Abstract
Although the Chlamydia trachomatis genome is predicted to encode 50 inclusion membrane proteins, only 18 have been experimentally localized in the inclusion membrane of C. trachomatis-infected cells. Using fusion proteins and anti-fusion protein antibodies, we have systematically evaluated all 50 putative inclusion membrane proteins for their localization in the infected cells, distribution patterns, and effects on subsequent chlamydial infection when expressed ectopically, as well as their immunogenicity during chlamydial infection in humans. Twenty-two of the 50 proteins were localized in the inclusion membrane, and 7 were detected inside the inclusions, while the location of the remaining 21 was not defined. Four (CT225, CT228, CT358, and CT440) of the 22 inclusion membrane-localized proteins were visualized in the inclusion membrane of Chlamydia-infected cells for the first time in the current study. The seven intra-inclusion-localized proteins were confirmed to be chlamydial organism proteins in a Western blot assay. Further characterization of the 50 proteins revealed that neither colocalization with host cell endoplasmic reticulum nor inhibition of subsequent chlamydial infection by ectopically expressed proteins correlated with the inclusion membrane localization. Interestingly, antibodies from women with C. trachomatis urogenital infection preferentially recognized proteins localized in the inclusion membrane, and the immunodominant regions were further mapped to the region predicted to be on the cytoplasmic side of the inclusion membrane. These observations suggest that most of the inclusion membrane-localized proteins are both expressed and immunogenic during C. trachomatis infection in humans and that the cytoplasmic exposure may enhance the immunogenicity.
Chlamydia trachomatis, a species of obligate intracellular bacterial pathogen, has more than 15 different serovars with some (serovars A to C) infecting the human ocular epithelium, potentially leading to preventable blindness (48), and others (D to K) infecting human urogenital tissues, which if left untreated can cause severe complications, such as ectopic pregnancy and infertility (23, 41). Serovars L1 to L3 that cause lymphogranuloma venereum are more invasive and can occasionally cause outbreaks of systemic infections in humans (5, 32, 43). Nevertheless, all C. trachomatis organisms share a nearly identical genome (25, 26, 45) and common intracellular biphasic growth cycle (17, 18). A typical chlamydial infection starts with the entry of elementary bodies (EBs), the infectious form, into host cells via endocytosis. The internalized EBs within the endosomal vacuole can rapidly differentiate into reticulate bodies, the metabolically active but noninfectious form of chlamydial organisms. After numerous rounds of replication, the reticulate bodies can differentiate back into EBs for spreading to adjacent cells. C. trachomatis organisms can accomplish their biosynthesis, replication, and differentiation within the cytoplasmic vacuole, also termed inclusion. The inclusion membrane serves as both a barrier for protecting the intravacuolar organisms and a gate for C. trachomatis interactions with host cells. To establish and maintain its intravacuolar growth, C. trachomatis must exchange both materials and signals with the host cells across the inclusion membrane. C. trachomatis is not only able to import nutrients and metabolic intermediates from host cells (7, 19, 20, 33, 46) but also secretes chlamydial factors into host cells (57). Furthermore, C. trachomatis can actively manipulate host signal pathways (12, 16, 46, 50). Despite the frequent exchanges of both materials and information between C. trachomatis and host cells, the mechanisms of these exchanges across the inclusion membrane are largely unknown. Since proteins localized in the inclusion membrane can potentially play important roles in chlamydial interactions with host cells, the identification and characterization of chlamydial inclusion membrane proteins have become an area of intensive investigation.
In the past decade, significant progress has been made in identifying chlamydial inclusion membrane proteins, designated as Inc. Since Rockey et al. (28) reported the first chlamydial inclusion membrane protein, designated as IncA, from C. caviae GPIC in 1995, many Inc homologues have been described for C. trachomatis. For example, the regions of the C. trachomatis genome covering open reading frames (ORFs) CT115 to -119 (4, 35) and CT222 to -233 (2, 3, 38) contain numerous inc genes, although not every protein encoded in these regions has been experimentally demonstrated to be in the chlamydial inclusion membrane (2). Several other C. trachomatis proteins encoded by genes outside the above genomic regions were also found in the chlamydial inclusion membrane, including CT050 (42), CT089 (14), CT147 (6), CT249 (22), CT442 (2, 44), CT529 (15), CT618 (42), and CT813 (8). As the chlamydial genome sequences became available and in an attempt to search for more inclusion membrane proteins, both Bannantine et al. (2) and Toh et al. (49) used computer-based methods to predict chlamydial inclusion membrane proteins. Although about 50 C. trachomatis and 100 Chlamydia pneumoniae proteins were predicted to localize in the inclusion membrane (2, 49), these computer prediction results have not been validated by sufficient experimental evidence. Indeed, some of the predicted inclusion membrane proteins were determined to be not in the inclusion membrane (2, 24). Therefore, it is necessary to use experimental approaches to identify and characterize these predicted inclusion membrane proteins. Due to the lack of genetic tools for manipulating the chlamydial genome, chlamydial researchers have been forced to use cell-free or surrogate/heterologous systems to characterize chlamydial proteins (40, 42, 52, 53, 58). For example, the expression of chlamydial proteins in Saccharomyces cerevisiae has led to the identification of novel inclusion membrane proteins (42). Alternatively, characterizing chlamydial proteins in Chlamydia-infected cells has also been productive (8, 27, 28, 31, 34, 57).
Since chlamydial protein intracellular localization is a phenotype that can be experimentally tracked with specific reagents, we have initiated an effort to use antibodies raised with chlamydial fusion proteins to localize the endogenous chlamydial proteins. To our surprise, only 22 of the 50 putative C. trachomatis inclusion membrane proteins were visualized in the inclusion membrane, with another 7 inside the inclusions and the remaining 21 undefinable. We further found that the inclusion membrane localization of a given protein in C. trachomatis-infected cells did not correlate with the protein's ability to colocalize with host cell endoplasmic reticulum (ER) or to inhibit subsequent chlamydial infection when expressed via a transgene. Interestingly, antibodies from C. trachomatis-infected women predominantly recognized the C termini of the proteins localized in the inclusion membrane, suggesting that most of the inclusion membrane protein C termini are immunogenic during chlamydial infection in humans.
MATERIALS AND METHODS
(i) Chlamydial infection.
C. trachomatis serovars D or L2 were grown, purified, and titrated as previously described (16). Aliquots of the organisms were stored at −80°C till use. HeLa cells (ATCC, Manassas, VA) maintained in Dulbecco's modified Eagle's medium (GIBCO BRL, Rockville, MD) with 10% fetal calf serum (GIBCO BRL) at 37°C in an incubator supplied with 5% CO2 were used in the present study. For immunofluorescence assays, HeLa cells grown on glass coverslips were inoculated with chlamydial organisms diluted in Dulbecco's modified Eagle's medium with 10% fetal calf serum and 2 μg/ml of cycloheximide (Sigma, St. Louis, MO). The infection dose was pretitrated, and an infection rate of ∼50% was applied. The cell samples were cultured at 37°C in a CO2 incubator and processed at various time points after infection as indicated for individual experiments.
(ii) Prokaryotic expression of chlamydial fusion proteins and production of anti-fusion protein antibodies.
The ORFs coding for 50 putative inclusion membrane proteins from the C. trachomatis serovar D genome (http://stdgen.northwestern.edu) were cloned into pGEX vectors (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) and expressed as fusion proteins with glutathione-S-transferase (GST) fused to the N termini of the chlamydial proteins. Most of the 50 ORFs were cloned as full-length, with the following exceptions: CT227 was cloned from the lysine amino acid residue at the 46th position (K46) to the stop codon, CT300 from T28 to stop, and CT365 from the start codon to N260. These cloning-position variations were made to overcome difficulties in fusion protein expression. The following 15 ORFs were also cloned in fragments for mapping immunodominant regions: the CT089 N-terminal fragment (CT089N) was cloned from the start codon (M1) to residue S211 (M1-S211), CT089C from S212 to stop (S212-stop), CT115N (M1-A71), CT115C (S72-stop), CT116N (M1-A67), CT116C (L68-stop), CT118N (M1-L84), CT118C (L85-F165), CT119N (M1-F138), CT119C (Y139-stop), CT147N (M1-A509), CT147M (M510-D1024), CT147C (M1025-stop), CT223N (M1-L164), CT223C (L165-stop), CT226N (M1-S88), CT226C (A89-stop), CT228N (M1-E98), CT228C (A99-stop), CT229N (M1-S107), CT229C (M108-stop), CT442N (M1-A75), CT442C (I76-stop), CT484N (M1-E164), CT484C (M165-stop), CT529N (M1-T149), CT529C (F150-stop), CT618N (M1-F142), CT618C (M143-stop), CT813N (M1-E133), and CT813C (V134-stop). The expression of the fusion proteins was induced with isopropyl-β-d-thiogalactopyranoside (IPTG; Invitrogen, Carlsbad, CA), and the fusion proteins were extracted by lysing the bacteria via sonication in a Triton X-100 lysis buffer (1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 75 units/ml of aprotinin, 20 mM leupeptin, and 1.6 mM pepstatin). After a high-speed centrifugation to remove debris, the fusion protein-containing supernatants were either used directly or further purified using glutathione-conjugated agarose beads (Pharmacia). The bead-bound fusion proteins were also used to deplete antigen-specific antibodies from antiserum samples (see below). For antibody production, the purified fusion proteins were used to immunize mice (54, 55, 59-61). The sera were collected and stored at −20°C till use.
(iii) Transient transfection of mammalian cells.
The ORFs coding for the 50 putative inclusion membrane proteins from the C. trachomatis serovar D genome were also cloned into the pDsRed Monomer C1 (BD Biosciences Clontech, San Jose, CA) mammalian expression vector system with the red fluorescent protein (RFP) gene fused to the 5′ end of the target genes. Most of the 50 ORFs were cloned as full-length, with the following exceptions: CT147 (M1-A509), CT227 (K46-stop), and CT300 (T28-stop). The recombinant plasmids were transfected into HeLa cells using Lipofectamine 2000 transfection reagent, following the protocol recommended by the manufacturer (Invitrogen, Carlsbad, CA). At various time points after transfection, as indicated for individual experiments, the RFP fusion protein expression was visualized via the RFP fusion tag. In some cases, the transfected cultures were used for analyzing colocalization between the RFP fusion proteins and host cell ER, while in others, the transfected cultures were subsequently infected with C. trachomatis serovar D organisms for analyzing the effect of prior expression of RFP fusion proteins on chlamydial infection.
(iv) Immunofluorescence staining.
HeLa cells grown on coverslips were fixed with 2% paraformaldehyde for 30 min at room temperature, followed by permeabilization with 1% saponin (Sigma) for an additional 1 h. After washing and blocking, the cell samples were subjected to various combinations of antibody and chemical staining. Hoechst stain (Sigma) was used to visualize nuclear DNA. A rabbit anti-chlamydial organism antibody (R1L2, raised with C. trachomatis serovar L2 organisms; unpublished data) or anti-CT395 (CT395 is a GrpE-related chaperonin with >70% amino acid sequence identity among all chlamydial species) plus a goat anti-rabbit immunoglobulin G (IgG) secondary antibody conjugated with Cy2 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used to visualize chlamydial inclusions. The mouse antibodies raised with GST-chlamydial ORF fusion proteins plus a goat anti-mouse IgG conjugated with Cy3 (Jackson ImmunoResearch) were used to visualize the corresponding antigens. In some cases, the mouse primary antibodies were preabsorbed with either the corresponding or heterologous fusion proteins prior to staining cell samples. The preabsorption was carried out by incubating the antibodies with bead-immobilized fusion proteins for 1 h at room temperature or overnight at 4°C, followed by pelleting the beads. The remaining supernatants were used for immunostaining. For the transfected cell samples, the C. trachomatis proteins were visualized via the RFP fusion tag. In some experiments, the transfected cells were costained with the mouse anti-GST chlamydial ORF fusion proteins plus a goat anti-mouse IgG conjugated with Cy2 antibody (Jackson ImmunoResearch). In others, the costaining was with a rabbit antibody against the ER marker calnexin plus a goat anti-rabbit IgG conjugated with Cy2 (Jackson ImmunoResearch).
After the appropriate immunolabeling, the cell samples were used for image analysis and acquisition with an Olympus AX-70 fluorescence microscope equipped with multiple filter sets (Olympus, Melville, NY) as described previously (12, 16, 51, 57). Briefly, the multicolor-labeled samples were exposed under a given filter set at a time and the single-color images were acquired by using a Hamamatsu digital camera. The single-color images were then superimposed with SimplePCI software to display multiple colors. All microscopic images were processed using the Adobe Photoshop program (Adobe Systems, San Jose, CA).
(v) Western blot assay.
The Western blot assay was carried out as described elsewhere (10-12, 37, 56). Briefly, either fusion protein, transfected cell, purified EB, or C. trachomatis-infected cell samples were solubilized in 2% sodium dodecyl sulfate (SDS) sample buffer and loaded onto SDS-polyacrylamide gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes and the blots were detected with primary antibodies. The primary antibody binding was probed with a horseradish peroxidase (HRP)-conjugated secondary antibody and visualized with an enhanced chemiluminescence kit (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The Western blot assay was used both for assessing the mouse anti-fusion protein antibody specificity and titrating human antibody reactivity. For measuring the pooled human serum reactivity with chlamydial GST fusion proteins, the purified fusion proteins were loaded in equal amounts onto the corresponding lanes of SDS-polyacrylamide gels in multiple sets. One set was stained with Coomassie blue (Sigma) for visualizing the total amount of protein in each lane, and the rest of the sets were transferred onto nitrocellulose membrane for assessing human antibody reactivity with the chlamydial fusion proteins.
(vi) ELISA.
Seventeen human sera collected from women diagnosed with C. trachomatis urogenital infections (designated as positive sera) and eight human sera collected from women without chlamydial infection (negative sera) were used in the current study. The reactivity of these human sera with chlamydial fusion proteins was measured by using a protein array enzyme-linked immunosorbent assay (ELISA) as described elsewhere (36, 60, 62). Briefly, bacterial lysates containing the GST fusion proteins were added to the 96-well microplates precoated with glutathione (Pierce, Rockford, IL). After the microplates were washed and blocked, individual human serum samples diluted 1:500 and pooled sera 1:200 were added to the antigen-immobilized microplates. The serum antibody binding was detected with a goat anti-human IgG conjugated with HRP (Jackson ImmunoResearch Laboratories) in combination with the soluble substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulforic acid) diammonium salt (ABTS; Sigma) and quantitated by reading the absorbance (optical density [OD]) at 405 nm using a microplate reader (Molecular Devices Corporation, Sunnyvale, CA). To reduce background binding, all human serum samples were preabsorbed with lysates made from XL-1-Blue bacteria expressing GST alone. In some experiments, the human serum samples were preabsorbed with either C. trachomatis-infected HeLa or normal HeLa cell lysates.
RESULTS
(i) Localization of 50 C. trachomatis proteins in Chlamydia-infected cells.
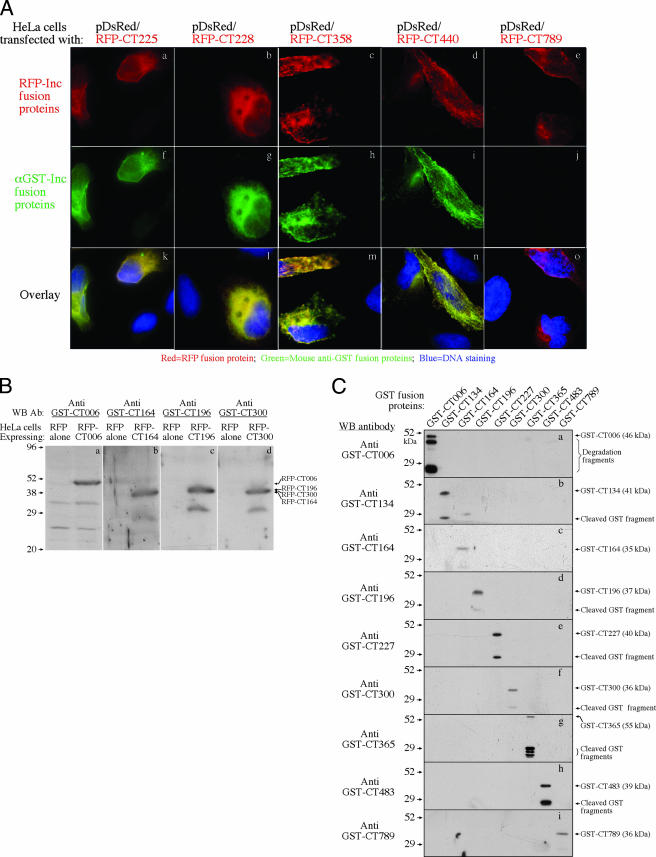
To track the intracellular location of the 50 C. trachomatis proteins in the infected cells, we expressed the 50 proteins as GST-tagged fusion proteins and raised antibodies with the fusion proteins in mice. The mouse anti-fusion protein antibodies were analyzed for their specificities using fusion proteins tagged with either RFP or GST (Table 1 and Fig. 1). Forty-one of the 50 antibodies recognized the corresponding RFP fusion proteins in an immunofluorescence assay, and the remaining 9 failed to do so (Fig. 1A). When the nine antibodies were reacted with the RFP fusion proteins in a Western blot assay, four (anti-CT006, anti-CT164, anti-CT196, and anti-CT300) picked up the corresponding bands but the remaining five (anti-CT134, anti-CT227, anti-CT365, anti-CT483, and anti-CT789) still failed to pick up any specific bands (Fig. 1B). The observation that a total of 45 anti-GST fusion protein antibodies reacted positively with both GST and RFP fusion proteins suggested that these antibodies specifically recognized the corresponding chlamydial proteins. The remaining five antibodies only reacted with the immunogen GST fusion proteins, but not the RFP fusion proteins (Table 1 and Fig. 1), suggesting that these five antibodies cannot recognize chlamydial antigens. Since these five antibodies detected the corresponding immunogens, but not the unrelated GST fusion proteins (Fig. 1C, panels b, e, g, h, and i), we speculated that these antibodies might recognize the epitopes created as a result of the fusion between the GST tag and the chlamydial proteins but not existing in either the fusion tag or chlamydial proteins alone.
TABLE 1.
Summary of anti-fusion protein antibody reactivity
| Antibodya | Reactivity with tagged fusion protein in indicated assayb
|
||
|---|---|---|---|
| RFP
|
GST (WB) | ||
| IFA | WB | ||
| Anti-CT005 | + | ND | + |
| Anti-CT006 | − | + | + |
| Anti-CT036 | + | ND | + |
| Anti-CT058 | + | ND | + |
| Anti-CT089 | + | ND | + |
| Anti-CT101 | + | ND | + |
| Anti-CT115 | + | ND | + |
| Anti-CT116 | + | ND | + |
| Anti-CT117 | + | ND | + |
| Anti-CT118 | + | ND | + |
| Anti-CT119 | + | ND | + |
| Anti-CT134 | − | − | +c |
| Anti-CT135 | + | ND | + |
| Anti-CT147 | + | ND | + |
| Anti-CT164 | − | + | + |
| Anti-CT179 | + | ND | + |
| Anti-CT192 | + | ND | + |
| Anti-CT195 | + | ND | + |
| Anti-CT196 | − | + | + |
| Anti-CT214 | + | ND | + |
| Anti-CT222 | + | ND | + |
| Anti-CT223 | + | ND | + |
| Anti-CT224 | + | ND | + |
| Anti-CT225 | + | ND | + |
| Anti-CT226 | + | ND | + |
| Anti-CT227 | − | − | +c |
| Anti-CT228 | + | ND | + |
| Anti-CT229 | + | ND | + |
| Anti-CT232 | + | ND | + |
| Anti-CT233 | + | ND | + |
| Anti-CT249 | + | ND | + |
| Anti-CT288 | + | ND | + |
| Anti-CT300 | − | + | + |
| Anti-CT345 | + | ND | + |
| Anti-CT357 | + | ND | + |
| Anti-CT358 | + | ND | + |
| Anti-CT365 | − | − | +c |
| Anti-CT383 | + | ND | + |
| Anti-CT440 | + | ND | + |
| Anti-CT442 | + | ND | + |
| Anti-CT449 | + | ND | + |
| Anti-CT483 | − | − | +c |
| Anti-CT484 | + | ND | + |
| Anti-CT529 | + | ND | + |
| Anti-CT565 | + | ND | + |
| Anti-CT618 | + | ND | + |
| Anti-CT728 | + | ND | + |
| Anti-CT789 | − | − | +c |
| Anti-CT813 | + | ND | + |
| Anti-CT850 | + | ND | + |
Mouse antibodies raised with GST fusion proteins.
RFP and GST were tagged to the N termini of chlamydial proteins. IFA, immunofluorescence assay; WB, Western blot assay; +, positive reaction; −, negative reaction; ND, not detected.
These five antibodies only reacted with the corresponding GST, but not RFP, fusion proteins, suggesting that these antibodies cannot recognize C. trachomatis-specific proteins.
FIG. 1.
Specificity of antibodies raised with GST chla myd i al fusion proteins. A total of 50 anti-GST fusion protein antibodies were reacted with RFP fusion proteins in immunofluorescence assay (A) or Western blot (WB) assay (B) and with GST fusion proteins in Western blot assay (C). Panel A shows examples of the reactivity of five anti-GST fusion protein antibodies (green; f to j) with HeLa cells expressing corresponding RFP fusion proteins (red; a to e). The DNA was labeled with a Hoechst dye (blue). Anti-GST-CT225, anti-GST-CT228, anti-GST-CT358, and anti-GST-CT440, together with 37 other antibodies (data not shown; see list in Table 1), reacted positively with the corresponding RFP fusion protein-expressing cells (k to n), but anti-GST-CT789, along with 8 other antibodies (images not shown; see list in Table 1), failed to do so (o). Overall, a total of 41 anti-GST fusion protein antibodies reacted positively with their corresponding RFP fusion proteins, and the remaining 9 did not. Panel B shows the binding of four anti-GST fusion protein antibodies to the corresponding RFP fusion protein bands (right lane of each panel), but not RFP-only bands (left lanes). The remaining five antibodies (anti-GST-CT134, anti-GST-CT227, anti-GST-CT365 anti-GST-CT483, and anti-GST-CT789) were also tested, but no specific bands were detected (data not shown). Panel C shows the reactivity of the nine antibodies with the corresponding GST fusion proteins, but not heterologous GST fusion proteins, under the experimental condition used. These anti-GST fusion protein antibodies were diluted highly enough to minimize the cross-reactivity. All 50 antibodies specifically reacted with their corresponding immunogens (GST fusion proteins) in the Western blot assay (images not shown). Ab, antibody; α, anti.
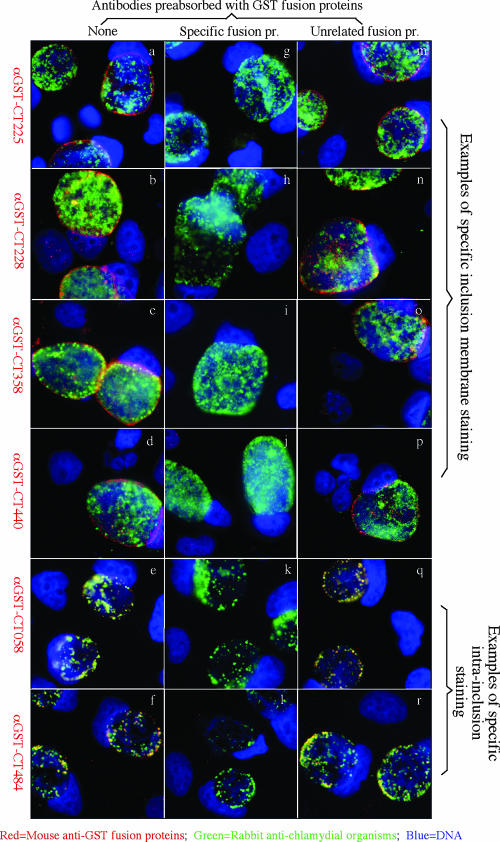
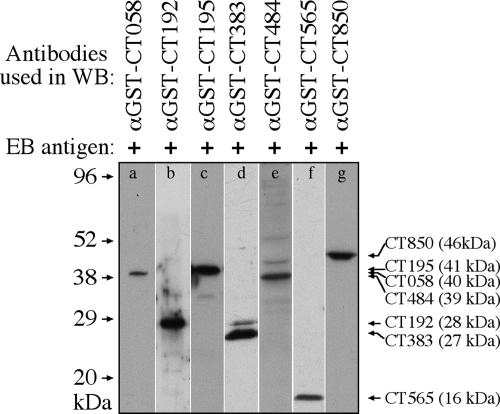
When the 50 anti-GST fusion protein antibodies were used to localize the endogenous proteins in C. trachomatis-infected cells (Table 2 and Fig. 2), 29 antibodies detected specific signals, with 22 visualizing the corresponding endogenous proteins in the inclusion membrane and 7 proteins inside the inclusions. These antibody-detected endogenous protein signals were removed by preabsorption of the antibodies with corresponding GST fusion proteins. Among the 22 inclusion membrane-localized proteins, 18 have been previously demonstrated (the corresponding references are listed in footnote a of Table 2) and four (CT225, CT228, CT358, and CT440) were identified for the first time in the current study. Despite the signature secondary structure of bilobed hydrophobic domains shared by most of the 50 putative inclusion membrane proteins, we clearly detected 7 proteins inside the C. trachomatis inclusions, suggesting that the bilobed hydrophobicity is not unique to inclusion membrane proteins. Furthermore, the seven antibodies recognized the corresponding chlamydial proteins from purified EBs in a Western blot assay (Fig. 3), suggesting that these proteins are associated with chlamydial organisms. Unfortunately, 21 of the 50 anti-GST fusion protein antibodies detected no specific signals in the C. trachomatis-infected cells in the immunofluorescence assay. Although 5 of them were known to lack the ability to recognize C. trachomatis-specific proteins, the remaining 16 recognized chlamydial proteins in the form of RFP fusion proteins (Table 1). We further used a Western blot assay to detect the reactivity of the 16 antibodies with C. trachomatis-infected cell lysates and found that three (anti-CT006, anti-CT164, and anti-CT300) recognized the corresponding endogenous proteins (data not shown). Thus, lack of recognition of endogenous proteins in immunofluorescence assays by these 16 antibodies may be due to low affinity of the anti-fusion protein antibodies to the endogenous proteins and/or insufficient amounts of the endogenous proteins expressed in the infected cells when the samples were processed for detection.
TABLE 2.
Summary of immunofluorescence detection of 50 C. trachomatis proteins in infected cells using mouse anti-fusion protein antibodies in comparison with previously known information
| Location of protein and ORF | Previous evidencea | Computer predictionb
|
|
|---|---|---|---|
| Ban | Toh | ||
| Inclusion membrane | |||
| CT089 | IM+ | − | − |
| CT115 | IM+ | + | + |
| CT116 | IM+ | + | + |
| CT117 | IM+ | + | + |
| CT118 | IM+ | + | + |
| CT119 | IM+ | + | + |
| CT147 | IM+ | − | + |
| CT223 | IM+ | + | + |
| CT225 | − | + | + |
| CT226 | IM+ | + | + |
| CT228 | − | + | + |
| CT229 | IM+ | + | + |
| CT232 | IM+ | + | + |
| CT233 | IM+ | + | + |
| CT249 | IM+ | − | + |
| CT288 | IM+ | + | + |
| CT358 | − | + | + |
| CT440 | − | + | + |
| CT442 | IM+ | + | − |
| CT529 | IM+ | − | − |
| CT618 | IM+ | + | − |
| CT813 | IM+ | + | + |
| Intrainclusion | |||
| CT058 | − | + | + |
| CT192 | − | + | + |
| CT195 | − | + | + |
| CT383 | − | + | + |
| CT484 | IB+ | + | + |
| CT565 | − | + | − |
| CT850 | − | + | − |
| Undefinedc | |||
| CT005 | − | + | − |
| CT006 | − | + | − |
| CT036 | − | + | + |
| CT101 | − | + | − |
| CT134 | − | + | − |
| CT135 | − | + | − |
| CT164 | − | + | − |
| CT179 | − | − | + |
| CT196 | − | + | + |
| CT214 | − | + | + |
| CT222 | − | − | + |
| CT224 | − | + | + |
| CT227 | − | + | + |
| CT300 | − | + | + |
| CT345 | − | + | + |
| CT357 | − | + | + |
| CT365 | − | − | + |
| CT449 | − | + | + |
| CT483 | − | + | + |
| CT728 | − | + | − |
| CT789 | − | + | − |
Previous experimental evidence for intracellular location of the 50 proteins is listed in this column. Previously described proteins and the references in which their localizations are described are CT089 (14); CT115 to -119 (35); CT147 (6); CT223, -229, -233, -288, -442, and -484 (2); CT226 (38); CT229 (30); CT232 (IncB) and CT233 (IncC) (3); CT249 (22); CT442 (44); CT529 (15); CT618 (42); and CT813 (8). IM+, positive localization in the inclusion membrane; IB+, positive localization inside the inclusions; −, published experimental evidence for the intracellular location of the corresponding proteins is lacking.
Ban and Toh stand for references 2, Bannantine et al., and 49, Toh et al., respectively. +, positive prediction for inclusion membrane localization; −, negative prediction for inclusion membrane localization.
No specific signals were detected using the corresponding antibodies under the conditions in which the experiments were carried out.
FIG. 2.
Intracellular localization of the 50 putative inclusion membrane proteins in C. trachomatis-infected cells. HeLa cells infected with C. trachomatis organisms were immunostained with mouse anti-GST fusion protein antibodies plus a goat anti-mouse IgG conjugated with Cy3 (red) as listed along the left side and a rabbit anti-chlamydial organism antibody plus a goat anti-rabbit IgG conjugated with Cy2 (green) and a DNA dye (blue). The antibodies whose results are shown in panels a to d all detected a dominant signal in the inclusion membrane, while the antibodies whose results are shown in panels e and f detected signals mainly inside the inclusions and the staining overlapped with the chlamydial organisms. These antibody labelings were removed by preabsorption of the antibodies with the corresponding GST fusion proteins (g to l), but not an unrelated control GST-CT858 fusion protein (m to r). Overall, a total of 22 anti-GST fusion protein antibodies labeled the inclusion membrane and 7 labeled the inclusions, while no significant specific staining signals were observed with the other 21 anti-GST fusion protein antibodies (images not shown; see list in Table 2). α, anti.
FIG. 3.
Detection of EB proteins with seven anti-fusion protein antibodies. The purified EBs were resolved in SDS-polyacrylamide gel, and the protein bands were transferred onto nitrocellulose membrane for measuring antibody reactivity in a Western blot (WB) assay. Seven mouse anti-GST fusion protein antibodies as listed on top of the figure were used to react with strips of the nitrocellulose membrane (a to g). The molecular masses in kDa are marked along the left, while the antibody-recognized EB protein bands are listed along the right side of the figure. The sizes of the EB proteins are also indicated, in parentheses following each protein's name. Clearly, the seven anti-fusion protein antibodies recognized the corresponding endogenous proteins from the purified EBs. α, anti.
(ii) Correlates of inclusion membrane localization.
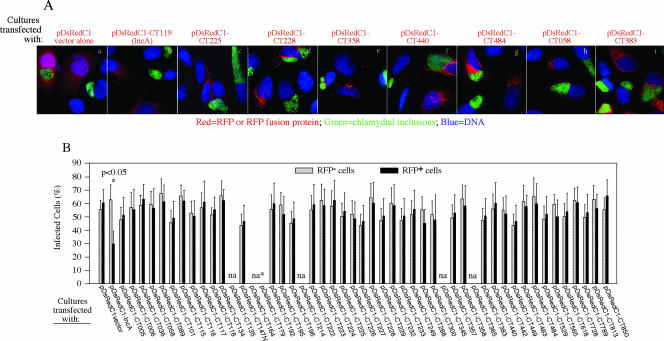
In addition to the computer program-based prediction of secondary structures potentially shared by the chlamydial inclusion membrane proteins, inclusion membrane proteins have also been correlated with their ability to colocalize with host cell ER structure (9). We expressed the 50 chlamydial proteins in HeLa cells as RFP fusion proteins and assessed their colocalization with the ER membrane marker calnexin (Table 3 and data not shown). We found that these 50 proteins displayed various degrees of overlap with the ER marker regardless of the location of their corresponding endogenous proteins in C. trachomatis-infected cells, suggesting that the ER localization test is not efficacious for the determination of proteins localized in the chlamydial inclusion membrane. It has also been reported that prior expression of inclusion membrane proteins significantly inhibited subsequent chlamydial infection (9). We assessed the effects of expression of the 50 proteins on C. trachomatis infection (Fig. 4). We found that although IncA expression significantly reduced the subsequent chlamydial infection in the transfected cells (P < 0.05), none of the other proteins, including the remaining 21 inclusion membrane-localized proteins, did so.
TABLE 3.
Summary of colocalization of ectopically expressed RFP fusion proteins with host cell ER
| Degree of colocalizationa | Location of endogenous protein
|
||
|---|---|---|---|
| Inclusion membrane | Inclusion | Undefined | |
| Most | CT115 | CT195 | CT005 |
| CT116 | CT484 | CT036 | |
| CT117 | CT565 | CT101 | |
| CT118 | CT850 | CT164 | |
| CT119 | CT179 | ||
| CT223 | CT214 | ||
| CT225 | CT345 | ||
| CT226 | CT357 | ||
| CT232 | CT728 | ||
| CT442 | CT789 | ||
| CT813 | |||
| Partial | CT222 | CT058 | CT224 |
| CT228 | CT192 | CT227 | |
| CT229 | CT365 | ||
| CT288 | |||
| CT529 | |||
| CT618 | |||
| Least | CT089 | CT383 | CT006 |
| CT147 | CT134 | ||
| CT233 | CT135 | ||
| CT249 | CT196 | ||
| CT358 | CT300 | ||
| CT440 | CT449 | ||
| CT483 | |||
The degrees of overlap between the RFP fusion proteins and the host cell ER marker calnexin were categorized as “most,” with almost-complete overlapping; “partial,” with some overlapping; and “least,” with no clear overlapping.
FIG. 4.
Effect of RFP-chlamydial fusion protein expression on chlamydial infection. HeLa cells grown on coverslips in 24-well plates were transfected with recombinant pDsRedC1 plasmids encoding each of the 50 RFP fusion proteins, and 12 h after transfection, the cultures were infected with C. trachomatis organisms. Forty hours after infection, the cell samples were processed for immunostaining with a rabbit anti-chlamydial organism antibody plus a goat anti-rabbit IgG conjugated with Cy2 (green) and a DNA dye (blue). The RFP fusion proteins were visualized via the RFP (red). Panel A shows examples of cultures transfected with pDsRedC1 vector alone and eight pDsRedC1/CT recombinant plasmids. Overall, all 50 C. trachomatis proteins were similarly evaluated (images not shown). Panel B shows the quantitative results for 100 RFP-positive (RFP+) and 100 RFP-negative (RFP−) cells which were counted from each coverslip culture that was transfected with each of the 50 plasmids as listed along the x axis. The rates of chlamydial infection in these two different cell populations from the same cultures were calculated separately, and the results are displayed along the y axis. The data are from three independent experiments. A statistically significant difference in infection rate (P value of <0.05 by two-tailed t test) was found between the RFP-positive and RFP-negative cells in the cultures transfected with pDsRedC1-CT119 (IncA). No other cultures showed any significant differences. It is clear that the expression of IncA, but not other chlamydial proteins, inhibited the subsequent chlamydial infection. Error bars show standard deviations. na, not analyzed due to insufficient number of cells counted.
(iii) Proteins localized in the inclusion membrane are more dominantly recognized by antibodies from women with C. trachomatis urogenital tract infection.
The 50 GST-C. trachomatis fusion proteins were analyzed for their reactivity with 17 human antisera, and we found that all but one (CT357) of the fusion proteins reacted positively with one or more human antisera (Fig. 5), suggesting that the 49 proteins were at least expressed during chlamydial infection in humans. The fusion proteins that reacted with human antibodies with both positive titers (average OD, ≥0.2) and high frequency (≥8 antisera) were defined as immunodominant antigens. A total of 17 proteins met the criteria, of which 15 (CT089, CT115, CT116, CT118, CT119, CT147, CT223, CT225, CT226, CT228, CT229, CT442, CT529, CT618, and CT813) were inclusion membrane localized (15/22, 68%) and 2 (CT484 and CT850) were chlamydial organism-associated proteins (2/7, 29%). None of the 17 immunodominant antigens belonged to the location-undefined group (0%). In fact, most of the 21 location-undefined proteins displayed low reactivity with the human sera. We defined the fusion proteins recognized by three or fewer of the 17 human antisera as the least antigenic proteins. A total of 26 fusion proteins met the criteria, of which 16 (CT006, CT134, CT135, CT164, CT179, CT196, CT224, CT227, CT300, CT345, CT357, CT365, CT449, CT483, CT728, and CT789) belong to the location-undefined (76%), 5 (CT058, CT192, CT195, CT383, and CT565) to chlamydial organism-associated (71%), and 5 (CT117, CT232, CT233, CT358, and CT440) to inclusion membrane-localized (23%) groups. It seemed that the group of inclusion membrane-localized proteins had the highest percentage of the most-immunodominant proteins (68% versus 29% or 0%) and the lowest percentage of the least-dominant proteins (23% versus 71% or 76%) compared with the chlamydial organism-associated or undefinable groups.
FIG. 5.
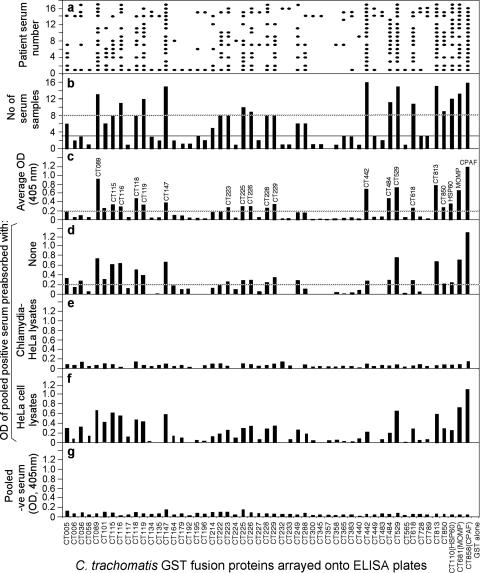
Reactivity of 17 human antibodies with 50 GST-chlamydial fusion proteins. Each of the 17 human antibodies (displayed along the y axis of panel a) was reacted after 1:500 dilution with each of the 50 GST fusion proteins (listed along the x axis) immobilized onto the 96-well microplates. The human antibody binding was detected with a secondary goat anti-human IgG antibody conjugated with HRP plus a soluble substrate. The results were expressed as OD readings obtained at the wavelength of 405 nm. Any given reaction with an OD reading of fourfold more than the value from the control well (GST-coated well) was designated positive and is represented with a filled oval in panel a. The total number of human serum samples that positively recognized a given fusion protein are summarized in panel b. The dashed line marks those fusion proteins recognized by eight or more human antisera, while the thin solid line marks the fusion proteins recognized by three or fewer human sera. The average OD readings, calculated by dividing the total OD values by 17, are displayed in panel c. The dashed line marks those fusion proteins with an average OD of 0.2 or above. The 17 human serum samples were pooled at an equal ratio and assayed against each of the fusion proteins at a dilution of 1:200, and the OD values are displayed along the y axis of panel d. The dashed line marks those fusion proteins with an OD of 0.2 or more. Please note that the OD reading patterns obtained with the pooled serum are similar to those obtained with the individual serum samples that are shown in panel c. In addition, the pooled serum was subjected to preabsorption with either C. trachomatis-infected (e) or HeLa cell-only (f) lysates prior to being reacted with the fusion proteins. Please note that the pooled serum reactivity with the fusion proteins was removed by the C. trachomatis-infected, but not HeLa cell-only, lysates. Finally, pooled negative (-ve) serum samples from eight individuals failed to react with the fusion proteins (g).
The specificity of the human antibody binding was confirmed by preabsorption of the human antisera with the endogenous antigens. For convenience of detection, the 17 human antisera were pooled at an equal ratio (designated as pooled positive serum). We found that the pooled positive serum and the individual serum samples reacted similarly with the GST fusion proteins (Fig. 5, compare panels d and c), and the reactivity was removed by preabsorption with C. trachomatis-infected cell lysates, but not HeLa-only lysates (e and f). A pooled negative human serum sample failed to react significantly with any of the GST fusion proteins (Fig. 5, panel g).
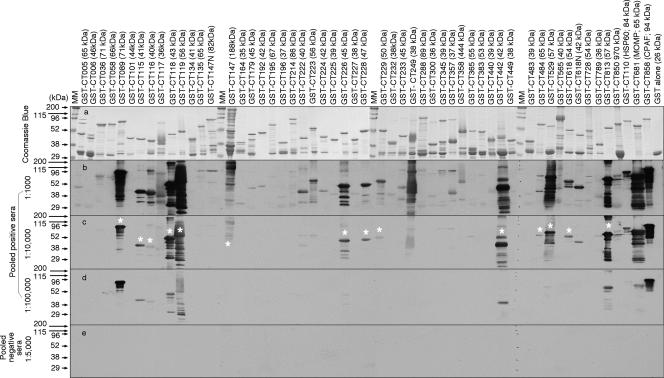
We further used a Western blot assay to measure the pooled positive human antibody reactivity with the GST fusion proteins (Fig. 6). All 50 GST fusion proteins and 3 control proteins, plus GST alone, were expressed and properly purified, as revealed in the results shown in Fig. 6, panel a. The pooled positive human antiserum sample reacted with most of the 50 GST fusion proteins when used at 1:1,000 dilution (Fig. 6, panel b) and still recognized 16 of the 17 immunodominant antigens (as identified in the ELISA described above) at 1:10,000 dilution (panel c). Interestingly, CT225, although identified as an immunodominant protein in the ELISA, was not recognized by the human antibody in the Western blot assay, suggesting that the linearization of CT225 in the Western blot assay altered the conformation required by the human antibody. In fact, careful comparison of the human antibody reactivity with the GST fusion proteins in the ELISA (Fig. 5) and the Western blot assay (Fig. 6) can help to determine the relative dependence on antigen conformation by the human antibodies. For example, human antibodies maintained a strong reactivity with GST-CT089, GST-CT119, GST-CT442, and GST-CT813 in Western blotting, suggesting that these fusion protein-reactive human antibodies are less dependent on antigen conformation. On the contrary, the human antibody reactivity with GST-CT147 and GST-CT529 was limited in the Western blot assay, suggesting that the corresponding human antibodies are more dependent on protein conformation.
FIG. 6.
Recognition of GST fusion proteins by the human antibodies in a Western blot. The 50 GST fusion proteins or fragments plus a few control fusion proteins were loaded onto an SDS gel, and after electrophoretic separation, one set of the gels was subjected to Coomassie blue dye staining for visualizing the total proteins (a). The parallel gels were used to transfer the resolved protein bands onto nitrocellulose membrane for measuring the human antibody reactivity. The pooled positive human serum used in the experiments whose results are shown in Fig. 5 was 10-fold serially diluted starting at 1:1,000 and then reacted with the membrane. Please note that at 1:1,000 dilution, many GST fusion proteins were picked up by the human antibodies (b). As the dilution increased to 1:10,000, fewer proteins were recognized (c), and only a few proteins were detectable when the serum was diluted another 10-fold (d). No significant reactivity was detected when the pooled negative serum was measured at 1:5,000 dilution (e). Among the 50 GST fusion proteins, those recognized by the human pooled positive serum at 1:10,000 were considered immunodominant and thus marked with white stars and subjected to further mapping of immunodominant regions. Please note that the results of Western blotting largely confirmed the ELISA results shown in Fig. 5, with the exception of the results for CT225. MM, molecular mass.
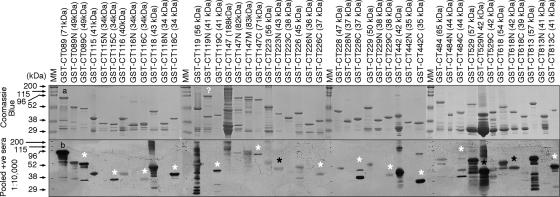
Finally, we compared the relative immunodominance of the N- and C-terminal portions of the immunodominant antigens using a Western blot assay (Fig. 7). We found that the C-terminal portions were consistently more immunodominant than the N termini in most proteins, with the exception of CT223, CT529, and CT618. In contrast, these three proteins displayed a more-immunodominant N-terminal region.
FIG. 7.
Mapping immunodominant regions. The immunodominant GST fusion proteins were further expressed as N- and C-terminal fragments, and these fragments, along with the corresponding full-length fusion proteins, were subjected to SDS gel and Western blot analysis as described in the Fig. 6 legend. The Coomassie blue-stained gel (a) showed the total protein levels, and a dominant protein band that migrated at the expected position was detected in most of the protein samples. The extra high-molecular-mass band in the IncA N-terminal fragment (CT119N) sample may represent an SDS-resistant aggregate, since the IncA N-terminal transmembrane region is known to form oligomer. The pooled positive (+ve) human serum detected both the full-length and one or more fragment bands from each protein. The fragments that were predominantly recognized by the human serum are marked with either a white (C-terminal fragment) or black (N-terminal fragment) star. Please note that most immunodominant fragments belong to the C terminus, with the exception of CT223, CT529, and CT618 that display an N-terminal immunodominant region. MM, molecular mass.
DISCUSSION
We have systematically characterized 50 C. trachomatis proteins that are predicted to localize in the inclusion membrane. By using the anti-fusion protein antibody approach, we were able to define the intracellular location of 29 such proteins in Chlamydia-infected cells, with 22 in the inclusion membrane and 7 associating with chlamydial organisms. The location of the remaining 21 proteins was undefined by this approach due to either poor quality of the corresponding anti-fusion protein antibodies or low levels of the corresponding endogenous proteins. Among the 22 inclusion membrane-localized proteins, 18 have been previously demonstrated (2-4, 6, 8, 15, 35, 39, 42). The current study has presented the first experimental evidence for the chlamydial inclusion membrane localization of CT225, CT228, CT358, and CT440. Although CT225 and CT228 belong to the CT222-CT233 gene cluster and are widely believed to be in the inclusion membrane (30), no experimental evidence has been presented in any prior publications. Although in Bannantine et al. (2) the Fig. 4 legend mentioned “p228,” it may be erroneous since the authors only described antibodies to CT288, but not CT228, in the paper (2). The four newly identified inclusion membrane proteins vary in size and charge, with 133 amino acids with a pI of 8.66 for CT225, 196 amino acids with a pI of 4.93 for CT228, 178 amino acids with a pI of 4.62 for CT358, and 113 amino acids with a pI of 10.17 for CT440. Nevertheless, all four proteins possess two or more transmembrane domains at their N termini, which provides them with the so called N-terminal bilobed hydrophobic domain, a common structural feature shared by the predicted inclusion membrane proteins (2, 49). However, not all proteins with the bilobed hydrophobic domain structure feature can be detected in the inclusion membrane. For example, the seven proteins that were determined to localize inside inclusions and associate with chlamydial organisms in the current study all contain the bilobed hydrophobic domain. Furthermore, not all experimentally demonstrated inclusion membrane proteins, such as CT529 (29) and CT089 (14), are predictable by the existing computer programs (2, 15, 49). Thus, there is an urgent need to identify new inclusion membrane proteins using experiments.
In the past decades, a tremendous amount of effort has been made to characterize inclusion membrane proteins, to search for correlates of inclusion membrane localization, and to probe the functions of the inclusion membrane proteins. Some inclusion membrane proteins, when expressed in the host cell cytosol via transgenes, were colocalized with host cell ER markers (9). However, we have presented evidence that colocalization with the ER is not a reliable correlate of inclusion membrane localization. Although IncA expression in host cell cytosol was shown to inhibit the subsequent chlamydial infection (1, 9), we have found that this IncA ability is not shared by any other C. trachomatis proteins tested. IncA may be unique in its ability to oligomerize, to mediate interaction between facing membranes (9), and to mediate fusion between inclusions in the same infected cells (13, 21, 47). The research interest in inclusion membrane proteins stems mainly from the potential roles of inclusion membrane proteins in mediating the interactions between C. trachomatis and host cells. Indeed, some Incs have been shown to be modified by (27) and to interact with (31, 34) host cell components. For example, IncG (CT118) and CT229 can interact with the host adaptor molecule 14-3-3 and Rab GTPases, as well as their effectors, respectively. The search for interaction partners of other inclusion membrane proteins is also underway in our laboratory.
It has been suggested that animals infected with live chlamydial organisms can develop higher titers of antibodies against inclusion membrane proteins than animals immunized with dead organisms (28). In fact, the first inclusion membrane protein, IncA, was identified by taking advantage of the animal antisera raised with live-organism infection (28). Since antibodies from women with urogenital-tract C. trachomatis infection are likely generated in response to live infection (36), we analyzed the reactivity of the 50 putative C. trachomatis inclusion membrane proteins with 17 human sera. We found that human antibodies predominantly recognized inclusion membrane-localized proteins, suggesting that the inclusion membrane proteins are immunodominant during natural infection in humans. Although multiple factors could affect the immunogenicity of a given protein, the inclusion membrane localization may enhance immunogenicity by presenting/exposing portions of the proteins to the host cell cytosol. The C-terminal fragments of many inclusion membrane proteins are believed to be exposed at the host cell cytoplasmic surface (27; data not shown) and are dominantly recognized by the human antibodies (Fig. 7). The correlation of cytoplasmic exposure and immunodominance is also true when the N-terminal fragment is exposed to the cytoplasm. For example, CT529 with its N-terminal region exposed to the cytoplasm displayed dominant immune recognition of its N-terminal epitopes both by antibodies (current study) and T cells (15, 44). The N-terminal portions of CT223 and CT618 were also more dominantly recognized by human antibodies. It will be interesting to test whether these two inclusion membrane proteins have their N termini exposed to the host cell cytoplasm.
Acknowledgments
This work was supported in part by grants (to G. Zhong) from the U.S. National Institutes of Health.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Alzhanov, D., J. Barnes, D. E. Hruby, and D. D. Rockey. 2004. Chlamydial development is blocked in host cells transfected with Chlamydophila caviae incA. BMC Microbiol. 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 235-47. [DOI] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., D. D. Rockey, and T. Hackstadt. 1998. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 281017-1026. [DOI] [PubMed] [Google Scholar]
- 4.Bannantine, J. P., W. E. Stamm, R. J. Suchland, and D. D. Rockey. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 666017-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauwens, J. E., H. Orlander, M. P. Gomez, M. Lampe, S. Morse, W. E. Stamm, R. Cone, R. Ashley, P. Swenson, and K. K. Holmes. 2002. Epidemic lymphogranuloma venereum during epidemics of crack cocaine use and HIV infection in the Bahamas. Sex. Transm. Dis. 29253-259. [DOI] [PubMed] [Google Scholar]
- 6.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 1008478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carabeo, R. A., D. J. Mead, and T. Hackstadt. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. USA 1006771-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C., D. Chen, J. Sharma, W. Cheng, Y. Zhong, K. Liu, J. Jensen, R. Shain, B. Arulanandam, and G. Zhong. 2006. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect. Immun. 744826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delevoye, C., M. Nilges, A. Dautry-Varsat, and A. Subtil. 2004. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 27946896-46906. [DOI] [PubMed] [Google Scholar]
- 10.Dong, F., M. Pirbhai, Y. Xiao, Y. Zhong, Y. Wu, and G. Zhong. 2005. Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect. Immun. 731861-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, F., Y. Zhong, B. Arulanandam, and G. Zhong. 2005. Production of a proteolytically active protein, chlamydial protease/proteasome-like activity factor, by five different Chlamydia species. Infect. Immun. 731868-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in Chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, K. A., E. Fischer, and T. Hackstadt. 2002. Inhibition of fusion of Chlamydia trachomatis inclusions at 32°C correlates with restricted export of IncA. Infect. Immun. 703816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 381048-1060. [DOI] [PubMed] [Google Scholar]
- 15.Fling, S. P., R. A. Sutherland, L. N. Steele, B. Hess, S. E. D'Orazio, J. Maisonneuve, M. F. Lampe, P. Probst, and M. N. Starnbach. 2001. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 981160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene, W., Y. Xiao, Y. Huang, G. McClarty, and G. Zhong. 2004. Chlamydia-infected cells continue to undergo mitosis and resist induction of apoptosis. Infect. Immun. 72451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackstadt, T. 1998. The diverse habitats of obligate intracellular parasites. Curr. Opin. Microbiol. 182-87. [DOI] [PubMed] [Google Scholar]
- 18.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5288-293. [DOI] [PubMed] [Google Scholar]
- 19.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15964-977. [PMC free article] [PubMed] [Google Scholar]
- 20.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 924877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1119-130. [DOI] [PubMed] [Google Scholar]
- 22.Jia, T. J., D. W. Liu, J. H. Luo, and G. M. Zhong. 2007. Localization of the hypothetical protein CT249 in the Chlamydia trachomatis inclusion membrane. Wei Sheng Wu Xue Bao 47645-648. (In Chinese.) [PubMed] [Google Scholar]
- 23.Kinnunen, A. H., H. M. Surcel, M. Lehtinen, J. Karhukorpi, A. Tiitinen, M. Halttunen, A. Bloigu, R. P. Morrison, R. Karttunen, and J. Paavonen. 2002. HLA DQ alleles and interleukin-10 polymorphism associated with Chlamydia trachomatis-related tubal factor infertility: a case-control study. Hum. Reprod. 172073-2078. [DOI] [PubMed] [Google Scholar]
- 24.Luo, J., G. Liu, Y. Zhong, T. Jia, K. Liu, D. Chen, and G. Zhong. 2007. Characterization of hypothetical proteins Cpn0146, 0147, 0284 & 0285 that are predicted to be in the Chlamydia pneumoniae inclusion membrane. BMC Microbiol. 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 312134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24217-228. [DOI] [PubMed] [Google Scholar]
- 28.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15617-626. [DOI] [PubMed] [Google Scholar]
- 29.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4333-340. [DOI] [PubMed] [Google Scholar]
- 30.Rzomp, K. A., A. R. Moorhead, and M. A. Scidmore. 2006. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 745362-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rzomp, K. A., L. D. Scholtes, B. J. Briggs, G. R. Whittaker, and M. A. Scidmore. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 715855-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schachter, J., and J. Moncada. 2005. Lymphogranuloma venereum: how to turn an endemic disease into an outbreak of a new disease? Start looking. Sex. Transm. Dis. 32331-332. [DOI] [PubMed] [Google Scholar]
- 33.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 391638-1650. [DOI] [PubMed] [Google Scholar]
- 35.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33753-765. [DOI] [PubMed] [Google Scholar]
- 36.Sharma, J., A. M. Bosnic, J. M. Piper, and G. Zhong. 2004. Human antibody responses to a Chlamydia-secreted protease factor. Infect. Immun. 727164-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma, J., F. Dong, M. Pirbhai, and G. Zhong. 2005. Inhibition of proteolytic activity of a chlamydial proteasome/protease-like activity factor by antibodies from humans infected with Chlamydia trachomatis. Infect. Immun. 734414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma, J., Y. Zhong, F. Dong, J. M. Piper, G. Wang, and G. Zhong. 2006. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect. Immun. 741490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37913-925. [DOI] [PubMed] [Google Scholar]
- 40.Shen, L., M. Li, and Y. X. Zhang. 2004. Chlamydia trachomatis sigma28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae. Microbiology 150205-215. [DOI] [PubMed] [Google Scholar]
- 41.Sherman, K. J., J. R. Daling, A. Stergachis, N. S. Weiss, H. M. Foy, S. P. Wang, and J. T. Grayston. 1990. Sexually transmitted diseases and tubal pregnancy. Sex. Transm. Dis. 17115-121. [DOI] [PubMed] [Google Scholar]
- 42.Sisko, J. L., K. Spaeth, Y. Kumar, and R. H. Valdivia. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 6051-66. [DOI] [PubMed] [Google Scholar]
- 43.Spaargaren, J. 2005. Slow epidemic of lymphogranuloma venereum l2b strain. Emerg. Infect. Dis. 111787-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starnbach, M. N., W. P. Loomis, P. Ovendale, D. Regan, B. Hess, M. R. Alderson, and S. P. Fling. 2003. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J. Immunol. 1714742-4749. [DOI] [PubMed] [Google Scholar]
- 45.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282754-759. [DOI] [PubMed] [Google Scholar]
- 46.Su, H., G. McClarty, F. Dong, G. M. Hatch, Z. K. Pan, and G. Zhong. 2004. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 2799409-9416. [DOI] [PubMed] [Google Scholar]
- 47.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, H. R., S. L. Johnson, J. Schachter, H. D. Caldwell, and R. A. Prendergast. 1987. Pathogenesis of trachoma: the stimulus for inflammation. J. Immunol. 1383023-3027. [PubMed] [Google Scholar]
- 49.Toh, H., K. Miura, M. Shirai, and M. Hattori. 2003. In silico inference of inclusion membrane protein family in obligate intracellular parasites chlamydiae. DNA Res. 109-17. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, Y., Y. Zhong, W. Greene, F. Dong, and G. Zhong. 2004. Chlamydia trachomatis infection inhibits both Bax and Bak activation induced by staurosporine. Infect. Immun. 725470-5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, Y., Y. Zhong, H. Su, Z. Zhou, P. Chiao, and G. Zhong. 2005. NF-kappa B activation is not required for Chlamydia trachomatis inhibition of host epithelial cell apoptosis. J. Immunol. 1741701-1708. [DOI] [PubMed] [Google Scholar]
- 52.Yu, H. H., D. Kibler, and M. Tan. 2006. In silico prediction and functional validation of σ28-regulated genes in Chlamydia and Escherichia coli. J. Bacteriol. 1888206-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, H. H., and M. Tan. 2003. Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 50577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, G., J. Berry, and R. C. Brunham. 1994. Antibody recognition of a neutralization epitope on the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 621576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, G., and R. C. Brunham. 1992. Antibody responses to the chlamydial heat shock proteins hsp60 and hsp70 are H-2 linked. Infect. Immun. 603143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, G., F. Castellino, P. Romagnoli, and R. N. Germain. 1996. Evidence that binding site occupancy is necessary and sufficient for effective major histocompatibility complex (MHC) class II transport through the secretory pathway redefines the primary function of class II-associated invariant chain peptides (CLIP). J. Exp. Med. 1842061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J. Exp. Med. 1911525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong, G., C. Reis e Sousa, and R. N. Germain. 1997. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc. Natl. Acad. Sci. USA 9413856-13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, G., I. Toth, R. Reid, and R. C. Brunham. 1993. Immunogenicity evaluation of a lipidic amino acid-based synthetic peptide vaccine for Chlamydia trachomatis. J. Immunol. 1513728-3736. [PubMed] [Google Scholar]
- 61.Zhong, G. M., and R. C. Brunham. 1991. Antigenic determinants of the chlamydial major outer membrane protein resolved at a single amino acid level. Infect. Immun. 591141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong, G. M., and R. C. Brunham. 1990. Immunoaccessible peptide sequences of the major outer membrane protein from Chlamydia trachomatis serovar C. Infect. Immun. 583438-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]