Abstract
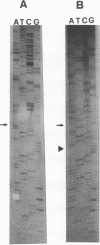
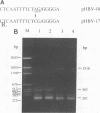
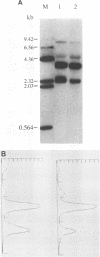
The transcriptional map of hepatitis B virus (HBV) has been expanded recently by the discovery of a singly spliced transcript in hepatoma cell lines transfected with cloned viral DNA and a doubly spliced one in naturally infected human liver tissues. By the use of reverse transcription and a subsequent polymerase chain reaction, the two spliced HBV RNAs were shown to be present in both types of cells. As further evidence, an HBV mutant was constructed and found to exclusively express the singly spliced RNA. This mutant was also used to quantitate the two spliced species in transfected HepG2 cells; they were found to be equally abundant, and each represented about 30% of the pregenomic RNA. The HBV mutant could still produce replication-competent HBV virions when transfected into HepG2 cells, indicating that the doubly spliced transcript, just like the singly spliced one, was not essential for HBV replication. However, the two abundant spliced HBV transcripts were detected in most naturally infected human liver tissues, suggesting that they may have biologic functions in vivo.
Full text
PDF

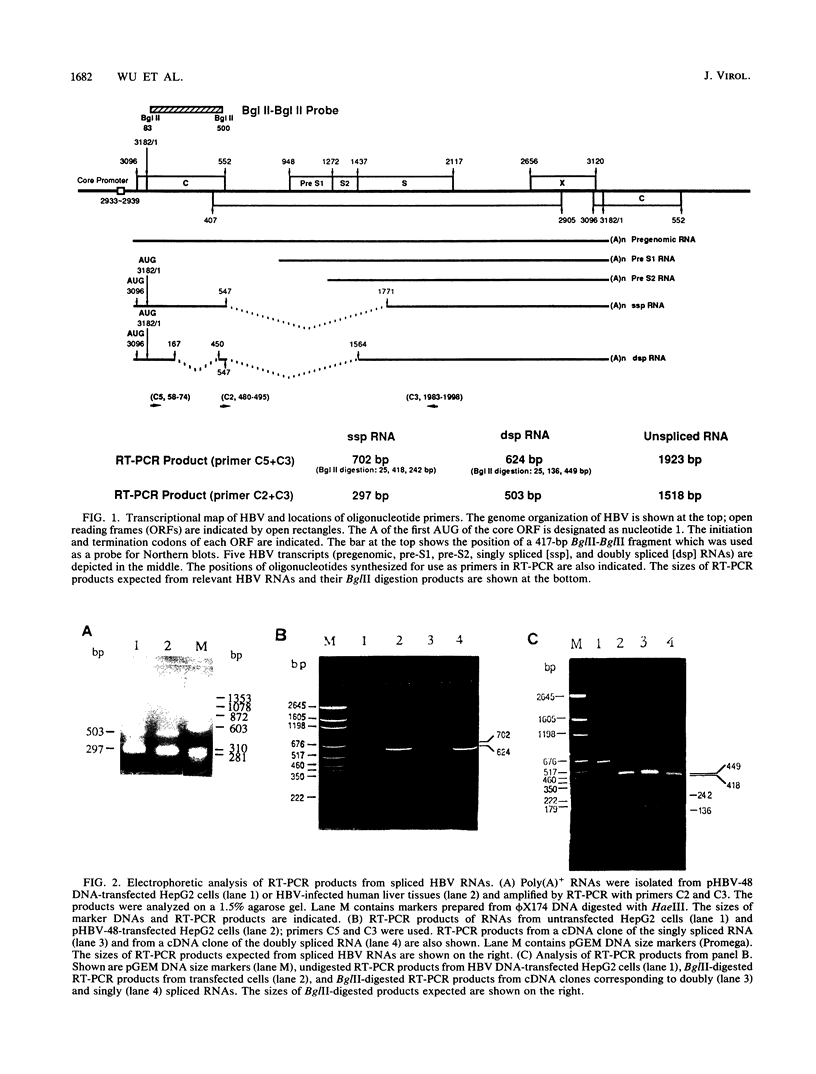
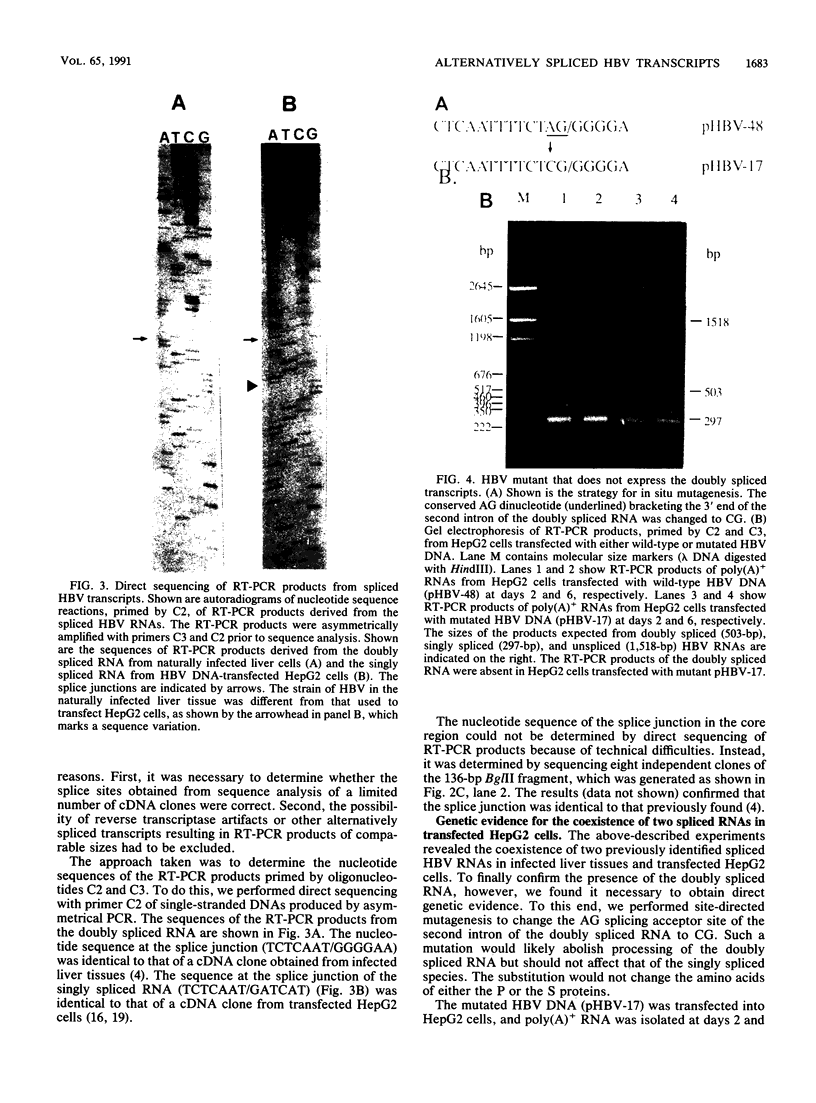



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartenschlager R., Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988 Dec 20;7(13):4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. M., Jeng K. S., Hu C. P., Lo S. J., Su T. S., Ting L. P., Chou C. K., Han S. H., Pfaff E., Salfeld J. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987 Mar;6(3):675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. J., Pryciak P., Ganem D., Varmus H. E. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature. 1989 Jan 26;337(6205):364–368. doi: 10.1038/337364a0. [DOI] [PubMed] [Google Scholar]
- Chen P. J., Chen C. R., Sung J. L., Chen D. S. Identification of a doubly spliced viral transcript joining the separated domains for putative protease and reverse transcriptase of hepatitis B virus. J Virol. 1989 Oct;63(10):4165–4171. doi: 10.1128/jvi.63.10.4165-4171.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng-Mayer C., Iannello P., Shaw K., Luciw P. A., Levy J. A. Differential effects of nef on HIV replication: implications for viral pathogenesis in the host. Science. 1989 Dec 22;246(4937):1629–1632. doi: 10.1126/science.2531920. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan P. M., Greenman R. L., Gerin J. L., Purcell R. H., Robinson W. S. DNA polymerase associated with human hepatitis B antigen. J Virol. 1973 Nov;12(5):995–1005. doi: 10.1128/jvi.12.5.995-1005.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987 Aug 20;196(4):947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Radziwill G., Tucker W., Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990 Feb;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht H. J., Radziwill G., Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989 Jan 13;56(1):85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- Schlicht H. J., Salfeld J., Schaller H. The duck hepatitis B virus pre-C region encodes a signal sequence which is essential for synthesis and secretion of processed core proteins but not for virus formation. J Virol. 1987 Dec;61(12):3701–3709. doi: 10.1128/jvi.61.12.3701-3709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. S., Lai C. J., Huang J. L., Lin L. H., Yauk Y. K., Chang C. M., Lo S. J., Han S. H. Hepatitis B virus transcript produced by RNA splicing. J Virol. 1989 Sep;63(9):4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. S., Lui W. Y., Lin L. H., Han S. H., P'eng F. K. Analysis of hepatitis B virus transcripts in infected human livers. Hepatology. 1989 Feb;9(2):180–185. doi: 10.1002/hep.1840090203. [DOI] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Masui N., Kajino K., Saito I., Miyamura T. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Will H., Reiser W., Weimer T., Pfaff E., Büscher M., Sprengel R., Cattaneo R., Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987 Mar;61(3):904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma K., Shirakata Y., Kobayashi M., Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci U S A. 1987 May;84(9):2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]