Abstract
Although many bacterial pathogens use specialized secretion systems for virulence, no such systems have been described for Mycobacterium tuberculosis, a major pathogen of humans that proliferates in host macrophages. In a screen to identify genes required for virulence of M. tuberculosis, we have discovered three components and two substrates of the first Sec-independent secretion pathway described in M. tuberculosis, which we designate the Snm pathway. Here we demonstrate that the proteins Snm1, -2, and -4 are required for the secretion of ESAT-6 and CFP-10, small proteins previously identified as major T cell antigens. Snm2, a member of the AAA ATPase family, interacts with substrates and with Snm1, another AAA ATPase. We show that M. tuberculosis mutants lacking either the Snm system or these substrates exhibit defects in bacterial growth during the acute phase of a mouse infection and are attenuated for virulence. Strikingly, snm mutants fail to replicate in cultured macrophages and to inhibit macrophage inflammatory responses, two well established activities of wild-type M. tuberculosis bacilli. Thus, the Snm secretion pathway works to subvert normal macrophage responses and is a major determinant of M. tuberculosis virulence.
The etiologic agent of human tuberculosis, Mycobacterium tuberculosis, infects one-third of the world's population and can survive within an infected individual for decades (1, 2). During infection, M. tuberculosis resides primarily within macrophages, myeloid cells whose function is to phagocytose and destroy invading microorganisms. Numerous experiments using in vitro infection of macrophages have demonstrated that live M. tuberculosis cells assert a profound inhibitory influence on their cellular host. One well studied example of M. tuberculosis-mediated manipulation of macrophage function is the bacterium's ability to alter macrophage signaling required for the production of immunostimulatory cytokines and effector molecules (3, 4). For example, M. tuberculosis actively suppresses the transcriptional induction of the p40 subunit of IL-12, a cytokine critical for control of mycobacterial infection (5, 6). The production of the proinflammatory cytokine tumor necrosis factor α (TNF-α) and the antimicrobial effector nitric oxide (NO) is also critical for controlling M. tuberculosis infection. Interestingly, avirulent mycobacterial strains elicit significantly more TNF-α and NO from infected macrophages than M. tuberculosis, suggesting that suppression of these responses is important for virulence (3).
Many bacterial pathogens influence immune responses by secreting effector proteins that directly manipulate host cell function (7). Although most secreted proteins are exported via the classical Sec pathway (8), virulence factors are often secreted by specialized Sec-independent systems (9). To date, no alternative secretion pathways have been described in M. tuberculosis. Many of the proteins secreted by M. tuberculosis bacilli during growth in culture have been identified, some of which lack identifiable N-terminal signal sequences that would target them to the Sec pathway (10). However, the reliability of these approaches to identify secreted proteins is confounded by the high rates of cell autolysis in mycobacterial cultures (11).
ESAT-6 and CFP-10 are two secreted proteins of unknown function originally identified as immunodominant antigens of M. tuberculosis. Several recent studies have suggested that these proteins are important for virulence. Deletion of the genes encoding ESAT-6 and CFP-10 from the virulent Mycobacterium bovis strain results in a diminution of virulence (12). Furthermore, all strains of the attenuated vaccine strains of bacillus Calmette–Guérin have deletions encompassing the esat-6 locus, also known as the RD1 region (13). Importantly, deletion of RD1 from M. tuberculosis attenuates the organism and, conversely, incorporation of the RD1 region from M. tuberculosis into bacillus Calmette–Guérin restores ESAT-6 and CFP-10 expression and increases virulence and immunogenicity (14–16). Interestingly, ESAT-6 and CFP-10 lack characteristic signal sequences that would target them to the Sec system of secretion and are secreted by an unknown mechanism.
Analyses of the M. tuberculosis genome have suggested that genes neighboring the esat-6-cfp-10 operon may be important for ESAT-6 and CFP-10 secretion (see Fig. 1A) (17–19). This speculation is strengthened by the observation that 4 of 10 homologues of the esat-6-cfp-10 operon scattered within the M. tuberculosis genome are also flanked by homologues of these neighboring genes. These genes encode a variety of proteins including two AAA ATPases (Rv3870 and Rv3871) and a 12-transmembrane domain protein (Rv3877). However, experimental evidence for the role of these proteins in ESAT-6 and CFP-10 secretion is lacking.
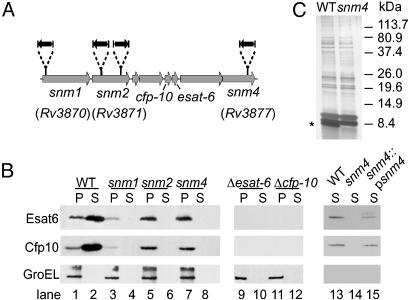
Fig. 1.
M. tuberculosis snm mutants fail to secrete ESAT-6 and CFP-10. (A) Schematic representation of the transposon insertion sites (black arrows) from three snm mutants within the M. tuberculosis genome. (B) Western blot detection of ESAT-6, CFP-10, and GroEL from cell supernatant (S) and cell pellet lysate (P) fractions of wild-type and mutant cells. Loading was normalized by OD280, and efficiency of transfer was confirmed by Ponceau S staining of the membrane. The decreased level of ESAT-6 and CFP-10 detected in the pellet of the snm1 mutant was not observed in subsequent experiments. (C) Silver stain of wild-type and mutant (snm4) cell culture supernatants separated by SDS/PAGE. The protein band absent in the snm4 mutant is marked with an asterisk.
Here we describe the identification of avirulent mutants of M. tuberculosis with lesions in Rv3870, Rv3871, and Rv3877. We show that these genes encode components of a secretion system that work together to export ESAT-6 and CFP-10. These mutants fail to inhibit macrophage cytokine responses and are attenuated in mice and macrophages. Our studies describe a previously uncharacterized alternative secretion system identified in M. tuberculosis and show that this pathway is a major determinant of M. tuberculosis virulence.
Materials and Methods
Strains and Plasmids. All strains, plasmids (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), and methods for genetic manipulation of M. tuberculosis used in this study are published as supporting information on the PNAS web site. M. tuberculosis (Erdman) and bacillus Calmette–Guérin (Pasteur) were cultivated as described (20). Sauton's medium was used for supernatant preparations, and yeast media were purchased from Qbiogene (Carlsbad, CA).
Protein Preparation and Analysis. Concentrated culture supernatants were prepared by growing M. tuberculosis in Sauton's medium supplemented with 0.05% Tween-80 to midlogarithmic phase. Cells were inoculated into Sauton's medium without Tween-80 at OD600 = 0.05, incubated in roller bottles for 5 d, harvested by centrifugation, and supernatants were concentrated. Cell lysates and supernatants were separated by SDS/PAGE by using 10–20% polyacrylamide gels. Proteins were visualized by silver stain or immunoblotting by using antibodies against Esat-6 (Hyb 76–8), CFP-10 (K8493), or GroEL (HAT5), all kind gifts of P. Andersen (Statens Serum Institut, Copenhagen, Denmark).
Yeast Two-Hybrid. Bait and prey vectors are listed in Table 1, and primers used to amplify genes are listed in Table 2, which is published as supporting information on the PNAS web site. For directed two-hybrid analysis, genes were amplified, sequenced, and inserted into both bait (pEG202) and prey (pjsc401) vectors. The resulting plasmids were used to transform yeast strains EGY48 and W303–1a, and their expression was verified by Western blot. All possible bait–prey combinations were tested by mating and replica plating to 5-bromo-4-chloro-3-indolyl β-d-galactoside + galactose plates (21).
Bacterial Infections. Mice were infected i.v., and samples were processed exactly as described (20). To normalize to inoculum size, total colony-forming units (cfu) at each time point were divided by total cfu at day 1. Bone marrow-derived macrophages were infected in triplicate wells by using DMEM containing 5% horse serum and 5% FCS at an multiplicity of infection of 1, incubated for 2 h, washed, and fresh medium was added. Media were changed 36 h after infection. Infected cells were lysed with 0.5% Triton X-100 and plated on 7H10 agar.
ELISAs and Nitrite Measurements. Cultured macrophage supernatants were assayed for cytokine levels by using ELISA kits (BD Biosciences, Palo Alto, CA). Nitrite levels were measured by using the Griess reaction.
Quantitative PCR. Three micrograms of total macrophage RNA was reverse transcribed in a 20-μl reaction, diluted to 100 μl with water, and 2.5 μl was used in a quantitative real-time PCR reaction with oligonucleotides specified in Table 2 (22) by using SYBR green as label. Results shown are from two separate infection experiments, with each PCR reaction performed in triplicate. All values reported were in the linear range of the experiment and were normalized to actin values. A relative standard curve for actin was generated from serial dilution of a pooled reference of all of the cDNA samples.
Results
Isolation of Attenuated M. tuberculosis Signature-Tagged Mutagenesis (STM) Mutants Defective for ESAT-6/CFP-10 Secretion. With a previously described STM methodology in which mice are infected with pools of 48 randomly generated transposon mutants (20), we identified four attenuated mutants, each containing a single transposon insertion within the RD1 region of the M. tuberculosis genome. Although this locus has been implicated in virulence, the individual genes within the region have not been studied (Fig. 1 A). We found that the transposons had inserted within the unknown genes Rv3870, Rv3871 (two separate isolates), and Rv3877, which flank the esat-6 and cfp-10 genes in the genome of M. tuberculosis (Fig. 1 A). For reasons described below, we have renamed these genes snm1, snm2, and snm4 (for secretion in mycobacteria). Each of the four mutants was well represented in its inoculum pool at the time of infection but was drastically under-represented in pools from the lungs of two infected mice after 3 wk of infection (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the proximity of the snm genes to one another in the genome and the severity of the growth defects of these four mutants suggest strongly that these genes function together to promote M. tuberculosis growth during the early stages of infection.
To test the hypothesis that Snm1, -2, and -4 are each required for ESAT-6 and CFP-10 secretion, we examined cell supernatants from exponentially growing M. tuberculosis cultures. With antibodies specific for ESAT-6 and CFP-10, we detected both proteins in the cell supernatant fraction from wild-type cultures as well as in extracts from cell pellets (Fig. 1B, lanes 1 and 2). To verify that the presence of ESAT-6 and CFP-10 in the cell supernatant was not due to cell lysis, we probed the blots with antibodies specific for GroEL, an intracellular chaperone. As expected, GroEL was found exclusively in cell pellets. Remarkably, the mutations in each of the snm genes blocked ESAT-6 and CFP-10 secretion into the cell culture supernatant because neither protein was detected in cell supernatants from the mutants (Fig. 1B, lanes 3–8). Importantly, ESAT-6 and CFP-10 were still detected in cell pellets of the mutants, demonstrating that synthesis of the proteins was not abrogated by the mutations. The decreased level of ESAT-6 and CFP-10 detected in the pellet of the snm1 mutant was not observed in subsequent experiments. The secretion defect in snm4 mutant cells was complemented to nearly wild-type levels by introduction of a single copy of the wild-type gene into the genome (lanes 13–15). Furthermore, the block in ESAT-6 and CFP-10 secretion was not due to a general defect in total protein export, because SDS/PAGE analysis of supernatants from both wild-type and snm4 mutant cell cultures revealed nearly identical protein profiles except for one of three prominent bands in the low molecularweight range (Fig. 1C). The protein profiles of cell supernatants from the snm1 and snm2 mutants appeared identical to that from the snm4 mutant (data not shown).
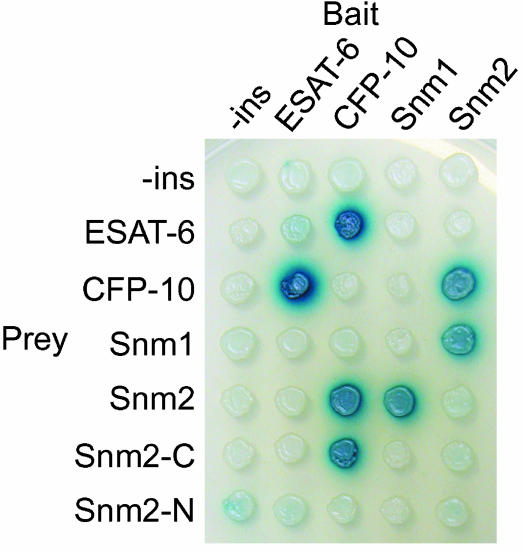
Components and Substrates of the Snm Secretory Pathway Interact. The observation that each of the three Snm proteins is required for ESAT-6 and CFP-10 secretion suggested that these proteins work together in the same pathway. This notion was strengthened by the results of a yeast two-hybrid screen in which we identified both ESAT-6 and Snm2 in a search for M. tuberculosis proteins that interacted with CFP-10. No other interactors were identified in this screen, suggesting that CFP-10 makes specific contacts with both proteins. To verify these results and identify other Snm interactions, we constructed both “bait” and “prey” vectors to test for interactions among ESAT-6, CFP-10, Snm2, and the soluble C-terminal domain of Snm1 (Fig. 2). Snm4 was not compatible for yeast two-hybrid analysis, because it is predicted to contain mostly transmembrane domains. As expected, an interaction between CFP-10 and ESAT-6 was detected, confirming biochemical evidence that these proteins interact (23). In addition to the CFP-10 and Snm2 interaction, we also identified interactions between Snm2 and Snm1. Thus, we were able to construct a simple interaction map connecting all four of these proteins, consistent with the model in Fig. 5. In particular, Snm2 appears to be a central player, because it interacts with both the CFP-10 substrate as well as Snm1, a component likely anchored in the cytoplasmic membrane in vivo. Deletion analysis showed that the C-terminal domain of Snm2, which includes the second AAA ATPase domain, is sufficient to interact with CFP-10, whereas full-length Snm2 is required for Snm1 binding (Fig. 2). Taken together, these data support our genetic data that the snm genes encode for components of a secretion pathway that functions to directly secrete ESAT-6 and CFP-10 from M. tuberculosis cells.
Fig. 2.
Components and substrates of the Snm secretion pathway interact. Diploid yeast strains harboring the indicated bait and prey yeast–two-hybrid constructs were replica plated to galactose + 5-bromo-4-chloro-3-indolyl β-d-galactoside indicator plates and allowed to develop overnight at 30°C. ESAT-6, CFP-10, and Snm2 were expressed as full-length fusion proteins and the Snm1 constructs expressed the N-terminal nontransmembrane domain portion of the protein (amino acids 252–747). Strains containing plasmids expressing only the C-terminal (Snm2-C, amino acids 248–591) or N-terminal (Snm2-N, amino acids 1–241) portions of Snm2 were also tested. Expression of all fusion proteins was confirmed by Western blotting by using antibodies that recognize LexA (baits) or the hemagglutinin epitope tag (preys).
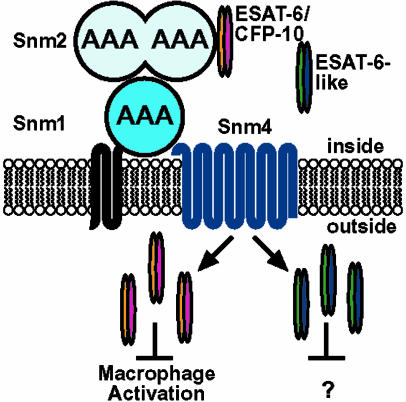
Fig. 5.
Hypthetical model of Snm-mediated secretion of ESAT-6 and CFP-10; see text for details.
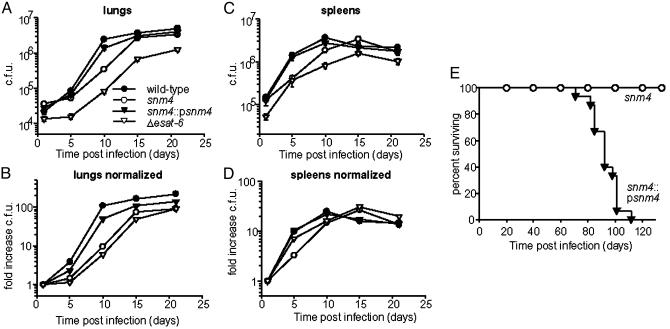
The Snm Secretory Pathway Is Required for Virulence. The identification of the three snm genes in our STM screen suggested that the Snm pathway is critical for M. tuberculosis growth in vivo. To determine whether the snm mutants were defective for growth in vivo when infected as a clonal population, we injected C57BL/6 mice with 1 × 106 cfu of either the wild-type Erdman strain or the snm4 mutant strain. Wild-type cells replicated extensively in the lungs during the first 10 d of infection, leading to a 100-fold increase in cfu (Fig. 3A). In contrast, snm4 mutant cells were defective for growth during the first 10 d after infection, resulting in only 10-fold growth at this time point. Interestingly, replication of the mutant progressed after 10 d of infection, reaching near wild-type levels 15 d after infection. Due to variations in seeding of the lungs, the difference in growth between wild-type and snm4 mutant cells is most clearly seen when the data are normalized to bacterial burdens immediately after infection (Fig. 3B). Histopathological examination of tissues 10 d after infection revealed that the snm4 mutant strain induced a less robust inflammatory immune response than wild-type cells, a finding consistent with a decreased bacterial burden at this time point (Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, the maximal in vivo growth rates of the mutant cells [doubling time (td) = 1.84 d for snm4] were significantly slower than wild-type bacilli (td = 1.03 d). Accordingly, the growth defect of snm4 mutant cells resulted in a lower total fold increase in cfu in the lung by 3 wk. Attenuated growth was also observed in the spleen at early time points (Fig. 3C). Defects in growth observed in vivo were not a result of an intrinsic growth defect, because growth rates of wild-type and mutant cells in liquid culture were identical (wild-type td = 0.93 d, snm4 td = 0.91 d). The in vivo growth defects of the snm4 mutant were rescued by expression of the snm4 gene because the complemented strain was able to replicate at nearly wild-type levels (Fig. 3 A and C). Importantly, snm1 and snm2 mutant cells displayed in vivo growth phenotypes identical to the snm4 mutant, demonstrating that all three genes are required for normal growth kinetics during infection (data not shown).
Fig. 3.
Snm-mediated secretion is required for M. tuberculosis virulence. (A) C57BL/6 mice were injected with 1 × 106 cfu of each strain, and bacilli were harvested from lungs at 1, 5, 10, 15, and 21 d after infection. Error bars represent the SEM from two combined experiments by using five mice per timepoint per experiment. (B) cfu data from A normalized to initial inoculum (see Materials and Methods). (C) cfu isolated from spleens of mice infected in A.(D) cfu data from C normalized to initial inoculum. (E) Survival of BALB/c mice (n = 15 per group) infected with 106 cfu of indicated strains.
To determine whether esat-6 and cfp-10 are required for growth in vivo, we made precise deletions of each gene from the genome by replacing each ORF with the hygromycin resistance marker. As shown in Fig. 1C (lanes 9–12), extracts prepared from either mutant strain lack ESAT-6 and CFP-10. Because cfp-10 is the upstream gene in an operon with esat-6 (24), transcriptional attenuation due to polarity can explain the absence of ESAT-6 in the Δcfp-10 mutant. However, the observation that CFP-10 protein is undetectable in the Δesat-6 mutant suggests that ESAT-6 is required for stable CFP-10 accumulation. In support of this notion, Renshaw et al. (23) have demonstrated that ESAT-6 and CFP-10 adopt fully folded states only when present together. Although the role of the individual proteins in virulence is therefore difficult to test, we sought to determine whether ESAT-6 and/or CFP-10 is required for replication in vivo. Like snm mutant cells, Δesat-6 mutant cells grow more slowly than wild-type cells during infection and fail to attain high levels during the later stages of infection (Fig. 3 A and B). The phenotype of the Δesat-6 mutant is unlikely to be the result of a polar effect on snm4, because there is a strong terminator downstream of the esat-6/cfp-10 operon. The slight increase in growth rate of snm4 mutant cells compared with Δesat-6 mutant cells could be due to partial complementation resulting from the release of intracellular ESAT-6 and CFP-10 from lysed cells. Overall, the similarity of the in vivo growth curves of the two mutants indicates that the growth defect of snm mutants results from the failure to secrete ESAT-6 and/or CFP-10. However, we cannot rule out the possibility that other substrates secreted by the Snm pathway may also be important during infection.
To determine whether snm4 is required for virulence of M. tuberculosis, BALB/c mice were infected with 1 × 106 cfu of snm4 mutant cells or the complemented strain. All of the mice infected with the complemented strain succumbed to infection by 112 d after infection, with a mean survival time of 90 d (Fig. 3E). These kinetics are very similar to those reported in the literature for infection with the wild-type Erdman strain under the same conditions (25). In contrast, none of the mice infected with snm4 mutant cells succumbed to infection during the course of the experiment (140 d). Measurement of bacterial burden on day 100 after infection revealed no significant difference in the total number of bacteria in the lungs (5.6 ± 1.0 × 106 for wild-type and 6.2 ± 1.7 × 106 for snm4). Therefore, despite the fact that snm4 mutant cells eventually attain wild-type levels in the tissues, they are strikingly less virulent than wild-type M. tuberculosis cells. Although it is not well understood, decreased virulence does not always correlate with a decrease in M. tuberculosis burden (26). Whether the modest growth defect of the snm4 mutant early after infection causes the decrease in virulence at later stages of infection or whether Snm4 acts during both phases of infection is impossible to determine at this time. However, our results demonstrate that the Snm pathway is required not only for normal growth kinetics in vivo but also for overall virulence of M. tuberculosis.
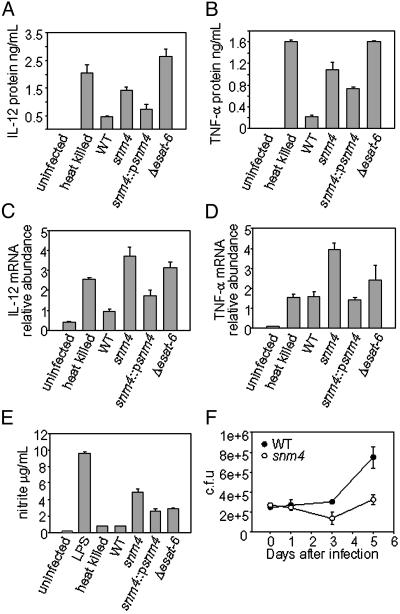
The Snm Pathway Alters Innate Responses of Macrophages. Macrophages infected with avirulent mycobacterial species produce greater amounts of cytokines and reactive nitrogen species than macrophages infected with virulent M. tuberculosis. Thus, it has been postulated that M. tuberculosis has evolved mechanisms for suppressing macrophage responses. To determine whether Snm components and substrates are required to dampen cytokine responses, we measured the amount of proinflammatory cytokines elicited by wild-type, snm4, and Δesat-6 mutant cells 24 h postinfection by ELISA. As has been shown for infection with human monocytes (5, 6), murine macrophages infected with heat-killed M. tuberculosis cells elicit substantially more IL-12 p40 than macrophages infected with live bacilli (Fig. 4A). Like heat-killed bacteria, live snm4 mutant cells elicited high amounts of IL-12 p40 secretion, yet the bacilli were viable at this time point (Fig. 4F). Importantly, Δesat-6 mutant cells also induced high levels of IL-12 p40. Although it has been suggested that IL-12 suppression by M. tuberculosis results from increased induction of the antiinflammatory cytokine IL-10 (5), no IL-10 was detected in any of the infections (data not shown). TNF-α induction was also enhanced in macrophages infected with both snm4 and Δesat-6 mutant cells (Fig. 4B). As expected, the differences in IL-12 cytokine levels were reflected in differences in IL-12 mRNA levels (Fig. 4C). Although this pattern of transcriptional activation generally holds true for TNF-α, heat-killed bacilli elicited large amounts of TNF-α cytokine release, yet TNF-α mRNA was induced only partially and was similar to that elicited by live cells (Fig. 4D). TNF-α expression in macrophages is controlled by multiple mechanisms, including transcriptional and posttranscriptional regulation (27). Although we do not yet understand the basis for this inconsistency, differences in posttranscriptional regulation may explain the discrepancy between TNF-α mRNA and protein at this timepoint.
Fig. 4.
Snm pathway mutants induce enhanced macrophage inflammatory responses. Bone marrow-derived macrophages were infected, and culture supernatants were collected and the concentration of IL-12 p40 (A) and TNF-α (B) was measured by ELISA. Total RNA was harvested from macrophage monolayers, and IL-12 p40 (C) and TNF-α (D) mRNA levels were measured by quantitative PCR and normalized to actin mRNA levels. Each sample was assayed in triplicate; error bars represent standard deviation from at least two experiments. (E) Nitrite concentration from culture supernatants was measured by using the Griess reaction. (F) M. tuberculosis-infected macrophages were infected at a multiplicity of infection of 1 and harvested immediately after the 2-h phagocytosis period (“0h”) and at the indicated time points, and bacterial cfu were determined by plating.
Macrophage recognition of M. tuberculosis molecules is also sufficient to elicit NO production in resting macrophages (28). As with the cytokine response, all of the mutants elicited a higher amount of NO (as determined by measuring nitrite concentrations) than that elicited by wild-type cells 24 h postinfection (Fig. 4E). Importantly, snm1 and snm2 mutant cells also elicited higher levels of NO as well as IL-12 p40 and TNF-α production compared with wild-type cells (data not shown). Taken together, these results suggest that the Snm pathway and ESAT-6/CFP-10 function to inhibit macrophage signaling, thus suppressing the proinflammatory and effector responses normally elicited on contact with bacteria.
The Snm Pathway Is Required for Growth in Macrophages. The inability of snm4 mutant cells to suppress macrophage activation could result from loss of a specific suppressive activity or from a decrease in viability soon after phagocytosis. To distinguish between these possibilities, we examined the ability of the snm4 mutant to replicate within cultured macrophages. Bone marrow-derived macrophages from C57BL/6 mice were infected at a multiplicity of infection of 1 with wild-type or snm4 mutant cells. As shown in Fig. 4F, viability of snm4 cells is unaffected 24 h after infection, indicating that the failure of snm4 to suppress cytokine production at 24 h does not simply result from a loss of viability. However, at later timepoints, snm4 mutant cells fail to replicate at wild-type levels, resulting in 3-fold fewer bacilli at 5 d after infection. Therefore, the Snm pathway is responsible for multiple suppressive effects on macrophages and is required for bacterial replication after phagocytosis.
Discussion
In this study, we have identified the first Sec-independent protein secretion system in M. tuberculosis, and we have shown that three Snm components are required for the transport of the ESAT-6 and CFP-10 substrates from the cell. The Snm secretion system is a major determinant of M. tuberculosis virulence, because mutants lacking either components or substrates are profoundly attenuated in a mouse model of infection. The reduced virulence of snm mutant cells displayed during in vivo infection is likely due to their inability to limit macrophage responses. Although a number of studies have described the immunosuppressive effects M. tuberculosis cells exert on infected macrophages, we have identified specific mycobacterial gene products that are required for manipulating macrophage activation during infection.
What Is the Function of the Snm Secretion System? Snm4 mutants fail to limit both cytokine and effector responses early after infection of cultured macrophages and ultimately fail to replicate after phagocytosis. We hypothesize that the Snm system also functions to inhibit initial macrophage responses to infection in vivo, leading to reduced amounts of direct antimicrobial effectors such as NO and reduced levels of cytokines required for communication with other cells of the immune system. The phenotype of snm4 mutants is specific and not simply a consequence of attenuation, because other mutants isolated from the STM screen are able to suppress macrophage responses (unpublished observations). Consistent with our macrophage results, the Snm secretion system is required for normal growth kinetics during the early stages of infection with M. tuberculosis. Although the defective growth of snm4 mutants in cultured macrophages is likely a result of elevated NO production, the attenuated growth observed in vivo could result from the elevated levels of both effector molecules and cytokines.
Despite an early growth delay in vivo, snm4 mutants ultimately reach bacterial numbers similar to wild type in the lungs and other tissues. Interestingly, this modest difference in growth stands in contrast to the marked attenuation of the snm4 mutant in our virulence studies. Others have reported that there is not a strict correlation between mycobacterial burden and virulence in a time-to-death assay (26). Indeed, mutants have recently been studied that have no apparent growth or persistence defects but are attenuated for virulence (29, 30). Because a decrease in virulence is not always accompanied by a decrease in cfu in the lungs, it is difficult to know whether the decrease in virulence of the snm4 mutant is related to the early growth defect. There are at least two possibilities. First, the Snm pathway is required only during the initial 2 wk of infection, but the delayed growth of snm4 mutant cells influences subsequent immune responses such that the animal is better able to cope with chronic infection. Alternatively, the virulence defect of snm4 mutant cells may be the result of the Snm system functioning during the chronic stage of infection. Although it will be interesting to determine whether the growth and virulence defects are separable, the tools required to experimentally regulate M. tuberculosis gene expression during infection do not currently exist.
Snm Proteins Constitute an Evolutionarily Conserved Protein Translocation System. Our data indicate that the Snm proteins interact to form a pathway to promote ESAT-6 and CFP-10 secretion and release into the extracellular milieu. Although we cannot rule out an indirect role for the Snm proteins in substrate secretion, the specific two-hybrid interactions and operon organization strongly suggest a direct role in secretion. Our results are consistent with a model in which snm1, snm2, and snm4 encode part of a previously unrecognized secretion apparatus that allows for the export of ESAT-6 and CFP-10 (Fig. 5). Primary sequence analysis has placed Snm1 and -2 within the SpoIIIE/FtsK subfamily of AAA ATPases (18, 19). Because members of this family transduce chemical energy into force (31), it is possible that Snm1 and Snm2 may push ESAT-6 and CFP-10 through a channel that includes Snm4. This activity would be analogous to other AAA ATPases, such as Cdc48 in eukaryotic cells and FtsH in E. coli, that interact with and translocate protein substrates across cellular membranes. Alternatively, the role of Snm1 and -2 may be akin to that of signal recognition particle (SRP) and its membrane-bound receptor (SR) in the general secretion pathway, where GTP binding and hydrolysis by cytoplasmic (SRP/Snm2) and membrane-bound proteins (SR/Snm1) endow fidelity and linearity to the pathway rather than drive substrate translocation (32). However, biochemical experiments in more tractable Mycobacterial species will be required for a more thorough mechanistic understanding of the Snm system.
Although the evolution of prokaryotic alternative secretion pathways to interact with and directly inhibit eukaryotic cell functions is a common theme among Gram-negative bacterial pathogens, equivalent systems in Gram-positive pathogens have not been as clearly defined. Although direct evidence for the secretion of ESAT-6/CFP-10 or their homologues directly into host cells is lacking, the Snm pathway likely represents a new mode of host–pathogen interaction. The presence of snm and esat-6/cfp-10 homologues in the genomes of a large number of Gram-positive bacteria, including pathogens such as Bacillus anthracis, suggests that the Snm system represents an evolutionarily conserved secretion pathway used by many different prokaryotes (18).
Supplementary Material
Acknowledgments
We thank W. R. Jacobs, Jr. (Albert Einstein College of Medicine, Bronx, NY) for phages and the transposon; M. Glickman for help with construction of the genomic library; P. Andersen and J. Gruenberg (University of Geneva, Geneva) for antibodies; C. Sihlbom and A. Burlingame for MS; L. Woo for technical assistance; A. Sil, P. Walter, H. Madhani, C. Gross, J. Engel, J. Ernst, R. Locksley, K. Cox, and members of the Cox laboratory for helpful comments throughout the course of this work; and K. Schneider, K. Jones, S. Kogan, and J. McKerrow for assistance with histopathology. This work was supported by National Institutes of Health Grant AI68540 (to J.S.C.). J.S.C. gratefully acknowledges the support of the Pew Scholars Program in the Biomedical Sciences and the Sandler Family Supporting Foundation.
Abbreviations: TNF-α, tumor necrosis factor α; cfu, colony-forming unit.
References
- 1.Dye, C., Scheele, S., Dolin, P., Pathania, V. & Raviglione, M. C. (1999) J. Am. Med. Assoc. 282 677-686. [DOI] [PubMed] [Google Scholar]
- 2.McKinney, J. D., Jacobs, J., W. R. & Bloom, B. R. (1998) in Emerging Infections, eds. Fauci, A. & Krause, R. (Academic, London), pp. 51-146.
- 3.Beltan, E., Horgen, L. & Rastogi, N. (2000) Microb. Pathog. 28 313-318. [DOI] [PubMed] [Google Scholar]
- 4.Falcone, V., Bassey, E. B., Toniolo, A., Conaldi, P. G. & Collins, F. M. (1994) FEMS Immunol. Med. Microbiol. 8 225-232. [DOI] [PubMed] [Google Scholar]
- 5.Giacomini, E., Iona, E., Ferroni, L., Miettinen, M., Fattorini, L., Orefici, G., Julkunen, I. & Coccia, E. M. (2001) J. Immunol. 166 7033-7041. [DOI] [PubMed] [Google Scholar]
- 6.Nau, G. J., Richmond, J. F., Schlesinger, A., Jennings, E. G., Lander, E. S. & Young, R. A. (2002) Proc. Natl. Acad. Sci. USA 99 1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, V. T. & Schneewind, O. (2001) Genes Dev. 15 1725-1752. [DOI] [PubMed] [Google Scholar]
- 8.Economou, A. (1999) Trends Microbiol. 7 315-320. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. B. & Falkow, S. (1997) Microbiol. Mol. Biol. Rev. 61 136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenberg, M. G. & Belisle, J. T. (1997) Infect. Immun. 65 4515-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tullius, M. V., Harth, G. & Horwitz, M. A. (2001) Infect. Immun. 69 6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wards, B. J., de Lisle, G. W. & Collins, D. M. (2000) Tuber. Lung Dis. 80 185-189. [DOI] [PubMed] [Google Scholar]
- 13.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178 1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46 709-717. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, K. N., Liao, R., Guinn, K. M., Hickey, M. J., Smith, S., Behr, M. A. & Sherman, D. R. (2003) J. Infect. Dis. 187 117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pym, A. S., Brodin, P., Majlessi, L., Brosch, R., Demangel, C., Williams, A., Griffiths, K. E., Marchal, G., Leclerc, C. & Cole, S. T. (2003) Nat. Med. 9 533-539. [DOI] [PubMed] [Google Scholar]
- 17.Tekaia, F., Gordon, S. V., Garnier, T., Brosch, R., Barrell, B. G. & Cole, S. T. (1999) Tuber. Lung Dis. 79 329-342. [DOI] [PubMed] [Google Scholar]
- 18.Pallen, M. J. (2002) Trends Microbiol. 10 209-212. [DOI] [PubMed] [Google Scholar]
- 19.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biol. 2, RESEARCH0044. [DOI] [PMC free article] [PubMed]
- 20.Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R., Jr. (1999) Nature 402 79-83. [DOI] [PubMed] [Google Scholar]
- 21.Golemis, E., Serebriiskii, I., Finley, R. L. J., Kolonin, M. G., Gyuris, J. & Brent, R. (1999) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), 20.1.1-20.1.28.
- 22.Overbergh, L., Valckx, D., Waer, M. & Mathieu, C. (1999) Cytokine 11 305-312. [DOI] [PubMed] [Google Scholar]
- 23.Renshaw, P. S., Panagiotidou, P., Whelan, A., Gordon, S. V., Hewinson, G. R., Williamson, R. A. & Carr, M. D. (2002) J. Biol. Chem. 277 21598-21603. [DOI] [PubMed] [Google Scholar]
- 24.Berthet, F. X., Rasmussen, P. B., Rosenkrands, I., Andersen, P. & Gicquel, B. (1998) Microbiology 144 3195-3203. [DOI] [PubMed] [Google Scholar]
- 25.McKinney, J. D., Honer zu Bentrup, K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, W. T., Swenson, D., Sacchettini, J. C., Jacobs, W. R., Jr., & Russell, D. G. (2000) Nature 406 735-738. [DOI] [PubMed] [Google Scholar]
- 26.North, R. J., Ryan, L., LaCource, R., Mogues, T. & Goodrich, M. E. (1999) Infect. Immun. 67 5483-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, J. J., Xue, Q., Papasian, C. J. & Morrison, D. C. (2001) J. Immunol. 166 6855-6860. [DOI] [PubMed] [Google Scholar]
- 28.Thoma-Uszynski, S., Stenger, S., Takeuchi, O., Ochoa, M. T., Engele, M., Sieling, P. A., Barnes, P. F., Rollinghoff, M., Bolcskei, P. L., Wagner, M., et al. (2001) Science 291 1544-1547. [DOI] [PubMed] [Google Scholar]
- 29.Kaushal, D., Schroeder, B. G., Tyagi, S., Yoshimatsu, T., Scott, C., Ko, C., Carpenter, L., Mehrotra, J., Manabe, Y. C., Fleischmann, R. D. & Bishai, W. R. (2002) Proc. Natl. Acad. Sci. USA 99 8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steyn, A. J., Collins, D. M., Hondalus, M. K., Jacobs, W. R., Jr., Kawakami, R. P. & Bloom, B. R. (2002) Proc. Natl. Acad. Sci. USA 99 3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vale, R. D. (2000) J. Cell Biol. 150 F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. (2001) Annu. Rev. Biochem. 70 755-775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.