Abstract
Reactive oxygen species (ROS) and oxidative stress have been considered in a variety of disease models, and cytochrome P450 (P450) enzymes have been suggested to be a source of ROS. Induction of P450s by phenobarbital (PB), β-naphthoflavone (βNF), or clofibrate in a mouse model increased ROS parameters in the isolated liver microsomes, but isoniazid treatment did not. However, when F2-isoprostanes (F2-IsoPs) were measured in tissues and urine, PB showed the strongest effect and βNF had a measurable but weaker effect. The same trend was seen when an Nfr2-based transgene reporter sensitive to ROS was analyzed in the mice. This pattern had been seen earlier with F2-IsoPs both in vitro and in vivo with rats (Dostalek, M., Brooks, J. D., Hardy, K. D., Milne, G. L., Moore, M. M., Sharma, S., Morrow, J. D., and Guengerich, F. P. (2007) Mol. Pharmacol. 72, 1419–1424). One possibility for the general in vitro-in vivo discrepancy in oxidative stress found in both mice and rats is that PB treatment might attenuate protective systems. One potential candidate suggested by an mRNA microarray was nicotinamide N-methyltransferase. PB was found to elevate nicotinamide N-methyltransferase activity 3- to 4-fold in mice and rats and to attenuate levels of NAD+, NADP+, NADH, and NADPH in both species (20–40%), due to the enhanced excretion of (N-methyl)nicotinamide. PB also down-regulated glutathione peroxidase and glutathione reductase, which together constitute a key enzymatic system that uses NADPH in protecting against oxidative stress. These multiple effects on the protective systems are proposed to be more important than P450 induction in oxidative stress and emphasize the importance of studying in vivo models.
Oxidative stress is a consideration in many diseases, including atherosclerosis, cancer, diabetes, neurodegeneration, and renal disease, as well as aging (1). In addition, ROS2 are a part of normal physiology and have beneficial roles as well, including roles in cell signaling (3–6) and protection against environmental insults, both biological and chemical (6, 7). The biochemistry of oxidative stress includes several issues. One is the nature of the damage to proteins, DNA, and lipids caused by ROS (8, 9). Another aspect is the nature of the biochemical processes involved in the production of ROS. Several enzyme systems have been considered in the production of ROS, including NADPH oxidase, xanthine oxidase, (uncoupled) mitochondrial electron transport, and P450 (1, 4, 10).
P450 enzymes are widely studied because of their roles in the metabolism of steroids, fat-soluble vitamins, fatty acids, eicosanoids, drugs, and carcinogens, plus other xenobiotic chemicals (11). In addition, the uncoupled oxidation of NADPH to generate H2O2 was demonstrated in rat liver microsomes in 1957 (12). Further research in the 1970s confirmed and further characterized the uncoupling (13, 14). Since the late 1980s (15), there has been growing interest in the generation of ROS by P450s. In some of the literature, the toxic actions of certain chemicals (e.g. 2,3,7,8-tetrachlorodibenzo-p-dioxin) have even been suggested to be the result of ROS generated due to the induction of P450s (16, 17). Different chemicals, including some drugs, selectively induce P450s and subfamilies of P450s, and P450 families 1, 2, 3, and 4 have all been proposed to have roles in the generation of ROS (18–26).
However, the literature is often inconsistent about the contribution of individual P450s to generation of ROS (22, 24, 27, 28). Moreover the great majority of reports in the area involve in vitro systems, i.e. enzymes, microsomes, and cultured cells. Few studies have been done in animals. One of the limitations has been the limited choice of appropriate biomarkers of oxidative stress in vivo.F2-IsoPs have been judged to be the most reliable in vivo measure of oxidative stress in a multicenter study organized by the NIEHS, National Institutes of Health (29). In our previous work with a rat model, we confirmed that several of the classic P450 inducers raise the level of ROS in liver microsomal assays but PB (and a polychlorinated biphenyl mixture associated with a barbiturate-type response) selectively elevated F2-IsoP levels in vivo (28). These and related experiments implicated the P450 Subfamily 2B enzymes, with some caveats (28). However, an open question raised in that study was why the discrepancy exists between the in vitro and in vivo patterns of oxidative stress produced by the various inducers.
In the present study we turned our attention to several mouse models (mostly derived from a C57BL/6 background), because several transgenics were of potential use, including P450 2e1-null (30, 31) and liver NPR-null (32, 33) mice and mice containing an Nrf2 reporter system to sense ROS (34, 35). The in vitro and in vivo results were qualitatively similar to those previously seen with rats, i.e. selective in vivo oxidative stress in animals treated with PB (28). The availability of mRNA microarray data for C57BL/6 mice treated with these inducers (36) led to new hypotheses about the basis of the patterns of in vivo oxidative stress. We analyzed NNMT and pyridine nucleotide levels, and the results led to an examination of the effect of PB and the other inducers on protective enzyme systems.
We conclude that the variable effects of P450 inducers on the regulation of pyridine nucleotide-dependent systems contribute, along with some P450 increases, to oxidative stress. In addition, the results with the transgenic mice define a role for constitutively expressed mouse P450s in oxidative stress in the liver, in vivo, and show that P450 2e1 is not involved. The results emphasize the importance of in vivo models.
EXPERIMENTAL PROCEDURES
Chemicals—βNF, CLOF, INH, and PB were purchased from Sigma. All other reagents and solvents were obtained from general commercial suppliers. All chemicals were used without further purification.
Animals—All experimental procedures involving the use of experimental animals were performed in accordance with Guiding Principles in the Care and Use of Laboratory Animals, the National Research Council Guide, and the Office of Research, Vanderbilt University Medical Center, Wadsworth Center-New York State Department of Health, and NCI, National Institutes of Health. The animals were fed a commercial solid diet (Purina 5001 rodent diet) and water ad libitum. Lighting was maintained on a 12-h light/dark cycle (lights on from 7:00 a.m. to 7:00 p.m.); the ambient temperature was maintained between 21 and 24 °C. Studies with rat liver were done with samples obtained in a previous study (28) and stored at –80 °C.
The ARE-reporter mice were developed as described previously (34). The reporter construct contained a 51-bp segment of the rat NAD(P)H:quinone oxidoreductase (NQOR1) promoter, with the core ARE inserted into a TATA-Inr minimal promoter attached to the human placental alkaline phosphatase reporter gene. Male mice expressing the transgene were mated with female C57BL/6 mice. Male pups of those matings were genotyped (34) (with the assistance of R. F. Burk and K. E. Hill, Division of Gastroenterology, Vanderbilt University), and those carrying the transgene (homozygotes) were used for studies. ARE mice used for studies were males weighing 20–23 g each.
The liver-Npr-null mice (32) used in the study were maintained in the animal facility at the Wadsworth Center, New York State Department of Health. The liver samples were collected from individual liver-Npr-null mice (on a B6N10 background) and wild-type C57BL/6 mice (female, 2 months old, 6 mice/strain); tissues were kept at –80 °C until use. The urine samples were collected over a 24-h period from individual liver-Npr-null and wild-type C57BL/6 mice (female, 6 months old, 8 mice/strain) using metabolic cages (one mouse in each metabolic cage); samples were kept at –80 °C until analysis. A second set of urine samples were collected 1 week later from the same groups of mice, and the two samples from the same mouse were pooled for analysis.
The Cyp2e1-null mouse line (Cyp2e1–/–) has been described previously (30). Male wild-type (Cyp2e1+/+) and Cyp2e1-null mice (on the 129/Sv strain background, from 2–3 months of age) were used in this study.
The following treatments were used for the ARE-reporter mice: βNF (40 mg/kg, once daily for 3 days, intraperitoneal, in corn oil), CLOF (250 mg/kg, once daily for 3 days, intraperitoneally, in corn oil), PB (continuously for 10 days, as a 0.1% solution (w/v) in drinking water), and INH (continuously for 10 days, as a 0.1% solution (w/v) in drinking water) (28). Each test group was compared with the control group, i.e. corn oil injections to match the βNF- and CLOF-treated groups and drinking water for the PB- and INH-treated groups. The animals were housed in metabolic cages (Nalgene® Metabolic Cage System, Braintree Scientific, Braintree, MA) with free access to food and water. Urine was collected for 12 h after the last administration of each compound until sacrifice. Tissue samples were harvested after mice were anesthetized with isoflurane (12 h after last administration of each substance) and exsanguinated by withdrawing blood from the inferior vena cava. Pieces of livers and brains and the right kidneys were used for in vivo measurements. The remainder of the livers and brains and the left kidneys were used for in vitro assays. Tissues were immediately frozen and stored at –80 °C until processing. Microsomes were prepared from mouse liver samples as described (37) and stored at –70 °C. Protein concentrations were estimated using a bicinchoninic acid method (Pierce-Fisher) with bovine serum albumin as a standard.
Assay of F2-IsoPs—Liver, kidney, brain, and urinary levels of F2-IsoPs were determined using a gas chromatography-mass spectrometry method, as described previously (38).
In Vitro Assays—Concentrations of total P450 in liver microsomal samples were determined as described by Omura and Sato (39). Microsomal NADPH-cytochrome c reduction was measured as described previously (37, 40). Microsomal NADPH oxidation was measured as described previously (41). Microsomal H2O2 formation was measured using a protocol adapted from Hildebrandt et al. (42). The formation of malondialdehyde and other thiobarbituric acid-reactive substrates in liver microsomes were measured with a thiobarbituric acid assay (43). Immunoelectrophoretic blotting assays were done to estimate P450 induction using rabbit polyclonal antibodies raised against rat P450s 1A1, 2B1, 2E1, and 3A1 and have been described previously (44, 45). Rabbit anti-rat P450 4A1 was a generous gift from J. Capdevila (Vanderbilt University).
Measurement of Enzyme Activities—Catalase activity was measured as the decomposition of H2O2 (46). Total superoxide dismutase activity was measured in a cytochrome c-xanthine-xanthine oxidase assay (47). NNMT activity was measured as described elsewhere (48, 49). Pyridine nucleotide concentrations were determined as described previously (50, 51). GSH peroxidase activity was measured as described previously (52). GSSG reductase activity was measured as the rate of NADPH reduction in the presence of GSSG, as described before (53).
Human placental alkaline phosphatase activity was measured using a chemiluminescence assay (34, 35). Briefly, 5% tissue homogenates (w/v) were prepared from frozen liver in 50 mm Tris-HCl buffer (pH 7.5) containing 5 mm MgCl2, 0.1 m NaCl, and 4% CHAPS. The homogenates, diluted to 1% in the same buffer devoid of CHAPS, were mixed with 0.2 m diethanolamine (1:3, v/v) and heated at 65 °C for 15 min. Samples were then incubated with a chemiluminescent substrate (Tropix Ready-To-Use CSPD Chemiluminescent Substrate with Emerald-II (Applied Biosystems, Foster City, CA)) for 20 min at room temperature in the dark. The chemiluminescent signal was detected using an Lmax Luminometer (Molecular Devices Corp., Sunnyvale, CA).
Statistical Evaluation—Data were analyzed by analysis of variance (one-way analysis of variance) followed by Dunnett's test for comparison of groups against control groups, the Student-Newman-Keul test was used for comparison of all groups pairwise, and Kruskal-Wallis tests were used for non-parametric data. The Kolmogorov-Smirnov test was used to test normality. SPSS version 13 software for Windows (SPSS Headquarters, Chicago, IL) was used for all steps of the analysis. Prism 5 software for Windows (GraphPad Software, San Diego, CA) was used to prepare graphs. Results in graphs are expressed as means ± S.E. Values of p < 0.05 were considered to be statistically significant.
RESULTS
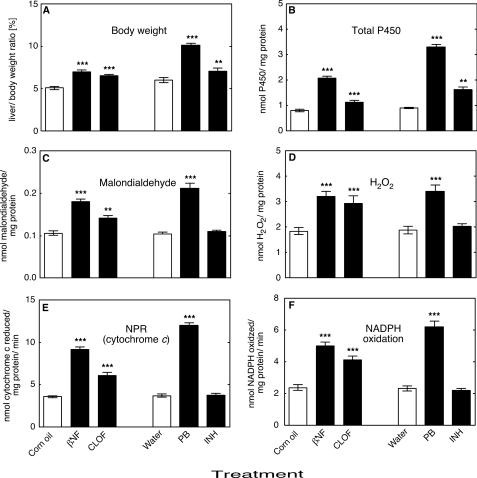
Cytochrome P450 Induction—No mortality or significant change in the body weight of different groups of ARE-reporter mice was recorded during the treatments. Treatment of animals with all of the classic P450 inducers βNF, CLOF, PB, or INH resulted in a significant enhancement in the liver/body weight ratios when compared with the control group (Fig. 1A). No significant changes were observed in kidney/body and brain/body weight ratio (results not presented).
FIGURE 1.
Effects of treatments on liver/body weight ratio and in vitro liver microsomal parameters. A, liver/body weight ratio; B, total microsomal P450; C, formation of malondialdehyde; D, microsomal H2O2 production; E, microsomal NADPH oxidation; F, microsomal NADPH-cytochrome c reduction (NPR) (specific activity). All values are presented as means ± S.E. (n = 5) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (**, p < 0.01; ***, p < 0.001).
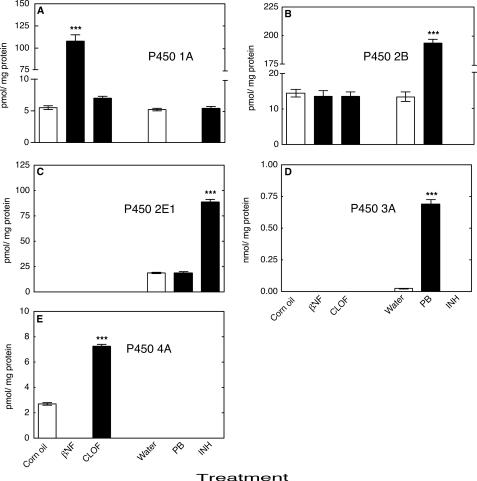
Treatment of animals with βNF, CLOF, PB, or INH significantly increased the total concentration of total P450 in mouse liver (Fig. 1B). The effects of inducers on individual liver P450s were determined by immunochemical analyses as described previously (44, 45) with the orthologous rat proteins used as standards for the mouse liver P450s (54). Subfamily 1A P450s were induced only by βNF (Fig. 2A); the Subfamily 2B P450s were induced only by PB (Fig. 2B); P450 2E1 was induced only by INH (Fig. 2C); the Subfamily 3A P450s were induced only by PB (Fig. 2D); and the Subfamily 4A P450s were induced only by CLOF (Fig. 2E).
FIGURE 2.
Induction of mouse liver P450s. Levels were estimated immunochemically and expressed in terms of comparison with the indicated rat P450s. P450 1A1 and 1A2 proteins were not resolved, whereas multiple P450 2B, 3A, or 4A proteins were detected. The values represent combined levels of all proteins detected for each P450 Subfamily. A, P450 1A Subfamily (rat P450 1A1 used as standard); B, P450 2B Subfamily (rat P450 2B1 used as standard); C, P450 2E1 (rat P450 2E1 used as standard); D, P450 3A Subfamily (rat P450 3A1 used as standard); E, P450 4a Subfamily (rat P450 4A1 used as standard). All values are presented as means ± S.E. (n = 5) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (***, p < 0.001). Measurements were not made in the blank sections of parts C–E.
In Vitro Measurements—Lipid peroxidation in mouse liver microsomes was significantly increased by treatment with PB, βNF, or CLOF, as indicated by the rates of formation of malondialdehyde (and any other thiobarbituric acid-reactive products) (Fig. 1C) as well as H2O2 (Fig. 1D). The rates of both NADPH oxidation (Fig. 1E) and NADPH-cytochrome c reduction (Fig. 2F) were increased by treatment with PB, βNF, and CLOF. None of these in vitro parameters were significantly changed by treatment of the mice with INH.
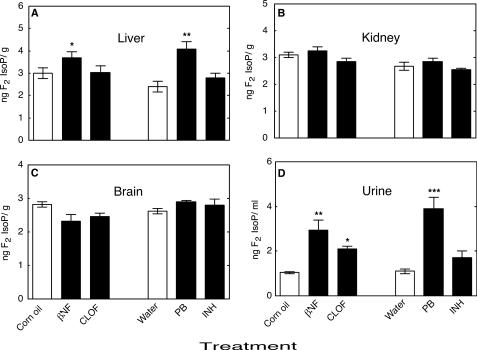
In Vivo F2-IsoP Measurements in Mice—Levels of liver F2-IsoPs were significantly increased by both PB and βNF, but no significant changes were found for CLOF or INH treatment (Fig. 3A). None of the treatments significantly increased F2-IsoPs levels in kidney (Fig. 3B) or brain (Fig. 3C) samples. Urinary F2-IsoP levels were significantly increased by treatment with PB or βNF (and less by CLOF) but not by INH (Fig. 3D).
FIGURE 3.
Measurements of F2-isoP levels in tissue and urine samples from ARE-reporter mice. A, liver F2-isoPs; B, kidney F2-isoPs; C, brain F2-isoPs; D, urinary F2-isoPs. All values are presented as means ± S.E. (n = 5), with statistical significance indicated (*, p < 0.05; **, p < 0.01; and ***, p < 0.001).
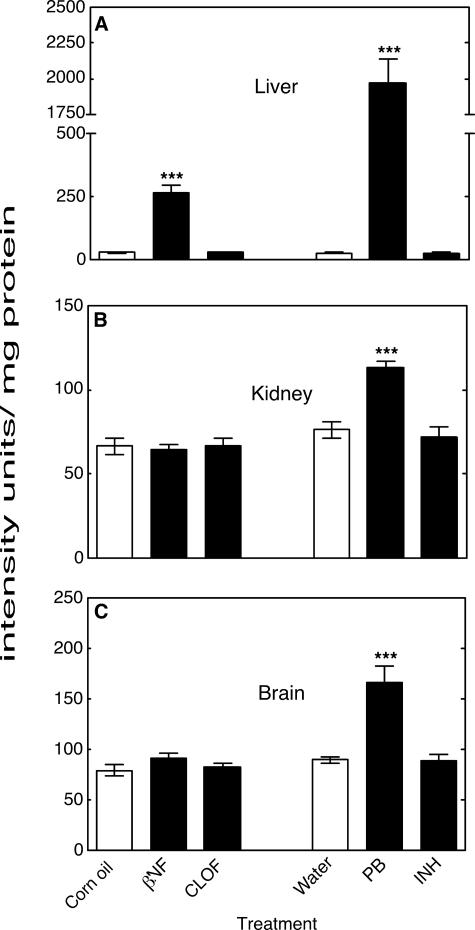
Effects of Treatments on ARE Response in a Transgenic Mouse System—The activity of an ARE-reporter enzyme (human placental alkaline phosphatase) in mouse liver homogenates was strongly increased by PB (75-fold) and also by βNF, but to a lesser extent (9-fold), when compared with the control group (Fig. 4A). The activity of the ARE-reporter in kidney and brain homogenates was increased only by PB but not by βNF. No significant changes in liver (Fig. 4A), kidney (Fig. 4B), or brain (Fig. 4C) homogenates were described after CLOF or INH treatment.
FIGURE 4.
Measurements of ARE-reporter enzyme activity in mouse tissue samples. A, liver ARE-reporter activity; B, kidney ARE-reporter activity; C, brain ARE-reporter activity. All values are presented as means ± S.E. (n = 5), with statistical significance indicated (***, p < 0.001).
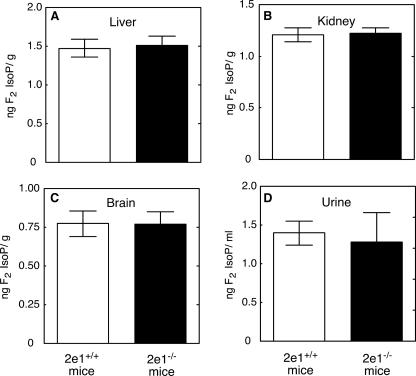
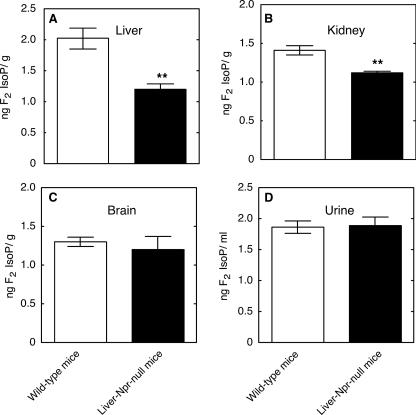
In Vivo F2-IsoP Measurements in Liver-Npr-null and Cyp2e1–/– Mice—F2-IsoPs levels in urine, liver, kidney, and brain samples from Cyp2e1–/– mice were not significantly different from Cyp2e1+/+ mice (Fig. 5). F2-IsoPs levels in liver and kidney samples from liver-Npr-null mice were significantly decreased compared with those from wild-type mice; however, no significant changes of F2-IsoPs level were found in brain or urine samples (Fig. 6).
FIGURE 5.
Measurements of F2-isoP levels in tissue and urine samples from Cyp2e1–/– and Cyp2e1+/+ mice. A, liver F2-isoPs; B, kidney F2-isoPs; C, brain F2-isoPs; D, urinary F2-isoPs. All values are presented as means ± S.E. (n = 5). No statistical significance was found in any case.
FIGURE 6.
Measurements of F2-isoPs levels in tissue and urine samples from liver-Npr-null and wild-type mice. A, liver F2-isoPs; B, kidney F2-isoPs; C, brain F2-isoPs; D, urinary F2-isoPs. All values are presented as means ± S.E. (n = 5), with statistical significance indicated (**, p < 0.01).
NNMT Activity, Pyridine Nucleotides, and Protective Enzymes in Mouse Liver—The previous measurements in mice (see above) and rats (28) indicated elevation of ROS parameters by PB, βNF, and CLOF in liver microsomes but selective enhancement of oxidative stress in vivo by PB. Information about mRNA microarray results of treatment of C57BL/6 mice with common enzyme inducers is available (C. A. Bradfield, University of Wisconsin, Madison, WI, edge.oncology.wisc.edu/) (36) (supplemental Figs. S1 and S2). Examination of these data for genes that corresponded differentially to PB compared with βNF, another fibrate (ciprofibrate), and other P450 inducers revealed some possible candidates, involving several P450 enzymes, fatty acyl CoA desaturase, carboxylesterase, and NNMT. The latter gene was of interest in that the increase in activity might deplete pyridine nucleotide stores and attenuate the systems that protect against ROS.
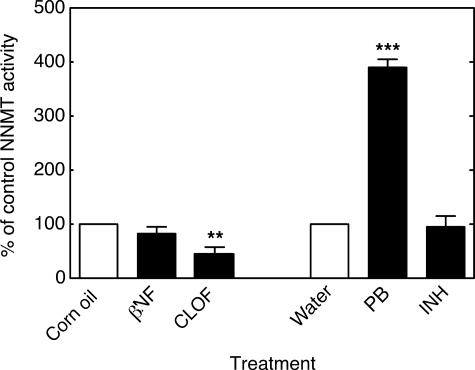
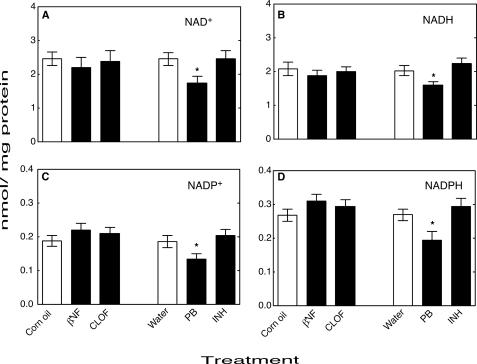
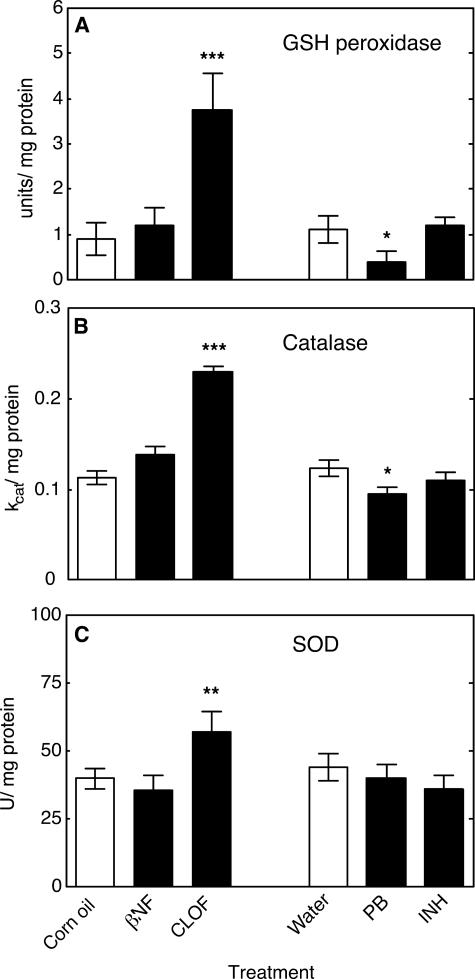
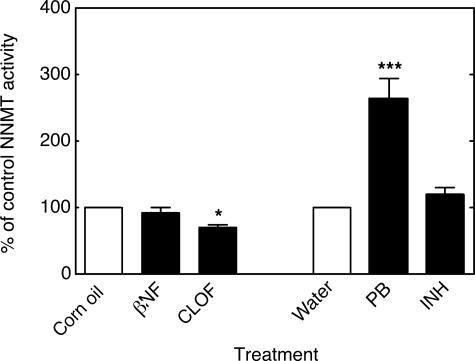
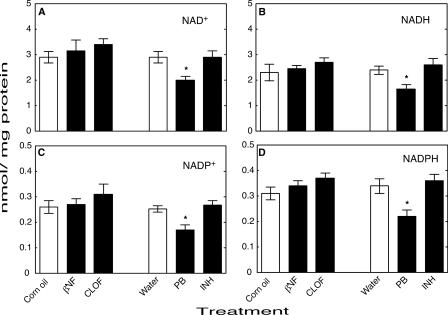
NNMT activity in mouse liver homogenates was significantly increased by PB treatment and significantly decreased by CLOF (Fig. 7). Levels of the pyridine nucleotides NAD+ (Fig. 8A), NADH (Fig. 8B), NADP+ (Fig. 8C), and NADPH (Fig. 8D) were all significantly decreased by treatment with PB compared with the control group. Neither βNF, INH, nor CLOF treatment significantly changed NNMT activity or the levels of pyridine nucleotides. GSH peroxidase was significantly increased by CLOF and decreased by PB (Fig. 9A). Treatment of ARE-reporter mice with CLOF caused a significant increase in catalase activity (Fig. 9B) and SOD activities (Fig. 9C) in liver homogenate compared with controls. No differences in catalase or SOD activities were observed in animals treated with INH, PB, or βNF.
FIGURE 7.
Effects of treatments on NNMT activity in mouse liver. All values are presented as means ± S.E. (n = 5) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (**, p < 0.01; ***, p < 0.001).
FIGURE 8.
Effects of treatments on levels of pyridine nucleotides in mouse liver. A, NAD+; B, NADH; C, NADP+; D, NADPH. All values are presented as means ± S.E. (n = 5) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (*, p < 0.05).
FIGURE 9.
Effects of treatments on activity of protective enzyme systems in mouse liver. A, GSH peroxidase (one unit of GSH peroxidase activity is defined as that amount of enzyme catalyzing oxidation of 1 μmol min–1 of GSH (52)); B, catalase (activity was calculated based on kcat = 2.3/dt(C0/Ct), dt = 1 min) (46); C, SOD. One unit of SOD activity is defined as inhibition of xanthine oxidase-dependent cytochrome c reduction by 50% in this assay. All values are presented as means ± S.E. (n = 5) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (**, p < 0.01; ***, p < 0.001).
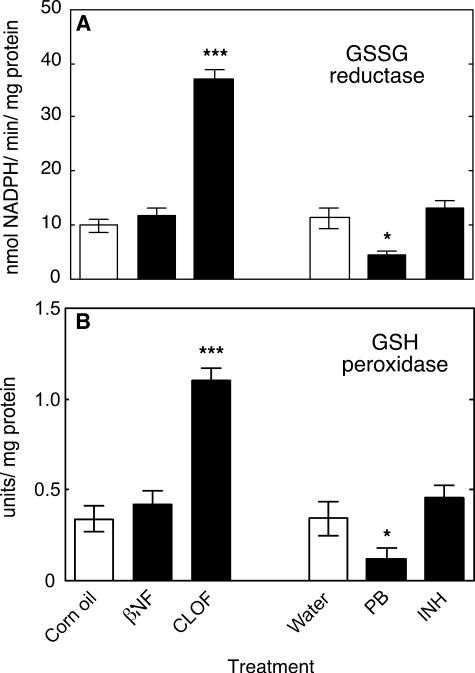
Measurements in Rat Liver—The studies with ARE-reporter mice were extended to rats, in that a complete set of samples from treated animals was available from a previous study (28). NNMT activity was significantly and selectively increased by PB treatment (Fig. 10); levels of pyridine nucleotides NAD+ (Fig. 11A), NADH (Fig. 11B), NADP+ (Fig. 11C), and NADPH (Fig. 11D) were all significantly decreased only by PB treatment. GSSG reductase and GSH peroxidase activities were both selectively decreased by PB treatment and increased by CLOF treatment (Fig. 12, A and B).
FIGURE 10.
Effects of treatments on NNMT activity in rat liver. All values are presented as means ± S.E. (n = 6) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (*, p < 0.05; ***, p < 0.001).
FIGURE 11.
Effects of treatments on levels of pyridine nucleotides in rat liver. A, NAD+; B, NADH; C, NADP+; D, NADPH. All values are presented as means ± S.E. (n = 6) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (*, p < 0.05).
FIGURE 12.
Effects of treatments on activities of protective enzymes in rat liver. A, GSSG reductase. Activity was calculated using Δε340 = 6.22 mm–1 cm–1 for NADPH oxidation; B, GSH peroxidase (one unit of GSH peroxidase is activity defined as that amount of enzyme catalyzing the oxidation of 1 μmol of GSH min–1 in the presence of H2O2 (52)). All values are presented as means ± S.E. (n = 6) and statistical significance relative to the appropriate vehicle control (within each group, indicated by the open bar in each set) (*, p < 0.05; ***, p < 0.001).
DISCUSSION
P450s have previously been found to produce ROS in vitro, going back to at least 1957 (12). The present study was designed to examine whether P450 induction alters oxidative stress using ARE-reporter mice, liver-Npr-null mice, and Cyp2e1–/– mice, particularly in vivo. We found that P450 induction by PB treatment was selectively associated with oxidative stress both in vitro and in vivo, supporting results found with rats in our previous study (28). An explanation for in vitro-in vivo oxidative stress discrepancy in the variously treated animals may involve enhancement of NNMT activity, which leads to depletion of pyridine nucleotides in the system, and attenuation of the NADPH-dependent antioxidative enzymes GSH peroxidase and GSSG reductase as well (Figs. 13 and 14).
FIGURE 13.
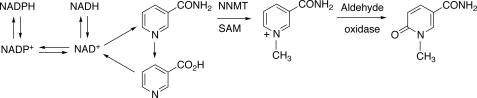
Competition of NNMT methylation and utilization of nicotinamide for pyridine nucleotide synthesis. SAM, S-adenosylmethionine.
FIGURE 14.
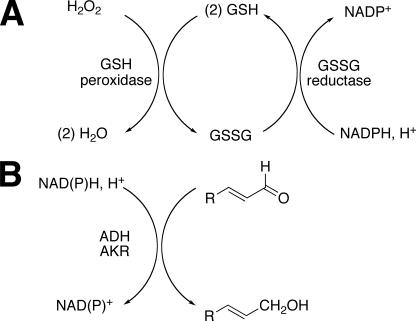
Detoxication of reactive species. A, H2O2 and GSH peroxidase system; B, reduction of reactive aldehydes by NAD(P)H. ALDH, aldehyde dehydrogenase; AKR, aldo-keto reductase.
The liver-Npr-null mouse model has previously been used for estimation of relative contributions of P450 and/or extrahepatic tissues in the biotransformation of endogenous as well as exogenous compounds (32, 33, 55). In our studies, the liver-Npr-null mouse model was used to confirm a role for P450 in “basal” oxidative damage. Male liver-Npr-null mice had a significant decrease in F2-IsoP levels in liver (and kidney) tissue samples compared with wild-type mice with normal NPR expression; no significant change was found in urine or brain tissue samples (Fig. 6). These results indicate that under physiological conditions some endogenous P450s contribute to the process of oxidative stress in mice, a conclusion similar to that found with the effect of 1-aminobenzotriazole on hepatic and renal F2-IsoP levels in rats (28). Notably, the decrease in oxidative stress in the liver occurred despite the substantial compensatory expression of multiple hepatic P450 enzymes in the liver-Npr-null mice (32, 56, 57), an observation consistent with the view that P450-mediated oxidative stress requires electron transfer from NPR. The decrease in kidney F2-isoP levels in the liver-Npr-null mice, in which the expression of neither P450 nor NPR was altered in the kidney (32), indicates that endogenous oxidants generated in the liver can be transported to the kidney.
Homozygous Cyp2e1-null mice were used to determine the role of P450 2E1 in oxidative stress processes. Cyp2e1–/– mice did not show significant differences in F2-IsoP levels in any tissues or urine samples when compared with control Cyp2e1+/+ mice. Moreover, INH, an established inducer of P450 2E1 activity (Fig. 2C), did not alter any of the oxidative stress markers in the ARE-reporter mice in vitro or in vivo. These results with both Cyp2e1–/– and ARE-reporter mice are in full agreement with results from our previous study with rats (28) and indicate that P450 2E1 does not play crucial role in oxidative damage in rats or mice.
A large majority of the studies on P450 and ROS have been done in vitro. In previous work with rats we found that most of the P450 inducers (except INH) enhanced in vitro ROS parameters, i.e. in assays using liver microsomes. However, F2-IsoP levels were selectively increased by PB in vivo (28). The F2-IsoP results obtained with the ARE-reporter mice in the present study were quantitatively similar, in that PB and βNF were the best inducers (Figs. 1 and 3). The in vivo patterns were reinforced by the results of assays with the ARE-driven reporter (Fig. 4), where the selective effect of PB was quite dramatic. The magnitude of the response was much stronger than for endogenous ARE-dependent genes, but such large increases have been seen in this system (35), possibly due to the lack of negative regulatory elements in the transgene.
At least three major explanations can be considered to explain the selective oxidative stress observed in vivo in rats (28) and mice (Figs. 3 and 4) following treatment with PB and barbiturate-like inducers, in contrast to the increased in vitro ROS parameters observed with several P450 inducers (28) (Fig. 1, C, D, and F): (i) the in vivo oxidative stress is due to (PB) induction of a non-P450 system that generates ROS; (ii) all of the P450 inducers raise levels of ROS (in microsomes), but βNF and CLOF also induce protective systems while PB does not; and (iii) all of the P450 inducers raise levels of ROS species but PB selectively attenuates systems that protect against oxidative stress. In evaluating these general possibilities, the available results with rats (28) argue against the first possibility, in that administration of the P450 mechanism-based inactivator 1-aminobenzotriazole (58) to rats ablated the PB-induced increase in F2-IsoP levels (28). The possibility that PB induces NPR, which in turn stimulates P450-mediated oxidative stress, is largely ruled out by the 1-aminobenzotriazole results with rats: 1-aminobenzotriazole attenuated the level of (PB-induced) P450 by 87% and the level of liver F2-IsoPs by 71%, while having an effect of lowering NPR activity by ≤20% (28). In contrast, the second possibility (see above) can be considered viable, at least in part. CLOF treatment elevated GSH peroxidase, catalase, and SOD activities in mouse liver (Fig. 9), and these results may contribute to the lack of in vivo stress induced by CLOF (Figs. 3 and 4). However, the limited enhancement of in vivo oxidative stress by βNF cannot be explained by induction of ROS-destroying enzymes (Fig. 9).
Mechanistic work on the problem is limited in the sense that more in vitro studies are of limited use in explaining an in vivo phenomenon already known to differ from the in vitro. Lipid peroxidation products (e.g. 4-hydroxynonenal) are already known to induce the ARE system in cultured cells (59–62) but this information does not explain which products drive the ARE system response seen with PB (Fig. 4). One approach was to examine available in vivo microarray results, which are available for C57BL/6 mice treated with the compounds of interest (36) (www.edge.oncology.wisc.edu) (supplemental Figs. S1 and S2). Of the genes differentially regulated by PB, βNF (also 3-methylcholanthrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin), and fibrates (ciprofibrate and Wyeth 14,643), many are either P450s or do not have obvious connections to oxidative stress (e.g. carboxyesterase, fatty acyl-CoA desaturase). One potential candidate considered was NNMT, which is unaffected by βNF, down-regulated by fibrates, and up-regulated by PB (supplemental Figs. S1–S3).
We hypothesized that up-regulation of NNMT could lead to a decrease in the pool of pyridine nucleotides available for defense against ROS (Figs. 13 and 14), in that 1-methylnicotinamide has long been known to be a major urinary excretion product (63). Precedent exists for the importance of the total level of NADP(H) in protection against oxidative ROS in cellular systems (64). We found that NNMT activity was selectively elevated 3- to 4-fold by PB treatment in both mouse (Fig. 7) and rat (Fig. 10) liver. We also found selective attenuation of levels of NAD+, NADH, NADP+, and NADPH in both mouse (Fig. 8) and rat (Fig. 11) liver, which we propose are linked to the elevated NNMT activities. The amount of NNMT activity in liver is already considerable in the absence of induction; e.g. it has long been known that administration of large amounts of nicotinamide to rats will lead to enough depletion of methyl groups (from S-adenosylmethionine) to produce fatty livers (63). The decrease in pyridine nucleotide supply (Figs. 8 and 11), which we propose is related to NNMT induction (Figs. 7 and 10), could decrease the ability of cells to reduce ROS-derived hydroperoxides and aldehydes (Fig. 14). The literature has limited information on the in vivo effects of barbiturate treatment on pyridine nucleotide supplies, and what is available is focused largely on the NADPH/NADP+ ratio (65, 66). Our interpretation of our results is that the total pyridine nucleotide levels may be a major issue in protection, in that these limit the function of the protective systems.
GSH peroxidase is a key enzyme involved in protection against oxidative stress (67) (Fig. 14). Analysis of the C57BL/6 mouse liver microarray data base showed increased mRNA (GSH peroxidase 1) following fibrate treatment (supplemental Fig. S3), and we also found increased enzymatic activity in both mouse and rat liver following CLOF treatment (Figs. 9 and 12). Although the C57BL/6 mouse data base did not show an effect of PB on GSH peroxidase mRNA levels (supplemental Fig. S3), a major decrease in GSH peroxidase activity was seen following PB treatment in both mouse (Fig. 9) and rat (Fig. 12) livers. GSSG reductase activity was also attenuated in rat liver following PB treatment (Fig. 12A) (measurements were not done in mouse liver).
The pyridine nucleotide levels we measured in rat and mouse liver (Figs. 8 and 11) are similar to literature values (68, 69). Most of the attention in the literature has been given to reduced versus oxidized ratios, often determined indirectly, in situations involving P450 induction (65, 66). The literature apparently contains only limited information on the effects of P450 inducers on GSH peroxidase and GSSG reductase (in rats). In one study (70), liver GSSG reductase was elevated 2-fold by treatment with PB or trans-stilbene oxide (and the latter compound did not alter GSH peroxidase). In another study, PB treatment was reported to elevate activities of both enzymes (71). However, our results clearly showed decreases in the activities of both GSH peroxidase and GSSG reductase (Figs. 9 and 12).
Our results show PB-induced increases in NNMT activity (Figs. 7 and 10), decreases in all pyridine nucleotide levels (Figs. 8 and 11), and decreases in both GSH peroxidase and GSSG reductase (Figs. 9 and 12). The increased level of NNMT activity is proposed to account for the decreased pyridine nucleotide levels (Fig. 13), although direct evidence is not available. The decreased pyridine nucleotide levels may limit the enzymatic defense systems (Fig. 14), although a direct relationship cannot be proven in light of the decreased activities of both GSH peroxidase and GSSG reductase (Figs. 9 and 12). Nevertheless, all of these changes observed following PB treatment could compromise rodents against the effects of the ROS and lead to elevated parameters of oxidative stress (Figs. 3 and 4).
At this time we do not understand the details of the regulation of the NNMT, GSH peroxidase, and GSSG reductase genes by barbiturates. The regulation of GSH peroxidases (actually a family of enzymes) is complex (67), involving selenium levels, and at this time we do not know which of the GSH peroxidases is most relevant here (Figs. 9 and 12). One possibility that can be considered is the use of transgenic mice devoid of CAR, a factor involved in PB induction of P450 2B enzymes (72). However, distinguishing between effects of CAR on induction of P450s and other genes may not be trivial. We do not know if the NNMT gene contains functional CAR (or PXR) elements (73). NNMT is up-regulated not only by PB (Figs. 7 and 10 and supplemental Fig. S3) but also in neoplasia (74) and a model of hepatitis (75). The regulation may be indirect in that Stat3 has been found to activate NNMT expression in a human cancer cell line (76).
In conclusion, we extended our previous findings on the roles of P450s in rats (28) to mice, again demonstrating a role of P450s in the basal hepatic state (Fig. 6) and no role of P450 2E1 in in vivo oxidative stress (Fig. 5). The selective ability of PB treatment to produce oxidative stress in vivo was observed. Similar patterns were observed with F2-IsoPs (Fig. 3), an established biomarker of in vivo oxidative stress (29), and a more sensitive transgenic reporter of the ARE system (34) (Fig. 4). In rats we concluded that one (or more) of the Subfamily 2B P450s is probably involved (28), but which of the five Subfamily 2B P450s (or another PB-inducible P450) in mice is involved is unclear. We propose that the ability of PB to selectively induce in vivo oxidative stress (Fig. 3) is not related only to the questions of which P450s produce ROS (in that several do this well, e.g. Figs. 1, C, D, and F) but also to the PB-induced changes in the levels of the enzyme GSH-peroxidase (Figs. 9 and 12) and pyridine nucleotides (Figs. 8 and 11), which can protect against ROS; these latter changes result from the selective induction of NNMT (Figs. 7 and 10), an enzyme involved in degradation of nicotinamide, a key intermediate in the pyridine nucleotide cycle (Fig. 13) (64). Finally, we emphasize the importance of in vivo models in studying oxidative stress, in that many components of the overall system are lacking in work with isolated systems (e.g. Fig. 1) or are not necessarily regulated in the same way in cultured cells.
Supplementary Material
Acknowledgments
We thank R. F. Burk, K. E. Hill, and L. M. Austin for their assistance with the ARE-reporter mice, C. A. Bradfield for access to the mouse liver microarray data base, S. Sanchez for technical assistance with F2-IsoP measurements, and K. Trisler for assistance in preparation of the manuscript. We also thank the late D. M. Ziegler (University of Texas, Austin), who often made the point to one of us (F. P. G.) that in vitro studies overestimate the extent of oxidative stress and encouraged study of the factors involved in vivo.
This work was supported, in whole or in part, by National Institutes of Health Grants R37 CA090426 (to F. P. G.), P30 ES000267 (to F. P. G., J. D. M., and R. F. B.), P50 GM15431, R01 DK48831, and P01 ES13125 (to J. D. M.), R01 ES07462 (to X. D.), and R01 ES10042 and R01 ES08089 (to J. A. J.) from the United States Public Health Services. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; ARE, antioxidant response element; βNF, β-naphthoflavone (5,6-benzoflavone); CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; CLOF, clofibrate; F2-isoP, F2-isoprostane; GSSG, oxidized glutathione; INH, isoniazid; NNMT, nicotinamide N-methyltransferase; NPR, NADPH-P450 reductase; Nrf2, nuclear factor erythroid 2-related factor 2; P450, cytochrome P450 (also termed heme-thiolate P450 (2)); PB, phenobarbital; SOD, superoxide dismutase.
References
- 1.Valko, M., Leibfritz, D., Moncol, J., Cronin, M. T., Mazur, M., and Telser, J. (2007) Int. J. Biochem. Cell Biol. 39 44–84 [DOI] [PubMed] [Google Scholar]
- 2.Palmer, G., and Reedijk, J. (1992) J. Biol. Chem. 267 665–6771309757 [Google Scholar]
- 3.Jabs, T. (1999) Biochem. Pharmacol. 57 231–245 [DOI] [PubMed] [Google Scholar]
- 4.Finkel, T., and Holbrook, N. J. (2000) Nature 408 239–247 [DOI] [PubMed] [Google Scholar]
- 5.Lee, S. R., Yang, K. S., Kwon, J., Lee, C., Jeong, W., and Rhee, S. G. (2002) J. Biol. Chem. 277 20336–20342 [DOI] [PubMed] [Google Scholar]
- 6.West, J. D., and Marnett, L. J. (2006) Chem. Res. Toxicol. 19 173–194 [DOI] [PubMed] [Google Scholar]
- 7.Nguyen, T., Sherratt, P. J., and Pickett, C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43 233–260 [DOI] [PubMed] [Google Scholar]
- 8.Collins, A. R. (1999) BioEssays 21 238–246 [DOI] [PubMed] [Google Scholar]
- 9.Liebler, D. C. (2006) Chem. Res. Toxicol. 19 610–613 [DOI] [PubMed] [Google Scholar]
- 10.Halliwell, B., and Gutteridge, J. M. (1990) Methods Enzymol. 186 1–85 [DOI] [PubMed] [Google Scholar]
- 11.Ortiz de Montellano, P. R. (ed) (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd Ed., KluwerAcademic/Plenum Publishers, New York, NY
- 12.Gillette, J. R., Brodie, B. B., and La Du, B. N. (1957) J. Pharmacol. Exp. Ther. 119 532–540 [PubMed] [Google Scholar]
- 13.Thurman, R. G., Ley, H. G., and Scholz, R. (1972) Eur. J. Biochem. 25 420–430 [DOI] [PubMed] [Google Scholar]
- 14.Nordblom, G. D., and Coon, M. J. (1977) Arch. Biochem. Biophys. 180 343–347 [DOI] [PubMed] [Google Scholar]
- 15.Ekström, G., and Ingelman-Sundberg, M. (1989) Biochem. Pharmacol. 38 1313–1319 [DOI] [PubMed] [Google Scholar]
- 16.Park, J. Y. K., Shigenaga, M. K., and Ames, B. N. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 2322–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shertzer, H. G., Nebert, D. W., Puga, A., Ary, M., Sonntag, D., Dixon, K., Robinson, L. J., Cianciolo, E., and Dalton, T. P. (1998) Biochem. Biophys. Res. Commun. 253 44–48 [DOI] [PubMed] [Google Scholar]
- 18.Persson, J. O., Terelius, Y., and Ingelman-Sundberg, M. (1990) Xenobiotica 20 887–900 [DOI] [PubMed] [Google Scholar]
- 19.Cederbaum, A. I., Wu, D., Mari, M., and Bai, J. (2001) Free Radic. Biol. Med. 31 1539–1543 [DOI] [PubMed] [Google Scholar]
- 20.Nieto, N., Friedman, S. L., and Cederbaum, A. I. (2002) J. Biol. Chem. 277 9853–9864 [DOI] [PubMed] [Google Scholar]
- 21.Imaoka, S., Osada, M., Minamiyama, Y., Yukimura, T., Toyokuni, S., Takemura, S., Hiroi, T., and Funae, Y. (2004) Cancer Lett. 203 117–125 [DOI] [PubMed] [Google Scholar]
- 22.Liu, L., Bridges, R. J., and Eyer, C. L. (2001) Mol. Cell. Biochem. 223 89–94 [DOI] [PubMed] [Google Scholar]
- 23.Robertson, G., Leclercq, I., and Farrell, G. C. (2001) Am. J. Physiol. 281 G1135–G1139 [DOI] [PubMed] [Google Scholar]
- 24.Dalton, T. P., Puga, A., and Shertzer, H. G. (2002) Chem-Biol. Interact. 141 77–95 [DOI] [PubMed] [Google Scholar]
- 25.Tong, V., Chang, T. K., Chen, J., and Abbott, F. S. (2003) Free Radic. Biol. Med. 34 1435–1446 [DOI] [PubMed] [Google Scholar]
- 26.Caro, A. A., and Cederbaum, A. I. (2004) Annu. Rev. Pharmacol. Toxicol. 44 27–42 [DOI] [PubMed] [Google Scholar]
- 27.Isayama, F., Froh, M., Bradford, B. U., McKim, S. E., Kadiiska, M. B., Connor, H. D., Mason, R. P., Koop, D. R., Wheeler, M. D., and Arteel, G. E. (2003) Free Radic. Biol. Med. 35 1568–1581 [DOI] [PubMed] [Google Scholar]
- 28.Dostalek, M., Brooks, J. D., Hardy, K. D., Milne, G. L., Moore, M. M., Sharma, S., Morrow, J. D., and Guengerich, F. P. (2007) Mol. Pharmacol. 72 1419–1424 [DOI] [PubMed] [Google Scholar]
- 29.Kadiiska, M. B., Gladen, B. C., Baird, D. D., Graham, L. B., Parker, C. E., Ames, B. N., Basu, S., Fitzgerald, G. A., Lawson, J. A., Marnett, L. J., Morrow, J. D., Murray, D. M., Plastaras, J., Roberts, L. J., 2nd, Rokach, J., Shigenaga, M. K., Sun, J., Walter, P. B., Tomer, K. B., Barrett, J. C., and Mason, R. P. (2005) Free Radic. Biol. Med. 38 711–718 [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. S. T., Buters, J. T. M., Pineau, T., Fernandez-Salguero, P., and Gonzalez, F. J. (1996) J. Biol. Chem. 271 12063–12067 [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez, F. J. (2005) Mutat. Res. 569 101–110 [DOI] [PubMed] [Google Scholar]
- 32.Gu, J., Weng, Y., Zhang, Q. Y., Cui, H., Behr, M., Wu, L., Yang, W., Zhang, L., and Ding, X. (2003) J. Biol. Chem. 278 25895–25901 [DOI] [PubMed] [Google Scholar]
- 33.Gu, J. C., H., Behr, M., Zhang, L., Zhang, Q.-Y., Yang, W., Hinson, J. A., and Ding, X. (2005) Mol. Pharmacol. 67 623–630 [DOI] [PubMed] [Google Scholar]
- 34.Johnson, D. A., Andrews, G. K., Xu, W., and Johnson, J. A. (2002) J. Neurochem. 81 1233–1241 [DOI] [PubMed] [Google Scholar]
- 35.Burk, R. F., Hill, K. E., Nakayama, A., Mostert, V., Levander, X. A., Motley, A. K., Johnson, D. A., Johnson, J. A., Freeman, M. L., and Austin, L. M. (2008) Free Radic. Biol. Med. 44 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, R. S., Rank, D. R., Penn, S. G., Zastrow, G. M., Hayes, K. R., Pande, K., Glover, E., Silander, T., Craven, M. W., Reddy, J. K., Jovanovich, S. B., and Bradfield, C. A. (2001) Mol. Pharmacol. 60 1189–1194 [DOI] [PubMed] [Google Scholar]
- 37.Guengerich, F. P., and Bartleson, C. J. (2007) in Principles and Methods of Toxicology, 5th Ed. (Hayes, A. W., ed) pp. 1981–2048, CRC Press, Boca Raton, FL
- 38.Morrow, J. D., and Roberts, L. J., II (1999) Methods Enzymol. 300 3–12 [DOI] [PubMed] [Google Scholar]
- 39.Omura, T., and Sato, R. (1964) J. Biol. Chem. 239 2370–2378 [PubMed] [Google Scholar]
- 40.Phillips, A. H., and Langdon, R. G. (1962) J. Biol. Chem. 237 2652–2660 [PubMed] [Google Scholar]
- 41.Yun, C.-H., Kim, K.-H., Calcutt, M. W., and Guengerich, F. P. (2005) J. Biol. Chem. 280 12279–12291 [DOI] [PubMed] [Google Scholar]
- 42.Hildebrandt, A. G., Roots, I., Tjoe, M., and Heinemeyer, G. (1978) Methods Enzymol. 52 342–350 [DOI] [PubMed] [Google Scholar]
- 43.Ernster, L., and Nordenbrand, K. (1967) Methods Enzymol. 10 574–580 [Google Scholar]
- 44.Guengerich, F. P., Wang, P., and Davidson, N. K. (1982) Biochemistry 21 1698–1706 [DOI] [PubMed] [Google Scholar]
- 45.Guengerich, F. P., Dannan, G. A., Wright, S. T., Martin, M. V., and Kaminsky, L. S. (1982) Biochemistry 21 6019–6030 [DOI] [PubMed] [Google Scholar]
- 46.Cohen, G., Dembiec, D., and Marcus, J. (1970) Anal. Biochem. 34 30–38 [DOI] [PubMed] [Google Scholar]
- 47.Crapo, J. D., McCord, J. M., and Fridovich, I. (1978) Methods Enzymol. 53 382–393 [DOI] [PubMed] [Google Scholar]
- 48.Sano, A., Endo, N., and Takitani, S. (1992) Chem. Pharm. Bull. (Tokyo) 40 153–156 [DOI] [PubMed] [Google Scholar]
- 49.Musfeld, C., Biollaz, J., Belaz, N., Kesselring, U. W., and Docesterd, L. A. (2001) J. Pharm. Biomed. Anal. 24 391–404 [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Z., Yu, J., and Stanton, R. C. (2000) Anal. Biochem. 285 163–167 [DOI] [PubMed] [Google Scholar]
- 51.Griffiths, M. M., and Bernofsky, C. (1972) J. Biol. Chem. 247 1473–1478 [PubMed] [Google Scholar]
- 52.Paglia, D. E., and Valentine, W. N. (1967) J. Lab. Clin. Med. 70 158–169 [PubMed] [Google Scholar]
- 53.Wheeler, C. R., Salzman, J. A., Elsayed, N. M., Omaye, S. T., and Korte, D. W. (1990) Anal. Biochem. 184 193–199 [DOI] [PubMed] [Google Scholar]
- 54.Kaminsky, L. S., Dannan, G. A., and Guengerich, F. P. (1984) Eur. J. Biochem. 141 141–148 [DOI] [PubMed] [Google Scholar]
- 55.Gu, J., Chen, C. S., Wei, Y., Fang, C., Xie, F., Kannan, K., Yang, W., Waxman, D. J., and Ding, X. (2007) J. Pharmacol. Exp. Ther. 321 9–17 [DOI] [PubMed] [Google Scholar]
- 56.Weng, Y., DiRusso, C. C., Reilly, A. A., Black, P. N., and Ding, X. (2005) J. Biol. Chem. 280 31686–31698 [DOI] [PubMed] [Google Scholar]
- 57.Henderson, C. J., Otto, D. M., Carrie, D., Magnuson, M. A., McLaren, A. W., Rosewell, I., and Wolf, C. R. (2003) J. Biol. Chem. 278 13480–13486 [DOI] [PubMed] [Google Scholar]
- 58.Ortiz de Montellano, P. R., and Mathews, J. M. (1981) Biochem. J. 195 761–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishii, T., Itoh, K., Ruiz, E., Leake, D. S., Unoki, H., Yamamoto, M., and Mann, G. E. (2004) Circ. Res. 94 609–616 [DOI] [PubMed] [Google Scholar]
- 60.Chen, Z. H., Saito, Y., Yoshida, Y., Sekine, A., Noguchi, N., and Niki, E. (2005) J. Biol. Chem. 280 41921–41927 [DOI] [PubMed] [Google Scholar]
- 61.Gong, P., and Cederbaum, A. (2006) Hepatology 43 144–153 [DOI] [PubMed] [Google Scholar]
- 62.Osburn, W. O., Wakabayashi, N., Misra, V., Nilles, T., Biswal, S., Trush, M. A., and Kensler, T. W. (2006) Arch. Biochem. Biophys. 454 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, A., Handler, P., and Smith, E. L. (1964) Principles of Biochemistry, 3rd Ed., McGraw-Hill, New York, pp. 954–957
- 64.Pollak, N., Niere, M., and Ziegler, M. (2007) J. Biol. Chem. 282 33562–33571 [DOI] [PubMed] [Google Scholar]
- 65.Kauffman, F. C., Evans, R. K., and Thurman, R. G. (1977) Biochem. J. 166 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kauffman, F. C., Evans, R. K., Jerkins, A. A., Reinke, L. A., Conway, J. G., and Thurman, R. G. (1987) Biochem. Pharmacol. 36 1083–1090 [DOI] [PubMed] [Google Scholar]
- 67.Burk, R. F. (1997) in Biotransformation, Vol. 3, Comprehensive Toxicology, 1st Ed. (Guengerich, F. P., ed) pp. 229–242, Elsevier Science Ltd., Oxford
- 68.Conway, J. G., Kauffman, F. C., and Thurman, R. G. (1983) J. Biol. Chem. 258 3825–3831 [PubMed] [Google Scholar]
- 69.Morgan, W. A., Prins, B., and Koster, A. S. (1997) Arch. Toxicol. 71 582–587 [DOI] [PubMed] [Google Scholar]
- 70.Carlberg, I., Depierre, J. W., and Mannervik, B. (1981) Biochim. Biophys. Acta 677 140–145 [DOI] [PubMed] [Google Scholar]
- 71.Torres, M., Jarvisalo, J., and Hakim, J. (1981) Enzyme 26 129–135 [DOI] [PubMed] [Google Scholar]
- 72.Sueyoshi, T., and Negishi, M. (2001) Annu. Rev. Pharmacol. Toxicol. 41 123–143 [DOI] [PubMed] [Google Scholar]
- 73.Yan, L., Otterness, D. M., and Weinshilboum, R. M. (1999) Pharmacogenetics 9 307–316 [DOI] [PubMed] [Google Scholar]
- 74.Seifert, R., Hoshino, J., and Kroger, H. (1984) Biochim. Biophys. Acta 801 259–264 [DOI] [PubMed] [Google Scholar]
- 75.Dong, H., Toyoda, N., Yoneyama, H., Kurachi, M., Kasahara, T., Kobayashi, Y., Inadera, H., Hashimoto, S., and Matsushima, K. (2002) Biochem. Biophys. Res. Commun. 298 675–686 [DOI] [PubMed] [Google Scholar]
- 76.Tomida, M., Ohtake, H., Yokota, T., Kobayashi, Y., and Kurosumi, M. (2008) J. Cancer Res. Clin. Oncol. 134 551–559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.