Abstract
We analyzed interleukin (IL) 12 and IL-23 production by monocyte-derived dendritic cells (mono-DCs). Mycobacterium tuberculosis H37Rv and zymosan preferentially induced IL-23. IL-23 but not IL-12 was efficiently induced by the combination of nucleotide-binding oligodimerization domain and Toll-like receptor (TLR) 2 ligands, which mimics activation by M. tuberculosis, or by the human dectin-1 ligand β-glucan alone or in combination with TLR2 ligands, mimicking induction by zymosan. TLR2 ligands inhibited IL-12 and increased IL-23 production. DC priming with interferon (IFN) γ strongly increased IL-12 production, but was not required for IL-23 production and inhibited IL-23 production induced by β-glucan. The pattern of IL-12 and IL-23 induction was reflected in accumulation of the IL-12p35 and IL-23p19 transcripts, respectively, but not IL-12/23p40. Although IL-23, transforming growth factor β, and IL-6 contained in the supernatants of activated mono-DCs played a role in the induction of IL-17 by human CD4+ T cells, IL-1β, in combination with one or more of those factors, was required for IL-17 production, and its production determined the differential ability of the stimuli used to elicit mono-DCs to produce soluble factors directing IL-17 production. Thus, the differential ability of pathogens to induce antigen-presenting cells to produce cytokines regulates the immune response to infection.
IL-12 and IL-23, two heterodimeric cytokines produced by APCs, are composed of a specific polypeptide, namely p35 for IL-12 and p19 for IL-23, disulfide linked to a common p40 chain to form the biologically active molecules (1, 2). Although p19 and p35 are not secreted in the absence of the p40 chain, a consistent amount of free p40, whose biological function is still debated (3), is produced, often in large excess over the complete IL-12 and IL-23 heterodimeric proteins (4). The function of IL-12 is pivotal in both innate and acquired immunity, because it induces IFN-γ production from NK and NKT cells in the early phases of the immune response, and induces the differentiation of Th1 cells, relevant in protection against infectious agents and tumor surveillance (5). A protective role for the Th1 response generated by IL-12/IL-23 has been suggested based on Mendelian susceptibility to mycobacterial and other infectious diseases in children with genetic defects of the IL-12/23–IFN-γ circuit (6). Studies using p19−/− or p35−/− mouse models of autoimmunity (7–9), tumors (10), and inflammatory bowel disease (11–14) have identified IL-23, rather than IL-12, as the major factor responsible for lesions caused by chronic inflammation, acting through the induction of a CD4+ T lymphocyte subset producing IL-17, and through other less-characterized mechanisms of innate immunity. In humans, a role for IL-23 in chronic inflammation has been proposed based on the association of polymorphisms in the IL-23 receptor gene with Crohn's disease (15).
Products from microorganisms are strong inducers of IL-12 heterodimer production from APCs that have been exposed to cytokines such as IFNs or IL-4, whereas in the absence of these mediators, IL-12 production is minimal, although high levels of free p40 are induced (5). Until now, very little has been known about the stimuli inducing p19 expression and IL-23 production. Cells of the innate immune system recognize microorganisms through a limited number of germline-encoded pattern recognition receptors (PRRs), (16, 17). The simultaneous engagement of distinct PRRs often cooperates in the innate response to microorganisms or their products, influencing both the magnitude and the quality of the immune response (18, 19). The synergistic effect of the engagement of cooperating Toll-like receptor (TLR) 7/8 and TLR3 or TLR4 (20, 21) is, however, particularly dramatic for the production of the biologically active heterodimeric form of IL-12.
In the present study, we observed that although PRR ligand–activated DCs represent the major producers of IL-12 and IL-23 among peripheral APCs, there are striking differences in the conditions leading to the expression of either cytokine because of the selective engagement of distinct combinations of PRRs and to the differential effect of IFN-γ priming of the DCs. We also found that the ability of the supernatants of activated DCs to induce IL-17 production from naive CD4+ T cells is caused by the differential secretion of IL-1β, which acts in cooperation with IL-23 and other cytokines.
RESULTS
Identification of IL-12, IL-23, and free IL-12 p40 by immunoprecipitation
Immunoprecipitation experiments in IFN-γ–primed, LPS-stimulated monocyte-derived DCs (mono-DCs) and monocyte-enriched PBMCs biosynthetically labeled with [35S]methionine with anti–IL-23 p19, anti–IL-12 p35, and anti–IL-12/23 p40 mAbs (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071450/DC1) allowed clear identification of the IL-12 p40 secreted alone or in association with p35 and p19 to form the heterodimeric cytokines IL-12 and IL-23. IL-23 appears as three major bands of 61, 57, and 51 kD, corresponding to the p19 chain disulfide linked to all of the glycosylation variants of the IL-12/23 p40 chain (22). Inhibition of glycosylation by tunicamycin (Fig. S2) demonstrated that N-glycosylation of IL-23 is not needed for secretion of this cytokine, unlike the fundamental role of N-glycosylation for secretion of IL-12 p75 (22).
Differential regulation of IL-12 and IL-23 production by inactivated Mycobacterium tuberculosis alone or in combination with other stimuli
IL-12 and IL-23 production by human DCs stimulated with heat-killed M. tuberculosis (H37Rv strain) was evaluated by immunoprecipitation and by ELISA. Because IL-12 is preferentially induced by a combination of TLR ligands, DCs were also stimulated with the TLR7/8-ligand R848 alone or in combination with H37Rv.
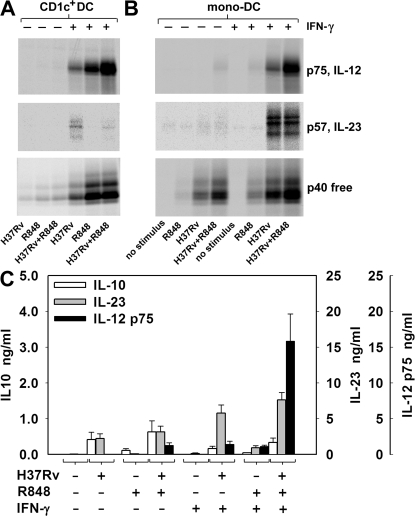
H37Rv induced IL-12 p75 and IL-23 production in IFN-γ–primed peripheral blood CD1c+ DCs, as detected by immunoprecipitation (Fig. 1 A). R848 synergized with H37Rv for IL-12 p75 production but consistently decreased IL-23 production. In contrast, LPS and R848 synergistically induced IL-12 p75 and IL-23 in either untreated or IFN-γ–pretreated monocytes and, at higher levels, CD1c+ DCs (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20071450/DC1).
Figure 1.
Differential regulation of IL-12 and IL-23 production in CD1c+ DCs and mono-DCs stimulated by H37Rv. CD1c+ DCs (A) and mono-DCs (B and C) were treated with IFN-γ or left untreated. Cells were labeled with [35S]methionine (A and B) or unlabeled (C) and stimulated with heat-killed H37Rv, alone or together with R848. After 18 h, supernatants were immunoprecipitated with anti–IL-12 p35, anti–IL-23 p19, or anti–IL-12/23 p40 mAbs and resolved by nonreducing SDS-PAGE (A and B) or tested for IL-12 p75, IL-23, and IL-10 production by ELISA (C). In A, supernatants were pooled from cells purified from three different donors. Results in B are representative of six independent experiments. Data in C are mean ± SE values for mono-DCs derived from 20 different donors.
In mono-DCs, H37Rv induced IL-23 but not IL-12 p75, as detected by immunoprecipitation (Fig. 1 B). In IFN-γ–primed mono-DCs, H37Rv alone induced relevant production of both IL-23 and IL-12 p75. The combination of H37Rv and R848 did not increase the amount of IL-23 production but synergistically induced IL-12 p75 production in both IFN-γ–primed and unprimed mono-DCs.
ELISA using mono-DC preparations obtained from 20 different donors confirmed the immunoprecipitation data (Fig. 1 C). H37Rv alone was unable to induce production of IL-12 p75 in mono-DCs but was effective when added either together with R848 (H37Rv+R848 versus R848; P = 0.005) or to IFN-γ–primed mono-DCs (H37Rv+IFN-γ vs. IFN-γ; P = 0.004). Significantly higher levels of IL-12 were produced when IFN-γ–primed mono-DCs were co-stimulated with H37Rv and R848 (H37Rv+R848+IFN-γ vs. H37Rv+IFN-γ; P = 0.001). Unlike IL-12, IL-23 was significantly induced by H37Rv alone (P = 0.005), and its production was only modestly increased by the combination of H37Rv and R848. IFN-γ priming increased IL-23 production (H37Rv+IFN-γ vs. H37Rv; P = 0.001), with no further sizable increase upon combination with R848. Like IL-23, IL-10 was induced in response to H37Rv alone (P = 0.052), and its production was moderately increased by R848 but reduced by IFN-γ priming.
Overall, these results indicate that M. tuberculosis efficiently induces IL-23 production but is only a modest stimulus for IL-12; however, it provides signals to DCs that efficiently synergize with the TLR7/8 ligand R848 in inducing IL-12 production. Because DCs produced IL-12 and IL-23 much more efficiently than did monocytes, and because mono-DCs closely mimicked the responses observed with purified blood CD1c+ DCs, all subsequent experiments in this study were performed using mono-DCs.
Combination of ligands for nucleotide-binding oligodimerization domain (NOD2) and TLR2 mimics IL-23 and IL-12 induction by M. tuberculosis
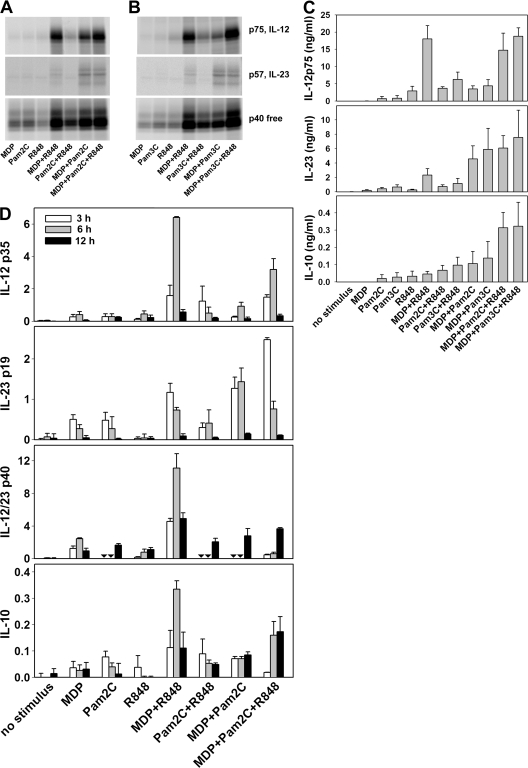
DCs reportedly recognize M. tuberculosis primarily through TLR2, TLR9, NOD2, and possibly TLR4 (23–27). Because human mono-DCs do not express functional levels of TLR9, we tested whether the association of the NOD2 ligand muramyl dipeptide (MDP) with ligands for TLR2 might mimic the stimulation observed with M. tuberculosis. Indeed, MDP together with the TLR2/6 ligand Pam2C (Fig. 2 A) or the TLR1/2 ligand Pam3C (Fig. 2 B) induced production of IL-23, IL-12 p75, and free p40 in IFN-γ–primed mono-DCs. Treatment with each stimulus alone did not induce IL-23 and induced only barely detectable amounts of IL-12 p75 and free p40. The combination of MDP with R848 induced much higher levels of IL-12 p75 but lower levels of IL-23 than did the combination of MDP with Pam2C or with Pam3C that preferentially induced IL-23. R848 and Pam2C (Fig. 2 A) or Pam3C (Fig. 2 B) modestly increased IL-12 but not IL-23 production. The association of MDP+Pam2C or MDP+Pam3C with R848 (Fig. 2, A and B, respectively) further increased IL-12 p75 and p40, whereas IL-23 remained highly expressed and almost unmodified, a pattern consistent with that observed in mono-DCs stimulated by H37Rv and H37Rv+R848. These results were confirmed by ELISA quantitation of cytokine secretion (Fig. 2 C) and by QuantiGene multiplex assay to measure mRNA accumulation (Fig. 2 D). No IL-12 production and lower production of IL-23 and IL-10 in all conditions were observed when non–IFN-γ–primed mono-DCs were used (unpublished data).
Figure 2.
A combination of NOD2 and TLR2 ligands mimics IL-23 and IL-12 induction by M. tuberculosis. mono-DCs treated with IFN-γ were labeled with [35S]methionine (A and B) or unlabeled (C and D) and stimulated with different combinations of MDP, Pam2C, and Pam3C, with or without R848. After 18 h, supernatants were immunoprecipitated with anti–IL-12 p35, anti–IL-23 p19, or anti–IL-12/23 p40 mAbs and resolved in nonreducing SDS-PAGE (A and B), or were evaluated for IL-12 p75, IL-23, and IL-10 by ELISA (C). In D, cells were lysed at 3, 6, and 12 h, and mRNA accumulation was determined using the QuantiGene multiplex assay. Results in A and B are representative of those obtained in three independent experiments with cells derived from different donors. Data in C are mean ± SE values from mono-DCs derived from three different donors. Results in D were obtained with cells from two different donors and are expressed as the mean ± SD of triplicate cultures. Inverted triangles indicate not done.
These data suggest that the combination of ligands for NOD2 and TLR2, known to be present in H37Rv, closely mimics the ability of M. tuberculosis to induce robust IL-23 production and, less efficiently, IL-12 production. The essential role of NOD2 in response to H37Rv was confirmed by the complete absence of production of IL-12, both p75 and p40, in IFN-γ–primed mono-DCs from three Crohn's disease patients homozygous for a frameshift mutation (1,007 fs) of the NOD2 gene (NOD2−/−) (28) stimulated by H37Rv alone (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20071450/DC1).
Although ligands for TLR7/8 have not been identified in M. tuberculosis, mycobacterial DNA is a ligand for TLR9 (23), an MyD88-coupled receptor with signaling properties similar to those of TLR7/8 and expressed on mouse but not human DCs. Because the combination of a NOD2 ligand with the TLR7/8 ligand R848 was much more efficient than the combination with a TLR2 ligand in inducing IL-12 production, the combination of NOD2 ligands with mycobacterial DNA may represent a strong stimulus for IL-12 induction by M. tuberculosis in mouse DCs.
Differential regulation of IL-12 and IL-23 production by zymosan
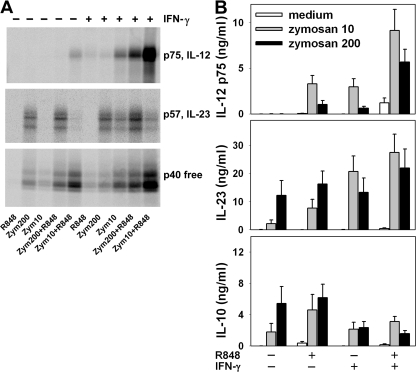
High concentrations of zymosan (200 μg/ml) induced mono-DCs to produce IL-23 and low levels of free p40 but no IL-12 p75, as detected by immunoprecipitation (Fig. 3 A). Lower amounts of zymosan (10 μg/ml) stimulated only free p40 production in mono-DCs. The association of zymosan with R848 induced IL-12 p75 and increased the amount of IL-23 and free p40. Note that the amount of IL-12 p75 detected in response to zymosan at 10 μg/ml combined with R848 was much higher than that observed in response to zymosan at 200 μg/ml combined with R848, whereas the opposite was observed with IL-23. Priming of mono-DCs with IFN-γ enhanced IL-12 p75, IL-23, and free p40 production in response to zymosan, either in the presence or absence of R848. Again, the amount of IL-12 p75 induced by zymosan at 10 μg/ml was consistently higher than that induced by zymosan at 200 μg/ml.
Figure 3.
Differential regulation of IL-12 and IL-23 in mono-DCs stimulated by zymosan. mono-DCs treated with or without IFN-γ were labeled with [35S]methionine (A) or unlabeled (B) and stimulated with 10 or 200 μg/ml zymosan alone or associated with R848. After 18 h, supernatants were immunoprecipitated with anti–IL-12 p35, anti–IL-23 p19, or anti–IL-12/23 p40 mAbs and resolved in nonreducing SDS-PAGE (A) or were tested by ELISA (B) for IL-12 p75, IL-23, and IL-10. Results in A are representative of three independent experiments. Results in B are mean ± SE values from mono-DCs derived from 10 different donors.
Evaluation of cytokine production by ELISA (Fig. 3 B) confirmed that zymosan induced IL-12 p75 production only when used in association with R848 or when mono-DCs were primed with IFN-γ. The amount of IL-12 p75 induced by zymosan at 10 μg/ml was significantly higher than that induced by zymosan at 200 μg/ml, either associated with R848 (P = 0.012) or in IFN-γ–primed cells (P = 0.01). Maximal IL-12 production was observed when IFN-γ–primed mono-DCs were stimulated with both R848 and low-concentration zymosan (10 vs. 200 μg/ml zymosan; P = 0.011). Unlike the observation for IL-12, high levels of IL-23 were induced by zymosan alone at 200 μg/ml, and these levels were not significantly modified by R848 or by IFN-γ priming. At 10 μg/ml, zymosan induced much lower levels of IL-23 than at 200 μg/ml (P = 0.026), but these were significantly increased by R848 co-stimulation (P = 0.01) or IFN-γ priming (P = 0.002). IL-10 production was induced in mono-DCs by zymosan alone at 200 μg/ml or by zymosan at 10 μg/ml in association with R848; production induced at the high concentration of zymosan was reduced when mono-DCs were primed with IFN-γ either in the absence (P = 0.047) or presence (P = 0.016) of R848.
Thus, zymosan is a potent inducer of IL-23 and IL-10 in a dose-dependent way. IFN-γ priming of DCs inhibited IL-10 production induced by zymosan, whereas it had no effect on the induction of IL-23 by high-dose zymosan. Unlike IL-23, IL-12 was poorly induced by zymosan, which paradoxically induced the latter cytokine in IFN-γ–primed mono-DCs only when used at low concentrations. However, zymosan was strongly synergistic with R848 in inducing IL-12, an effect that was again more efficient at lower concentrations of zymosan and was amplified in IFN-γ–primed mono-DCs.
Combination of ligands for dectin-1 and TLR2 mimics IL-23 and IL-12 induction by zymosan
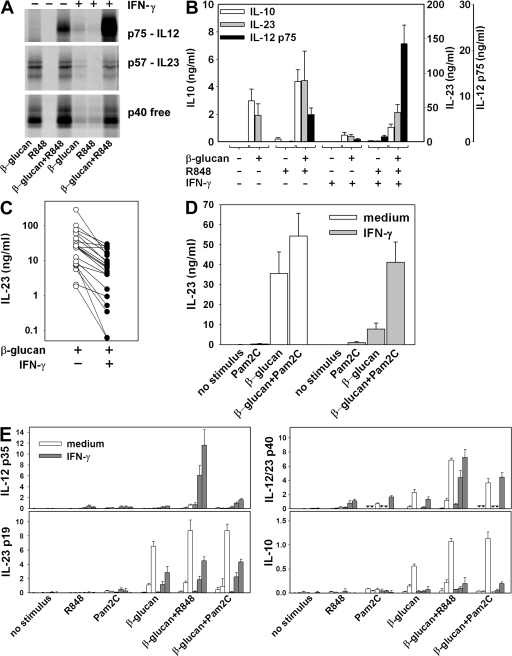
TLR2 and the β-glucan receptor (human dectin-1) have been previously described as the two major PRRs involved in the stimulation of DCs by zymosan (29, 30). To assess the relative contribution of these PRRs, we tested the ability of purified ligands of these receptors to mimic zymosan induction of IL-12 and IL-23 production in mono-DCs. As observed by both immunoprecipitation (Fig. 4 A) and ELISA (Fig. 4 B), β-glucan alone induced production of IL-23 and free p40, but not IL-12 p75, analogous to the pattern observed in zymosan-stimulated mono-DCs. IL-12 p75 production was observed only when mono-DCs were stimulated by β-glucan in association with R848, a condition in which IL-23 and free p40 production were also increased (Fig. 4, A and B). Stimulation of mono-DCs with β-glucan also induced production of IL-10, as previously reported by others (31, 32), and this production was increased by R848 (Fig. 4 B).
Figure 4.
Role of dectin-1 and TLR2 in IL-12 and IL-23 production in mono-DCs. mono-DCs treated with IFN-γ or left untreated were labeled with [35S]methionine (A) or unlabeled (B–E) and stimulated with β-glucan or Pam2C, or with a combination of β-glucan+Pam2C, with or without R848. After 18 h, supernatants were immunoprecipitated with anti–IL-12 p35, anti–IL-23 p19, or anti–IL-12/23 p40 mAbs and resolved in nonreducing SDS-PAGE (A) and tested for IL-23, IL-12 p75, and IL-10 production by ELISA (B–D) and for mRNA accumulation using the QuantiGene multiplex assay (E). Results in A are representative of four independent experiments. Results in B are mean ± SE values from mono-DCs derived from 18 donors. Results in C represent individual IL-23 levels from mono-DCs derived from 21 different donors. Results in D are mean ± SE values from five experiments. Results in E are mean ± SD (n = 3) mRNA accumulation in cells derived from three separate donors and lysed at 3 h (right column of each triplet), 6 h (middle column of each triplet), and 12 h (right column of each triplet). Inverted triangles indicate not done.
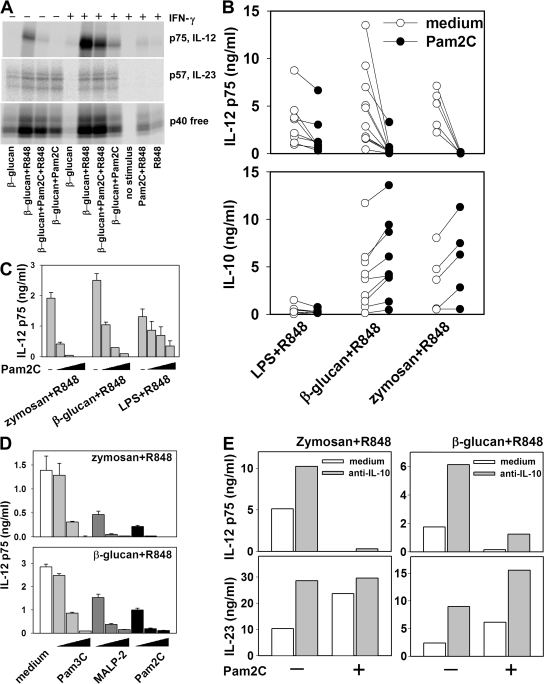
IFN-γ priming of mono-DCs significantly enhanced IL-12 p75 production induced by β-glucan (P = 0.018; Fig. 4, A and B), similar to the observations with zymosan. However, priming of mono-DCs with IFN-γ drastically inhibited IL-23 and IL-10 expression induced by β-glucan (Fig. 4, A and B). IL-10 production was significantly (P < 0.02) lower in IFN-γ–primed DCs in response to β-glucan alone or in combination with either R848 (Fig. 4 B) or Pam2C (not depicted). IL-23 induction by β-glucan was significantly lower in IFN-γ–primed cells from a large panel of donors (P = 0.02; Fig. 4 C). Dectin-1 expression detected by immunofluorescence did not differ between IFN-γ–primed and unprimed mono-DCs (unpublished data), indicating that the reduced IL-23 and IL-10 production in IFN-γ–primed mono-DCs was not dependent on dectin-1 downmodulation. Note that co-stimulation of mono-DCs with β-glucan and Pam2C, a ligand for TLR2 that is the other major receptor involved in the recognition of zymosan, reversed the inhibition of IL-23 production observed after IFN-γ priming (Fig. 4 D).
mRNA accumulation closely paralleled the pattern of protein expression (Fig. 4 E). Transcripts of the gene encoding the IL-12/23 common p40 chain accumulated in response to either single or combined PRR ligands tested. Accumulation of IL-23 p19 and IL-10 transcripts was similarly regulated, with strong accumulation induced by β-glucan and enhanced when either R848 or Pam2C was associated with β-glucan; IFN-γ priming of mono-DCs decreased both IL-23 p19 and IL-10 mRNA accumulation in response to these ligands. Accumulation of IL-12 p35 transcripts was strictly dependent on IFN-γ priming of mono-DCs, with maximal levels in IFN-γ–primed mono-DCs stimulated with a combination of β-glucan and R848. The mRNA accumulation data show that the pattern of IL-12 and IL-23 induction was reflected in accumulation of the IL-12p35 and IL-23p19 transcripts, respectively, but not IL-12/23p40. However, the analysis of mRNA at three different time points showed that the mRNA accumulation peaks earlier when DCs are stimulated with TLR ligands (Fig. 4 E and not depicted) than when they are stimulated or co-stimulated with β-glucan. Although the amount of mRNA accumulation provides important information on the degree and kinetics of the expression of the genes encoding the heterodimeric cytokines, even when analyzed at different time points it may not give a full account of the total amount of transcripts available for translation during the period of stimulation. Thus, the analysis of p35 and p19 mRNA accumulation that has often been used in past reports to evaluate IL-12 and IL-23 production is less representative than the analysis of protein accumulation in the supernatant fluids at the end of the stimulation for evaluating the transcriptional and posttranscriptional regulation of cytokine production.
These results show that unlike high-dose zymosan, stimulation with β-glucan alone induces IL-23 production in nonprimed mono-DCs but does so poorly in IFN-γ–primed DCs. However, the association of β-glucan with Pam2C enhanced IL-23 production by nonprimed mono-DCs of most donors and significantly reversed the inhibition of IL-23 production observed in the IFN-γ–primed cells. Thus, like high doses of zymosan, the combination of dectin-1 and TLR2 ligands results in elevated IL-23 production independent of IFN-γ priming.
TLR2 ligands inhibit IL-12 production by mono-DCs induced by combinations of other PRR ligands
When the combination of β-glucan and R848 was further associated with Pam2C, IL-12 p75 production by untreated or IFN-γ–primed mono-DCs was drastically inhibited and IL-12 p40 was minimally decreased, whereas IL-23 was consistently increased (Fig. 5 A). To further investigate the inhibitory effect of TLR2 ligands on IL-12 p75 production, mono-DCs from several donors were incubated with LPS, β-glucan, or low-dose zymosan in association with R848, three ligand combinations that strongly induced IL-12 p75 production in the presence or absence of Pam2C (Fig. 5 B). Pam2C strongly inhibited IL-12 p75 production induced by zymosan and R848 (P = 0.006) or by β-glucan and R848 (P = 0.007), whereas IL-12 p75 production induced by LPS and R848 was moderately reduced (P = 0.012). In contrast, IL-10 was increased (zymosan+R848, P = 0.019; β-glucan+R848, P = 0.002) or unaffected (LPS+R848) by treatment with Pam2C. The lower sensitivity of IL-12 p75 induced by LPS+R848 to inhibition by TLR2-ligands was confirmed by the observation that 50 ng/ml Pam2C was required for 75% inhibition of the IL-12 p75 induced by LPS+R848, whereas 5 and 0.5 ng/ml were sufficient for >75% inhibition of the IL-12 p75 production induced by β-glucan+R848 and by zymosan+R848, respectively (Fig. 5 C). The three synthetic ligands 2-kD macrophage-activating lipopeptide (MALP-2), Pam2C (both TLR2/TLR6 ligands), and Pam3C (TLR2/TLR1 ligand) all showed a strong, dose-dependent capacity to inhibit IL-12 p75 production induced by zymosan+R848 or by β-glucan+R848. MALP-2 and Pam2C, were, however, more potent than Pam3C (Fig. 5 D).
Figure 5.
TLR2-mediated inhibition of IL-12 p75. (A) mono-DCs treated with IFN-γ or left untreated were labeled with [35S]methionine and stimulated with β-glucan, Pam2C, R848, and their combinations. After 18 h, supernatants were immunoprecipitated with anti–IL-12 p35, anti–IL-23 p19, or anti–IL-12/23 p40 mAbs and resolved in nonreducing SDS-PAGE. (B) Cytokine production was measured by ELISA in mono-DCs 18 h after stimulation with 10 ng/ml LPS+R848, 10 μg/ml β-glucan+R848, or 10 μg/ml zymosan+R848 in the presence or absence of 50 ng/ml Pam2C (each symbol represents a single donor). (C) mono-DCs were stimulated as indicated in B in the presence or absence of 0.5, 5, and 50 ng/ml Pam2C, and IL-12 p75 was measured by ELISA (mean ± SD from triplicate cultures). (D) IL-12 p75 production was assessed by ELISA (mean ± SD from triplicate cultures) in mono-DCs stimulated with 10 μg/ml zymosan+R848 or 10 μg/ml β-glucan+R848 without or with 2, 20, and 200 ng/ml Pam3C; 1, 10, and 100 ng/ml MALP-2; or 0.5, 5, and 50 ng/ml Pam2C. (E) mono-DCs were stimulated with10 μg/ml zymosan+R848 (left) or with 0.1 μg/ml β-glucan+R848 (right) in the absence (open column) or presence (gray column) of 30 μg/ml of the neutralizing anti–IL-10 mAb 19F1, with or without 50 ng/ml Pam2C. Cytokines were measured by ELISA (mean values from duplicate cultures in a representative experiment of six performed with similar results).
The ability of TLR2 ligands to inhibit IL-12 p75 but not IL-23 production induced by β-glucan+R848 might explain the inverse dose–response observed in IL-12 p75 but not IL-23 production induced by zymosan. Indeed, this paradoxical dose dependence might reflect a differential contribution of dectin-1 and TLR2 ligands at different concentrations of zymosan. The dose-dependent production of IL-23 could reasonably be attributed to the presence in zymosan of the dectin-1 ligand β-glucan, an efficient stimulus for IL-23 that is augmented by the combination with TLR2 ligands. The inhibitory activity for IL-12 but not IL-23 production exerted by the TLR2 ligands also present in zymosan could account for the decreased IL-12 production when high-dose zymosan was used in association with R848.
IL-10 is induced by TLR2 ligands but does not account for the inhibition of IL-12 p75 production
Both zymosan and β-glucan induced production of IL-10 and accumulation of IL-10 transcripts in mono-DCs, with enhanced levels upon co-stimulation with either R848 or Pam2C (Fig. 3 B; Fig. 4, B and E; and not depicted). To determine whether autocrine IL-10 might underlie the inhibition of IL-12 p75 and IL-23 production, mono-DCs were stimulated with low-dose zymosan+R848 or β-glucan+R848 in the presence or absence of a neutralizing anti–IL-10 mAb (Fig. 5 E) and/or exogenously added recombinant IL-10 (not depicted). Both IL-23 and IL-12 p75 were inhibited by both exogenous IL-10 (not depicted) and by autocrine IL-10, as demonstrated by the increased production of IL-23 and IL-12 p75 in the presence of the neutralizing anti–IL-10 mAb (Fig. 5 E). The inhibitory effect of Pam2C on IL-12 p75 production was not altered by the neutralizing anti–IL-10 mAb, indicating that IL-10 was not responsible for the inhibition. This conclusion is further supported by the fact that although IL-10 was able to suppress both IL-12 and IL-23 production, Pam2C enhanced IL-23 production while suppressing IL-12 production. An anti–TGF-β neutralizing antibody, alone or together with the anti–IL-10 mAb, also did not prevent Pam2C inhibition of IL-12 p75 production (unpublished data).
Induction of IL-17 and IFN-γ in human naive CD4+ T cells by supernatants of stimulated human mono-DCs
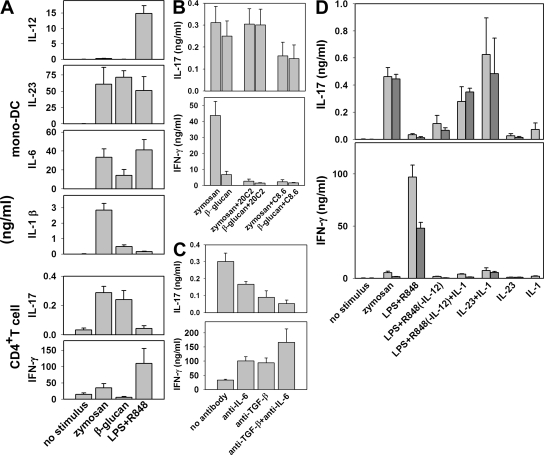
We next investigated whether the supernatants of mono-DCs stimulated in the conditions leading to maximum IL-23 production were able to induce IL-17 production in naive human CD4+ T cells stimulated with anti-CD3 and anti-CD28. Supernatants from zymosan-, β-glucan–, and LPS+R848-treated cells containing approximately equivalent concentrations of IL-23 were tested. Unlike IL-23, IL-12 was undetectable in the supernatant from β-glucan–stimulated cells; instead, IL-12 levels were highest in the LPS+R848 supernatant and low in the zymosan supernatants (Fig. 6 A). The induction of IFN-γ from naive human CD4+ T cells correlated with the IL-12 content in the supernatants, but only the supernatants from zymosan- and β-glucan–stimulated mono-DCs, and not that from LPS+R848-stimulated mono-DCs, induced IL-17 production, although all three supernatants contained IL-23. Antibody 20C2, which blocks the biological activity of IL-12 but not of IL-23, completely inhibited IFN-γ but not IL-17 production, whereas anti–IL-12 p40 antibody C8.6, which inhibits both IL-12 and IL-23, completely inhibited IFN-γ production but only half of IL-17 production (P = 0.0002 and 0.013 for zymosan and β-glucan, respectively; Fig. 6 B). We therefore turned to TGF-β and IL-6. Antibodies neutralizing TGF-β and IL-6, two cytokines shown to be essential for IL-17 production in mice (33), each partially inhibited IL-17 production and, in combination, induced profound inhibition of IL-17, whereas these antibodies, alone or in combination, resulted in increased production of IFN-γ (Fig. 6 C). However, IL-6, like IL-23, was present in all three supernatants. IL-1β, another cytokine potentially involved in IL-17 induction in human CD4+ T cells (34–36), was detected in the zymosan- and β-glucan–stimulated supernatants but at minimal levels in the LPS+R848-stimulated supernatant. The use of IL-1β together with recombinant IL-23 or addition of IL-1β to the supernatant from mono-DCs stimulated with LPS+R848 (and from which IL-12 was removed by affinity chromatography with anti–IL-12 p35 mAb beads) reconstituted IL-17 production in human naive CD4+ T cells (Fig. 6 D). Also, IL-1RA inhibited IL-17 production induced by the supernatants (unpublished data). Thus, although IL-23, TGF-β, and IL-6 all appeared to play a role in the induction of IL-17 by human CD4+ T cells, IL-1β, in combination with one or more of those factors, was essential in the induction of IL-17 production and determined the differential ability of the supernatants from activated mono-DCs to direct IL-17 production by CD4+ T cells.
Figure 6.
Induction of IL-17 and IFN-γ in human CD4+T cells by supernatants of stimulated mono-DCs. Supernatants from differently stimulated mono-DCs were used to induce IL-17 and IFN-γ production from human CD4+ CD45RO− T lymphocytes stimulated with anti-CD3 and anti-CD28. (A) Supernatants from IFN-γ–primed mono-DCs stimulated with 200 μg/ml zymosan or 1 μg/ml LPS+R848, or from unprimed mono-DCs stimulated with 10 μg/ml β-glucan, were evaluated by ELISA for IL-12, IL23, IL-6, and IL-1β production and for the capacity to induce IL-17 and IFN-γ in naive CD4+ T cells (mean ± SE from 10 independent experiments). (B) IL-17 and IFN-γ production was determined by ELISA in naive T cell cultures in the presence of supernatants from zymosan- or β-glucan–stimulated mono-DCs, and in the presence or absence of neutralizing anti–IL-12 p75 (20C2) or anti-p40 (C8.6) mAbs (mean ± SE from seven independent experiments). (C) Effect of neutralizing anti–TGF-β or anti–IL-6, or a mixture of both mAbs on IL-17 and IFN-γ production measured by ELISA in naive T cells cultured in the presence of supernatants from zymosan-stimulated mono-DCs. (D) Effect of 10 ng/ml IL-1β on IL-17 and IFN-γ production, as determined by ELISA in naive T cells cultured in the presence of IL-23 or an IL-12–depleted supernatant from LPS+R848–stimulated mono-DCs. Supernatants were diluted to contain IL-23 at a final concentration of 15 or 1.5 ng/ml (light and dark gray, respectively). Recombinant IL-23 was used at 15 or 1.5 ng/ml (light and dark gray, respectively). C and D show the mean ± SD from triplicate cultures. Similar results were obtained in three independent experiments.
DISCUSSION
The heterodimeric cytokine IL-12 is produced by cells of innate immunity, particularly DCs and macrophages stimulated through PRRs and interaction with NK cells or antigen-activated T cells (5). However, stimulation of individual PRRs is not sufficient to induce optimal production of the IL-12 heterodimer; instead, it induces secretion of a large excess of the free p40 chain (4, 20). In addition to individual PRR stimulation, exposure of the producer cells to cytokines (e.g., IFN-γ and IL-4) (37–39) or co-stimuli (e.g., CD40 ligand) (40), or the association of different TLR ligands (e.g., TLR7/8 or TLR9 ligands together with TLR3 or TLR4 ligands) (18, 20, 21) is required for efficient production of the functional IL-12 heterodimer. These findings indicate that different PRR ligands expressed by pathogens during an infection cooperate in inducing IL-12 production, which is further regulated by soluble products or surface receptor/ligand interaction generated during both the innate and immune response to the infection.
Like IL-12, the cytokine IL-23 is a heterodimer composed of a specific p19 polypeptide linked to the p40 chain shared with IL-12 (2). Previous studies of IL-23 have been hampered by the limited methods available to detect expression at the protein level. SDS-PAGE analysis of immunoprecipitates with the anti–IL-23 p19, anti–IL-12 p35, and anti-p40 mAbs allowed us to analyze the single components of the IL-12–IL-23 complex, including the free p40 chain, and of the assembly of the IL-23 heterodimer. We find that the p19 chain associates as a disulfide-linked heterodimer to the 34-, 41-, and 44-kD glycosylation variants of the IL-12/23 p40 chain, analogous to IL-12 p35 (22). However, our biochemical analysis revealed a different sensitivity to N-linked posttranslational modifications, which are required for IL-12 p75 secretion (22, 41) but are irrelevant for IL-23 secretion. Immunoprecipitation and ELISA analyses indicated some similarities in the regulation of IL-12 and IL-23 production. Both cytokines were efficiently produced by freshly purified peripheral blood DCs and by mono-DCs, whereas lower production was observed in monocytes. Many of the stimuli (e.g., LPS or poly-(I:C); unpublished data) associated with the TLR-7/8 ligand R848, which induced maximal expression of IL-12, also induced expression of IL-23. However, differential IL-12 and IL-23 expression in DCs was triggered by selected associations of PRR ligands that mimicked structures associated with microorganisms. Our data identify a crucial role for IFN-γ, TLR7/8, and TLR2 in determining preferential expression of either IL-12 or IL-23.
Early in our studies, we observed that activation of DCs with two complex microorganisms—heat-killed M. tuberculosis H37Rv strain and the yeast zymosan—efficiently induced IL-23 but little or no IL-12 p75. The association of the TLR7/8 ligand R848 or IFN-γ priming to stimulation of DCs with either H37Rv or zymosan induced strong expression of IL-12 p75, with no major effect on IL-23 production. Thus, the relative pattern of expression of the two cytokines appears to be strictly dependent on the PRRs engaged by different microorganisms, as depicted in the model presented in Fig. S5 (available at http://www.jem.org/cgi/content/full/jem.20071450/DC1).
A protective role for IL-23 in M. tuberculosis infection has recently been described based on its activity in inducing a Th17 response and facilitating a memory T cell response (6, 42–46). The emerging role of IL-23, together with the well-known and possibly predominant role of IL-12 and IFN-γ in the protection against M. tuberculosis, suggests that a balanced production of IL-23 and IL-12 might be crucial in the defense against this intracellular pathogen (46). M. tuberculosis H37Rv induced IL-12 p75 in mono-DCs only after IFN-γ priming and/or co-stimulation with R848, whereas IL-23 was induced by H37Rv alone and was modestly affected by co-stimulation. The differential regulation of IL-12 p75 and IL-23 was even more striking in IFN-γ–primed peripheral blood DCs stimulated by H37Rv, in which R848 co-stimulation not only strongly increased IL-12 p75 production but also inhibited IL-23 production. Among the PRRs known to recognize mycobacteria (23–27), the combined stimulation of TLR2 and NOD2 was described to be required for the production of TNF (25). Our results show that the association of the NOD2 ligand MDP with the TLR2 ligand Pam3C or Pam2C, or with the TLR7/8 ligand R848, induced both IL-12 and IL-23 in IFN-γ–primed mono-DCs with a striking and reproducible preferential activation of IL-12 by the combination of ligands for NOD2 and TLR7/8, and of IL-23 by the combination of ligands for NOD2 and TLR2. The expression pattern of these cytokines induced by the combinations MDP and Pam3C (or Pam2C) or MDP and Pam3C (or Pam2C) associated with R848 was similar to the pattern induced by H37Rv alone or associated with R848, respectively, suggesting that NOD2 and TLR2 acting together are crucial receptors for IL-12 and IL-23 expression in response to M. tuberculosis. DCs from NOD2−/− patients are impaired for IL-12 production in response to H37Rv. In addition to NOD2 and TLR2, M. tuberculosis expresses heat-labile TLR4 ligands that might be involved in cooperating with other PRRs for cytokine induction (27). It is also of interest that in the mouse, TLR9 plays an important role in IL-12 p40 production, whereas TLR2 is mostly responsible for production of TNF (23). Human DCs do not express TLR9 (47), possibly explaining the relative inefficiency of M. tuberculosis to induce IL-12 in human DC cultures. Co-stimulation with R848, a ligand for the well-expressed TLR8 on human DCs, may mimic the effect of the mycobacterial TLR9 ligands on mouse DCs.
Like M. tuberculosis, the yeast zymosan induced IL-23 production in the absence of IL-12 p75. Both M. tuberculosis and zymosan also required co-stimulation of DCs with R848 or IFN-γ, or their association, for IL-12 p75 induction. The combination of a TLR2 ligand (Pam2C) and the β-glucan receptor/dectin-1 ligand β-glucan, which are the most important PRR ligands known to be present in zymosan, allowed us to identify TLR2 as a key PRR that differentially regulates the expression of IL-12 and IL-23. β-glucan induced very high-level IL-23 production in DCs, consistent with results recently reported for mouse DCs (48), and production was increased two- to threefold by co-stimulation with TLR2 or TLR7/8 ligands. However, production of IL-12 p75 was only induced by β-glucan and R848 together and not by β-glucan alone or in combination with TLR2 ligands. Furthermore, the IL-12 production induced by β-glucan and R848, unlike that of IL-23, was drastically inhibited by the addition of TLR2 ligands. Interestingly, IL-23 was preferentially induced by high-dose zymosan, whereas IL-12 was more efficiently induced by low-dose zymosan in combination with R848 and/or IFN-γ. This paradoxical inverse dose dependence is likely explained by the differential contribution of dectin-1 and TLR2 ligands when zymosan was used at low or high doses.
IL-10, which inhibits IL-12 production (49), was also induced by zymosan (31, 32), and its production was enhanced by TLR2 ligands, consistent with previous reports (50–52). However, despite efficient inhibition of IL-23 production by IL-10, Pam2C inhibited IL-23 production even in the presence of a neutralizing anti–IL-10 mAb, excluding IL-10 as a major mechanism mediating the inhibition of IL-12 p75 by TLR2 ligands. Previous papers described the promotion of Th2 and T regulatory cell responses by TLR2 ligands via IL-10 production (52, 53). IL-12 p75 favors Th-1 responses and prevents the differentiation of other Th subsets, including the Th17 subset, promoted by IL-23. Thus, our results indicating the IL-10–independent inhibition of IL-12 p75 and enhancement of IL-23 production by TLR2 add a new mechanism by which engagement of this receptor can modulate the Th responses.
IFN-γ enhances transcription of genes encoding both p40 and p35, and it has a marked effect on production of the IL-12 p75 (54). Thus, it seemed reasonable to hypothesize a similar enhancing effect of IFN-γ on the IL-23 heterodimer. Indeed, IFN-γ was required for optimal IL-12 and IL-23 production in mono-DCs stimulated by LPS and in both freshly isolated DCs and mono-DCs stimulated by H37Rv alone or in association with the TLR7/8 ligand R848. However, the enhancing effect of IFN-γ on IL-23 production was dependent on the nature of the PRR triggered, because β-glucan–induced IL-23 production, unlike that of IL-12, was effectively inhibited at both the protein and mRNA levels by IFN-γ. On the other hand, IFN-γ enhanced IL-23 induction by low dose zymosan. Because TLR2 ligands increased IL-23 production in both IFN-γ–primed and unprimed cells stimulated by β-glucan, it is possible that the different results obtained using β-glucan or zymosan rest in the engagement of TLR2 or other PRRs by ligands present in the yeast. IFN-γ inhibited the production of IL-10, as previously reported by others using different inducers (55–57). The mechanism by which IFN-γ inhibits IL-10 involves the GSK3 and CREB/AP1 transcription factors (57). The finding that both IL-23 and IL-10 were inhibited by IFN-γ raises the possibility of similarities in the pathways by which dectin-1 induces the two genes.
A dissociated production of IL-12 and IL-23 was previously reported for Candida yeasts that induced both IL-12 and IL-23 compared with the hyphal form that induced only IL-23 (58). Those data are consistent with previous data in the mouse (59) but not with others reported for human DCs, indicating an inability of the hyphal form to induce IL-12 (60). Because β-glucan is exposed only in the yeast and not in the filamentous form (61), it is possible that receptors other than dectin-1 play a major role in IL-23 induction by Candida. However, the ability of zymosan to induce both IL-23 and IL-12 was blocked by laminarin (unpublished data), strongly suggesting a major role for the β-glucan receptor in the induction of both cytokines by the yeast zymosan. Evidence suggesting an important role for the IL-23–IL-17 axis, as well as for dectin-1, in antifungal resistance is rapidly emerging (58, 62–65).
The IL-23–containing supernatants from human DCs stimulated by zymosan or β-glucan induced production of IL-17 in naive human CD4+ T cells stimulated polyclonally. IL-17 production was not affected or slightly enhanced by an antibody neutralizing IL-12, whereas it was diminished by an antibody to IL-12 p40, neutralizing both IL-12 and IL-23. Surprisingly, the supernatant of cells stimulated with LPS+R848 that also contained IL-23 failed to induce IL-17 production. We found that this inability of the LPS+R848-induced supernatant to induce IL-17 production was caused by the absence of high IL-1β concentrations that were present in the zymosan- and β-glucan–induced supernatants. IL-1 was required for IL-17 induction in human CD4+ cells in combination with one or more of the cytokines IL-23, TGF-β, and IL-6, as also suggested by other studies (34–36), and the level of IL-1 produced by the various stimuli used in our study was responsible for the differential induction of IL-17 production. The ability of the supernatants to induce IFN-γ production in CD4+ T cells correlated with their content of IL-12, but not of IL-23, and was completely blocked by antibodies neutralizing IL-12 p75, thus excluding a major role for IL-23 in IFN-γ production. IL-1β is also required for production of IFN-γ by human T cells stimulated by IL-12 (49); however, much lower concentrations of IL-1β appear to be required for this effect than those required for IL-17 production, and sufficient IL-1 is probably present in all the supernatants tested or produced in the T cell cultures to allow IFN-γ production. The anti–TGF-β antibody inhibits IL-17 production. Previous studies (35, 36) failed to observe an effect of added TGF-β on the differentiation of human Th17, suggesting that endogenous TGF-β present in the cultures was sufficient for permitting IL-17 production, whereas additional exogenous TGF-β had no effect or even inhibited IL-17. The requirement for TGF-β in IL-17 production might be partially caused by its inhibiting effect on IFN-γ production, in addition to a direct effect on T cell differentiation.
The separate regulation of IL-12 and IL-23 through different associations of PRRs might lead to a preferential production of IL-23 responsible for the chronic inflammation and the Th17 response associated with this cytokine. In particular, we found that production of IL-23 is either enhanced or inhibited by IFN-γ depending on the receptor involved in the induction of IL-23. This may have important consequences in different types of infections, e.g., in a mycobacterial infection in which the presence of a strong Th1 response with high IFN-γ levels may not prevent the simultaneous or successive production of IL-23, although it would likely prevent a full-fledged Th17 response by acting at the T cell level. In fungal infections, the presence of a strong Th1 response may also prevent the effective dectin-1–mediated stimulation of IL-23 production and the generation of Th17 cells acting at the APC level. We also find that ligands for both TLR1/2 and TLR2/6, when associated with stimulation through NOD2 or dectin-1, preferentially induce IL-23 rather than IL-12, and when associated with several ligand combinations that induce IL-12 very efficiently, block IL-12 production. The very contrasting effect of the different TLR ligands is exemplified by β-glucan, which alone or in association with TLR2 ligands induces only IL-23 production, whereas its association with R848 represents the most powerful IL-12–inducing stimulus reported thus far, even more potent then the combination of LPS and R848. In addition to our data indicating that IL-23 and IL-12 are differentially regulated by various PRR ligands expressed by microorganisms, we also find that the differential induction of IL-1 production may profoundly affect the production of IL-17 and the Th17 responses. We conclude that both pathogen-intrinsic factors and regulatory components of the inflammatory and immune responses likely affect the nature of the initial innate and immune responses to the infection, as well as their evolution when the infection progresses or resolves.
MATERIALS AND METHODS
Antibodies.
The following antibodies were affinity purified on Sepharose–protein G (GE Healthcare) and covalently immobilized at 1–6 mg/g of CNBr-activated Sepharose 4B beads (GE Healthcare): C11.79 mAb and C8.6 mAb, recognizing the IL-12 β chain (p40) (38); 12H4 mAb, recognizing the IL-12 α chain (p35; provided by S. Wolf, Wyeth Pharmaceuticals, Cambridge, MA); and PAB 512 (7G10) and PAB 187 mouse anti–human IL-23 p19 mIgG1 and mIgG2a, respectively, recognizing the p19 of the IL-23 heterodimer (2). To increase binding capacity, C11.79 and C8.6 mAbs or 512 and 187 mAbs were coupled as a mixture. 30 μg/ml of the neutralizing anti–IL-10 19F1.1 mAb (provided by K. Moore, Schering-Plough Biopharma, Palo Alto, CA), 10 μg/ml anti–TGF-β 1D11 mAb (provided by F. Ruscetti, National Cancer Institute, Frederick, MD), and goat anti–human IL-6 antiserum (1:100) were used in naive T cell cultures.
Reagents.
The following reagents were used: 10 ng/ml LPS (or as indicated in the figure legends; Sigma-Aldrich), 1 μM R848 (InvivoGen), 10 μg/ml β-glucan (from baker's yeast; Sigma-Aldrich), 200 ng/ml Pam3CSK4 (or as indicated in the figure legends; InvivoGen), 50 ng/ml Pam2CSK4 (or as indicated in the figure legends; InvivoGen), MALP-2 (Qbiogene), 10 or 200 μg/ml zymosan (from Saccharomyces cerevisiae; InvivoGen), and 10 μg/ml MDP (InvivoGen). Heat-killed M. tuberculosis H37Rv was prepared as follows: bacteria were grown in standing cultures in Middlebrook 7H9 broth (BD Biosciences) supplemented with albumin–dextrose complex and 0.05% Tween 80 until the mid-log phase. Cultures were heated at 80°C for 1 h in a water bath, and bacteria were harvested by centrifugation. After three washes in RPMI 1640 medium, the pellet was resuspended at a concentration of 5 mg/ml (wet weight) in RPMI 1640. The bacterial suspension was divided into aliquots and stored frozen at −80°C until use at a 1:15 dilution.
Cells and cell cultures.
PBMCs obtained from healthy human donors were separated by Ficoll-Paque density gradient centrifugation. Leukopaks of peripheral blood from healthy donors were collected according to the National Institutes of Health approved institutional review board protocols, or were obtained from the blood bank of the University of Verona as discarded material after preparation of therapeutic blood products. Monocytes were enriched from PBMCs by depletion of T cells using E-rosetting with neuraminidase-treated sheep red blood cells and density gradient separation on Ficoll-Paque. The enriched cell suspension contained >70% monocytes, as evaluated by direct immunofluorescence with anti-CD14 mAb (Becton Dickinson). Cells were resuspended in RPMI 1640 medium supplemented with 10% heat-inactivated, low-endotoxin FCS and l-glutamine, and were treated for 24 h with 12 ng/ml IL-4 (108 U/mg; Schering-Plough Biopharma) and for 18 h with 1,000 U/ml IFN-γ (provided by Roussel Uclaf). mono-DCs were obtained by a 5-d culture of plastic-adherent PBMCs in medium with 12 ng/ml IL-4 and 50 ng/ml GM-CSF (107 U/mg; Schering-Plough Biopharma) and by an 18-h culture with or without 1,000 U/ml IFN-γ. CD1c+ DCs were obtained from PBMCs by depletion of CD19+ B lymphocytes with CD19 microbeads, followed by positive selection with CD1c-biotin and antibiotin microbeads, according to the manufacturer's procedure (Miltenyi Biotec). Purified cells (>98% CD1c+), as evaluated by labeling cells with streptavidin-PE, were treated with or without IFN-γ for 4 h.
CD4+, CD45RO− naive T lymphocytes were obtained from PBMCs by negative depletion using a naive CD4+ T cell enrichment kit, according to the manufacture's procedure (StemCell Technologies Inc.). CD4+, CD45RO− (>99% CD4+ and <2% CD45RO+) cells were stimulated with plastic-bound anti-CD3 mAb (eBioscience) and anti-CD28 in the presence of supernatants from variously stimulated mono-DCs. After 5 d, T cells were washed and stimulated with soluble anti-CD3 mAb and 10 ng/ml PMA. Supernatants were collected after 18 h.
NOD2 genotyping.
Three Crohn's disease patients homozygous for the NOD2 L1007fs variant were selected among the patient group described in Giachino et al. (28). In healthy controls, the presence of the major Crohn's disease–associated variants R702W, G908R, and L1007fs was excluded, as previously described (28). The study was conducted under the approval of the ethical review board of San Giovanni Battista Hospital of Torino. Written informed consent in accordance with the Declaration of Helsinki was obtained from each participant.
[35S]Methionine radiolabeling.
For continuous labeling with [35S]methionine, monocyte-enriched PBMCs, mono-DCs, and CD1c+ DCs were resuspended in methionine-free RPMI 1640 supplemented with 10% FCS and 7% normal RPMI 1640 as a source of cold methionine, and were incubated for 18 h with 60 μCi/ml [35S]methionine (1,200 Ci/mmol; NEN-DuPont) and stimuli. In some experiments, IFN-γ–primed mono-DCs were preincubated for 1 h with tunicamycin (TM) (22) at 2 μg/ml in methionine-free medium, followed by incubation for 17 h with [35S]methionine and LPS in the presence of 2 μg/ml TM. The viability of TM-treated cells, as determined by Trypan blue exclusion, did not differ from that of control cells and was always >95%. Radiolabeled antigens were isolated and purified as described in the following paragraphs.
Immunoprecipitation of radiolabeled IL-12 and IL-23.
All procedures (66) were performed at 4°C. Cell-culture supernatants were recovered, centrifuged to eliminate residual cells, and immunoprecipitated after the addition of a 10% TBS solution (10 mM Tris-HCl [pH 8.2], 150 mM NaCl, and 0.02% NaN3) containing 1% Nonidet P-40 and 10 μg/ml leupeptin (Sigma-Aldrich), 10 μg/ml antipain (Sigma-Aldrich), 2 mM EDTA, and 2 mM iodoacetamide as protease inhibitors. Radiolabeled cells were washed by centrifugation in PBS, and the cell pellet was resuspended in TBS solution containing 1% Nonidet P-40 and protease inhibitors. After a 1-h incubation, lysates were centrifuged at 15,000 g for an additional 10 min. Supernatants and cell lysates were precleared with Sepharose–protein A and incubated with Sepharose-immobilized mAbs for 2 h with shaking. mAb-coupled Sepharose beads were recovered by centrifugation and washed sequentially with TBS/0.1% Nonidet P-40 (four times) and with 10 mM Tris-HCl (pH 8.2) containing 0.1% Nonidet P-40 (twice). Immunoprecipitated materials were eluted from the beads by heating at 100°C for 4 min in SDS-PAGE sample buffer.
SDS-PAGE and isolation of proteins.
[35S]methionine-labeled specific immunoprecipitates were separated by SDS-PAGE, conducted essentially as previously described (67). Relevant bands were identified by autoradiography (24–48 h), the appropriate gel region was cut out, and single polypeptides were eluted for 24–36 h in three steps with 250 μl PBS containing 0.2% SDS and 50 mM dithiothreitol. Sample glycoproteins were boiled for 4 min, alkylated in the dark with 130 mM iodoacetamide at 37°C for 45 min, precipitated (66) with trichloroacetic acid (final concentration = 12% wt/vol) for 4 h at 4°C, and recovered by centrifugation at 14,000 g for 5 min on a microfuge. Protein pellets were incubated with cold acetone at −20°C for 2 h, washed three times with cold acetone in a microfuge (14,000 g for 5 min), and vacuum dried and resolubilized in SDS-PAGE or two-dimensional peptide mapping sample buffer (68).
Two-dimensional peptide mapping.
This was conducted essentially as previously described (68), with minor modifications (22). Silica gel plates were dried, exposed to a phosphor screen (Kodak) for 25–45 d, and developed on a PhosphorImager (Molecular Dynamics).
Detection of cytokine production by ELISA.
50 × 103 IFN-γ–primed and unprimed mono-DCs per well were treated with different stimuli or left untreated. After 18 h, supernatants were collected and tested in ELISA for IL-12p75 (Bender), IL-23 (eBioscience), and IL-10 (Bender) production. Supernatants from T cells were evaluated for IFN-γ (B133.1 and B133.5 mAbs) and IL-17 (Invitrogen) production.
Quantitation of mRNA accumulation.
mRNA accumulation was analyzed using the QuantiGene multiplex assay (Panomics). mono-DCs were lysed 3, 6, and 12 h after treatment in lysis mixture. Transcript expression in cell lysates was detected and quantitated according to the manufacturer's instructions. Expression of target-specific RNA molecules was calculated as the mean values from triplicate cultures and normalized against peptidylprolyl isomerase B (cyclophilin B).
Statistical analysis.
Results were compared using a two-tailed paired Student's t test. Differences were considered significant at P < 0.05.
Online supplemental material.
The supplemental figures provide information on the assembly of IL-12 and IL-23 (Fig. S1), the effect of glycosylation on the secretion of IL-12 and IL-23 (Fig. S2), the production of IL-12 and IL-23 in CD1c+ DCs and monocyte-enriched PBMCs (Fig. S3), the production of IL-12 p75 and p40 in NOD2−/− Crohn's disease patients (Fig. S4), and a model for the differential regulation of IL-12 and IL-23 production in mono-DCs by M. tuberculosis or zymosan (Fig. S5). Supplemental results and discussion provides further information regarding Figs. S1 and S2. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071450/DC1.
Supplementary Material
Acknowledgments
We thank NOD2−/− Crohn's disease patients for donating blood and Ms. Marina Hoffman for editing.
This work was supported in part by grants from Fondazione Cariverona (bandi 2004 and 2006) and by the intramural program of the Nation Cancer Institute.
Schering-Plough Biopharma is supported by the Schering-Plough Corporation. The authors have no other conflicting financial interests.
Abbreviations used: MALP-2, 2-kD macrophage-activating lipopeptide; MDP, muramyl dipeptide; mono-DC, monocyte-derived DC; NOD2, nucleotide-binding oligodimerization domain; PRR, pattern recognition receptor; TLR, Toll-like receptor.
References
- 1.Kobayashi, M., L. Fitz, M. Ryan, R.M. Hewick, S.C. Clark, S. Chan, R. Loudon, F. Sherman, B. Perussia, and G. Trinchieri. 1989. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 170:827–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oppmann, B., R. Lesley, B. Blom, J.C. Timans, Y. Xu, B. Hunte, F. Vega, N. Yu, J. Wang, K. Singh, et al. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 13:715–725. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, A.M., and S.A. Khader. 2007. IL-12p40: an inherently agonistic cytokine. Trends Immunol. 28:33–38. [DOI] [PubMed] [Google Scholar]
- 4.D'Andrea, A., M. Rengaraju, N.M. Valiante, J. Chehimi, M. Kubin, M. Aste, S.H. Chan, M. Kobayashi, D. Young, E. Nickbarg, et al. 1992. Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med. 176:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133–146. [DOI] [PubMed] [Google Scholar]
- 6.Filipe-Santos, O., J. Bustamante, A. Chapgier, G. Vogt, L. de Beaucoudrey, J. Feinberg, E. Jouanguy, S. Boisson-Dupuis, C. Fieschi, C. Picard, and J.L. Casanova. 2006. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin. Immunol. 18:347–361. [DOI] [PubMed] [Google Scholar]
- 7.Cua, D.J., J. Sherlock, Y. Chen, C.A. Murphy, B. Joyce, B. Seymour, L. Lucian, W. To, S. Kwan, T. Churakova, et al. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748. [DOI] [PubMed] [Google Scholar]
- 8.Langrish, C.L., Y. Chen, W.M. Blumenschein, J. Mattson, B. Basham, J.D. Sedgwick, T. McClanahan, R.A. Kastelein, and D.J. Cua. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy, C.A., C.L. Langrish, Y. Chen, W. Blumenschein, T. McClanahan, R.A. Kastelein, J.D. Sedgwick, and D.J. Cua. 2003. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 198:1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langowski, J.L., X. Zhang, L. Wu, J.D. Mattson, T. Chen, K. Smith, B. Basham, T. McClanahan, R.A. Kastelein, and M. Oft. 2006. IL-23 promotes tumour incidence and growth. Nature. 442:461–465. [DOI] [PubMed] [Google Scholar]
- 11.Hue, S., P. Ahern, S. Buonocore, M.C. Kullberg, D.J. Cua, B.S. McKenzie, F. Powrie, and K.J. Maloy. 2006. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 203:2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullberg, M.C., D. Jankovic, C.G. Feng, S. Hue, P.L. Gorelick, B.S. McKenzie, D.J. Cua, F. Powrie, A.W. Cheever, K.J. Maloy, and A. Sher. 2006. IL-23 plays a key role in Helicobacter hepaticus–induced T cell–dependent colitis. J. Exp. Med. 203:2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlig, H.H., B.S. McKenzie, S. Hue, C. Thompson, B. Joyce-Shaikh, R. Stepankova, N. Robinson, S. Buonocore, H. Tlaskalova-Hogenova, D.J. Cua, and F. Powrie. 2006. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 25:309–318. [DOI] [PubMed] [Google Scholar]
- 14.Yen, D., J. Cheung, H. Scheerens, F. Poulet, T. McClanahan, B. McKenzie, M.A. Kleinschek, A. Owyang, J. Mattson, W. Blumenschein, et al. 2006. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 116:1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duerr, R.H., K.D. Taylor, S.R. Brant, J.D. Rioux, M.S. Silverberg, M.J. Daly, A.H. Steinhart, C. Abraham, M. Regueiro, A. Griffiths, et al. 2006. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 314:1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135–145. [DOI] [PubMed] [Google Scholar]
- 17.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. [DOI] [PubMed] [Google Scholar]
- 18.Whitmore, M.M., M.J. DeVeer, A. Edling, R.K. Oates, B. Simons, D. Lindner, and B.R. Williams. 2004. Synergistic activation of innate immunity by double-stranded RNA and CpG DNA promotes enhanced antitumor activity. Cancer Res. 64:5850–5860. [DOI] [PubMed] [Google Scholar]
- 19.Trinchieri, G., and A. Sher. 2007. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7:179–190. [DOI] [PubMed] [Google Scholar]
- 20.Gautier, G., M. Humbert, F. Deauvieau, M. Scuiller, J. Hiscott, E.E. Bates, G. Trinchieri, C. Caux, and P. Garrone. 2005. A type I interferon autocrine–paracrine loop is involved in Toll-like receptor–induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med. 201:1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Napolitani, G., A. Rinaldi, F. Bertoni, F. Sallusto, and A. Lanzavecchia. 2005. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carra, G., F. Gerosa, and G. Trinchieri. 2000. Biosynthesis and posttranslational regulation of human IL-12. J. Immunol. 164:4752–4761. [DOI] [PubMed] [Google Scholar]
- 23.Bafica, A., C.A. Scanga, C.G. Feng, C. Leifer, A. Cheever, and A. Sher. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202:1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brightbill, H.D., D.H. Libraty, S.R. Krutzik, R.B. Yang, J.T. Belisle, J.R. Bleharski, M. Maitland, M.V. Norgard, S.E. Plevy, S.T. Smale, et al. 1999. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 285:732–736. [DOI] [PubMed] [Google Scholar]
- 25.Ferwerda, G., S.E. Girardin, B.J. Kullberg, L. Le Bourhis, D.J. de Jong, D.M. Langenberg, R. van Crevel, G.J. Adema, T.H. Ottenhoff, J.W. Van der Meer, and M.G. Netea. 2005. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 1:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, S., S. Uematsu, S. Akira, and P. Salgame. 2004. IL-6 and IL-10 induction from dendritic cells in response to Mycobacterium tuberculosis is predominantly dependent on TLR2-mediated recognition. J. Immunol. 173:3392–3397. [DOI] [PubMed] [Google Scholar]
- 27.Means, T.K., S. Wang, E. Lien, A. Yoshimura, D.T. Golenbock, and M.J. Fenton. 1999. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. 163:3920–3927. [PubMed] [Google Scholar]
- 28.Giachino, D., M.M. van Duist, S. Regazzoni, D. Gregori, M. Bardessono, P. Salacone, N. Scaglione, R. Sostegni, N. Sapone, F. Bresso, et al. 2004. Analysis of the CARD15 variants R702W, G908R and L1007fs in Italian IBD patients. Eur. J. Hum. Genet. 12:206–212. [DOI] [PubMed] [Google Scholar]
- 29.Gantner, B.N., R.M. Simmons, S.J. Canavera, S. Akira, and D.M. Underhill. 2003. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197:1107–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, G.D., J. Herre, D.L. Williams, J.A. Willment, A.S. Marshall, and S. Gordon. 2003. Dectin-1 mediates the biological effects of β-glucans. J. Exp. Med. 197:1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi, H., T.L. Denning, and L. Soong. 2003. Differential induction of interleukin-10 and interleukin-12 in dendritic cells by microbial toll-like receptor activators and skewing of T-cell cytokine profiles. Infect. Immun. 71:3337–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers, N.C., E.C. Slack, A.D. Edwards, M.A. Nolte, O. Schulz, E. Schweighoffer, D.L. Williams, S. Gordon, V.L. Tybulewicz, G.D. Brown, and C. Reis e Sousa. 2005. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 22:507–517. [DOI] [PubMed] [Google Scholar]
- 33.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 34.van Beelen, A.J., Z. Zelinkova, E.W. Taanman-Kueter, F.J. Muller, D.W. Hommes, S.A. Zaat, M.L. Kapsenberg, and E.C. de Jong. 2007. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 27:660–669. [DOI] [PubMed] [Google Scholar]
- 35.Acosta-Rodriguez, E.V., G. Napolitani, A. Lanzavecchia, and F. Sallusto. 2007. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 8:942–949. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, N.J., K. Boniface, J.R. Chan, B.S. McKenzie, W.M. Blumenschein, J.D. Mattson, B. Basham, K. Smith, T. Chen, F. Morel, et al. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957. [DOI] [PubMed] [Google Scholar]
- 37.Hayes, M.P., J. Wang, and M.A. Norcross. 1995. Regulation of interleukin-12 expression in human monocytes: selective priming by interferon-gamma of lipopolysaccharide-inducible p35 and p40 genes. Blood. 86:646–650. [PubMed] [Google Scholar]
- 38.D'Andrea, A., X. Ma, M. Aste-Amezaga, C. Paganin, and G. Trinchieri. 1995. Stimulatory and inhibitory effects of interleukin (IL)-4 and IL-13 on the production of cytokines by human peripheral blood mononuclear cells: priming for IL-12 and tumor necrosis factor α production. J. Exp. Med. 181:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, X., J.M. Chow, G. Gri, G. Carra, F. Gerosa, S.F. Wolf, R. Dzialo, and G. Trinchieri. 1996. The interleukin 12 p40 gene promoter is primed by interferon γ in monocytic cells. J. Exp. Med. 183:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz, O., D.A. Edwards, M. Schito, J. Aliberti, S. Manickasingham, A. Sher, and C. Reis e Sousa. 2000. CD40 triggering of heterodimeric IL-12 p70 production by dendritic cells in vivo requires a microbial priming signal. Immunity. 13:453–462. [DOI] [PubMed] [Google Scholar]
- 41.Murphy, F.J., M.P. Hayes, and P.R. Burd. 2000. Disparate intracellular processing of human IL-12 preprotein subunits: atypical processing of the P35 signal peptide. J. Immunol. 164:839–847. [DOI] [PubMed] [Google Scholar]
- 42.Fletcher, H.A. 2007. Correlates of immune protection from tuberculosis. Curr. Mol. Med. 7:319–325. [DOI] [PubMed] [Google Scholar]
- 43.Khader, S.A., G.K. Bell, J.E. Pearl, J.J. Fountain, J. Rangel-Moreno, G.E. Cilley, F. Shen, S.M. Eaton, S.L. Gaffen, S.L. Swain, et al. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8:369–377. [DOI] [PubMed] [Google Scholar]
- 44.Umemura, M., A. Yahagi, S. Hamada, M.D. Begum, H. Watanabe, K. Kawakami, T. Suda, K. Sudo, S. Nakae, Y. Iwakura, and G. Matsuzaki. 2007. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis Bacille Calmette-Guerin infection. J. Immunol. 178:3786–3796. [DOI] [PubMed] [Google Scholar]
- 45.Wozniak, T.M., A.A. Ryan, and W.J. Britton. 2006. Interleukin-23 restores immunity to Mycobacterium tuberculosis infection in IL-12p40-deficient mice and is not required for the development of IL-17-secreting T cell responses. J. Immunol. 177:8684–8692. [DOI] [PubMed] [Google Scholar]
- 46.Khader, S.A., J.E. Pearl, K. Sakamoto, L. Gilmartin, G.K. Bell, D.M. Jelley-Gibbs, N. Ghilardi, F. deSauvage, and A.M. Cooper. 2005. IL-23 compensates for the absence of IL-12p70 and is essential for the IL-17 response during tuberculosis but is dispensable for protection and antigen-specific IFN-gamma responses if IL-12p70 is available. J. Immunol. 175:788–795. [DOI] [PubMed] [Google Scholar]
- 47.Kadowaki, N., S. Ho, S. Antonenko, R.W. Malefyt, R.A. Kastelein, F. Bazan, and Y.J. Liu. 2001. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 194:863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeibundGut-Landmann, S., O. Gross, M.J. Robinson, F. Osorio, E.C. Slack, S.V. Tsoni, E. Schweighoffer, V. Tybulewicz, G.D. Brown, J. Ruland, and C. Reis e Sousa. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 8:630–638. [DOI] [PubMed] [Google Scholar]
- 49.D'Andrea, A., M. Aste-Amezaga, N.M. Valiante, X. Ma, M. Kubin, and G. Trinchieri. 1993. Interleukin-10 inhibits human lymphocyte IFN-γ production by suppressing natural killer cell stimulatory factor/interleukin-12 synthesis in accessory cells. J. Exp. Med. 178:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Re, F., and J.L. Strominger. 2004. IL-10 released by concomitant TLR2 stimulation blocks the induction of a subset of Th1 cytokines that are specifically induced by TLR4 or TLR3 in human dendritic cells. J. Immunol. 173:7548–7555. [DOI] [PubMed] [Google Scholar]
- 51.Netea, M.G., R. Sutmuller, C. Hermann, C.A. Van der Graaf, J.W. Van der Meer, J.H. van Krieken, T. Hartung, G. Adema, and B.J. Kullberg. 2004. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J. Immunol. 172:3712–3718. [DOI] [PubMed] [Google Scholar]
- 52.Dillon, S., A. Agrawal, T. Van Dyke, G. Landreth, L. McCauley, A. Koh, C. Maliszewski, S. Akira, and B. Pulendran. 2004. A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J. Immunol. 172:4733–4743. [DOI] [PubMed] [Google Scholar]
- 53.Sutmuller, R.P., M.H. den Brok, M. Kramer, E.J. Bennink, L.W. Toonen, B.J. Kullberg, L.A. Joosten, S. Akira, M.G. Netea, and G.J. Adema. 2006. Toll-like receptor 2 controls expansion and function of regulatory T cells. J. Clin. Invest. 116:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, J., S. Cao, L.M. Herman, and X. Ma. 2003. Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-γ–primed IL-12 production by IFN regulatory factor 1. J. Exp. Med. 198:1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donnelly, R.P., S.L. Freeman, and M.P. Hayes. 1995. Inhibition of IL-10 expression by IFN-gamma upregulates transcription of TNF-alpha in human monocytes. J. Immunol. 155:1420–1427. [PubMed] [Google Scholar]
- 56.Flores, R.R., K.A. Diggs, L.M. Tait, and P.A. Morel. 2007. IFN-gamma negatively regulates CpG-induced IL-10 in bone marrow-derived dendritic cells. J. Immunol. 178:211–218. [DOI] [PubMed] [Google Scholar]
- 57.Hu, X., P.K. Paik, J. Chen, A. Yarilina, L. Kockeritz, T.T. Lu, J.R. Woodgett, and L.B. Ivashkiv. 2006. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 24:563–574. [DOI] [PubMed] [Google Scholar]
- 58.Acosta-Rodriguez, E.V., L. Rivino, J. Geginat, D. Jarrossay, M. Gattorno, A. Lanzavecchia, F. Sallusto, and G. Napolitani. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646. [DOI] [PubMed] [Google Scholar]
- 59.d'Ostiani, C.F., G. Del Sero, A. Bacci, C. Montagnoli, A. Spreca, A. Mencacci, P. Ricciardi-Castagnoli, and L. Romani. 2000. Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans: implications for initiation of T helper cell immunity in vitro and in vivo. J. Exp. Med. 191:1661–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romagnoli, G., R. Nisini, P. Chiani, S. Mariotti, R. Teloni, A. Cassone, and A. Torosantucci. 2004. The interaction of human dendritic cells with yeast and germ-tube forms of Candida albicans leads to efficient fungal processing, dendritic cell maturation, and acquisition of a Th1 response-promoting function. J. Leukoc. Biol. 75:117–126. [DOI] [PubMed] [Google Scholar]
- 61.Gantner, B.N., R.M. Simmons, and D.M. Underhill. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang, W., L. Na, P.L. Fidel, and P. Schwarzenberger. 2004. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J. Infect. Dis. 190:624–631. [DOI] [PubMed] [Google Scholar]
- 63.Kleinschek, M.A., U. Muller, S.J. Brodie, W. Stenzel, G. Kohler, W.M. Blumenschein, R.K. Straubinger, T. McClanahan, R.A. Kastelein, and G. Alber. 2006. IL-23 enhances the inflammatory cell response in Cryptococcus neoformans infection and induces a cytokine pattern distinct from IL-12. J. Immunol. 176:1098–1106. [DOI] [PubMed] [Google Scholar]
- 64.Saijo, S., N. Fujikado, T. Furuta, S.H. Chung, H. Kotaki, K. Seki, K. Sudo, S. Akira, Y. Adachi, N. Ohno, et al. 2007. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat. Immunol. 8:39–46. [DOI] [PubMed] [Google Scholar]
- 65.Taylor, P.R., S.V. Tsoni, J.A. Willment, K.M. Dennehy, M. Rosas, H. Findon, K. Haynes, C. Steele, M. Botto, S. Gordon, and G.D. Brown. 2007. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 8:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson, D.J., and G. Blobel. 1983. Immunoprecipitation of proteins from cell-free translations. Methods Enzymol. 96:111–120. [DOI] [PubMed] [Google Scholar]
- 67.Maizel, J.V. 1971. Polyacrylamide gel electrophoresis of viral protein. In Methods in Virology. Vol. 5. K. Maramarosch and H. Koprowski, editors. Academic Press, New York. 179–191.
- 68.Mole, L.R. 1975. A genetic marker in the variable region of rabbit immunoglobulin heavy chain. Biochem. J. 151:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.