. [DOI] [PubMed] [Google Scholar]
- 38.McVean, G., P. Awadalla, and P. Fearnhead. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melotto, M., W. Underwood, J. Koczan, K. Nomura, and S. Y. He. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126:969-980. [DOI] [PubMed] [Google Scholar]
- 40.Mindrinos, M., F. Katagiri, G. L. Yu, and F. M. Ausubel. 1994. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78:1089-1099. [DOI] [PubMed] [Google Scholar]
- 41.Narra, H. P., and H. Ochman. 2006. Of what use is sex to bacteria? Curr. Biol. 16:R705-R710. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Losada, M., E. B. Browne, A. Madsen, T. Wirth, R. P. Viscidi, and K. A. Crandall. 2006. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect. Genet. Evol. 6:97-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitman, A. R., R. W. Jackson, J. W. Mansfield, V. Kaitell, R. Thwaites, and D. L. Arnold. 2005. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 15:2230-2235. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 45.Ronald, P. C., J. M. Salmeron, F. M. Carland, and B. J. Staskawicz. 1992. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174:1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 47.Rosebrock, T. R., L. Zeng, J. J. Brady, R. B. Abramovitch, F. Xiao, and G. B. Martin. 2007. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarkar, S. F., J. S. Gordon, G. B. Martin, and D. S. Guttman. 2006. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174:1041-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scally, M., E. L. Schuenzel, R. Stouthamer, and L. Nunney. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 71:8491-8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shenge, K. C., R. B. Mabagala, C. N. Mortensen, D. Stephan, and K. Wydra. 2007. First report of bacterial speck of tomato caused by Pseudomonas syringae pv. tomato in Tanzania. Plant Dis. 91:462. [DOI] [PubMed] [Google Scholar]
- 52.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 53.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spratt, B. G., W. P. Hanage, and E. J. Feil. 2001. The relative contribution of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 4:602-606. [DOI] [PubMed] [Google Scholar]
- 55.Swofford, D. L. 2003. PAUP* phylogenetic analysis using parsimony (and other methods), 4th ed. Sinauer Associates, Sunderland, MA.
- 56.Vinatzer, B. A., and C. T. Bull. The impact of genomics approaches on our understanding of diversity and taxonomy of plant pathogenic bacteria. In R. W. Jackson (ed.), Plant pathogenic bacteria: genomics and molecular biology, in press. Horizon Scientific Press, Norfolk, United Kingdom.
- 57.Whalen, M. C., R. W. Innes, A. F. Bent, and B. J. Staskawicz. 1991. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiebe, W. L., and R. N. Campbell. 1993. Characterization of Pseudomonas syringae pv. maculicola and comparison with P. s. tomato. Plant Dis. 77:414-419. [Google Scholar]
- 59.Wildermuth, M. C., J. Dewdney, G. Wu, and F. M. Ausubel. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562-565. [DOI] [PubMed] [Google Scholar]
- 60.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Z. 1997. A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 62.Zaccardelli, M., A. Spasiano, C. Bazzi, and M. Merighi. 2005. Identification and in planta detection of Pseudomonas syringae pv. tomato using PCR amplification of hrpZPst. Eur. J. Plant Pathol. 111:85-90. [Google Scholar]
- 63.Zhao, Y., J. P. Damicone, D. H. Demezas, V. Rangaswamy, and C. L. Bender. 2000. Bacterial leaf spot of leafy crucifers in Oklahoma caused by Pseudomonas syringae pv. maculicola. Plant Dis. 84:1015-1020. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, N., T. L. Tootle, F. Tsui, D. F. Klessig, and J. Glazebrook. 1998. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.