Abstract
Little is currently known about proteins that make contact with the pre-mRNA in the U12-dependent spliceosome and thereby contribute to intron recognition. Using site-specific cross-linking, we detected an interaction between the U11-48K protein and U12-type 5′ splice sites (5′ss). This interaction did not require branch point recognition and was sensitive to 5′ss mutations, suggesting that 48K interacts with the 5′ss during the first steps of prespliceosome assembly in a sequence-dependent manner. RNA interference-induced knockdown of 48K in HeLa cells led to reduced cell growth and the inhibition of U12-type splicing, as well as the activation of cryptic, U2-type splice sites, suggesting that 48K plays a critical role in U12-type intron recognition. 48K knockdown also led to reduced levels of U11/U12 di-snRNP, indicating that 48K contributes to the stability and/or formation of this complex. In addition to making contact with the 5′ss, 48K interacts with the U11-59K protein, a protein at the interface of the U11/U12 di-snRNP. These studies provide important insights into the protein-mediated recognition of the U12-type 5′ss, as well as functionally important interactions within the U11/U12 di-snRNP.
Most metazoans and some unicellular eukaryotes contain two distinct spliceosomes (reference 28 and references therein). The majority of introns are excised by the major, U2-dependent spliceosome, while a subset (<0.5%) with highly conserved 5′ splice sites (5′ss) and branch point sequences (BPS) are removed by the minor, U12-dependent spliceosome (for a review, see reference 24). Both spliceosomes consist of five small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP proteins. The two spliceosomes share the U5 snRNP, while the remaining four snRNPs in each spliceosome are distinct but functionally analogous, with U11, U12, U4atac, and U6atac of the minor spliceosome being the counterparts of U1, U2, U4, and U6 in the major spliceosome, respectively (12, 14, 16, 34, 35, 42).
Intron recognition is achieved via multiple, dynamic RNA- and protein-mediated interactions. RNA-RNA interactions in the two spliceosomes are highly analogous. In the major spliceosome, the first assembly step (generating the E complex) involves the recognition of the 5′ss by U1, while the non-snRNP protein factors SF1, U2AF65, and U2AF35 bind to the BPS, the polypyrimidine tract, and the 3′ss, respectively (for a review, see reference 26). During this stage, U1 base pairs with the pre-mRNA's 5′ss, while U1-associated proteins facilitate 5′ss recognition or stabilize the U1-5′ss complex (for a review, see reference 37). During prespliceosome (A complex) formation, U2 associates stably with the BPS (17), while non-snRNP proteins bridge U1 and U2 snRNPs (40, 41). The U4/U6/U5 tri-snRNP then joins the spliceosome (generating the B complex), which triggers the displacement of U1 from the 5′ss by U6 and additional rearrangements that lead to catalytic core formation (for a review, see references 22 and 33).
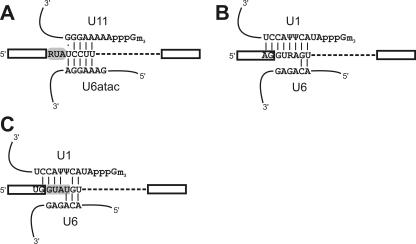
The overall assembly pathway of the minor spliceosome is similar. Initially, U11 base pairs with the 5′ss and U12 base pairs with the BPS (16, 35, 42). U11 is displaced from the 5′ss by U6atac after the association of the U4atac/U6atac/U5 tri-snRNP, and this displacement is followed by additional rearrangements that result in a catalytic core with an architecture highly similar to that of the major spliceosome (9, 14, 32). However, intriguing differences during the initial intron recognition step are observed. For example, U12-type E complexes have not been detected. Instead, the 5′ss and BPS are recognized cooperatively by a preformed U11/U12 di-snRNP (10, 36), suggesting that components connecting the 5′ss and the BPS are already present in the di-snRNP prior to prespliceosome formation. The initial base-pairing interactions at the 5′ss in the two spliceosomes are also different. The U1-5′ss interaction spans nucleotides (nt) −2 or −1 through +6 relative to the 5′ss both in mammals and in Saccharomyces cerevisiae (see Fig. 1B and C). In contrast, the first three nucleotides of U12-type introns (RUA) do not engage in base pairing with either U11 or U6atac snRNAs (see Fig. 1A), but their identities are almost 100% conserved (31). Mutations in the +1 and +2 positions block U12-dependent splicing and lead to the activation of cryptic splice sites (2). This observation suggests that a protein interacts with the RUA motif in a sequence-specific manner during intron recognition.
FIG. 1.
RNA-RNA interactions at the 5′ss. (A) Base-pairing interactions between the U12-type 5′ss and the human U11 and U6atac snRNAs. The putative protein binding site is shaded. (B and C) Base pairing of the human (B) and yeast (C) U2-type 5′ss with U1 and U6. The sequence recognized by the yeast U1 C protein is shaded. Ψ denotes a pseudouridine residue, and 3m Gppp indicates the trimethylguanosine cap.
In addition to Sm proteins, the human 18S U11/U12 di-snRNP comprises SF3b, a heptameric protein complex also present in U2 snRNPs, plus seven proteins not found in the major spliceosome (38). Four of the latter (59K, 48K, 35K, and 25K) are associated with the 12S U11 snRNP and thus potentially play an important role in 5′ss recognition and/or prespliceosome assembly. The 59K protein, via its interaction with the U12-associated 65K protein, appears to be involved in di-snRNP formation (1). The role of the other U11-associated proteins is currently unknown.
Here, we show that the 48K protein makes direct contact with the +2 position of the U12-type 5′ss, demonstrating for the first time an interaction between a U11 protein and the pre-mRNA. In addition, we provide evidence that 48K is essential for the splicing of U12-type introns in vivo and also contributes to U11/U12 di-snRNP formation and stability, potentially by interacting with the di-snRNP interface protein U11-59K.
MATERIALS AND METHODS
Oligonucleotides.
DNA oligonucleotides were purchased from Sigma-Aldrich or from Oligomer Oy. 2′-O-Methyl (2ome) RNA oligonucleotides and 2ome DNA chimeric oligonucleotides were purchased from the Keck Oligonucleotide Synthesis Facility at Yale University or from Proligo. Small interfering RNA (siRNA) oligonucleotides were synthesized in-house or obtained from Qiagen. The 2ome oligonucleotides used for the pretreatment of nuclear extracts were as follows: U2b, used to block U2-dependent splicing (18); U6atac1-20, used to arrest U12-dependent spliceosome assembly at the A complex stage (34); and U12a, used to block the U12-BPS interaction (9, 35). DNA oligonucleotides used for cloning intron 9 of the mouse Vps16 gene were Vps16-1 (AGGAAAGGGCTGTTGTTGTG) and Vps16-2 (CTTCATGCAGGAACTCATGG), and those for generating the template for the in vitro transcription of the Vps16 substrates were Vps16-7 (GCGAAGCTTAATACGACTCACTATAGGGAAAGGGCTGTTGTTGTGG) and Vps16-8 (TACTTACCTTCATGCAGGAACTCATGG). DNA oligonucleotides used for RNase H assays were P120-13 and P120-26 (10), which are complementary to the 5′ss (nt −4 through +10) and intron (nt +90 through +103) of the wild-type (WT) P120 construct, and P120-182 and P120-193, which are essentially similar to P120-13, with appropriate changes to complement the mutations CC+5+6GG and A+3G in the P120 mutant substrates, respectively. The following 21-nt siRNA duplexes with 2-nt deoxyribosylthymine (dT) 3′-end overhangs were used for RNA interference (RNAi; only the sense strand sequence is shown) and were annealed as described previously (7): BB1 (targeting the fly luciferase gene; CGTACGCGGAATACTTCGA-dT-dT), 139-1 (targeting the 48K open reading frame; CGGTTTGTTTGTGATCTAA-dT-dT), and CT2 (targeting the human Prp8 [hPrp8] gene 3′ untranslated region; GGCCGCTGACATTCAGCAG-dT-dT). Quantitative real-time PCR (qRT-PCR) analysis of siRNA-targeted mRNAs was carried out using the following gene-specific primers: 48Kfor (specific for exons 7 and 8; GATGCCGAAAAGAATGAAGAAAG), 48Krev (specific for exon 8; TGGGCTTCTACTCCTATCCCTTC), GLUD2for (TCGTGGAGGACAAGTTGGTG), and GLUD2rev (TTGCAGGGCTTGATGATCCG). The RT-PCR primers used to detect the effect of siRNA-induced knockdown on splicing are described in Table S1 in the supplemental material, along with their target introns.
Plasmids.
For the construction of pVps16-i9, the last 70 nt of exon 9, intron 9, and the first 89 nt of exon 10 of the mouse Vps16 gene (Ensembl release 36 accession no. ENSMUSG00000027411; www.ensembl.org) were amplified by PCR from genomic mouse (strain Sv129) DNA. This fragment was then inserted into pCRII-TOPO. For the production of RNA site-specifically labeled with 4-thiouridine (4SU), plasmid pVps16-e9m was constructed, in which nt −70 to +2 (relative to the 5′ss) of pVps16 were modified to contain a T residue solely at position +2, similar to the construction of the previously described P120 cross-linking substrate (8, 9). The mutations CC+5+6GG and A+3G were introduced into pP120 (35) and pVps16-i9 by PCR and verified by sequencing.
Splicing substrates.
Capped pre-mRNA substrates uniformly labeled with 32P were produced by in vitro transcription as described previously (9). For cross-linking, substrates containing a 4SU residue and/or a 32P phosphate group at a specific site were constructed using the two-piece RNA ligation protocol (9). Substrates lacking the BPS were truncated 3 nt upstream of the BPS.
In vitro spliceosome assembly and splicing.
In vitro splicing was performed as described previously (21), except that the concentration of the U2b oligonucleotide was 2.4 μM. Polyvinyl alcohol (1.0%) was used in all reaction mixtures except those subjected to native-gel analyses and RNase H protection assays. The concentrations of splicing substrates were 2 nM for native-gel analyses, RNase H protection assays, and splicing assays and 10 nM for cross-linking experiments. To deplete ATP, ATP and creatine phosphate were omitted and the reaction mixture was preincubated for 30 min before the addition of the substrate. To deplete Mg2+, MgCl2 was omitted and 2.5 mM EDTA was added. Blocking with 2ome oligonucleotides, RNase H assays, and native-gel analyses were performed as described in reference 10, and Northern blot analyses were performed as described in reference 35.
Cross-linking.
For 4SU cross-linking, 5 to 20 μl of a splicing reaction mixture was irradiated for 3 min with a 337-nm N2 excimer laser (PSX-100; Neweks Ltd., Estonia) and then subjected to treatment with a nuclease mixture (0.01 mg of micrococcal nuclease/ml, 0.02 U of RNase T1/μl, 0.1 mg of RNase A/ml) at 37°C for 30 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; on a 4 to 12% NuPAGE or 8 to 16% Precise protein gel) and detected by autoradiography or phosphorimager (Fuji FLA-5010) analysis.
IPs.
Rabbit antibodies against the U11/U12 25K, 35K, 48K, 59K, and 65K proteins and SF3b49 were raised as described previously (6, 38). Twenty-microliter aliquots of affinity-purified antibody or serum (in the case of anti-48K and anti-SF3b49) were preabsorbed to 20 μl of protein A-Sepharose beads in 200 μl of 100 mM Tris-HCl (pH 8.0)-phosphate-buffered saline (PBS) overnight at 4°C. Beads were washed three times with PBS, and 20 μl of a UV-treated splicing reaction mixture was added. Samples were either left in their native state or treated with RNases and denatured at 95°C for 2 min in the presence of 0.15% SDS prior to immunoprecipitation (IP) (see Fig. 3C). IPs were performed overnight in 0.1% Nonidet P-40 (NP-40)-PBS, after which the beads were washed five times with buffer (10 mM Tris-HCl [pH 8.0], 0.1% NP-40, 150 mM NaCl). Proteins were eluted by adding 20 μl of SDS-PAGE loading buffer supplemented with 4 μl of nuclear extract as a carrier and incubating at 95°C for 10 min. IP of U11/U12 snRNPs from nuclear extracts with SF3b155 antibodies was performed essentially as described previously (39).
FIG. 3.
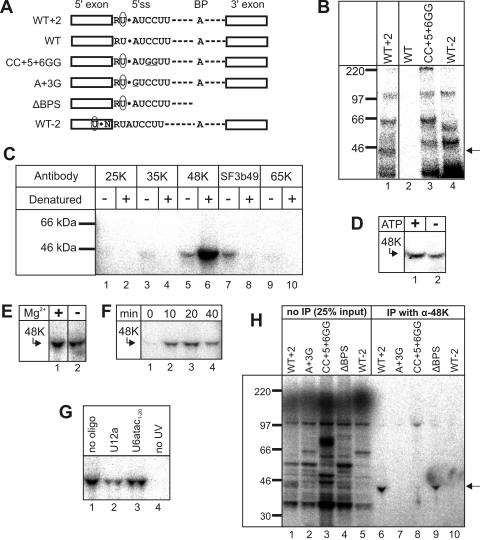
The U11-48K protein interacts with the +2 position of U12-type introns. (A) Schematic representation of the cross-linking substrates used in this study. 4SU is indicated by a circled U, and the position of the 32P-labeled phosphate group is indicated by a black dot. Underlining indicates mutated nucleotides. BP, branch point; WT−2, substrate with 4SU at the −2 position; ΔBPS, substrate truncated directly upstream of the BPS. (B) Cross-linking was performed after 40 min of splicing with the P120 substrate indicated above each lane. Cross-linked proteins were separated by SDS-PAGE and visualized by autoradiography. Molecular size markers are indicated on the left. The 45-kDa cross-linked protein is indicated by an arrow. (C) Identification of the cross-linked 45-kDa protein. Cross-linking was performed with the WT+2 P120 substrate. Samples were immunoprecipitated with the indicated antibody either directly (−) or after RNase digestion and denaturing with SDS (+) and were analyzed as described in the legend to panel B. (D to G) Effect of ATP depletion (D) or Mg2+ depletion (E) on 48K-5′ss cross-link formation. Reactions were performed with (+) or without (−) exogenously added ATP or Mg2+. For panels D to G, cross-linking was performed with the WT+2 P120 substrate and samples were denatured and immunoprecipitated with anti-48K antibody. (F) Time course of 48K-5′ss cross-link formation. Samples were removed from the reaction mixture at the indicated times after the start of splicing and placed on ice prior to cross-linking. (G) Effects of anti-snRNA 2ome oligonucleotides on 48K cross-link formation. Splicing was performed in the presence of a 2ome oligonucleotide against the indicated snRNA. For lane 4, UV irradiation was omitted. (H) Cross-linking was performed after 40 min of splicing with the P120 substrate indicated above each lane. Cross-linked proteins immunoprecipitated with anti-48K antibody (α-48K) were separated by SDS-PAGE (lanes 6 to 10) along with 25% of the input (lanes 1 to 5). Molecular weight markers are shown on the left. The 48K band is indicated by an arrow.
Far-Western overlays.
Coupled transcription and translation of the 48K protein and mutant forms thereof and a 59K mutant protein comprising amino acids 137 to 336 (59K137-336) were performed using the TNT system (Promega) in the presence of [35S]methionine or [14C]leucine, or alternatively, in vitro-transcribed mRNA was isolated and translated in wheat germ extract (Zoegene, Japan) essentially as described in reference 29. Far-Western overlay analyses were performed with 35S-labeled protein, and purified U11/U12 proteins were immobilized on nitrocellulose as described previously (1).
Yeast two-hybrid assays.
Yeast two-hybrid analyses were performed with the Matchmaker 3 two-hybrid system (Clontech). cDNAs encoding full-length or truncated proteins were cloned into pGBKT7 (the bait vector) or pGADT7 (the prey vector). S. cerevisiae strain AH109 was cotransformed with bait and prey plasmids, and the plasmids were selected on agarose plates prepared with minimal synthetic dropout medium lacking leucine and tryptophan by incubation at 30°C for 3 to 5 days. To identify protein-protein interactions, cotransformants were then replicated on plates additionally lacking histidine and adenine (plates containing synthetic dropout medium without Ade, His, Leu, or Trp).
StrepII tag pulldown assays.
For pulldown assays, cDNA encoding 59K137-336 was cloned into pEU3-NII-StrepII via PCR-based techniques and 59K137-336 containing a C-terminal StrepII tag was translated in wheat germ extract. Forty microliters of the translated 59K137-336 was incubated with 40 μl of translated 48K for 90 min at 0°C, prior to the addition of 15 μl of Strep-Tactin Sepharose (IBA, Göttingen, Germany) and 200 μl of IPP150 buffer (20 mM HEPES-KOH, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.05% NP-40). The mixture was incubated with end-over-end rotation for 1 h, and after the beads were washed four times with IPP150 buffer, bound protein was recovered and analyzed by SDS-PAGE.
Cell culture, RNAi, and qRT-PCR analysis of siRNA-targeted mRNAs.
Culturing of HeLa S6 cells and the transfection of cells with siRNA duplexes by using Oligofectamine (Invitrogen) were performed essentially as described in reference 38. The targeted regions were not found in any other known genes via BLAST searches of the human genome. To assay for effects on cell growth, HeLa cells were harvested 24, 48, 72, and 96 h after transfection with siRNA and counted with a CASY model TT cell counter according to the instructions of the manufacturer (Schaerfe System, Germany). For 48K rescue experiments, the 48K cDNA with four mutations (silent on the amino acid level) in the region bound by the 139-1 siRNA was cloned into the pcDNA4 expression vector (Invitrogen), generating pcDNA-48K. Twelve hours after siRNA addition, cells were transfected with pcDNA-48K (75 ng per 2.25 × 105 cells) by using Transfectin lipid reagent according to the instructions of the manufacturer (Bio-Rad). Total cellular RNA was isolated using an RNeasy mini kit (Qiagen) and digested with RQ1 DNase (Promega) to remove any contaminating DNA. The level of 48K mRNA remaining after transfection with siRNA BB1 versus siRNA 139-1 was determined by qRT-PCR with an Opticon continuous-fluorescence detector (MJ Research). As an internal control, the level of mRNA encoding glutamate dehydrogenase 2 (which is not spliced) was also determined by qRT-PCR. Small-scale nuclear extracts were prepared essentially as described in reference 19. Nuclear extracts were fractionated on 10 to 30% glycerol gradients, and snRNAs were detected by Northern blotting as described previously (38).
RT-PCR analysis of endogenous pre-mRNA in 48K knockdown cells.
The removal of U12- and U2-type introns from pre-mRNA was analyzed by RT-PCR with total cellular RNA from control and 48K knockdown cells after 72 h. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) with oligo(dT) primers, followed by 26 cycles (32 cycles with RCD8 and PPP2R2A primers) of amplification using Phusion DNA polymerase (Finnzymes). For details of the analyzed introns and the primers used, see Table S1 in the supplemental material. The RT-PCR products were separated on 2.5 to 3.0% Metaphor agarose (Cambrex) gels.
RESULTS
The mutation of the conserved +3 position of U12-type introns blocks prespliceosome assembly.
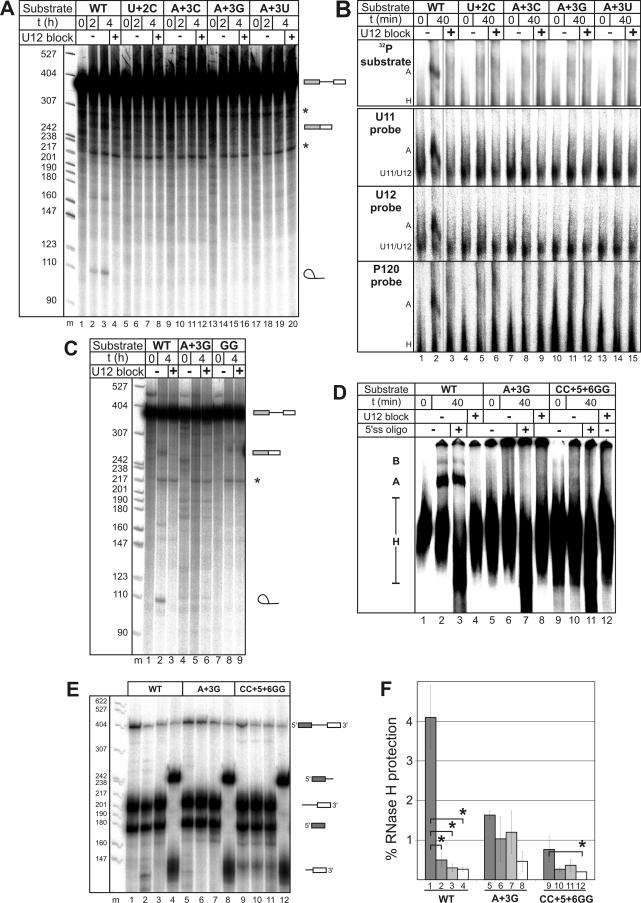
The RUA motif at the 5′ss of U12-type introns is nearly 100% conserved (31) (Fig. 1A), and the identities of the first two nucleotides of this motif are critical for splicing in vivo (2). Here, we initially tested whether mutations at the +3 position affect spliceosome assembly and/or the splicing of a pre-mRNA substrate (the P120 substrate) containing a U12-type intron. The P120 substrate with a U+2C mutation was used as a positive control. The WT P120 substrate was spliced in vitro, as indicated by the accumulation of excised intron products (Fig. 2A, lanes 2 and 3), and formed a stable A complex (Fig. 2B, lane 2) whose formation could be blocked by the addition of an anti-U12 oligonucleotide (Fig. 2B, lane 3). In contrast, no splicing or A complex formation was observed when A+3 was mutated to C, G, or U (Fig. 2A and B). Thus, the identity of the third intron nucleotide is also critical for splicing.
FIG. 2.
5′ss mutations block spliceosome assembly. (A) In vitro splicing of 32P-labeled P120 splicing substrates. Substrates, incubation times, and the presence or absence of anti-U12 2ome oligonucleotide used to block A complex formation are indicated at the top. After splicing, RNA was separated on a 7.5 M urea-5% acrylamide gel and visualized by autoradiography. DNA molecular weight markers (m) are indicated on the left, and the positions of pre-mRNAs (bars and line), ligated exons (bars), and the excised intron (lariat) are indicated on the right. Asterisks indicate unspecific products. t, time; +, present; −, absent. (B) Splicing complex formation with P120 splicing substrates. For the top panel, complexes with 32P-labeled pre-mRNA were separated by nondenaturing PAGE and visualized by autoradiography. For the remaining panels, complexes with unlabeled pre-mRNA were separated by nondenaturing PAGE and visualized by Northern blotting with the 32P-labeled probes indicated on the left. The positions of the U11/U12 di-snRNP and A and H complexes are indicated. (C) In vitro splicing of 32P-labeled WT, A+3G, and CC+5+6GG P120 pre-mRNAs. Splicing was analyzed as described in the legend to panel A, and symbols are as defined in the legend to panel A. (D) Splicing complex formation with 32P-labeled WT, A+3G, and CC+5+6GG P120 pre-mRNAs. Complexes were separated by nondenaturing PAGE and visualized by autoradiography. “5′ss oligo” indicates a DNA oligonucleotide complementary to the 5′ss, used to induce the hydrolysis of pre-mRNA by endogenous RNase H. H, A, and B complexes are indicated on the left. (E) Splicing complex formation with WT and mutant P120 substrates as assayed by RNase H protection assays. A DNA oligonucleotide complementary to the 5′ss was used to induce site-specific cleavage by RNase H (lanes 1, 5, and 9). Background protection of the 5′ss was monitored either by blocking the A complex as described in the legend to panel A (lanes 2, 6, and 10) or by preannealing the DNA oligonucleotide to the substrate prior to A complex assembly (lanes 3, 7, and 11). An intron-specific DNA oligonucleotide was used to determine the general background level of protection (lanes 4, 8, and 12). RNA was subsequently separated on a 7.5 M urea-6% acrylamide gel. The positions of the protected P120 substrates and their hydrolysis products are indicated on the right, and the positions of molecular weight markers are shown on the left. (F) Quantitation of 5′ss protection. The amount of protected pre-mRNA is expressed as the percentage of the total pre-mRNA in each lane of the gel in panel E. The mean values obtained from three independent experiments are shown, with error bars indicating the standard deviations. Column pairs with paired-t-test P values below 0.05 are indicated by asterisks. Columns correspond to the identically numbered lanes in panel E.
In subsequent studies, we used the A+3G substrate, in which a conservative purine-to-purine change was made. For comparison, we also tested P120 pre-mRNA with a CC+5+6GG mutation, which has been shown to block U12-dependent splicing in vivo due to the disruption of the base pairing of both U11 and U6atac snRNAs (14). Like the A+3G substrate, the CC+5+6GG substrate was not spliced and did not form stable spliceosomal complexes (Fig. 2C and D). RNase H protection assays that allow the detection of less stable U11-5′ss interactions (10) confirmed that splicing complex formation with the A+3G and CC+5+6GG substrates was essentially abolished; no protection of the 5′ss (as indicated by the lack of protection values significantly over background protection values) was observed (Fig. 2E and F). In contrast, ca. 4% of the WT substrate (i.e., ca. 10-fold more than the background level) was protected (Fig. 2F), similar to previously published observations (10). Thus, mutations in the RUA motif block spliceosome assembly at a very early stage, suggesting that the motif is recognized in a sequence-specific manner by a functionally important component during prespliceosome assembly.
The U11/U12 48K protein makes contact with the 5′ss during prespliceosome assembly.
To detect proteins interacting with the RUA motif, we performed site-specific cross-linking with WT, A+3G, and CC+5+6GG P120 pre-mRNAs, in which a 4SU residue and a single 32P-labeled phosphate group were placed at the +2 position relative to the 5′ss. Control substrates with 4SU at the −2 position, without 4SU, or with a 3′ truncation were also analyzed (Fig. 3A). Splicing and splicing complex formation were not significantly hindered by introducing 4SU at positions +2 and −2 (9, 10) (data not shown).
Splicing was performed with HeLa nuclear extracts, and cross-linking was induced by UV irradiation. RNA was then degraded enzymatically, and cross-linked proteins were separated by SDS-PAGE and visualized by autoradiography. Four bands (corresponding to ∼35, 45, 100, and 220 kDa) were reproducibly observed after 40 min of splicing with the P120 substrate containing 4SU at the +2 position (WT+2) (Fig. 3B, lane 1) but not with the substrate lacking 4SU (the WT) (Fig. 3B, lane 2). Of these bands, only the 45-kDa cross-link band was not observed with either the CC+5+6GG substrate or the substrate with 4SU at the −2 position (Fig. 3B, cf. lanes 3 and 4 with lane 1), indicating that the 45-kDa cross-linked protein interacts specifically with the +2 position in a manner dependent on either U11 or U6atac base pairing with the 5′ss. In the absence of SDS, the cross-linked 45-kDa protein was precipitated to various degrees by antibodies against the U11/U12 35K, 48K, and 65K proteins and the SF3b49 protein (Fig. 3C), suggesting that it is a U11/U12 protein. In contrast, after denaturation with SDS, which disrupts all protein-protein and protein-RNA interactions, it was precipitated solely by the 48K antibody (Fig. 3C, lane 6), identifying it as the U11/U12 48K protein.
Similar to that of U11-5′ss cross-linking (42), the efficiency of 48K-5′ss cross-linking was not significantly affected when the splicing reaction mixture was depleted of ATP (Fig. 3D) or Mg2+ (Fig. 3E). However, unlike U11 (8, 42), 48K was not efficiently cross-linked at 0°C (Fig. 3F, lane 1). The 48K cross-linked protein was present in equal amounts after incubation at 30°C for 10 and 20 min (when only the A complex forms), but the amount started to decline by 40 min (when the B complex appears) (10) (Fig. 3F, lanes 2 to 4), suggesting that 48K interacts early during spliceosome assembly in a transient manner. Indeed, blocking the U6atac-5′ss interaction with a 2ome oligonucleotide against U6atac had no effect on 48K-5′ss cross-link formation, suggesting that 48K interacts prior to B complex formation (Fig. 3G). Blocking U12-BPS base pairing with a 2ome oligonucleotide against U12 (Fig. 3G) or truncating the splicing substrate directly upstream of the BPS (Fig. 3H, lane 9) led to a 3- to 4-fold or ∼2-fold reduction in cross-linking efficiency, respectively. Thus, the 48K-5′ss interaction is not dependent on BPS recognition but may be enhanced by the cooperative recognition of the U12-type 5′ss and the BPS (10). We conclude that 48K makes contact with the 5′ss during the early stages of intron recognition, apparently prior to BPS and 5′ss recognition by the U12 and U6atac snRNAs, respectively.
Strikingly, the A+3G mutation almost completely abolished cross-linking of the 48K protein (yielding a 30- to 60-fold reduction relative to the level observed with the WT+2 construct) (Fig. 3H, lane 7). As this mutation was also shown to inhibit prespliceosome formation (Fig. 2F), the loss of the 48K cross-linked protein correlated with impaired 5′ss recognition, suggesting that the cross-link is not lost due to merely a change in the local orientations of the RNA and the cross-linker. These data suggest that 48K interacts in a sequence-specific manner with the RUA motif either directly or indirectly via an unknown partner that specifically recognizes this motif (see Discussion).
The 48K-5′ss interaction is generally found in U12-dependent spliceosomes.
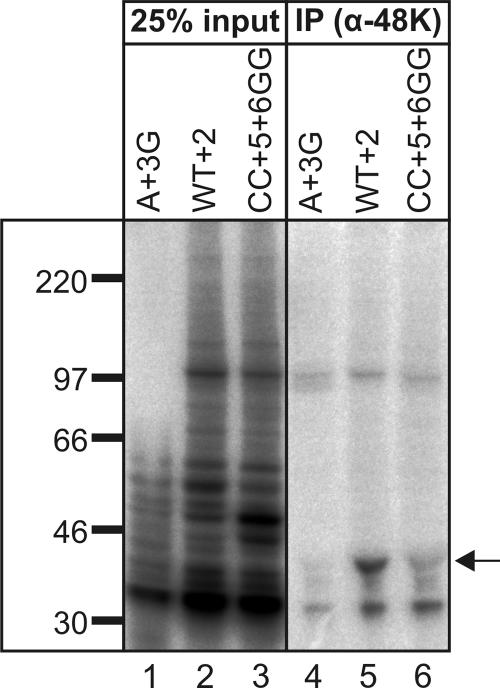
To address the generality of the 48K-5′ss interaction, we constructed a second set of splicing substrates containing a U12-type intron of the GT-AG subtype, derived from the ninth intron of the mouse Vps16 gene. Like the P120 substrates (Fig. 2C and D), the uniformly labeled Vps16 WT substrate was assembled into spliceosomal complexes and was spliced while the A+3G and CC+5+6GG mutant forms were not (data not shown). The efficiency of cross-link formation with the Vps16 substrate was somewhat higher than that with the P120 substrate, resulting in more background bands (Fig. 4). IP with anti-48K antibody revealed that 48K is efficiently cross-linked to the Vps16 WT+2 substrate (Fig. 4, lane 5). In contrast, a large reduction (ca. 50- and 10-fold) in 48K cross-linking was observed with the A+3G and CC+5+6GG mutant substrates, respectively (Fig. 4). Thus, 48K makes contact with the 5′ss of both AT-AC and GT-AG U12-type introns.
FIG. 4.
48K is cross-linked to the GU-AG U12-type intron of the Vps16 pre-mRNA. 48K cross-link formation with the Vps16 WT+2 and 5′ss mutant substrates was analyzed as described in the legend to Fig. 3H. α-48K, anti-48K antibody.
RNAi reveals an essential cellular function for the 48K protein.
To investigate whether the 48K protein carries out an essential function in vivo, we performed RNAi-mediated 48K knockdowns. We transfected HeLa cells with four different siRNA duplexes against 48K. Three led to either no or solely a moderate reduction in the level of 48K mRNA and correspondingly had no effect on cell growth (data not shown). With a fourth siRNA (139-1), cell growth was inhibited by 70% relative to that corresponding to the control siRNA BB1 (directed against fly luciferase) after 96 h (Fig. 5A). By comparison, the knockdown of the essential spliceosomal protein hPrp8 led to 90% growth inhibition. Due to the low cellular abundance of U11/U12 proteins and the relatively weak antibodies at our disposal, it was not possible to determine the amount of 48K protein in the control versus that in knockdown cells. However, qRT-PCR confirmed that only ∼10 to 15% of the 48K mRNA was present in cells transfected with siRNA 139-1 already after 24 h and at later time points but that the levels of mRNAs encoding other U11 proteins (e.g., 35K and 25K) were not affected by the addition of 139-1 siRNA (data not shown). These results indicate that 48K plays an essential role in the cell, consistent with the idea that it functions in U12-independent splicing.
FIG. 5.
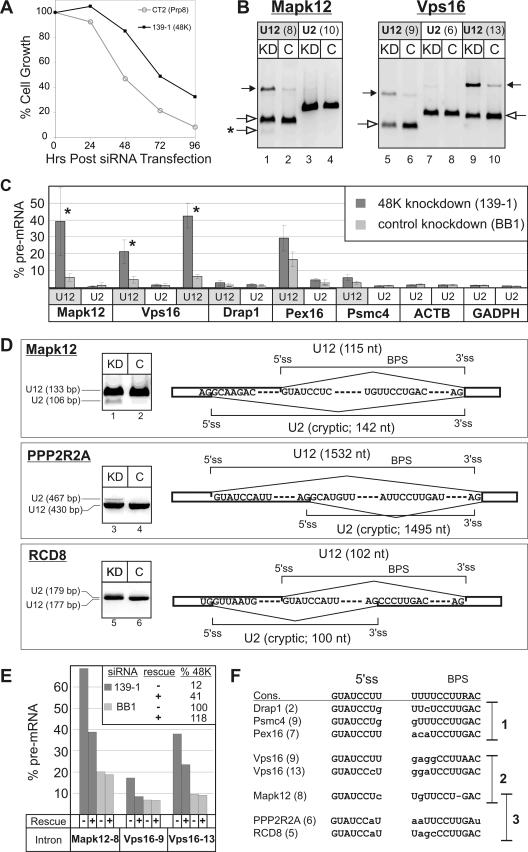
48K knockdown affects cell growth and U12-dependent splicing. (A) 48K knockdown inhibits cell growth. Cell growth, expressed as the percentages of cells present relative to the numbers of cells in the control siRNA BB1 knockdown culture, was determined 24, 48, 72, and 96 h after the transfection of cells with an siRNA duplex against the 48K protein or, as a positive control, against hPrp8. Values are the averages of triple determinations. (B and C) The splicing of a subset of U12-type pre-mRNAs is reduced after 48K knockdown. RNA from either 48K knockdown (KD) or control (C) cells was used for RT-PCR amplification across U12- or U2-type introns of the indicated transcripts. In panel B, the RT-PCR products of Mapk12 and Vps16 pre-mRNAs containing U12-type introns and spliced mRNA are indicated by closed and open arrows, respectively, and the cryptically spliced product is indicated by an asterisk. Intron numbers are indicated in parentheses. (C) The amount of pre-mRNA as a percentage of the total pre-mRNA and mRNA for all introns studied is shown. Mean values obtained from six (Mapk12 and Vps16 transcripts) or four experiments are shown, with error bars indicating standard deviations. Column pairs with paired-t-test P values below 0.01 are marked with asterisks. ACTB, beta-actin transcripts. (D) 48K knockdown leads to the activation of cryptic splice sites. The RT-PCR products of mRNA spliced by the U12- or U2-type spliceosomes are indicated on the left. Splice site usage is shown schematically on the right, with the intron lengths indicated in parentheses. The normal U12-type splice sites are indicated above, while the cryptic U2-type splice sites activated in the knockdown cells are indicated below. (E) The inhibition of U12-type splicing is partially reversed by transfection with the plasmid encoding 48K. The rescue plasmid (pcDNA-48K) was added 12 h after transfection with BB1 or 139-1 siRNA, as indicated, and RNA was isolated 72 h posttransfection with siRNA. The level of unspliced pre-mRNA was determined as described in the legend to panel C, and the averages from triplicate RT-PCRs are shown. The presence (+) or absence (−) of the rescue plasmid is indicated below the graph, and the level of 48K mRNA in each case is shown in the insert. (F) Comparison of the 5′ss and BPS of U12-type introns investigated by RT-PCR. The human consensus (cons.) sequences are shown at the top. The intron number is in parentheses. Deviations from the consensus are indicated by lowercase letters, and deletions are indicated by hyphens. The introns are divided into three groups according to the severity of the 48K knockdown effect, as follows: group 1, little or no effect; group 2, reduced U12-dependent splicing; group 3, activation of cryptic splice sites.
RNAi knockdown of 48K leads to the activation of cryptic splice sites.
To ascertain whether the observed reduction in cell viability was due to a defect in U12-dependent splicing, we performed RT-PCR with endogenous RNA isolated from cells 72 h after the addition of siRNA. The seven U12-type intron-containing transcripts studied here were recently described by Sheth et al. (31). We observed a modest reduction in the splicing of the U12-type introns of Mapk12 and Vps16 pre-mRNAs in the 48K knockdown cells, as evidenced by an increase in unspliced pre-mRNA and a corresponding reduction in mRNA (Fig. 5B and C). In contrast, splicing of U2-type introns of the same pre-mRNAs or of the beta-actin and glyceraldehyde-3-phosphate dehydrogenase (GADPH) pre-mRNAs, which lack U12-type introns, was not affected (Fig. 5C). However, the splicing efficiencies for some of the U12-type introns assayed were not significantly affected by 48K knockdown (a maximum difference of twofold in Drap1, Pex16, and Psmc4 pre-mRNA levels was detected) (Fig. 5C), presumably due to the residual amount of 48K present in the cells (see above), which for some U12-type introns may still be sufficient for splicing.
To exclude the possibility that the observed inhibition of U12-type splicing was due to off-target effects of the 48K 139-1 siRNA, cells were transfected 12 h after siRNA addition with a plasmid containing the 48K cDNA with silent point mutations in the region targeted by 139-1 (designated pcDNA-48K). The level of 48K mRNA (72 h after transfection with siRNA) increased in the 48K knockdown cells from 12% in the absence of pcDNA-48K to 41% in its presence, demonstrating the partial rescue of 48K levels (Fig. 5E). Significantly, the splicing of the U12-type introns Mapk12-8, Vps16-9, and Vps16-13 was partially restored upon the addition of pcDNA-48K, as evidenced by the significant decrease in the levels of pre-mRNAs containing these introns in 48K knockdown cells that received the plasmid compared to those in cells that did not (Fig. 5E). In contrast, U12-type splicing in BB1 control knockdown cells was not affected by the addition of pcDNA-48K (Fig. 5E). Taken together, these data demonstrate that 48K knockdown leads to a reduction in the splicing of at least a subset of U12-type introns.
For Mapk12, a faster-migrating band not present in the control reaction mixture was detected in the knockdown cells (Fig. 5B, lane 1). Sequence analysis of this band revealed that it arose by the activation of a cryptic, U2-type 5′ss (Fig. 5D). The activation of cryptic splice sites after 72 h of 48K knockdown was also observed with two other transcripts, namely, those for PPP2R2A and RCD8 (Fig. 5D), neither of which displayed a significant accumulation of pre-mRNA after 48K knockdown. RCD8 transcripts displayed the highest level of cryptic splicing, with ∼30% of the mRNA in the knockdown cells spliced at cryptic sites (data not shown).
Interestingly, the 5′ss and BPS of the U12-type introns that were most sensitive to 48K knockdown exhibited a number of deviations from the respective U12-type consensus sequences, suggesting that they may be weak sites, while those not affected by 48K knockdown had 5′ss and BPS closely matching the consensus sequences (Fig. 5F). The switch from U12- to U2-dependent splicing does not appear to be an alternative splicing event, as expressed-sequence-tag (EST) database searches revealed no Mapk12, PPP2R2A, or RCD8 ESTs generated from splicing at the observed U2-type splice sites. Thus, 48K plays a critical role in the recognition of U12-type introns in vivo.
The knockdown of 48K leads to a reduction in U11/U12 di-snRNP levels.
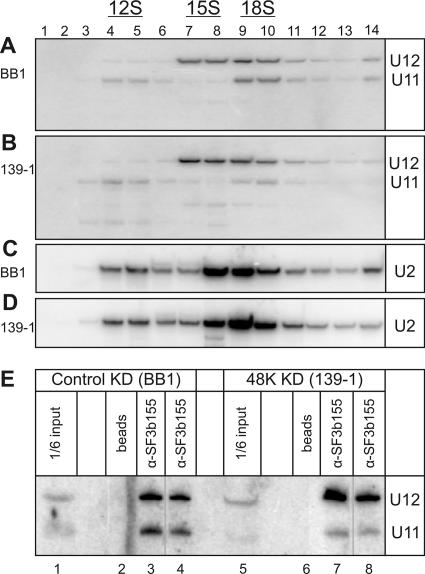
To test whether 48K contributes to di-snRNP stability and formation, we prepared nuclear extracts from control or 48K knockdown cells. Extracts were then subjected to glycerol gradient centrifugation, and after fractionation, the sedimentation behavior of snRNPs containing U11 or U12 was determined by Northern blot analysis with the corresponding 32P-labeled probe; as an internal control, the distribution of U2 snRNPs was also analyzed (Fig. 6A to D). No significant difference in the sedimentation behavior of U2-containing snRNPs (i.e., 17S and 12S particles) was observed in extracts from the control versus the 48K knockdown cells (Fig. 6C and D). In contrast, upon 48K knockdown, a smaller percentage of the total U11 snRNA present sedimented in 18S peak fractions 9 to 10 (20 versus 39% in the control as determined by phosphorimager analysis) and a larger percentage sedimented in 12S peak fractions 4 to 5 (27 versus 14% in the control) (Fig. 6A and B), suggesting that the knockdown cells contained ∼50% less U11/U12 di-snRNP than the control cells. Likewise, more U12 sedimented as a 15S monoparticle in the 48K knockdown cells; however, due to the close migration of 15S and 18S particles, a meaningful quantitation of U12 mono-snRNPs versus di-snRNPs could not be carried out. To provide additional evidence for a reduction in di-snRNP levels after 48K knockdown, we also performed co-IP assays with antibodies directed against the U12-associated SF3b155 protein. Significantly, on average only half as much (50% ± 7%) of the input U11 was precipitated together with U12 by anti-SF3b155 antibodies from the 48K knockdown extracts as from the control extracts (Fig. 6E, cf. lanes 3 and 4 with lanes 7 and 8), consistent with the conclusion that 48K knockdown cells contain ∼50% fewer U11/U12 di-snRNPs than control cells. Thus, 48K appears to contribute to the formation and/or the stability of the U11/U12 di-snRNP.
FIG. 6.
48K knockdown leads to a reduction in U11/U12 di-snRNPs. (A to D) Fewer U11 and U12 snRNPs sediment as an 18S di-snRNP upon the knockdown of 48K. Nuclear extracts isolated from cells treated with BB1 (A and C) or 139-1 (B and D) siRNA for 72 h were separated on a 10 to 30% glycerol gradient. RNA was recovered from the indicated gradient fraction, and Northern blotting was performed with 32P-labeled probes against U11 and U12 snRNAs (A and B) or subsequently with a probe against U2 (C and D). (E) Less U11 is coprecipitated with U12 snRNPs from 48K knockdown (48K KD) extracts than from control knockdown (control KD) extracts. IPs were performed with SF3b155 antibody (α-SF3b155) and nuclear extracts from BB1- or 139-1-treated cells, as indicated. RNA was recovered and analyzed by Northern blotting with probes against U11 and U12. In lanes 4 and 8, 20% less extract was analyzed.
The N terminus of 48K interacts with the central region of U11-59K.
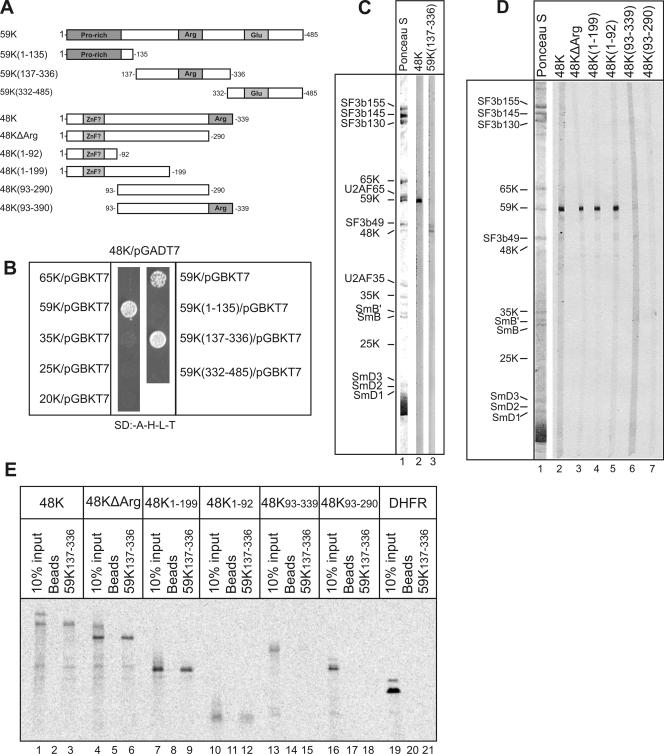
To identify protein interaction partners of the 48K protein within the U11/U12 di-snRNP, we first performed two-hybrid interaction assays with the yeast S. cerevisiae. When 48K was used as bait, it interacted strongly with U11-59K but not with any of the other U11/U12-specific proteins tested (Fig. 7B, left panel). As the 48K cDNA could not be cloned into the pGBKT7 vector, reciprocal two-hybrid screens with 48K as prey could not be performed. To further delineate the 48K-59K interaction, 59K was divided into its N-terminal proline-rich region (59K1-135), its arginine-rich region (59K137-336), and its C-terminal region rich in glutamic acid (59K332-485) (Fig. 7A). When these 59K deletion mutant proteins were used as prey, only 59K137-336 interacted with 48K (Fig. 7B, right panel). Significantly, no interaction between 59K137-336 and the U11/U12-specific 65K, 35K, 25K, or 20K protein used as either bait or prey was observed (data not shown). These results indicate that 48K interacts specifically with amino acids 137 to 336 of the 59K protein. Attempts to express full-length 48K as a fusion protein in Escherichia coli were not successful, apparently due to the arginine-rich C terminus of 48K. To provide additional evidence that 48K interacts with 59K, we performed far-Western overlay analyses with in vitro-translated, 35S-labeled 48K or 59K137-336 protein and nitrocellulose strips containing U11/U12 proteins. Consistent with the yeast two-hybrid results, 35S-labeled 48K interacted predominantly with 59K and 35S-labeled 59K137-336 interacted predominantly with 48K (Fig. 7C, lanes 2 and 3). The 48K-59K interaction appears to be specific, as other proteins that were equally abundant (as evidenced by Ponceau staining) (Fig. 7C, lane 1) were not efficiently bound by the 35S-labeled proteins.
FIG. 7.
The N terminus of the U11-48K protein interacts with the central region of the U11 59K protein. (A) Schematic of WT and truncated 48K and 59K proteins. Arginine (Arg)-, proline (Pro)-, and glutamic acid (Glu)-rich regions and a potential zinc finger (ZnF?) are shaded gray. (B) Results from yeast two-hybrid assays with the 48K protein and the U11/U12-associated 20K, 25K, 35K, 59K, and 65K proteins and deletion mutant forms of 59K, as indicated. SD:-A-H-L-T, synthetic dropout medium without Ade, His, Leu, or Trp. (C) 48K and 59K interact in far-Western overlays. U11/U12 proteins on nitrocellulose strips were visualized by staining with Ponceau S (lane 1) or incubated with 35S-labeled, in vitro-translated 48K protein or 59K137-336 and visualized by fluorography (lanes 2 and 3). U11/U12 proteins are indicated on the left. (D) The N-terminal region of 48K interacts with 59K in Far Western overlays. Overlay analyses were performed as described in the legend to panel C. (E) The N-terminal region of 48K interacts with 59K137-336. 14C-labeled 48K and deletion mutant forms thereof were incubated alone (lanes 2, 5, 8, 11, 14, 17, and 20) or with StrepII-tagged 59K137-336 (lanes 3, 6, 9, 12, 15, 18, and 21), and pulldowns were subsequently performed with Strep-Tactin Sepharose beads. Coprecipitated 48K protein or 10% of the input (lanes 1, 4, 7, 10, 13, 16, and 19) was analyzed by SDS-PAGE and visualized by fluorography. DHFR, dihydrofolate reductase.
To further delineate which region of 48K interacts with 59K, several 48K deletion mutant proteins were constructed (Fig. 7A), translated in vitro, and first tested in Far Western overlays with nitrocellulose strips containing U11/U12 proteins. Full-length 35S-labeled 48K, as well as all deletion mutant proteins containing the 92 N-terminal amino acids (i.e., 48KΔArg, 48K1-199, and 48K1-92), interacted strongly with 59K (Fig. 7D, lanes 2 to 5). In the absence of amino acids 1 to 92, no interaction was detected (Fig. 7D, lanes 6 and 7), indicating that the N terminus of 48K is necessary and sufficient for interaction with 59K. Interestingly, this region of 48K contains a potential CHHC zinc finger motif, which is highly conserved evolutionarily (see Fig. S1 in the supplemental material). To confirm this interaction, 59K137-336 containing a StrepII tag was translated in vitro and incubated with in vitro-translated 14C-labeled 48K and deletion mutant forms thereof. 59K was then precipitated with Strep-Tactin Sepharose beads, and coprecipitating proteins were analyzed by SDS-PAGE, followed by autoradiography. Consistent with the Far Western results, all 48K mutant proteins containing amino acids 1 to 92 bound to 59K137-336 with efficiencies nearly identical to that of the WT 48K protein (∼10%), whereas mutant proteins lacking this region did not (Fig. 7E). These results indicate that 48K not only makes contact with the 5′ss of U12-type introns but also interacts with the U11-59K protein and, thus, may possess dual binding affinities.
DISCUSSION
In this study, we detected a novel interaction between the U11-48K protein and the 5′ss of U12-type introns and provided evidence suggesting that this interaction is required for proper intron recognition. In addition, we demonstrated that 48K interacts with the U11-59K protein, a protein at the interface of the U11/U12 di-snRNP, and provided evidence that 48K contributes to di-snRNP formation and/or stability. Taken together, our data provide important insights into the protein-mediated recognition of the U12-type 5′ss, as well as functionally important interactions within the U11/U12 di-snRNP.
48K makes contact with the 5′ss in early spliceosomal complexes.
We detected an interaction between 48K and the 5′ss (Fig. 3 and 4), consistent with the idea that 48K participates in U12-type 5′ss recognition and that 48K knockdown leads to impaired U12-dependent splicing by preventing this interaction. Furthermore, several lines of evidence support the idea that the 48K-5′ss interaction takes place early during intron recognition. First, 48K could be cross-linked soon after the start of splicing in vitro (Fig. 3F) and seemed to require the U11-5′ss base pairing interaction (see below). Second, 48K-5′ss cross-link formation did not require U6atac-5′ss or U12-BPS base pairing (Fig. 3G) or the presence of the BPS (Fig. 3H), suggesting that the 48K-5′ss interaction precedes U12-BPS base pairing. Finally, 48K knockdown led to the activation of cryptic, U2-type splice sites (Fig. 5D). As commitment to either U2- or U12-type splicing is thought to occur at the time of A complex formation (for a discussion, see reference 14), 48K must act during or prior to this step. 48K is associated with the U11 moiety of the U11/U12 di-snRNP, and thus, it likely is displaced from the 5′ss at a later stage of spliceosome assembly, when the U11 snRNP is thought to dissociate. Indeed, 48K cross-linking was reduced after 40 min of splicing (Fig. 3F), which correlates with reduced U11-5′ss cross-link formation (8). However, at present, we cannot exclude the possibility that 48K remains bound to the 5′ss at later stages of spliceosome assembly.
The 48K-5′ss interaction appears to be greatly enhanced by U11-5′ss base pairing. The CC+5+6GG mutation, which abolishes U11-5′ss base pairing (14), greatly diminished 48K-5′ss cross-link formation (Fig. 3H and Fig. 4). Theoretically, this mutation could also hinder protein interactions with nt +5 and/or +6. However, as the splicing defect caused by the CC+5+6GG mutation could be eliminated by compensatory mutations in U11 and U6atac snRNAs in vivo (14), it appears that there are no critical RNA-protein interactions with these nucleotides. In summary, 48K interacts with the 5′ss during the very first steps of intron recognition, apparently together with or after U11 snRNA.
48K may recognize the 5′ss in a sequence-specific manner.
The mutation of the conserved RUA motif (A+3G), which prevents base pairing with the U11 snRNA, abolished both the stable interaction of U11 snRNP with the 5′ss (Fig. 2) and the 48K cross-link (Fig. 3 and 4). This finding suggests indirectly that 48K recognizes the 5′ss in a sequence-specific manner and, further, that the 48K-5′ss interaction helps to stabilize the U11-5′ss interaction. In the major spliceosome, the 5′ss is recognized by the U1-C protein, which can bind the 5′ss on its own and subsequently promotes the U1-5′ss interaction (4, 5, 27). It is not clear whether 48K and U1-C act in mechanistically similar manners. However, the different compositions of U1 and U11 proteins hint at mechanistic differences. Furthermore, in contrast to the U1-C-5′ss interaction, data presented here suggest that the 48K-5′ss interaction occurs either concomitantly with or after U11-5′ss base pairing.
Whether 48K can bind the U12-type 5′ss on its own is currently also unknown; due to the inability to overexpress the WT 48K protein in E. coli, RNA binding studies with purified 48K could not be performed. Recently, serine-arginine-rich (SR) proteins were shown to interact with a U12-type 5′ss (30), and they may possibly mediate the 48K-5′ss interaction either directly or indirectly by modulating U11-5′ss base pairing. Other U11/U12 proteins not found in the major spliceosome may also contribute to 5′ss recognition. Most notably, the U11/U12-20K protein shows some similarity to U1-C, including an N-terminal zinc finger, and it was thus proposed that the 20K protein may be involved in U12-type 5′ss recognition (38). However, 20K was not copurified with U11 monoparticles. Furthermore, we did not detect a 20K-5′ss interaction via cross-linking, at least at the −2 or + 2 position (data not shown). Thus, additional experiments are required to clarify whether proteins in addition to 48K uniquely contribute to 5′ss recognition in the minor spliceosome.
48K plays a critical role in the recognition of U12-type introns.
RNAi-induced knockdown of 48K leads to reduced cell growth, indicating that 48K has an essential cellular function (Fig. 5A). This function appears to be related to the recognition of U12-type introns, as 48K knockdown leads to reduced U12-dependent splicing and/or the activation of cryptic splice sites in certain pre-mRNAs containing U12-type introns (Fig. 5). These effects may be due partially to the reduction of U11/U12 di-snRNP levels in 48K knockdown cells (Fig. 6). However, as snRNPs are present in large excess in the cell, it is unlikely that a 50% reduction in di-snRNP levels alone would cause the observed effects. Indeed, residual amounts (<20%) of U4atac snRNA are sufficient to support the splicing of endogenous pre-mRNAs at WT levels (25), suggesting that the reduction of a single spliceosomal component by 80 to 90% does not a priori lead to splicing inhibition.
The removal of some U12-type introns was not significantly affected by 48K knockdown. As residual amounts of 48K mRNA remain after knockdown, some introns appear to be more sensitive to 48K levels than others. Interestingly, 48K knockdown had little or no effect on the removal of introns in which both the 5′ss and BPS matched the consensus sequences, whereas introns with deviations in their 5′ss or BPS were removed less efficiently or no longer recognized as U12-type introns after knockdown. It is likely that the formation and/or stability of the minor prespliceosome is dependent on the combined effects of several sequence-specific interactions at the 5′ss and BPS, including U11-5′ss base pairing, 48K-5′ss interaction, and U12-BPS base pairing, possibly augmented by SR proteins that recognize nearby regulatory elements (13, 30). Thus, 48K knockdown may lead to the defective formation of prespliceosomes solely on introns with suboptimal recognition sequences, especially in the absence of splicing enhancers.
Protein-protein interactions within the U11/U12 di-snRNP.
Our knockdown data indicated that 48K contributes to the stability of the U11/U12 di-snRNP (Fig. 6). Interestingly, direct interactions between 48K and integral components of U12 (e.g., SF3b or the 65K and 20K proteins) were not detected, suggesting that 48K may not be located at the interface between the U11 and U12 moieties of the di-snRNP. However, protein-protein interaction studies (Fig. 7) demonstrated that 48K interacts with another U11-associated protein, namely, 59K, and that this interaction involves the central arginine-rich region of 59K and the N-terminal region of 48K. The latter contains a putative zinc finger, a motif shown in several cases to mediate protein-protein interactions (11). Significantly, the C-terminal part of 59K interacts with the U12-65K protein, and thus, both of these proteins lie at the interface of the di-snRNP and likely play an important role in di-snRNP formation and stability (1). Thus, it is conceivable that 48K, via its interaction with 59K, may indirectly affect the association of the U11 and U12 snRNPs in the U11/U12 di-snRNP.
Our studies further suggest that 48K possesses dual binding activities, interacting not only with 59K but also with the 5′ss. At present, the regions of 48K responsible for RNA interactions are not known, as the inability to express WT 48K in significant amounts in E. coli has hampered studies to examine this issue. The sequence of 48K also offers few clues as to which amino acids may be involved in RNA binding, as it contains no known motifs apart from a putative zinc finger. However, there is an arginine-rich sequence at its C terminus (38) which, due to its overall positive charge, may exhibit general affinity for RNA. In addition, a centrally located region of 48K (i.e., amino acids 206 to 256 in humans) is highly conserved from plants to humans (20), suggesting that it may be involved in RNA and/or additional protein interactions.
Intron bridging in the U12-dependent prespliceosome.
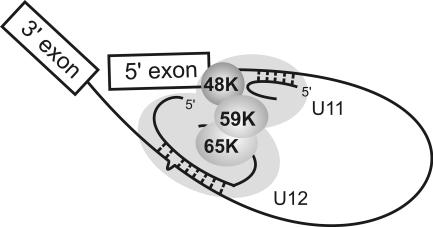
To enable splicing catalysis, the functional sites of the pre-mRNA, i.e., the 5′ss, the BPS, and the 3′ss, are brought into close proximity during the earliest assembly phases of both spliceosomes (3, 8, 15). For the major spliceosome, SR proteins and the DEAD box protein Prp5 have been implicated in bridging U1 at the 5′ss and U2 at the BPS in E and A complexes (40, 41). In contrast, in the minor spliceosome, U11 and U12 are bridged by integral proteins of the U11/U12 di-snRNP, namely, 59K and 65K, with 65K also directly interacting with the U12 snRNA (1). Our data revealing 48K interactions with the 5′ss and 59K extend this network such that interactions involving 48K, 59K, and 65K would bridge the 5′ss and U12 bound to the BPS (Fig. 8). The link between components interacting with the 5′ss and the BPS is also consistent with the idea that combinatorial interactions at these sites contribute to intron recognition, as discussed above.
FIG. 8.
Model of interactions in the U12-type spliceosomal A complex. The U11/U12 di-snRNP proteins are represented by gray shading, with 48K, 59K, and 65K highlighted. U12 snRNA is base paired to the BPS, and U11 snRNA is base paired to the 5′ss.
Implications for regulating the activity of the U12-dependent spliceosome.
Early protein-pre-mRNA interactions contributing to intron recognition may play a decisive role in determining which spliceosome ultimately forms on a particular intron. The posttranslational modification of proteins may affect U12-type intron recognition directly by either enhancing or interfering with splice site recognition or indirectly by affecting di-snRNP assembly or by changing the conformation of the complex. Intriguingly, based on the migration behavior of the 48K protein on polyacrylamide gels, it was previously suggested that the protein may be posttranslationally modified (38). The excision of U12-type introns appears to be the rate-limiting step in the splicing of genes containing both types of introns (23, 25). Therefore, the posttranslational modification of unique spliceosomal factors, such as 48K, may potentially allow for the rapid regulation of the U12-dependent spliceosome and thus affect the expression of genes with U12-type introns.
Supplementary Material
Acknowledgments
We are grateful to Marja-Leena Peltonen, Annukka Ruokolainen, Heike Behr, Tomma Eisbein, and Gabi Heyne for excellent technical assistance, Markus Hossbach for help with qRT-PCR assays, Dmitry Agafonov for helpful discussions regarding in vitro translation in wheat germ extracts, Helmut Pospiech for help with nuclear extract preparation, and members of the Frilander and Lührmann labs for critically reading the manuscript.
This work was supported by grants from the Academy of Finland and Sigrid Jusélius Foundation to M.J.F. and from the Deutsche Forschergruppe (Lu294/12-1), Fonds der Chemischen Industrie, and the Ernst Jung Stiftung to R.L. J.J.T. was supported by the Helsinki Graduate School in Biotechnology and Molecular Biology.
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Benecke, H., R. Lührmann, and C. L. Will. 2005. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J. 243057-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietrich, R. C., J. D. Fuller, and R. A. Padgett. 2005. A mutational analysis of U12-dependent splice site dinucleotides. RNA 111430-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dönmez, G., K. Hartmuth, B. Kastner, C. L. Will, and R. Lührmann. 2007. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell 25399-411. [DOI] [PubMed] [Google Scholar]
- 4.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 41986-90. [DOI] [PubMed] [Google Scholar]
- 5.Du, H., D. F. Tardiff, M. J. Moore, and M. Rosbash. 2004. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. Proc. Natl. Acad. Sci. USA 10114841-14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dybkov, O., C. L. Will, J. Deckert, N. Behzadnia, K. Hartmuth, and R. Lührmann. 2006. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 262803-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411494-498. [DOI] [PubMed] [Google Scholar]
- 8.Frilander, M. J., and X. Meng. 2005. Proximity of the U12 snRNA with both the 5′ splice site and the branch point during early stages of spliceosome assembly. Mol. Cell. Biol. 254813-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frilander, M. J., and J. A. Steitz. 2001. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell 7217-226. [DOI] [PubMed] [Google Scholar]
- 10.Frilander, M. J., and J. A. Steitz. 1999. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 13851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamsjaeger, R., C. K. Liew, F. E. Loughlin, M. Crossley, and J. P. Mackay. 2007. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 3263-70. [DOI] [PubMed] [Google Scholar]
- 12.Hall, S. L., and R. A. Padgett. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 2711716-1718. [DOI] [PubMed] [Google Scholar]
- 13.Hastings, M. L., and A. R. Krainer. 2001. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Incorvaia, R., and R. A. Padgett. 1998. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA 4709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent, O. A., and A. M. MacMillan. 2002. Early organization of pre-mRNA during spliceosome assembly. Nat. Struct. Biol. 9576-581. [DOI] [PubMed] [Google Scholar]
- 16.Kolossova, I., and R. A. Padgett. 1997. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA 3227-233. [PMC free article] [PubMed] [Google Scholar]
- 17.Krämer, A. 1988. Presplicing complex-formation requires 2 proteins and U2 snRNP. Genes Dev. 21155-1167. [DOI] [PubMed] [Google Scholar]
- 18.Lamond, A. I., B. S. Sproat, U. Ryder, and J. Hamm. 1989. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2′-OMe RNA. Cell 58383-390. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. A., and M. R. Green. 1990. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 18120-30. [DOI] [PubMed] [Google Scholar]
- 20.Lorković, Z. J., R. Lehner, R. Forstner, and A. Barta. 2005. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 111095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell, T. S., S. J. Cho, M. J. Frilander, and J. A. Steitz. 2002. Branchpoint selection in the splicing of U12-dependent introns in vitro. RNA 8579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsen, T. W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing, p. 279-307. In R. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Patel, A. A., M. McCarthy, and J. A. Steitz. 2002. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 213804-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel, A. A., and J. A. Steitz. 2003. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 4960-970. [DOI] [PubMed] [Google Scholar]
- 25.Pessa, H., A. Ruokolainen, and M. J. Frilander. 2006. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA 121883-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12340-345. [DOI] [PubMed] [Google Scholar]
- 27.Rossi, F., T. Forne, E. Antoine, J. Tazi, C. Brunel, and G. Cathala. 1996. Involvement of U1 small nuclear ribonucleoproteins (snRNP) in 5′ splice site-U1 snRNP interaction. J. Biol. Chem. 27123985-23991. [DOI] [PubMed] [Google Scholar]
- 28.Russell, A. G., J. M. Charette, D. F. Spencer, and M. W. Gray. 2006. An early evolutionary origin for the minor spliceosome. Nature 443863-866. [DOI] [PubMed] [Google Scholar]
- 29.Sawasaki, T., T. Ogasawara, R. Morishita, and Y. Endo. 2002. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 9914652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen, H., and M. R. Green. 2007. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat. Struct. Mol. Biol. 14597-603. [DOI] [PubMed] [Google Scholar]
- 31.Sheth, N., X. Roca, M. L. Hastings, T. Roeder, A. R. Krainer, and R. Sachidanandam. 2006. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 343955-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla, G. C., and R. A. Padgett. 1999. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA 5525-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92315-326. [DOI] [PubMed] [Google Scholar]
- 34.Tarn, W.-Y., and J. A. Steitz. 1996. Highly divergent U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 2731824-1832. [DOI] [PubMed] [Google Scholar]
- 35.Tarn, W.-Y., and J. A. Steitz. 1996. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84801-811. [DOI] [PubMed] [Google Scholar]
- 36.Wassarman, K. M., and J. A. Steitz. 1992. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 121276-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Will, C. L., and R. Lührmann. 2006. Spliceosome structure and function, p. 369-400. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Will, C. L., C. Schneider, M. Hossbach, H. Urlaub, R. Rauhut, S. Elbashir, T. Tuschl, and R. Lührmann. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10929-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Lührmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 204536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 751061-1070. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Y.-Z., C. M. Newnham, S. Kameoka, T. Huang, M. M. Konarska, and C. C. Query. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, Y.-T., and J. A. Steitz. 1997. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl. Acad. Sci. USA 946030-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.