Abstract
Seed development in flowering plants is a paradigm for the coordination of different tissues during organ growth. It requires a tight interplay between the two typically sexually produced structures: the embryo, developing from the fertilized egg cell, and the endosperm, originating from a fertilized central cell, along with the surrounding maternal tissues. Little is known about the presumptive signal transduction pathways administering and coordinating these different tissues during seed growth and development. Recently, a new signal has been identified emanating from the fertilization of the egg cell that triggers central cell proliferation without prior fertilization. Here, we demonstrate that there exists a large natural genetic variation with respect to the outcome of this signaling process in the model plant Arabidopsis thaliana. By using a recombinant inbred line population between the two Arabidopsis accessions Bayreuth-0 and Shahdara, we have identified two genetic components that influence the development of unfertilized endosperm. Exploiting this natural variation, we could further dissect the interdependence of embryo and endosperm growth during early seed development. Our data show an unexpectedly large degree of independence in embryo growth, but also reveal the embryo's developmental restrictions with respect to endosperm size. This work provides a genetic framework for dissection of the interplay between embryo and endosperm during seed growth in plants.
THE formation of seeds is an essential step in the life cycle of flowering plants (angiosperms). Seeds permit plants to survive at unfavorable conditions in a quiescent stage. They also serve as dispersal units and allow plants to spread out and colonize new territories. In addition, seeds are often nutritional units that support the germinating seedling, facilitating rapid growth during this crucial life phase. Thus, the proper formation of seeds is decisive for the reproductive success of a plant.
Seeds are highly elaborated structures composed of different tissues that represent different genetic systems: The embryo makes up the next plant generation and its growth is supported by the surrounding endosperm tissue. Usually, both the embryo and the endosperm are products of the so-called double fertilization that is unique for flowering plants. However, these two fertilization products are not genetically equivalent: In the case of diploid plants, one of the two haploid male gametes fertilizes the haploid egg cell and generates a diploid embryo whereas the second gamete fuses with the usually homodiploid central cell, giving rise to a triploid endosperm (Sitte et al. 2002). Embryo and endosperm are enclosed by a seed coat comprising several integument layers, which are contributed by the mother plant. Thus, there is an apparent need for a temporal and spatial coordination of growth of these different tissues.
It has been found that there is a strong maternal sporophytic effect from the integuments regulating seed size (Berger et al. 2006). Seed size is either increased, e.g., in apetala 2 (ap2) or auxin response factor 2/megaintegumenta (arf2/mnt) mutants (Jofuku et al. 2005; Ohto et al. 2005; Schruff et al. 2006), or reduced, e.g., in transparent testa glabra 2 (ttg2) mutants (Johnson et al. 2002; Garcia et al. 2005). These genes appear to act sporophytically, most likely in the integuments surrounding the seed, since the genotype of the mother determined seed size in a recessive manner. In addition, AP2 is likely to have a function in the gametophytes (see also below).
ARF2/MNT encodes a transcription factor that binds to auxin-responsive elements in the promotors of auxin-regulated genes (Ulmasov et al. 1999; Schruff et al. 2006). ARF2/MNT controls cell proliferation in the whole plant and mutants display a pleiotropic phenotype with enlarged organs containing extra cells. In particular, supernumerary cells in the integuments of still unfertilized ovules were observed (Schruff et al. 2006).
TTG2 codes for a WRKY transcription factor and, in addition to reduction of leaf hair (trichome) branching, mutants in ttg2 show differentiation defects of the endothelium, a layer of the inner integuments adjacent to the endosperm. In contrast to wild-type seeds, ttg2 mutants do not accumulate proanthocyanidins in the endothelium upon fertilization (Johnson et al. 2002). However, it is currently not clear how this defect is related to the reduced seed size in ttg2 mutants, especially since genetic ablation of the endothelium layer did not alter seed size (Debeaujon et al. 2003).
AP2 also acts as a transcription factor, belonging to the class of ethylene-responsive element-binding proteins, and was first identified as a regulator of floral organ identity and meristem determinacy (Koornneef et al. 1980; Bowman et al. 1989; Kunst et al. 1989; Drews et al. 1991). In the enlarged ap2 mutant seeds, a higher ratio of glucose to sucrose was found (Jofuku et al. 2005; Ohto et al. 2005), and from legume and other species, it has been reported that increased hexose concentrations, e.g., glucose, correlate with increased cell proliferation activity (Wobus and Weber 1999). However, no target genes involved in seed growth control have been identified for the above-mentioned transcription factors, and it is not clear how direct these genes control seed size.
In addition to the integuments, the endosperm size has been found to play a major role in seed size regulation (Berger et al. 2006). The endosperm in Arabidopsis develops in a syncytial manner with initially three highly synchronous division rounds (Boisnard-Lorig et al. 2001). In subsequent divisions, three mitotic domains become visible and ∼4 days after fertilization the endosperm reaches ∼200 nuclei and cellularizes (Boisnard-Lorig et al. 2001). In contrast to the endosperm of crops, e.g., barley or wheat, there is no proliferation activity in the Arabidopsis endosperm after cellularization is completed. In the following stages of seed development, the Arabidopsis endosperm is consumed, leaving only one single-celled tissue layer in the mature seed (Beeckman et al. 2000; Olsen 2004).
A number of recessively acting alleles that are thought to affect seed growth through endosperm size have been found, for example, the WRKY transcription factor MINISEED 3 (MINI3) or the leucine-rich repeat (LRR) kinase gene HAIKU2 (IKU2) (Garcia et al. 2003; Luo et al. 2005). Both MINI3 and IKU2 are expressed in the endosperm and display a loss-of-function phenotype with a premature stop of nuclear divisions associated with cellularization.
A similar phenotype was observed in interploidy crosses when mother plants had a higher ploidy level than father plants (Lin 1982, 1984; Scott et al. 1998). Conversely, raising the paternal contribution has been found to result in larger seeds displaying an increased number of endosperm nuclei that cellularized only at later stages of seed development. Since in both interploidy crosses the embryo has the same ploidy level, it was concluded that, in particular, gene dosage in the endosperm is crucial.
Moreover, these experiments have demonstrated that maternal and paternal alleles have different effects on seed size. Indeed, it has been found that, similar to mammals, the expression of certain genes, e.g., the homeodomain transcription factor FWA, in the Arabidopsis endosperm depends on the parental origin; i.e., they are imprinted (Kinoshita et al. 2004). In mammals, imprinted genes include growth factors, e.g., insulin-like growth factor (Igf2) (Haig and Graham 1991), and often growth-reducing factors are active only in the maternal genome while promoting factors are expressed only from the paternal allele (Morison et al. 2005). This expression pattern is consistent with the parental conflict theory (kinship theory) according to which mothers and fathers have a different interest in allocation of resources to their offspring (Haig and Westoby 1989, 1991).
In flowering plants, imprinting has been found to be controlled by the FERTILIZATION-INDEPENDENT SEED (FIS) complex, showing similarities to the Polycomb repressive complex 2 (PrC2) from mammals that mediates histone methylation. The FIS complex comprises at least four subunits in Arabidopsis: the SET domain protein MEDEA (MEA), the zinc-finger transcription factor FERTILIZATION-INDEPENDENT SEED 2 (FIS2), the WD40-repeat protein FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), and the WD40-repeat protein MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) (Ohad et al. 1996; Chaudhury et al. 1997; Grossniklaus et al. 1998; Kiyosue et al. 1999; Kohler et al. 2003).
Mutants for individual components display an overproliferation phenotype of the endosperm. The embryos of all these mutants arrest at late heart stage and are significantly bigger (overproliferated) than wild-type heart-stage embryos. Currently, the reason for this seed abortion and how the endosperm and the embryo defects are linked are not known. Thus, a major challenge is the unraveling of signal transduction routes that coordinate seed development.
Recently, we and others have characterized an Arabidopsis mutant in the key cell cycle regulator CDKA;1 (CYCLIN DEPENDENT KINASE A;1, the yeast cdc2a/CDC28 homolog) that allows a genetic dissection of seed development (Iwakawa et al. 2006; Nowack et al. 2006). cdka;1 mutant pollen contains only a single haploid gamete that exclusively fertilizes the egg cell, leaving the central cell unfertilized. Strikingly, we found that the central cell in these ovules autonomously undergoes a few rounds of free-nuclear divisions, revealing a previously unrecognized proliferation signal from the fertilization of the egg cell toward the central cell (Nowack et al. 2006). Moreover, we observed that this positive signal in a fis class mutant background was sufficient to induce the completion of seed development, giving rise to a viable embryo from which a fertile plant developed after germination (Nowack et al. 2007).
Here we report that there exists an unexpected large natural genetic variation in the autonomous proliferation of the central cell nucleus upon fertilization of the egg cell with cdka;1 mutant pollen. We have exploited this genetic variation to identify the principal genetic components that regulate the onset of endosperm proliferation that depends on this proliferation signal. Furthermore, this work also allowed us to dissect seed growth, in particular the relationship between embryo and endosperm during early stages of seed development, thus defining criteria for future genetic and molecular analyses of seed formation.
MATERIALS AND METHODS
Plant material and growth conditions:
Plants were germinated on soil or 1/2 MS medium and grown under standard greenhouse conditions or in a growth chamber. Arabidopsis plants used in this study were derived from the following accessions: Antwerp-1 (An-1; ABRC22626), Bayreuth-0 (Bay-0; ABRC22633), Burren-0 (Bur-0; ABRC22656), C24 (ABRC22620), Cape Verde Islands-0 (Cvi-0; ABRC22614), Columbia-0 (Col-0; ABRC22625), Estland-1 (Est-1; ABRC22629), Kashmir-1 (Kas-1; ABRC22638), Landsberg erecta-1 (Ler-1; ABRC22618), Martuba-0 (Mt-0; ABRC22642), Niederzenz-1 (Nd-1; ABRC22619), Nossen-0 (No-0; CS1394), Shahdara (Sha; ABRC22652), Wassilewskija-0 (WS-0; ABRC22623). In addition, a core set of 165 recombinant inbred lines (RIL) derived from a cross between Bay-0 and Sha was used (Loudet et al. 2002). Throughout this work the previously characterized cdka;1-1 allele in the Col-0 genetic background (SALK_106809.34.90.X) was used (Nowack et al. 2006). The endosperm marker line KS22 was contributed by F. Berger and is in the C24 genetic background (Ingouff et al. 2005). The ProFIS2:GUS reporter line was kindly provided by A. Chaudhury and is in the C24 genetic background (Luo et al. 2000).
Histology:
Pistils and siliques of different developmental stages were prepared as described previously (Grini et al. 2002). Dissected siliques were fixed and mounted on microscope slides in a chloral hydrate clearing solution. Gus staining was performed as described previously (Grini et al. 2002). Light microscopy was performed with a Zeiss Axiophot microscope using DIC optics. GFP fluorescence in seeds was analyzed with a Leica TCS SP2 AOBS confocal laser scanning microscope as described in Nowack et al. (2007). For vanillin staining, siliques were emasculated, hand pollinated, and harvested between 3 and 6 days after pollination (DAP). The silique walls were removed and dissected seeds were incubated in 6 n HCl solution containing 1% (w/v) vanillin (Sigma-Aldrich) at room temperature for 1 hr. Siliques were mounted on slides in a drop of vanillin containing acidic solution and directly inspected using an Axioplan 2 Carl Zeiss Microscope. Images were acquired with an AxioCam HRc Carl Zeiss camera and processed with AxioVs40 V 4.5.0.0 software.
QTL mapping:
For QTL mapping, three to five independent plants per RIL, each with 15–20 siliques, were emasculated and pollinated 3 days later with heterozygous cdka;1 mutant plants, giving rise, on average, to 50–60 analyzable single-fertilized seeds per RIL. The endosperm division value (EDV; for definition, see text) was determined at 3 DAP. QTL mapping was performed on the mean EDV of each RIL. QTL analysis was done using the software MapQTL 5.0 (van Ooijen 2004). A permutation test using 1000 permutations of the original data resulted in a genomewide 95% LOD threshold of ∼2.4. The automatic cofactor selection procedure was applied per chromosome to select markers to be used as cofactors for the composite interval mapping procedure (CIM). Markers most closely linked to QTL that appeared only after each round of CIM mapping were also selected as cofactors. The results of CIM mapping provided the variance explained by each and by all detected QTL as well as their additive allelic effect. The heritability was calculated by dividing the genetic variance by the sum of the genetic and the environmental variance.
RESULTS
The autonomously proliferating central cell nuclei in cdka;1 fertilized seeds display characteristics of genuine endosperm:
We have previously shown that embryo development can be successfully supported by an unfertilized diploid endosperm in sexually reproducing plants (Nowack et al. 2007). However, this was possible only in a fis class mutant background where the unfertilized proliferating central cell displays characteristics of endosperm differentiation (Chaudhury et al. 1997; Ingouff et al. 2006). We therefore asked whether, and if so, to what degree the central cell nuclei in a wild-type genetic background that autonomously divide upon fertilization with cdka;1 mutant pollen represent a developing endosperm.
In a fertilized endosperm, the proliferating nuclei follow a predictable migration pattern (Boisnard-Lorig et al. 2001): After the first divisions, usually one nucleus moves toward the chalazal pole (opposite the embryo) while the other nuclei are distributed at the micropylar pole (adjacent to the embryo). To better trace the migration pattern of endosperm nuclei, we used a promotor FIS2-GUS reporter that is expressed in the central cell prior to fertilization and persists in the initial five divisions of endosperm development (Luo et al. 2000). Both the cleared preparations of wild-type plants and the detection of β-glucuronidase (GUS) activity in the FIS2 promotor reporter lines revealed that the same migration pattern found in seeds fertilized with cdka;1 mutant pollen was found in seeds fertilized with wild-type pollen (Figure 1, A–F, data not shown).
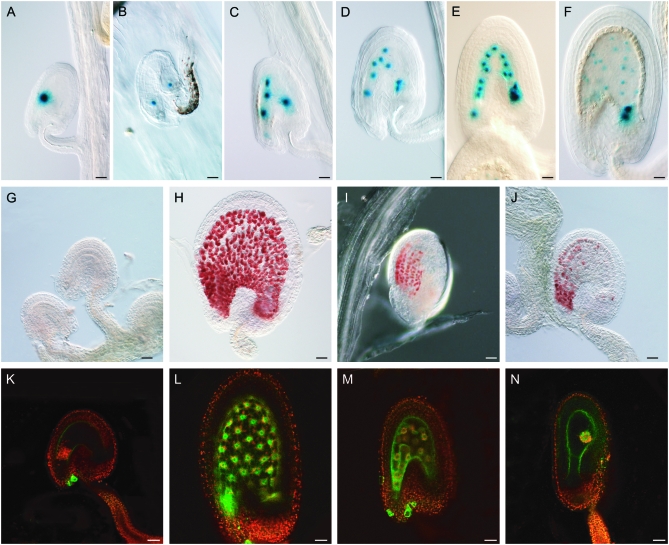
Figure 1.—
Endosperm characteristics of autonomously proliferating central cell nuclei. DIC light and confocal laser scanning (CLS) micrographs of developing double-fertilized (with wild-type pollen) and single-fertilized (with cdka;1 pollen) seeds. (A–F) Histochemical detection of GUS activity in seeds expressing a maternal FIS2-GUS construct (in the C24 ecotype) and pollinated with cdka;1 mutant pollen (A–E) or with wild-type pollen (F). The division pattern and nuclear migration of autonomous endosperm in cdka;1 fertilized seeds is similar to that seen in double-fertilized wild-type seeds. (A) FIS2-GUS expression in central cell nucleus (fused polar nuclei). (B) After the first division, one nucleus moves to the chalazal part of the seed. (C) Second division. (D) Third division. (E) Fourth division. (F) Fifth division cycle of an endosperm fertilized with wild-type pollen. (G–J) Detection of proanthocyanidin accumulation by vanillin staining in unpollinated Sha ovules or fertilized seeds at 3 DAP. (G) No proanthocyanidin can be detected in unfertilized Sha ovules. (H) Sha seeds fertilized with wild-type pollen accumulate proanthocyanidin in the endothelium. (I) Col-0 seeds fertilized with cdka;1 mutant pollen also start synthesizing proanthocyanidin. (J) Even seeds with little or nonproliferating central cell nuclei—as seen, for example, here in Sha fertilized with cdka;1 mutant pollen—are vanillin positive. (K–N) CLS micrographs of seeds or ovules containing the KS22 endosperm marker construct at 3 DAP, green GFP from the marker construct, red autofluorescence of plastids. (K) No GFP could be detected in an unpollinated wild-type C24 central cell. (L) Seeds fertilized with wild-type pollen showing expressing of the KS22 GFP reporter in the endosperm nuclei. (M) The autonomous endosperm in seeds fertilized with cdka;1 mutant pollen express GFP, even in cases where the central cell nucleus has not divided (N). All pictures are oriented such that the micropylar pole with the developing embryo points to the left and the chalazal pole of the seed to the right. Bars, 10 μm.
Next we tested by vanillin staining whether proanthocyanidins would accumulate in the endothelium layer of the seed coat upon cdka;1 fertilization (Debeaujon et al. 2003). A developmental trigger appears to be required to induce this differentiation process since the endothelium layer of unfertilized ovules or in seeds surrounding the proliferating nuclei of the central cell in retinoblastoma-related (rbr) mutants is not stained by vanillin (Ingouff et al. 2006). Proanthocyanidin accumulation can be induced by developing endosperm since, in addition to seeds of unfertilized msi plants, in the unfertilized seeds of mea and fis2 mutants without an embryo the differentiation of the endothelium layer also could be initiated (Ingouff et al. 2006; R. Shirzadi and P. E. Grini, unpublished data). In contrast to unfertilized seeds, we found that seeds fertilized with cdka;1 mutants were vanillin positive (Figure 1, G–J). Although previous experiments have suggested a key role for an endosperm in endothelium differentiation, we currently cannot distinguish whether this differentiation process can also be triggered by a developing embryo.
However, since the nuclei in cdka;1 fertilized seeds expressed the endosperm marker gene KS22 that is not active in the central cell of unfertilized seeds (Figure 1, K–M) (Ingouff et al. 2005), we conclude that the autonomously proliferating nuclei adopt at least some characteristics of wild-type endosperm.
Development of autonomous endosperm varies between different Arabidopsis accessions:
Within the species Arabidopsis thaliana, large genetic variation exists and many hundreds of different wild-type isolates, called accessions, have so far been collected (Koornneef et al. 2004; Nordborg et al. 2005; Bakker et al. 2006; Schmid et al. 2006). This natural variation represents a powerful tool for dissecting physiological or developmental processes.
We pollinated 14 accessions with pollen from heterozygous cdka;1 mutant plants and analyzed seed development 3 DAP. In these crosses, half of the ovules are fertilized by two wild-type sperm cells whereas in the other half only the egg cell is fertilized by mutant pollen containing only one sperm cell (Iwakawa et al. 2006; Nowack et al. 2006). In accordance with this, we found that half of the developing seeds in each tested accession followed the typical wild-type seed developmental program (Figure 2A; data not shown). In the other half of the seeds, we found the previously described autonomous proliferation of the central cell along with the development of an embryo that eventually leads to seed abortion (Figure 2, C–E) (Nowack et al. 2006). Remarkably, we observed a large variation in endosperm nuclei number between the different accessions ranging from 1 to 48 nuclei.
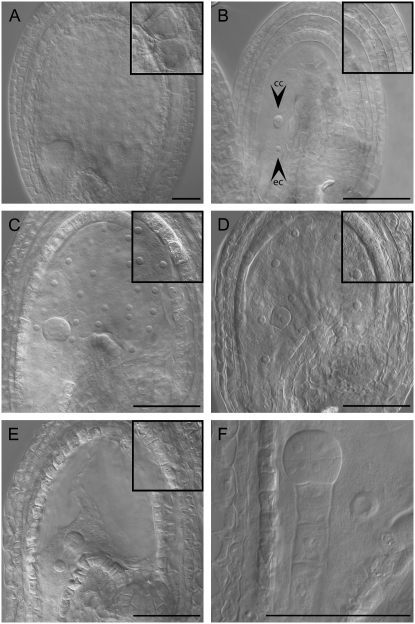
Figure 2.—
Autonomously proliferating central cell nuclei after single fertilization with cdka;1 mutant pollen. Light micrograph of cleared whole-mount ovules (unfertilized) or seeds originating from the pollination with wild-type pollen (double fertilized) or cdka;1 mutant pollen (only egg cell fertilized). (A) Col-0 seed pollinated with Col-0 wild-type pollen at 3 DAP. (B) Nonfertilized Col-0 ovules at 6 days after emasculation (ec and cc indicate egg cell and central cell, respectively) (C) Col-0 seed pollinated with cdka;1 mutant pollen at 3 DAP; the central cell nucleus has undergone five rounds of division and here 28 nuclei are visible. This represents one of the largest nuclei numbers found upon pollination with cdka;1 mutant pollen and is typical for seeds in the Col genetic background that have initiated autonomous proliferation. (D) Bay-0 seed pollinated with cdka;1 mutant pollen at 3 DAP. The central cell nucleus has undergone three rounds of division and eight nuclei are visible, characteristic for seeds in the Bay genetic background that have initiated autonomous proliferation. (E) Sha seed pollinated with cdka;1 mutant pollen at 3 DAP. The central cell contains only a single nucleus; the predominant number of seeds in the Sha genetic background do not initiate autonomous proliferation. (F) Close-up of a Sha seed pollinated with cdka;1 mutant pollen showing a single central cell nucleus and a 16-cell embryo at 3 DAP. Insets in A–E show the inner integument layers at the same magnification. The integument cells strongly enlarge and vacuolize in double-fertilized seeds (A), whereas in unfertilized ovules (B) no cell expansion is found. Upon single fertilization with cdka;1 mutant pollen (C–E), the integuments slightly expand. All pictures are oriented such that the micropylar pole with the developing embryo points to the left and the chalazal pole of the seed to the right. Bars, 50 μm.
To describe and to quantify this variation, we sought for a fast and robust way to present our data that would also facilitate a straightforward comparison between the different data sets. The mean number of nuclei per developing seed in an accession crossed with cdka;1 could serve as a measure of autonomous endosperm. However, this method has a number of disadvantages: First, it overemphasizes seeds with high nuclei numbers due to the exponential character of the largely synchronized nuclear divisions in the central cell. In addition, taking the average number of nuclei generates an increasingly larger standard deviation with growing nuclei numbers and thus complicates a simple classification of the observed cases. Finally, accurate determination becomes very challenging at high nuclei numbers, e.g., >30.
Therefore, we defined EDV as the number of anticipated division cycles per aborting seed, i.e., the number of divisions necessary to reach the corresponding nuclei numbers within the respective seed; e.g., 28 nuclei were scored as being in the fifth cycle (28 ≈ 25).
EDV overestimates the actual endosperm nuclei, especially after the first three highly synchronous division rounds, due to the occurrence of distinct mitotic domains (Boisnard-Lorig et al. 2001). However, larger nuclei numbers could still be assigned to division cycles 4–8 in previous studies (Boisnard-Lorig et al. 2001). Consistently, we found that the counted nuclei numbers of the autonomous endosperm were usually close to the expected value for a fully completed division cycle.
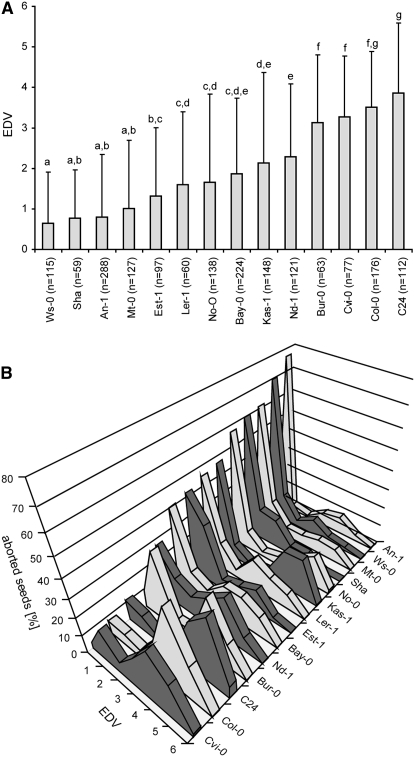
The obtained EDVs for the 14 different accessions ranged between 0.64 for Wassilewskija (Ws-0) and 3.84 for C24. Figure 3A lists groups of accessions that can be statistically separated with an error of 5%; i.e., accessions within one group are not significantly different. For example, our data set does not allow separating the autonomously developing endosperm in Sha from Est-1, while endosperm proliferation in Sha vs. Ler-1 is significantly different.
Figure 3.—
Natural variation in initiation of autonomous endosperm proliferation upon cdka;1 pollination. (A) EDVs for 14 Arabidopsis accessions. Mean values are shown with error bars representing the standard deviation. a–g indicate groups of significantly different subsets as shown by a Student–Newman–Keuls test with α = 0.5. (B) Distribution of autonomously proliferating endosperm nuclei of cdka;1-pollinated ovules in the Arabidopsis accessions An-1, Bay-0, Bur-0, C24, Cvi-0, Col, Est-1, Kas-1, Ler-1, Mt-0, Nd-1, No-0, Sha, and WS-0. The x-axis indicates EDV; the y-axis shows the wild-type ovules pollinated with cdka;1 mutant pollen in percentages. The graph shows that the observed natural variation in endosperm proliferation is mostly in the frequency of seeds that start proliferating endosperm nuclei. If nuclear proliferation has started, endosperm usually proceeds through four to five rounds of division.
General seed growth parameters contribute only a little to autonomous endosperm formation:
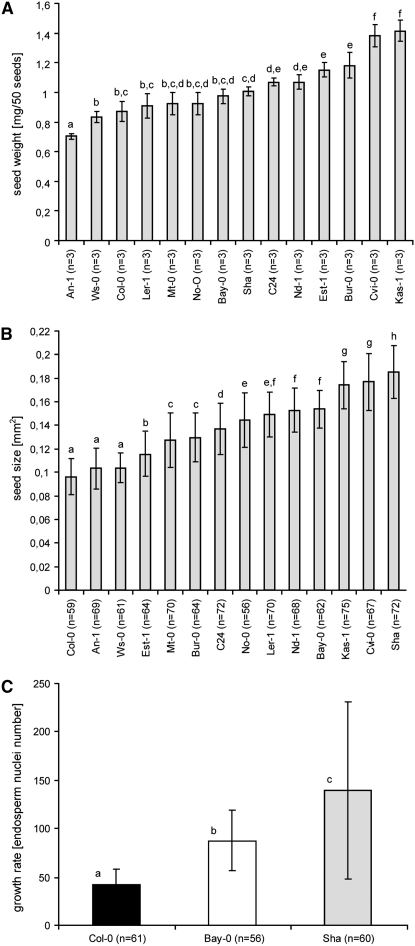
The variation of the autonomously forming endosperm could be due to natural differences in seed growth, i.e., cdka;1-pollinated seeds may develop more autonomous endosperm nuclei in accessions with larger or heavier seeds than in accessions with smaller or lighter seeds. To test this, we first determined the seed mass of the accessions used and observed a large variation in seed mass, ranging from 0.73 mg/50 seeds in An-1 to 1.41 mg/50 seeds in the accession Kas-1 (Figure 4A). Our measurements were in line with some previously reported seed masses for Ler-1 and Cvi-0 (Alonso-Blanco et al. 1999). We also found that, in addition to Kas-1 and Cvi-0, especially the accessions C24, Nd-1, Est-1, and Bur-0 have heavier seeds (Figure 4A).
Figure 4.—
Seed growth factors contribute only a little to the development of autonomous endosperm. (A) Natural variation in the seed mass (milligrams per 50 seeds) of double-fertilized ovules of the 14 accessions; seed mass could account for ∼20% of the observed variation in the autonomous endosperm (R2 = 0.20). (B) Natural variation in seed size in square millimeters of double-fertilized seeds; seed size factors have very little effect on the development of autonomous endosperm (R2 = 0.0043). (C) Natural variation in endosperm growth rate in nuclei numbers of double-fertilized endosperm at 3 DAP; factors controlling nuclear proliferation affect ∼30% of the variation seen in autonomous endosperm formation (R2 = 0.3189). Mean values are shown with error bars representing standard deviations. a–c indicate groups of significantly different subsets as shown by a Student–Newman–Keuls test with α = 0.5.
Refuting our initial hypothesis, large seed masses contributed only a little to high EDVs in cdka;1-pollinated seeds; i.e., ∼20% of the EDV might be accounted for by mass (linear regression R2 = 0.20) (Figure 3A, Figure 4A).
The effect of seed size on the formation of autonomous endosperm was even much lower (linear regression R2 = 0.0043; Figure 3A, Figure 4B).
To address putative differences in growth rate between the tested accessions, we counted endosperm nuclei in wild-type seeds of three representative accessions at 3 DAP (Figure 4C). Remarkably, we also found here differences in the proliferation rate of wild-type endosperm between the tested accessions. Among the seed growth parameters analyzed, proliferation rate had the largest effect on autonomous endosperm formation, contributing to ∼30% of the observed variation (linear regression R2 = 0.3189; Figure 3A, Figure 4C). Finally, an analysis of the distribution of nuclei numbers revealed that the natural variation found is not in the degree of autonomous endosperm formed, but rather in the frequency at which the central cell starts proliferating or not (Figure 3B). Once a central cell proliferates, even in an accession with a low EDV, it usually progresses to the fourth or fifth division cycle, i.e., roughly between 16 and 32 nuclei.
Taken together, we have detected natural variation in three major seed growth parameters in Arabidopsis, i.e., mass, size, and growth rate, consistent with previous reports (Krannitz et al. 1991; Alonso-Blanco et al. 1999). However, although factors that influence seed growth also have an effect on nuclear proliferation in the endosperm upon fertilization with cdka;1 mutant pollen, in particular proliferation rate, the development of autonomous endosperm relies largely on additional factors.
QTL analysis of the Sha–Bay RIL population reveals two new loci involved in autonomous endosperm development:
In addition to natural variation in seed growth, another possible explanation for the observed variation in autonomous endosperm development is a difference in the previously identified positive signal that links endosperm development with the fertilization of the egg cell. On the one hand, there could be natural differences in the signal generation and/or transmittance from the fertilization process. On the other hand, variation might exist on the perception side, i.e., in the central cell. Since we have previously found that fis-class mutants and cdka;1 can mutually restore their seed viability (Nowack et al. 2007), an obvious hypothesis was that natural variation in the expression or function of FIS genes might be responsible for the observed variation in autonomous endosperm formation.
To identify loci that influence the proliferation of the autonomous endosperm, we used an established RIL population between two accessions that differed significantly in their EDV: Bay-0 from southern Germany and Shahdara (Sha), originating from Tajikistan (Figure 3A) (Loudet et al. 2002).
Of the 165 lines of the core collection, we grew 160 lines and pollinated each of these RILs with pollen from heterozygous cdka;1 mutant plants. Between four and eight siliques derived from each cross were analyzed and the number of endosperm nuclei were determined at 3 DAP. EDVs varied among the RIL population and differed substantially from the Bay-0 and Sha parents (Figure 3A, supplemental Table 1).
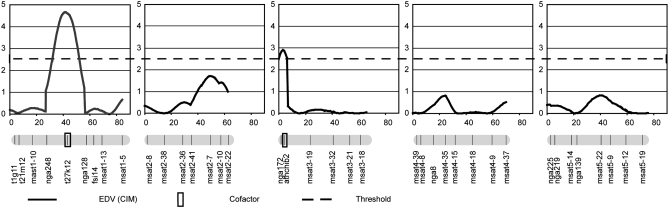
Two QTL explaining ∼20% of the total variance in EDVs with a heritability of 0.93 were detected and localized by CIM (Figure 5). We named the first QTL, located on chromosome I (peak at 40.9 cM) and associated with the marker T27K12 (at 43.6 cM or 15.9 Mb) KIRKE (KIR). This QTL was detected with a LOD of 4.66 and was found to be responsible for 13.7% of the observed phenotypic variation in the Bay-0 × Sha population. The alleles of Bay-0 at this position increased the number of divisions by a factor of 0.64.
Figure 5.—
QTL mapping of factors controlling the autonomous proliferation of the central cell after cdka;1 pollination. LOD traces of QTL mapping (in centimorgans) for EDV in the Bay-Sha RIL population are reported for each of the five Arabidopsis chromosomes represented by the shaded bars at the bottom of each chart (top on the left and bottom on the right of the bar). Marker names are indicated on each chromosome at their genetic position (Loudet et al. 2002). CIM (continuous line) is reported. The markers used as cofactors in CIM are boxed. The threshold of QTL detection (LOD = 2.4; dashed line) has been determined using a permutation test (see materials and methods).
The second QTL was mapped to the tip of chromosome III around the marker athcib2 (6.8 cM or 3.96 Mb) and was given the name KALYPSO (KAL). [In Greek mythology Kirke and Kalypso are two challenges the hero Odysseus had to master on his journey (Homer 2005). Both tried to seduce Odysseus and tried to distract him from his way home.] KAL had a LOD of 2.91 and explained 8.1% of the observed phenotypic variation in the Bay-0 × Sha population. Contrary to KIR, the presence of the alleles from Bay-0 at the KAL locus decreased the number of endosperm divisions. This resulted in a high transgression of the phenotype in the direction of higher EDVs, reaching up to 4.5 in some RILs. In contrast, no RILs that had an EDV lower than that determined for Sha were found. No genetic interaction between KAL and KIR was identified (data not shown).
The map positions of KIR and KAL were compared with the position of the FIS class genes and genes related to PRC2 in the Arabidopsis genome, i.e., MEA (AT1G02580), SWN/EZA1 (AT4G02020), FIS2 (AT2g35670), FIE (AT3G20740), the molecularly unidentified BORGIA (BGA) located in the vicinity of FIS2 (Guitton et al. 2004), and MULTICOPY SUPPRESSOR OF IRA 1 (MSI1; AT5G58230). However, none of these genes map closely to the two QTL identified here.
Furthermore, we compared the map position of other genes known to influence seed size: TTG2 (AT2G37260), IKU2 (AT3G19700), MINI3 (AT1G55600), AP2 (AT4G36920), and ARF2/MNT (AT5G62000). Again, neither KAL nor KIR mapped near any of these genes, indicating that they represent two new loci involved in endosperm development.
Uncoupling of endosperm development from proliferation:
The obtained genetic variation allowed us to dissect the relationship between endosperm and embryo development in detail. A first question was whether nuclear proliferation is required for the adaptation of endosperm fate and subsequent changes in seed development. Indeed, even in seeds fertilized with cdka;1 mutant pollen and containing only a single central cell nucleus, changes in seed morphology typical for fertilized ovules could be observed. For example, while in unfertilized ovules the cells of the endothelium layer remained small, in seeds with a single central cell the endothelium cells expanded and vacuolized as in wild-type seeds (Figure 2, A–E insets).
To further test the properties of seeds with a single nucleated endosperm, we analyzed the accumulation of proanthocyanidin in the endothelium layer in the accession Sha. In Sha, >60% of cdka;1 fertilized seeds display only a single nucleus in the central cell (Figure 4). Nonetheless, >85% of the cdka;1 fertilized Sha seeds were found to be vanillin positive (Figure 1J; data not shown). Therefore, we conclude that differentiation of the endothelium is independent of endosperm nuclei number and might be triggered by either an embryo and/or a single endosperm nucleus.
Finally, we found that in the endosperm marker line KS22, which is in the C24 background, cdka;1 fertilized seeds with only a single nucleus in the endosperm expressed the KS22 reporter (Figure 1N).
Thus, endosperm differentiation can be uncoupled from proliferation and even a single endosperm nucleus can adopt endosperm properties.
Relationship between endosperm and embryo development during seed growth:
Next we analyzed the effect of an underproliferated endosperm on embryo development. In Arabidopsis (and other plants) it has been found that the primary endosperm nucleus divides much earlier than the zygote; i.e., at the time of the first zygotic division there are already between 12 and 16 nuclei in the endosperm (Boisnard-Lorig et al. 2001). This might suggest a dependence of embryo development upon endosperm formation. However, we found that an embryo developed in all cases even if the central cell did not divide (n = 45) (Figure 6, Figure 7). Consistently, in the recently reported glauce (glc) mutant that is impaired in central cell fertilization, the embryo reaches a globular stage with a single central cell nucleus (Ngo et al. 2007). Thus, we conclude that the start of embryo development is independent of endosperm proliferation and likely reflects an autonomous program depending solely on the fertilization of the egg cell.
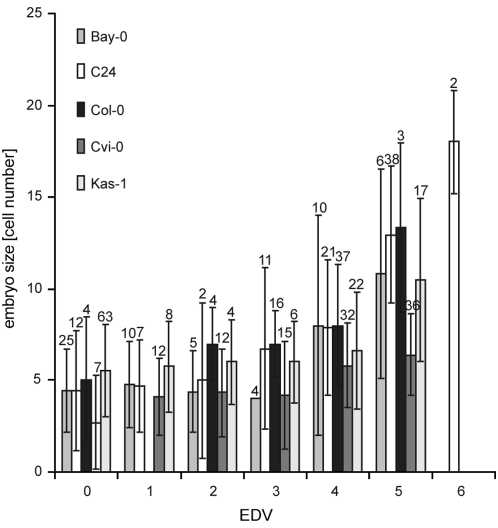
Figure 6.—
Early correlation of embryo development and endosperm proliferation. Correlation between the size of the autonomously formed endosperm (in EDV) with embryo size (in numbers of cells) at 3 DAP. In all five analyzed accessions, endosperm size correlated with embryo size [R2 (Bay-0) = 0.64; R2 (C24) = 0.82; R2 (Col-0) = 0.74; R2 (Cvi-0) = 0.90; R2 (Kas-1) = 0.61]. The sample size n is provided at the top of each column. Error bars indicate standard deviation.
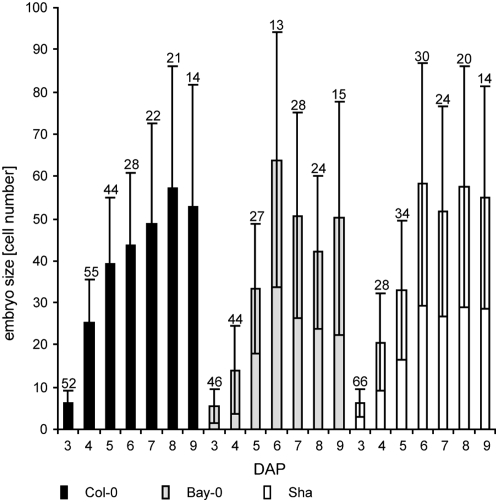
Figure 7.—
Independence and constraints of embryo development. Time course of embryo development in cdka;1-pollinated seeds of the three accessions, Col-0, Bay-0, Sha, from 3 to 9 DAP. After 3 DAP, no additional divisions in the endosperm were observed, yet embryos in all three accession continued to grow until 6–9 DAP, indicating a self-sufficient embryo developmental program. However, embryos reached a size of only 55–65 cells (globular stage), showing the limitations of this endosperm-independent growth. The sample size n is provided at the top of each column. Error bars indicate standard deviation.
This embryonic autonomy raised the question to what degree embryo development depends on endosperm size. Therefore, we plotted embryo growth as measured by cell number against number of endosperm nuclei in cdka;1 fertilized seeds at 3 DAP. Figure 6 shows that more nuclei in the endosperm are strongly correlated with more cells in the embryo, suggesting an early positive interaction between embryo and endosperm growth [R2 (Bay-0) = 0.64; R2 (C24) = 0.82; R2 (Col-0) = 0.74; R2 (Cvi-0) = 0.90; R2 (Kas-1) = 0.61]. Although currently we cannot exclude a more complicated interaction, for example, via maternal layers, the most likely explanation appears to be a positive and dose-dependent effect of developing endosperm on embryo growth.
Endosperm proliferation in cdka;1 fertilized seeds ceased ∼3 DAP. Independent of this, the embryo continued to grow for ∼3 more days, reaching on average a size of ∼50 cells at 9 DAP (Figure 7). On the one hand, this underlines the autonomous developmental program of the embryo. On the other hand, it shows the limits of embryo development without or with very little endosperm development, corroborating that further embryo growth depends on the presence of endosperm as first reported by Cooper and Brink (1942) in the middle of the last century.
DISCUSSION
Seed growth requires an intricate interplay between the different tissues that compose a seed. However, little is known about the coordination and integration of these parts into a functional whole. Analysis of seed development appears in particular to be difficult due to the observed coordinated growth behavior; i.e., small seeds also contain a reduced endosperm and large seeds a correspondingly larger endosperm (Berger et al. 2006). Here we have followed a genetic dissection of double fertilization and seed development and exploited natural variation found in Arabidopsis seed development to study embryo–endosperm interactions.
Since we have previously found that cdka;1 mutants can be rescued in combination with fis-class mutants, a simple hypothesis would be that natural variation in the imprinting machinery might be responsible for the observed difference in endosperm size upon cdka;1 pollination (Nowack et al. 2007). In fact, it has been observed that a mea mutant in an accession background that we identified to have a high EDV, i.e., Col, displayed greater numbers of autonomously formed endosperm nuclei than a mea mutant in an accession with a low EDV, i.e., Ws, while in Ler intermediate values were obtained for both traits (Kiyosue et al. 1999). One possible explanation is that both the increased autonomous endosperm formation and the high EDV values are due to natural variation in SWN that acts redundantly with MEA (Wang et al. 2006). Our data, however, do not substantiate such a hypothesis since the loci identified here do not map to any known FIS-class genes. Thus it remains to be seen whether the effect of the genetic background on the penetrance of the mea mutant has the same genetic source as the variation of autonomous endosperm formation observed upon fertilization with cdka;1 mutant pollen.
Interdependency of embryo and endosperm development:
The QTL identified in this study also did not correlate to any known loci affecting seed growth regulators. Moreover, we could show that factors regulating seed growth account only for a minor effect on the development of autonomous endosperm, pinpointing a new class of seed regulators. At the same time, variation in EDV is largely driven by genetic factors (h2 = 0.93) and the two QTL identified here together explain only ∼20% of the total variance. Thus, many minor QTL were presumably not detected due to a lack of statistical power, and among these minor QTL, we suspect seed growth regulators.
It currently seems likely that the two QTL identified here influence the signal transduction cascade between the fertilization of the egg cell and the central cell. The pending molecular identification of these loci offers a chance to obtain molecular insights into the signaling system that coordinates seed development.
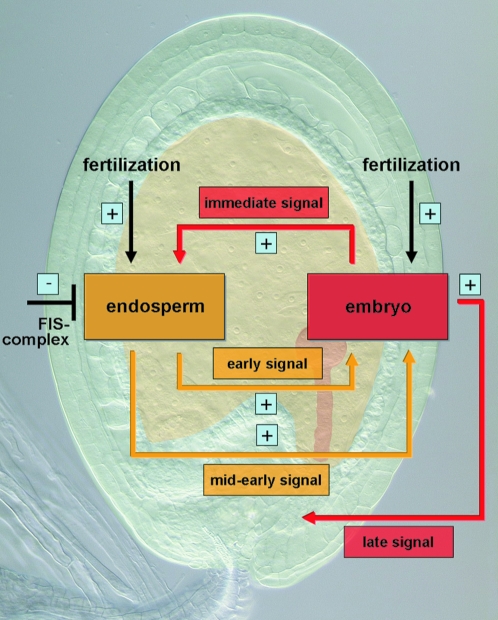
Data from this work and previous studies suggest that not only one but also multiple signals are at work and that information has to be exchanged repeatedly between the developing parts of a seed (Figure 8). Evidence for an immediate early signal from the zygote to the central cell comes from the observation of the autonomous endosperm proliferation observed upon fertilization of the egg cell (Nowack et al. 2006). Therefore, as presented in this study, there likely exists a second interaction phase during early endosperm development since at least ∼3 DAP embryo growth was found to correlate with the amount of endosperm formed (Figure 8). A similar correlation was also found for the maternal-effect capulet1 (cap1) mutant (Grini et al. 2002). Although the direction of a putative signaling pathway underlying this phenotype cannot unambiguously be clarified, it seems likely that the degree of endosperm influences embryo size and not vice versa. Consistent with this directionality is that, in experiments in which the embryo was genetically ablated, no obvious effects on endosperm development were reported (Baroux et al. 2001a,b).
Figure 8.—
Model of signal transduction pathways and cross talk between embryo and endosperm. On the basis of genetic evidence, four different signaling pathways between the embryo and the endosperm are postulated: an immediate early signal originating from the fertilization of the egg cell and stimulating endosperm fate adoption of the central cell; an early signal from the endosperm that stimulates in a quantitative manner embryo development; a mid-to-early action of the endosperm that is required for embryo development beyond a globular stage; and finally, a late signal stemming from the embryo and responsible for seed survival. For further explanation, see text.
For further embryo development, the endosperm has to reach a critical size as already suggested by Cooper and Brink (1942). It is possible that an endosperm size checkpoint at or beyond 64 nuclei exists since in all accessions pollinated with cdka;1 mutant pollen the embryo reached roughly the same size of ∼50 cells (Figure 8; see also below). Evidence for such a checkpoint also comes from the maternal-effect mutant capulet2 (cap2) in which the embryo developed to roughly the same stage as cdka;1 fertilized seeds (Grini et al. 2002).
Interestingly, such checkpoint behavior was previously not found in plants in which the endosperm was genetically ablated, and Weijers et al. (2003) found that embryogenesis could continue with very little endosperm although these embryos would eventually arrest. The driver line used for these ablation experiments becomes expressed ∼2 DAP, and a possible explanation is that this expression is already later than a putative seed size checkpoint. One hypothesis derived from this is that the establishment phase of endosperm might be especially crucial for seed development. A large number of recently identified gametophytic and seed-specific reporter and driver lines will enable a high-resolution dissection of the requirements of endosperm for embryo and seed development (Stangeland et al. 2005; Tiwari et al. 2006; Steffen et al. 2007).
The function of this putative endosperm checkpoint could be complex and could involve, for example, interaction with the mother plant. Cooper and Brink (1940) observed that in interspecies crosses the development of both the embryo and the endosperm were initiated but endosperm proliferation slowed down, followed by seed abortion. Interestingly, they found that the maternal layer, in particular the nucellar tissue, overproliferated preceding seed degeneration. Remarkably, we observed somewhat similar overproliferation and meristematization of maternal tissue at the chalazal pole in the aborting seeds (data not shown). On the one hand, this overproliferation could reflect a competition between the different seed tissues and an underdeveloped endosperm that might not be able to draw sufficient resource from the mother plant. On the other hand, this overproliferation could be an active execution mechanism of the mother to cut off developing seeds from nutrient supply. In any case, the successful establishment of endosperm as a sink tissue is likely to be a major step in seed development.
In addition, there appears to be a late signal coming from the embryo that is crucial for seed survival (Figure 8). Seeds in unpollinated fis mutants with a developing endosperm and no embryo will abort whereas fis mutants pollinated with cdka;1, which develop autonomous endosperm accompanied by an embryo, can complete embryogenesis and give rise to viable plants (Nowack et al. 2007).
Autonomy of embryo and endosperm development:
The observed natural variation also allowed us to address the developmental potential of endosperm and embryo. An unexpected finding was the relatively large degree of embryonic autonomy. Even without any divisions in the endosperm, embryo development was started and on a morphological basis was indistinguishable from wild-type embryo formation. At least one formal possibility is that nutrients could be provided from the mother plant through the suspensor. However, to our knowledge, the suspensor as a route for nutrients into the developing embryo has not been explored in much detail.
However, a single-nucleated endosperm appears to be functional, and differentiation of the central cell into endosperm along with morphological changes of the single-fertilized seeds was independent of cell divisions. For example, we observed that even in seeds with a single endosperm nucleus differentiation of the endothelium layer was induced. Notably, the accumulation of proanthocyanidins started from the micropylar pole of the seed in the surrounding area of the developing embryo and not in the immediate vicinity of the single endosperm nucleus. Similarly, the accumulation of proanthocyanidins started at the micropylar side in unfertilized seeds of the FIS-class mutants msi1, fis2, and mea (Ingouff et al. 2006; R. Shirzadi and P. E. Grini, unpublished results). Thus, it is possible that proanthocyanidin production is a readout of a rapidly polarizing central cell with a very distinct basal (micropylar) domain. However, no subcellular markers are currently available to follow this putative polarization.
The differentiation of the single central cell nucleus became most evident in the expression of endosperm marker gene KS22. Thus, the developmental potential for endosperm appears to be already programmed into the central cell as a part of the female gametophyte and neither fertilization nor proliferation of this cell is required for the adoption of this fate. However, the fertilization of the egg cell and the generation of subsequent signals appear to be required for a cell fate change. This is consistent with the hypothesis of a gametophytic evolutionary origin of endosperm in flowering plants and with Strasburger's (1900) hypothesis that the fertilization of the central cell is used in higher plants to trigger proliferation.
Acknowledgments
The authors thank Maarten Koornneef and Reidunn B. Aalen for critical reading and helpful comments on the manuscript. We are grateful to Frederic Berger and Abdul M. Chaudhury for providing marker lines used in this study. We are grateful to Hugues Barbier for help with statistical analyses. A.U. is a fellow of the Konrad-Adenauer-Stiftung. M.K. is a fellow of the International Graduate School in Genetics and Functional Genomics. P.E.G. and R.S. were supported by a grant from the Norwegian Research Council. This work was supported by grants of the Volkswagen-Stiftung, by the Deutsche Forschungsgemeinschaft SFB 572, by core funding of the Centre National de la Recherche Scientifique to A.S., and by an European Research Area in Plant Genomics grant to P.E.G. and A.S.
References
- Alonso-Blanco, C., H. Blankestijn-de Vries, C. J. Hanhart and M. Koornneef, 1999. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 4710–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, E. G., E. A. Stahl, C. Toomajian, M. Nordborg, M. Kreitman et al., 2006. Distribution of genetic variation within and among local populations of Arabidopsis thaliana over its species range. Mol. Ecol. 15 1405–1418. [DOI] [PubMed] [Google Scholar]
- Baroux, C., R. Blanvillain and P. Gallois, 2001. a Paternally inherited transgenes are down-regulated but retain low activity during early embryogenesis in Arabidopsis. FEBS Lett. 509 11–16. [DOI] [PubMed] [Google Scholar]
- Baroux, C., R. Blanvillain, I. R. Moore and P. Gallois, 2001. b Transactivation of BARNASE under the AtLTP1 promoter affects the basal pole of the embryo and shoot development of the adult plant in Arabidopsis. Plant J. 28 503–515. [DOI] [PubMed] [Google Scholar]
- Beeckman, T., R. De Rycke, R. Viane and D. Inze, 2000. Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113 139–148. [Google Scholar]
- Berger, F., P. E. Grini and A. Schnittger, 2006. Endosperm: an integrator of seed growth and development. Curr. Opin. Plant Biol. 9 664–670. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., A. Colon-Carmona, M. Bauch, S. Hodge, P. Doerner et al., 2001. Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. L., D. R. Smyth and E. M. Meyerowitz, 1989. Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A. M., L. Ming, C. Miller, S. Craig, E. S. Dennis et al., 1997. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D. C., and R. A. Brink, 1940. Somatoplastic sterility as a cause of seed failure after interspecific hybridization. Genetics 25 593–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D. C., and R. A. Brink, 1942. The endosperm as a barrier to interspecific hybridization in flowering plants. Science 95 75–76. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., N. Nesi, P. Perez, M. Devic, O. Grandjean et al., 2003. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G. N., J. L. Bowman and E. M. Meyerowitz, 1991. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002. [DOI] [PubMed] [Google Scholar]
- Garcia, D., V. Saingery, P. Chambrier, U. Mayer, G. Jurgens et al., 2003. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., J. N. Fitz Gerald and F. Berger, 2005. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini, P. E., G. Jurgens and M. Hulskamp, 2002. Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., J. P. Vielle-Calzada, M. A. Hoeppner and W. B. Gagliano, 1998. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton, A. E., D. R. Page, P. Chambrier, C. Lionnet, J. E. Faure et al., 2004. Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 2971–2981. [DOI] [PubMed] [Google Scholar]
- Haig, D., and C. Graham, 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64 1045–1046. [DOI] [PubMed] [Google Scholar]
- Haig, D., and M. Westoby, 1989. Parent-specific gene expression and the triploid endosperm. Am. Nat. 134 147–155. [Google Scholar]
- Haig, D., and M. Westoby, 1991. Genomic imprinting in endosperm: its effects on seed development in crosses between species and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333 1–13. [Google Scholar]
- Homer, 2005. Odyssee. Saur, Munich.
- Ingouff, M., J. Haseloff and F. Berger, 2005. Polycomb group genes control developmental timing of endosperm. Plant J. 42 663–674. [DOI] [PubMed] [Google Scholar]
- Ingouff, M., P. E. Jullien and F. Berger, 2006. The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., A. Shinmyo and M. Sekine, 2006. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Jofuku, K. D., P. K. Omidyar, Z. Gee and J. K. Okamuro, 2005. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. S., B. Kolevski and D. R. Smyth, 2002. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., A. Miura, Y. Choi, Y. Kinoshita, X. Cao et al., 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., N. Ohad, R. Yadegari, M. Hannon, J. Dinneny et al., 1999. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., L. Hennig, R. Bouveret, J. Gheyselinck, U. Grossniklaus et al., 2003. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., J. H. de Bruine and P. Goettsch, 1980. A provisional map of chromosome 4 of Arabidopsis. Arabidop. Inf. Serv. 17 11–18. [Google Scholar]
- Koornneef, M., C. Alonso-Blanco and D. Vreugdenhil, 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Krannitz, P. G., L. W. Aarssen and J. M. Dow, 1991. The effect of genetically based differences in seed size on seedling survival in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 78 446–450. [Google Scholar]
- Kunst, L., J. E. Klenz, J. Martinez-Zapater and G. W. Haughn, 1989. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y., 1982. Association of endosperm reduction with parental imprinting in maize. Genetics 100 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y., 1984. Ploidy barrier to endosperm development in maize. Genetics 107 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., S. Chaillou, C. Camilleri, D. Bouchez and F. Daniel-Vedele, 2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104 1173–1184. [DOI] [PubMed] [Google Scholar]
- Luo, M., P. Bilodeau, E. S. Dennis, W. J. Peacock and A. Chaudhury, 2000. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., E. S. Dennis, F. Berger, W. J. Peacock and A. Chaudhury, 2005. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison, I. M., J. P. Ramsay and H. G. Spencer, 2005. A census of mammalian imprinting. Trends Genet. 21 457–465. [DOI] [PubMed] [Google Scholar]
- Ngo, Q. A., J. M. Moore, R. Baskar, U. Grossniklaus and V. Sundaresan, 2007. Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134 4107–4117. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M. K., P. E. Grini, M. J. Jakoby, M. Lafos, C. Koncz et al., 2006. A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Nowack, M. K., R. Shirzadi, N. Dissmeyer, A. Dolf, E. Endl et al., 2007. Bypassing genomic imprinting allows seed development. Nature 447 312–315. [DOI] [PubMed] [Google Scholar]
- Ohad, N., L. Margossian, Y. C. Hsu, C. Williams, P. Repetti et al., 1996. A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, M. A., R. L. Fischer, R. B. Goldberg, K. Nakamura and J. J. Harada, 2005. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O. A., 2004. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16(Suppl.): S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., O. Torjek, R. Meyer, H. Schmuths, M. H. Hoffmann et al., 2006. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 112 1104–1114. [DOI] [PubMed] [Google Scholar]
- Schruff, M. C., M. Spielman, S. Tiwari, S. Adams, N. Fenby et al., 2006. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133 251–261. [DOI] [PubMed] [Google Scholar]
- Scott, R. J., M. Spielman, J. Bailey and H. G. Dickinson, 1998. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sitte, P., E. W. Weiler, J. W. Kadereit, A. Bresinsky and C. Körner, 2002. Strasburger. Lehrbuch der Botanik für Hochschulen. Spektrum Akademischer Verlag, Heidelberg, Germany/Berlin.
- Stangeland, B., R. Nestestog, P. E. Grini, N. Skrbo, A. Berg et al., 2005. Molecular analysis of Arabidopsis endosperm and embryo promoter trap lines: reporter-gene expression can result from T-DNA insertions in antisense orientation, in introns and in intergenic regions, in addition to sense insertion at the 5′ end of genes. J. Exp. Bot. 56 2495–2505. [DOI] [PubMed] [Google Scholar]
- Steffen, J. G., I. H. Kang, J. MacFarlane and G. N. Drews, 2007. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51 281–292. [DOI] [PubMed] [Google Scholar]
- Strasburger, E., 1900. Einige Bemerkungen zur Frage nach der “doppelten Befruchtung” bei den angiospermen. Bot. Zeit. 58 294–315. [Google Scholar]
- Tiwari, S., M. Spielman, R. C. Day and R. J. Scott, 2006. Proliferative phase endosperm promoters from Arabidopsis thaliana. Plant Biotechnol. J. 4 393–407. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., G. Hagen and T. J. Guilfoyle, 1999. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, J. W., 2004. MapQTL5: Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Kyazma, Wageningen, The Netherlands.
- Wang, D., M. D. Tyson, S. S. Jackson and R. Yadegari, 2006. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., J. P. Van Hamburg, E. Van Rijn, P. J. Hooykaas and R. Offringa, 2003. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133 1882–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and H. Weber, 1999. Sugars as signal molecules in plant seed development. Biol. Chem. 380 937–944. [DOI] [PubMed] [Google Scholar]