Abstract
Screening human sequence databases for endogenous retroviral elements with coding envelope genes has revealed 16 candidate genes that we assayed for their fusogenic properties. All 16 genes were cloned in a eukaryotic expression vector and assayed for cell–cell fusion by using a large panel of mammalian cells in transient transfection assays. Fusion was observed for two human endogenous retrovirus (HERV) envelopes, the previously characterized HERV-W envelope, also called syncytin, and a previously uncharacterized gene from the HERV-FRD family. Cells prone to env-mediated fusion were different for the two envelopes, indicating different receptor usage. A search for the FRDenv gene in primates indicated that the corresponding proviral element is present in all simians, from New World monkeys to humans, being absent only in prosimians. Cloning of the corresponding env genes in simians disclosed conservation of the fully coding status of the gene, and most remarkably, conservation of its fusogenic property. Finally, a Northern blot analysis for the expression of the FRD family among a series of human tissues demonstrated specific expression in the placenta, as previously demonstrated for the other fusogenic human envelope of the HERV-W family. Altogether, the present data have identified a previously uncharacterized envelope (that we propose to name syncytin 2 after renaming syncytin as syncytin 1) with a potential role in placenta formation, and the identification of the complete set of retroviral envelopes with fusogenic properties now allows a definite analysis of the possible role of HERV in this physiological process, via classical genetic approaches.
The human genome contains a large fraction (≈8%) of elements of retroviral origin, with thousands of sequences close to the integrated proviral form of infectious retroviruses, with two LTRs bordering internal regions homologous to the gag, pol, and env genes (1–3). These elements, named human endogenous retroviruses (HERV), are most probably the proviral remnants of ancestral germ-line infections by active retroviruses, which have thereafter been transmitted in a Mendelian manner. HERV can be grouped according to sequence homologies into ≈100 distinct families, each containing a few to several hundreds elements. Strong similarities between HERV and present-day retroviruses can be inferred from phylogenetic analyses on the reverse transcriptase domain of the pol gene or the transmembrane (TM) moiety of the env gene, which disclose interspersion of both classes of elements and suggest a common history and shared ancestors (4, 5). Similarities are also observed at the functional level. For instance, HERV-K elements express particles detected in cell lines established from human teratocarcinomas closely resembling retroviruses (6, 7), and some of them encode a regulatory protein (called Rec or cORF) functionally homologous to the Rev protein encoded by the human HIV retrovirus (8, 9). Similarly, the envelope protein of a HERV-W family member has retained the ability to interact with the receptor of the D-type retroviruses (10, 11) and can confer infectivity on pseudotyped retroviral particles (11, 12). As a consequence of the close relationship between HERVs and infectious retroviruses, and despite the fact that most HERVs have accumulated mutations, deletions, and/or truncations, it remains plausible that some elements still possess functions of infectious retroviruses that may have been diverted by the host to its benefit. Along this line, it has been proposed (reviewed in refs. 13 and 14) that the HERV envelopes could play a role in several processes including (i) protection against infection by present-day retroviruses through receptor interference (15), (ii) protection of the fetus against the maternal immune system via an immunosuppressive domain located in the envelope TM subunit (16, 17), and (iii) placenta morphogenesis through fusogenic effects, allowing differentiation of cytotrophoblast cells into the syncytiotrophoblast (10, 18). In accordance with a symbiotic role for HERVs, it has recently been shown that the HERV-W envelope gene product is a highly fusogenic glycoprotein that is specifically expressed in the placenta and can mediate cell–cell fusion ex vivo (10, 18). Involvement of HERV proteins in such physiological processes, however, remains a debated issue, and definite evidence is still lacking. Actually, we had previously demonstrated, via a classical human genetic approach, that one postulated candidate, namely the placenta highly expressed ERV-3/HERV-R envelope gene, is not necessary for any fundamental placental function as 1% of individuals of caucasian origin carried a homozygous stop mutation resulting in severe protein truncation (19). Due to the high number of HERV elements, it had been argued that this negative answer did not preclude other envelope genes from being involved. As an essential step to settle this issue, we made a systematic screen of the human genome for envelopes with fusogenic activity. This search revealed a previously uncharacterized fusogenic envelope gene belonging to the HERV-FRD family (4, 5, 20); this gene and the previously identified HERV-W envelope most probably constitute the only two candidate genes for a fusogenic function in vivo. We show that this previously uncharacterized envelope gene is expressed in the placenta and is functionally conserved on primate evolution over >40 million years, thus strongly suggesting positive selection. The down-sizing of an a priori unsolvable genetic problem (associated with highly reiterated elements) to a simple two-gene analysis should now allow a definite answer to be given as to the role of HERVs in placenta formation.
Materials and Methods
Cell Lines. The human TE671 rhabdomyosarcoma cells (ATCC CRL8805), 293T embryonal kidney cells (ATCC CRL11268), HeLa epithelioid carcinoma cells (ATCC CCL2), the Cos-7 African green monkey kidney cells (ECACC 87021302), the G355-5 feline astrocyte cells (ATCC CRL2033), and the NIH 3T3 mouse fibroblasts were grown in DMEM supplemented with 10% FCS; the Chinese hamster ovary cells (ATCC-CCL61) were grown in F12K nutrient medium (GIBCO) supplemented with 7% FCS. All cell culture media were supplemented with streptomycin (100 μg/ml) and penicillin (100 units/ml).
DNAs. Human bacterial artificial chromosome (BACs) were obtained from BACPAC Resources (Oakland, CA) and from the U.K. Human Genome Mapping Project Resource Centre (Cambridge, U.K.). The sources of the genomic DNAs are given in refs. 21 and 22 for human, chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), gibbon (Hylobates Lar Moloch), Rhesus macaque (Macaca cynomolgus), New World monkeys (Saguinus midas and Cebus capucinus), and prosimian (Eulemur fulvus). The marmoset New World monkey (Callithrix jacchus) DNA was from the European Collection of Cell Cultures (85011419). Mouse (Mus musculus) and cat (Felis cattus) DNAs were extracted from spleen tissue samples (provided by J. Richardson, Institut Cochin de Génétique Moléculaire, Paris).
Envelope Expression Vectors. The FBA-RlessSALF expression vector (23) encoding the A-Rless hyperfusogenic mutant amphotropic murine leukemia virus envelope glycoprotein and the phCMV-G expression plasmid (GenBank accession no. AJ318514) were gifts from F.-L. Cosset (Ecole Normale Supérieure, Lyon, France). The HERV-W envelope expression vector (phCMV-EnvpH74) was a gift from F. Mallet (Ecole Normale Supérieure, Lyon, France). The fifteen other endogenous envelope expression vectors were constructed as follows. The full-length envelope of each provirus was PCR-amplified from the corresponding BAC DNA by using a proofreading DNA polymerase and appropriate primers (sequences available on request). PCR were carried out for only 15 cycles (1 min at 94°C, 1 min at 60°C, 4 min at 72°C), in 50 μl, by using 100 ng of BAC DNA, 48 pmol of each primer, 200 μM of each dNTP, 2.5 μlof PfuTurbo Hotstart polymerase, and 1× Pfu reaction buffer (Stratagene). Each PCR product was then cloned into the ph-CMV-G vector, opened with EcoRI, and blunt-ended by Kleenow treatment, except for the HERV-Fc2 envelope, which was cloned into phCMV-G restricted with EcoRI and Bsu36I and blunt-ended.
Cell–Cell Fusion Assay. Cells were transfected by using Lipofectamine (Invitrogen, 2 μg of DNA for 5 × 105 cells), except for 293T and TE671 cells, which were transfected by using calcium phosphate precipitation (Invitrogen, 5 μg of DNA for 5 × 105 cells). Fusion activity of envelope glycoproteins was measured 12 to 36 h after transfection of the corresponding expression vectors. To visualize syncytia, cells were fixed in methanol and stained by adding May–Grünwald and Giemsa solutions (Sigma) according to the manufacturer's instructions. The fusion index, which represents the percentage of fusion events in a cell population is defined as [(N - S)/T] × 100, where N is the number of nuclei in the syncytia, S is the number of syncytia, and T is the total number of nuclei counted.
Cloning of the HERV-FRD Envelope Gene from Simians. The envelope genes orthologous to the human HERV-FRD gene were PCR-amplified from simian (and human as a control) genomic DNAs. PCR were carried out for 25 cycles (10 s at 93°C, 30 s at 56°C, and 4 min at 68°C), in 50 μl, by using 100 ng of genomic DNA, 48 pmol of each primer, 350 μM of each dNTP, 0.75 μl of Expand long template enzyme mix, and 1× reaction buffer (Roche Applied Science). XhoI-containing primers were ATCACCTCGAGCACCATGGGCCTGCTCCTGCTGGTTCTCATTC as forward primer and ATCACCTCGAGGCTTCAGTACAGGTGGATA as reverse primer. Each PCR product was then XhoI restricted and cloned into the phCMV-G vector opened with XhoI. Sequencing of the simian envelope genes was performed on the bulk of the PCR products before cloning (MWG Biotech, Ebersberg, Germany). Sequences have been deposited in the EMBL nucleotide sequence database under accession numbers AJ577595 to AJ577600 for the chimpanzee, gorilla, orangutan, gibbon, macaque, and marmoset envelope, respectively.
In Vitro Transcription/Translation. In vitro transcription/translation assays were performed by using the Promega TNT coupled reticulocyte lysate system, following the manufacturer's instructions. DNA templates were obtained by PCR with a forward primer designed with a T7 promoter (GCTAATACGACTCACTATAGGAACAGACCACCATGGGTCTGCTCCTGCTG CTTC for marmoset and GCTAATACGACTCACTATGGAACAGACCACCATGCTCCTGCTGGTTCTCATTC for all other species) and a reverse primer (ATCACCTCGAGG CTTCAGTACAGGTGGATA for marmoset and TTTGAGCAAGGGTGATTCAT for all other species). [35S]Methionine was from ICN. Posttranslational analyses were performed by SDS/PAGE, with the gels exposed to x-ray films (Fuji) for 6 h.
DNA Slot Blot. DNAs (3 μg) from each species were loaded on Hybond N+ membranes (Amersham Biosciences) by using a slot blot apparatus (Hoefer). Blots were hybridized overnight at 65°C under standard conditions (24), by using a 32P-labeled full-length FRD env gene (1,679-bp PCR fragment) as a probe. Membranes were then washed at 65°C, once with 2× SSC/0.1% SDS for 15 min and once with 1× SSC/0.1% SDS for 15 min. Labeling was detected by using a PhosphorImager (FLA-3000 scanner, Molecular Dynamics).
Northern Blot Analysis and RNA Probe. The full-length FRD env gene obtained by PCR as described above was cloned in the antisense orientation under the T7 promoter into the pBluescript (Stratagene) vector restricted with EcoRV. The construct was linearized by XhoI digestion and an antisense riboprobe was synthesized by using the Strip-EZ RNA probe system from Ambion (Austin, TX). A human Northern blot (first choice poly(A)+ Human Northern blot 2, Ambion) was used according to the manufacturer's instructions, with prehybridization at 68°C in the UltraHyb buffer for 1 h, riboprobe addition and overnight hybridization at 68°C. The membrane was washed at 68°C, twice with the NorthernMax low-stringency buffer and once with the NorthernMax high-stringency buffer (Ambion), and then exposed to x-ray film (Kodak) at -80°C for 12 h.
Results
Rationale of the Assay for HERV Envelopes with Fusogenic Properties. A systematic search for envelope genes of retroviral origin has been performed by screening human genome databases (5). An extensive search for fully coding sequences among these genes, based on the latest human genome releases (which included >95% of the human genome), has finally led to the identification of a total of 16 genes (Table 1) that potentially encode complete envelope proteins (45). Ten of these genes have been identified (21, 25–31), and six new genes emerged, including genes from the FRD, T, R(b), F(c)1, and F(c)2 HERV families. For each identified gene, a BAC was obtained from BACPAC Resources and used to clone the corresponding envelope gene in a plasmid vector for expression in eukaryotic cells. The genes were amplified by using a proofreading DNA polymerase and a limited number of PCR cycles and introduced into phCMV, a vector in which expression is under the control of the strong hCMV promoter, with the gene placed in between a β-globin intron and polyadenylation sequences. For each amplified and cloned gene, nucleotide sequencing was performed that disclosed amino acid identity with the sequences in the database, as well as an in vitro transcription/translation assay (as in Fig. 4B for envFRD) resulting in proteins of the expected size (data not shown). The assay for fusogenicity of the cloned envelopes then relies on the transfection of cells expected to carry a receptor for the corresponding envelope, with the expression vectors for the envelope proteins. Accordingly, fusion assays were performed for all 16 envelope genes on transfection of a large series of cells, including feline, murine, simian, and human cells. In the latter case, different cell lines were used, namely HeLa cells, 293T embryonic kidney cells, and TE671 cells that have been previously used because they possess the receptors for several retrovirus groups (32). In a standard assay, 5 × 105 cells were transfected with 2–5 μg of the Env-expression vector, and fusion was tested by screening the transfected cell culture for multinucleated giant cells (or syncytia) 12–36 h after transfection. Controls included the highly fusogenic A-Rless envelope from the amphotropic murine leukemia virus (33) and an “empty” vector.
Table 1. Assay for fusogenicity of the 16 coding endogenous envelope genes of the human genome.
| Gene name | Chromosome localization | Accession no. | Fusion assay* |
|---|---|---|---|
| envH1 | 2q24.3 | AJ289709 | — |
| envH2 | 3q26 | AJ289710 | — |
| envH3 | 2q24.1 | AJ289711 | — |
| envK1 | 12q14.1 | AC074261 | — |
| envK2 | 7p22.1 | AF164614 | — |
| envK3 | 19q12 | Y17833 | — |
| envK4 | 6q14.1 | AF164615 | — |
| envK5 | 19p13.11 | AY037928 | — |
| envK6 | 8p23.1 | AY037929 | — |
| envT | 19p13.11 | AC078899 | — |
| envW | 7q21.2 | AC000064 | + |
| envFRD | 6p24.1 | AL136139 | + |
| envR | 7q11.21 | AC073210 | — |
| envR(b) | 3p24.3 | AC018389 | — |
| envF(c)2 | 7q36.2 | AC016222 | — |
| envF(c)1 | Xq21.33 | AL354685 | — |
Fusion assay was performed with the following target cells: NIH3T3, CHO, G355-5, Cos-7, TE671, 293T, and HeLa. —, Negative result in all cell types; +, positive result in at least one cell type
Fig. 4.
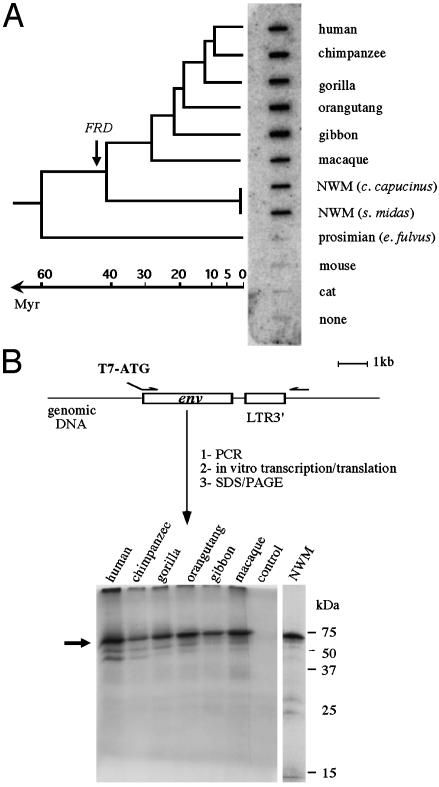
Conservation of the HERV-FRD env locus and ORF on primate evolution. (A) Slot-blot of genomic DNA from simian, prosimian, cat, and mouse species, probed with a full-length HERV-FRD env fragment. The inferred date of insertion of HERV-FRD is indicated with an arrow on the primate phylogenetic tree on the left. NWM, New World monkey. (B) In vitro transcription/ translation assay of the human HERV-FRD env gene and of the orthologous genes from the indicated simian species. Env genes were PCR-amplified as schematized, submitted to in vitro transcription/translation (“control” is without DNA template), run on a polyacrylamide gel, and autoradiographed. The arrow points to the bands of the expected size.
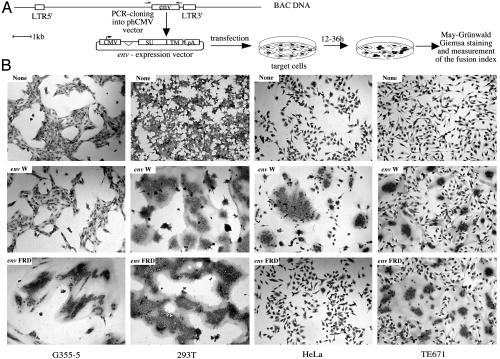
Identification of a Previously Uncharacterized Fusogenic HERV Envelope. As illustrated in Fig. 1 and Table 1, among all of the coding env genes, only two induced syncytia formation, namely the HERV-W and the HERV-FRD genes, the other genes resulting in no effect. The result obtained with HERV-W is confirmatory, as it has been demonstrated in refs. 10 and 18. Yet, an original outcome of the present extensive investigation is the sorting out of a previously uncharacterized envelope gene with fusogenic properties (the HERV-FRD env gene) and the evidence that these two genes most probably constitute the sole reservoir of fusogenic human endogenous retroviral envelopes.
Fig. 1.
Envelope-mediated cell–cell fusion. (A) Construction of envelope-expressing vectors and rationale of the fusion assay. Each of the 16 envelope genes was PCR-amplified from BAC DNA and cloned into the phCMV expression vector. The env genes inserted in between the β-globin intron and pA sequences are schematized with the putative cleavage site between the SU and the TM envelope subdomains. Cells were transfected with the env-expressing vectors and stained with May–Grünwald and Giemsa solutions 12 h (for the G355-5 cells) or 36 h (for the other cell types) after transfection. Fusion indices were calculated as indicated in Materials and Methods. (B) Syncytia formation by HERV envelope glycoproteins in various cell types. Cells were transfected with vectors expressing the HERV-FRD envelope (envFRD), the HERV-W envelope (envW), or an empty vector (none).
The properties of the HERV-FRD env gene were analyzed further and compared with that of HERV-W. First, as illustrated in Table 2, it can be observed that the syncytia-forming activity of both envelopes, although of quite related extent when using the syncytiaforming index defined in Materials and Methods, does not coincide for a given cell type: for instance, the HERV-FRD Env is highly fusogenic in feline cells whereas that of HERV-W has no activity, and within human cells whereas the HERV-W Env is fusogenic, that of HERV-FRD is not fusogenic in all cell types, with only limited effect in HeLa cells. Such differences can be simply accounted for by taking into consideration that retroviral envelope-mediated cell fusion depends on the presence of an appropriate receptor on the cell surface, which may be different for different envelope proteins, as currently observed for most exogenous infectious animal retroviruses (reviewed in refs. 34 and 35). In this respect, it has been demonstrated that the receptor for the HERV-W envelope protein is the receptor for the D-type retroviruses (10, 11). Clearly, the receptor for the HERV-FRD envelope is distinct because the cell specificity for fusion for both envelopes is different. This finding was further confirmed by performing an interference assay using the panel of cells derived in ref. 10, where no decrease in the fusogenic activity of the HERV-FRD Env could be detected, under conditions where the cells had been stably transfected with an expression vector for the envelope of a type-D retrovirus (RD114, not shown).
Table 2. Fusion host range of the HERV-FRD envelope.
| Fusion index* |
|||||
|---|---|---|---|---|---|
| Species | Target cells | HERV-FRD | HERV-W | A-Rless | None |
| Mouse | NIH3T3 | 1.0 ± 0.9 | 0.2 ± 0.5 | 41 ± 17 | 0.4 ± 0.3 |
| Hamster | CHO | 1.8 ± 0.6 | 1.3 ± 0.8 | 1.8 ± 0.4 | 1.0 ± 0.5 |
| Cat | G355—5 | 90 ± 2 | 2.2 ± 3.2 | 83 ± 3 | 0.7 ± 0.8 |
| Monkey | Cos-7 | 20 ± 4 | 62 ± 3 | 19 ± 4 | 13 ± 3 |
| Human | TE671 | 66 ± 6 | 56 ± 10 | 39 ± 8 | 3.1 ± 1.8 |
| 293T | 81 ± 7 | 90 ± 4 | 9 ± 3 | 0.3 ± 0.6 | |
| HeLa | 1.8 ± 0.7 | 65 ± 9 | 23 ± 6 | 1.0 ± 0.4 | |
Target cells were transfected with expression vectors for the HERV-FRD envelope, the HERV-W envelope, the hyperfusogenic mutant amphotropic MLV envelope (A-Rless), or no protein (none) as a negative control, and fusion indices were determined as indicated in Materials and Methods (means ± SDs; n = 5)
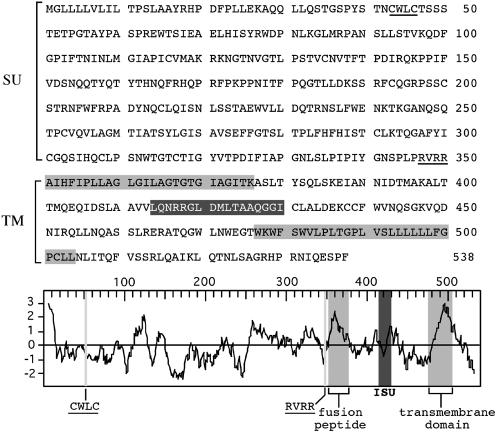
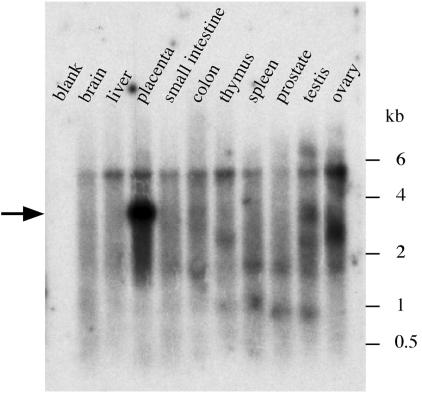
Consistent with its fusogenic property, analysis of the amino acid sequence of the FRD envelope together with its hydrophobic profile (Fig. 2) discloses the characteristic features of retroviral envelopes (reviewed in ref. 34), with a canonical cleavage site [consensus: R/K-X-R/K-R; (36)] between the surface (SU) and TM moieties of the protein, and the presence of hydrophobic domains corresponding to the fusion peptide and the transmembrane domain. A “CWLC” domain, involved in the interaction between the SU and TM moieties in retroviral envelopes (37), and an “immunosuppressive” domain (16) can also be identified in the SU and TM moieties, respectively. Finally, a Northern blot analysis was performed with a membrane containing poly(A)+ RNA from a panel of human tissues that we hybridized with a riboprobe for the HERV-FRD envelope. As can be observed in Fig. 3, a strong band is observed in the placenta, and actually almost exclusively in this organ, at the expected position for a spliced subgenomic retroviral transcript. A real-time RT-PCR analysis of the HERV-FRD env gene transcripts further disclosed high level expression in cytotrophoblast cells isolated from the placenta, not observed in the corresponding fibroblast cells (see Supporting Methods and Fig. 6, which are published as supporting information on the PNAS web site, www.pnas.org).
Fig. 2.
Primary sequence, hydrophobicity profile, and predicted features of the HERV-FRD envelope. The SU and TM moieties of the envelope are delineated, with the canonical RVRR cleavage site between the two subunits underlined (consensus: R/K-X-R/K-R); in the TM subunit, the hydrophobic fusion peptide and transmembrane domains are shaded in light gray, and the putative immunosuppressive domain (ISU) in dark gray; in the SU subunit, the canonical CWLC domain involved in SU-TM interaction is underlined.
Fig. 3.
Northern blot analysis of HERV-FRD env expression in human tissues. Poly(A)+ RNAs (Ambion, Human Northern Blot 2) were probed with an antisense-strand HERV-FRD env riboprobe. Exposure time was 12 h.
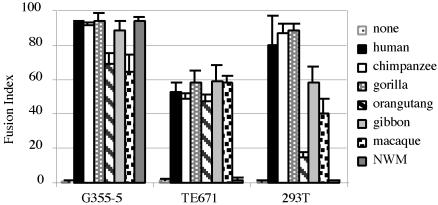
Conservation of the FRD Locus and Fusogenicity on Primate Evolution. The FRD family of endogenous retroviruses consists of ≈50 provirus copies per haploid genome (4, 20). This family is most probably a very ancient family whose members entered the primate genome after the divergence between prosimians and simians: this finding is demonstrated in the zoo-slot in Fig. 4A, where hybridization can be detected by using an envelope probe and genomic DNAs from humans to New World monkeys, with no signal for prosimians or rodents (or other mammals, not shown). Taking into consideration that conservation of the HERV-FRD envelope gene with an ORF might not be fortuitous but rather be the result of positive selection for a still unknown function, we analyzed the status of the orthologous locus in the course of primate evolution, by using a PCR approach with a “flanking” primer located 3′ to the proviral insertion, and an “internal” proviral primer 5′ to the env gene. Consistent with the zoo-blot analysis, PCR amplification was found positive for all simians, resulting in DNA fragments of the expected size. The identified putative envelope genes were characterized further, along three lines: first, the PCR products were sequenced for an unambiguous identification of the env genes and the determination of their level of divergence in the course of primate evolution; second, an in vitro transcription/translation assay was performed to determine whether the genes are fully coding; and third, a fusion assay was performed as above, to determine whether env gene function has been preserved. As illustrated in Table 3, sequencing of the PCR products disclosed very high sequence conservation among the primate ERV-FRD env genes, with values in the 95–100% range from human to macaque, and still 88% identity between human and New World monkey, which diverged at least 40 millions years ago. No stop codon could be found in the sequences, a result consistent with the in vitro transcription/translation assay performed directly on the PCR products (Fig. 4B), which demonstrates full-length products for all genes. To assay for the possible conservation of the fusogenic property of the identified genes, the PCR env fragments were cloned into the phCMV expression vector and tested in the syncytia-forming assay, as above. For each env gene, at least six clones were assayed to circumvent possible mutations introduced by the PCR amplification procedure (a feature with no consequence on both the sequencing and the transcription/translation assay above, directly performed on the bulk of the PCR product), by using both the highly fusogenic feline G355.5 cells, and human cells (see Fig. 5). Remarkably, as illustrated in the figure, fusion of the transduced feline cells could be observed for all env genes (with in each case at least four of six cloned PCR fragments positive, a result consistent with the expected rate of mutation of the DNA polymerase used for the corresponding genomic amplifications), with no significant difference in the extent of fusogenicity from human to New World monkey. A similar conclusion holds using human cells for the fusion assay, as expected, but with an exception for the most distantly related env gene (the New World monkey gene), which is not positive in human cells although clearly fusogenic when assayed in feline cells. This unexpected result is most probably relevant to subtle differences in the envelope amino acid sequence, which, together with expected differences among the human and feline receptors, result in the impairment of a productive interaction with the former. Such differences might be of interest for a future identification of the amino acids involved in the fusion process.
Table 3.
Percentage identity of amino acid sequences of envFRD in simians*
| Human | Chimpanzee | Gorilla | Orangutan | Gibbon | Macaque | NWM | |
|---|---|---|---|---|---|---|---|
| Human | - | ||||||
| Chimpanzee | 99.6 | - | |||||
| Gorilla | 99.1 | 99.1 | - | ||||
| Orangutan | 96.7 | 96.7 | 96.8 | - | |||
| Gibbon | 97.2 | 97.2 | 97.4 | 97.0 | - | ||
| Macaque | 95.2 | 95.2 | 95.4 | 95.7 | 96.5 | - | |
| NWM | 88.1 | 88.1 | 88.0 | 87.9 | 88.5 | 87.9 | - |
Orthologous envFRD genes were PCR-amplified and sequenced. Alignments were performed with the lalign program. Sequences are deposited in the EMBL nucleotide sequence database (see Materials and Methods). NWM, New World monkey
Fig. 5.
Fusion activity of the orthologous primate ERV-FRD envelope glycoproteins. The indicated target cells were transfected as indicated in Fig. 1 with vectors expressing the human or simian orthologous ERV-FRD envelopes, and fusion indices were determined as indicated in Table 2 (means ± SDs; n = 5).
Discussion
The present investigation has unraveled a fusogenic protein of retroviral origin that is encoded in primate genomes. This envelope protein promotes cell–cell fusion in a specific manner as it depends on the cell type used, with positive effects in both feline and human cells. Interestingly, analysis of the cell types prone to fusion discloses differences with the other fusogenic envelope identified so far, i.e., the HERV-W envelope (10, 18), indicating differences in receptor usage. In the case of the HERV-W envelope, interference assays had led to the identification of the corresponding receptor as being that for D-type retroviruses (10, 11). Similar assays demonstrate that this receptor is not that for the HERV-FRD envelope, and preliminary experiments suggest that it is none of the identified receptors for the present-day infectious retroviruses (not shown). Specific search for the HERV-FRD receptor will therefore be necessary to identify the cell membrane protein involved in the observed cell–cell fusion. Following the proposed nomenclature for the HERV-W envelope gene, that was named syncytin in relation with its ability to make syncytia via cell–cell fusion (18), we propose to name the fully coding HERV-FRD envelope gene the syncytin 2 gene, and accordingly to rename the syncytin gene as syncytin 1 in future studies.
An important issue of the present investigation is the discovery that the identified envelope gene has been conserved in primate evolution in a functional state over >40 million years, as the corresponding locus is present from New World monkeys to humans as a full-length coding gene, for which we further demonstrate that the fusogenic function is conserved in all primates tested. Conservation of the fusogenic function is a strong hint for a functional role of the gene in the physiology of the host and suggests a selective process by which the retroviral gene function has been diverted by the host to its own benefit. Similar diversions have already been described for “parasitic” genes, with examples of retroviral promoters that confer novel tissue specificities to nearby cellular genes (38–40), and examples of coding sequences of retroviral origin that, in the mouse, confer resistance to infections by exogenous retroviruses (reviewed in ref. 15).
Among the numerous functions that have been suggested for a possible physiological role of HERV envelopes, the fusogenic function has been frequently hypothesized as being plausibly involved in placenta formation, via the fusion of the cytotrophoblast cells into the syncytiotrophoblast (10, 18, 41). This hypothesis still remains elusive but would be consistent with the identified fusogenic activities of the HERV-W and –FRD envelope genes and their observed expression in the placenta and cytotrophoblast cells. A possible approach to assay the model in humans would be a genetic one, with the finding of possible “natural” mutants among the human population that could be associated with a pathological situation. Along these lines, one of the drawbacks that the present investigation relieves is the possible redundancy of genes with the same function. Indeed, we had previously shown, for the first envelope gene that had been unambiguously demonstrated to be highly expressed in the placenta (the HERV-R env gene), that it could not be involved in placenta formation, due to the occurrence of a severe polymorphism (a premature stop codon) found in 16% of caucasians in the heterozygote state, and in 1% in the homozygote state, and not resulting in any placenta-related phenotype (19). It was thereafter argued that this defect could be complemented by other genes with the same function, an interpretation that, indeed, could not be rejected. In fact, the present study, as a consequence of its exhaustivity, now provides the required genetic tools to unambiguously address the issue: there are two identified HERV envelope genes with fusogenic properties in the human genome, and these two genes are expressed in the placenta. A concomitant search for polymorphisms of these two genes among the human population and, in a more refined manner, among groups disclosing abnormalities in placenta formation, such as preeclampsia, Down's syndrome, or uncontrolled trophoblast invasion as observed in choriocarcinoma (42–44), should now provide hints for the possible involvement of these genes in placenta physiology. Finally, it is noteworthy that the identified HERV-FRD envelope gene carries a domain (the immunosuppressive domain) that could play a physiological role in protecting the fetus against the maternal immune system and, as such, could have also participated in the “positive selection” of the syncytin 2 gene in the course of primate evolution. Again, a genetic approach might help in answering these debated issues.
Supplementary Material
Acknowledgments
We thank A. Calteau for help with characterizing some of the envelope genes, F.-L. Cosset and A. Ruggieri for invaluable discussions and the gift of phCMV-G and FBA-RlessSALF plasmids and TE671 cells, J.-L Frendo and D. Evain-Brion for the gift of placenta cell-derived RNA samples, and F. Mallet for phCMV-EnvpH74 plasmid. We thank C. Lavialle for comments and critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique and the Ligue Nationale Contre le Cancer (Equipe “labellisée”).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HERV, human endogenous retrovirus; TM, transmembrane; SU, surface; BAC, bacterial artificial chromosome.
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (accession nos. AJ577595–AJ577600 for the chimpanzee, gorilla, orangutan, gibbon, macaque, and marmoset envelope, respectively).
References
- 1.Löwer, R., Löwer, J. & Kurth, R. (1996) Proc. Natl. Acad. Sci. USA 93 5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke, J. D. & Stoye, J. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-436. [PubMed]
- 3.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- 4.Tristem, M. (2000) J. Virol. 74 3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benit, L., Dessen, P. & Heidmann, T. (2001) J. Virol. 75 11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boller, K., König, H., Sauter, M., Mueller-Lantzsch, N., Löwer, R., Löwer, J. & Kurth, R. (1993) Virology 196 349-353. [DOI] [PubMed] [Google Scholar]
- 7.Löwer, R., Boller, K., Hasenmeier, B., Korbmacher, C., Mueller-Lantzsch, N., Löwer, J. & Kurth, R. (1993) Proc. Natl. Acad. Sci. USA 90 4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magin, C., Löwer, R. & Löwer, J. (1999) J. Virol. 73 9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, J., Bogerd, H. P., Peng, S., Wiegand, H., Truant, R. & Cullen, B. R. (1999) Proc. Natl. Acad. Sci. USA 96 13404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blond, J. L., Lavillette, D., Cheynet, V., Bouton, O., Oriol, G., Chapel-Fernandes, S., Mandrand, B., Mallet, F. & Cosset, F.-L. (2000) J. Virol. 74 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavillette, D., Marin, M., Ruggieri, A., Mallet, F., Cosset, F. L. & Kabat, D. (2002) J. Virol. 76 6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An, D. S., Xie, Y.-M. & Chen, I. S. Y. (2001) J. Virol. 75 3488-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venables, S., Brookes, M., Fan, W., Larsson, E., Maini, R. N. & Boyd, M. T. (1995) Virology 211 589-592. [DOI] [PubMed] [Google Scholar]
- 14.Harris, J. R. (1998) BioEssays 20 307-316. [DOI] [PubMed] [Google Scholar]
- 15.Best, S., Le Tissier, P. R. & Stoye, J. P. (1997) Trends Microbiol. 5 313-318. [DOI] [PubMed] [Google Scholar]
- 16.Cianciolo, G. J., Copeland, T., Orozlan, S. & Snyderman, R. (1985) Science 230 453-455. [DOI] [PubMed] [Google Scholar]
- 17.Mangeney, M. & Heidmann, T. (1998) Proc. Natl. Acad. Sci. USA 95 14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., LaVallie, E., Tang, X. Y., Edouard, P., Howes, S., Keith, J. C. J. & McCoy, J. M. (2000) Nature 403 785-788. [DOI] [PubMed] [Google Scholar]
- 19.de Parseval, N. & Heidmann, T. (1998) J. Virol. 72 3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seifarth, W., Skladny, H., Krieg-Schneider, F., Reichert, A., Hehlmann, R. & Leib-Mosch, C. (1995) J. Virol. 69 6408-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Parseval, N., Casella, J. F., Gressin, L. & Heidmann, T. (2001) Virology 279 558-569. [DOI] [PubMed] [Google Scholar]
- 22.Benit, L., Lallemand, J.-B., Casella, J.-F., Philippe, H. & Heidmann, T. (1999) J. Virol. 73 3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavillette, D., Maurice, M., Roche, C., Russell, S. J., Sitbon, M. & Cosset, F.-L. (1998) J. Virol. 72 9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Church, G. M. & Gilbert, W. (1984) Proc. Natl. Acad. Sci. USA 81 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbulescu, M., Turner, G., Seaman, M. I., Deinard, A. S., Kidd, K. K. & Lenz, J. (1999) Curr. Biol. 9 861-868. [DOI] [PubMed] [Google Scholar]
- 26.Blond, J.-L., Besème, F., Duret, L., Bouton, O., Bedin, F., Perron, H., Mandrand, B. & Mallet, F. (1999) J. Virol. 73 1175-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen, M., Powers, M., O'Connell, C. & Kato, N. (1985) Virology 147 449-458. [DOI] [PubMed] [Google Scholar]
- 28.Lindeskog, M., Mager, D. & Blomberg, J. (1999) Virology 258 441-450. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, J., Sauter, M., Racz, A., Scherer, D., Mueller-Lantzsch, N. & Meese, E. (1999) Nat. Genet. 21 257-258. [DOI] [PubMed] [Google Scholar]
- 30.Tönjes, R. R., Czauderna, F. & Kurth, R. (1999) J. Virol. 73 9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner, G., Barbulescu, M., Su, M., Jensen-Seaman, M. I., Kidd, K. K. & Lenz, J. (2001) Curr. Biol. 11 1531-1535. [DOI] [PubMed] [Google Scholar]
- 32.Porter, C. D., Collins, M. K., Tailor, C. S., Parkar, M. H., Cosset, F.-L., Weiss, R. A. & Takeuchi, Y. (1996) Hum. Gene Ther. 7 913-919. [DOI] [PubMed] [Google Scholar]
- 33.Rein, A., Mirro, J., Haynes, J. G., Ernst, S. M. & Nagashima, K. (1994) J. Virol. 68 1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter, E. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 71-119.
- 35.Overbaugh, J., Miller, A. D. & Eiden, M. V. (2001) Microbiol. Mol. Biol. Rev. 65 371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffin, J. M. (1986) Cell 46 1-4. [DOI] [PubMed] [Google Scholar]
- 37.Pinter, A., Kopelman, R., Li, Z., Kayman, S. C. & Sanders, D. A. (1997) J. Virol. 71 8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson, D. A., Mager, D. L. & Leong, J.-A. C. (1994) in The Retroviridae, ed. Levy, J. A. (Plenum, New York), Vol. 3, pp. 465-535. [Google Scholar]
- 39.Samuelson, L. C., Philips, R. S. & Swanberg, L. J. (1996) Mol. Biol. Evol. 13 767-779. [DOI] [PubMed] [Google Scholar]
- 40.Schulte, A. M., Lai, S., Kurtz, A., Czubayko, F. & Riegel, A. T. (1996) Proc. Natl. Acad. Sci. USA 93 14759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frendo, J.-L., Olivier, D., Cheynet, V., Blond, J.-L., Bouton, O., Vidaud, M., Rabreau, M., Evain-brion, D. & Mallet, F. (2003) Mol. Cell. Biol. 23 3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox, H. (1997) in Pathology of the Placenta, ed. Fox, H. (Saunders, Philadelphia), pp. 151-175.
- 43.Frendo, J. L., Vidaud, M., Guibourdenche, J., Luton, D., Muller, F., Bellet, D., Giovagrandi, Y., Tarrade, A., Porquet, D., Blot, P. & Evain-Brion, D. (2000) J. Clin. Endocrinol. Metab. 85 3700-3707. [DOI] [PubMed] [Google Scholar]
- 44.Baergen, R. N. (1997) Gen. Diagn. Pathol. 143 127-141. [PubMed] [Google Scholar]
- 45.de Parseval, N., Lazar, V., Casella, J. F., Benit, L. & Heidmann, T. (2003) J. Virol. 77 10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.