Abstract
Background
Peripheral blood mononuclear cells (PBMC) serve a sentinel role allowing the host to efficiently sense and adapt to the presence of danger signals. Herein we have directly compared the genome-level expression patterns (microarray) of a human PBMC model (THP-1 cells) subjected to one of two canonical danger signals, heat shock or lipopolysaccharide (LPS).
Results and Discussion
Based on sequential expression and statistical filters, and in comparison to control cells, we found that 3,988 genes were differentially regulated in THP-1 cells subjected to LPS stress, and 2,921 genes were differentially regulated in THP-1 cells subjected to heat shock stress. Venn analyses demonstrated that the majority of differentially regulated genes (≥ 70%) were uniquely expressed in response to one of the two danger signals. Functional analyses demonstrated that the two danger signals induced expression or repression of genes corresponding to unique pathways, molecular functions, biological processes, and gene networks. In contrast, there were 184 genes that were commonly upregulated by both stress signals, and 430 genes that were commonly downregulated by both stress signals. Interestingly, the 184 commonly upregulated genes corresponded to a gene network broadly related to inflammation, and more specifically to chemokine signaling.
Conclusion
These data demonstrate that the mononuclear cell responses to the canonical stress signals, heat shock and LPS, are highly divergent. However, there is a heretofore unrecognized common pattern of gene network expression corresponding to chemokine-related biology. The data also serve as a reference database for investigators in the field of stress signaling.
Background
Peripheral blood mononuclear cells serve a sentinel role by allowing rapid host adaptations to danger signals associated with various biological stresses such as infection, trauma, or drastic environmental changes [1]. Two of the more primitive and universal danger signals recognized by peripheral blood mononuclear cells, as well as other types of cells, are lipopolysaccharide (LPS) of gram negative bacteria and heat stress (a.k.a. heat shock). Previous studies in our laboratory, and that of others, have implied that the cellular responses to LPS and heat shock are mutually exclusive, particularly at the transcriptional level [2-6]. The transcriptional response to LPS has been traditionally viewed as one involving expression of immune- and inflammation-associated genes that serve to rid the host of infection. In contrast, the transcriptional response to heat shock has been traditionally viewed as a specific re-prioritization of gene expression toward "stress proteins" while other forms of gene expression are globally suppressed.
These two traditional paradigms involving the transcriptional responses to LPS or heat stress may represent oversimplifications. For example, transcriptional profiling experiments have suggested the existence of previously unrecognized genes that are expressed in response to LPS stimulation [7-12]. Our own studies have demonstrated that genes not traditionally regarded as "heat shock proteins" can be specifically regulated and expressed in response to heat shock [13-18].
In the current work, we sought to expand our understanding of the peripheral blood mononuclear cell transcriptional response to canonical stress/danger signals. We have exposed peripheral blood mononuclear cells to either LPS or heat shock, and have directly compared their respective genome-level responses using microarray technology. The focus of the analytical approach is coordinated expression/repression of genes corresponding to functional annotations and gene networks.
Methods
Cell model and experimental conditions
Human THP-1 mononuclear cells (American Type Culture Collection, Manassas, VA; ATCC#: TIB-202™) were used as a model of peripheral blood mononuclear cells as previously described [19]. Cells were maintained in a room air/5% CO2 incubator at 37°C using RPMI (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. One group of cells was exposed to LPS (1 μg/ml, E. coli serotype 055:B5, Sigma, St. Louis, MO) for 4 hours at 37°C. Heat shock was carried out by placing one group of cells in a dedicated room air/5% CO2 incubator set at 43°C for 1 hour. After heat shock, cells were returned to a 37°C, room air/5% CO2 incubator and allowed to recover for 4 hours. Control cells were maintained in basal growth media at 37°C. After exposure to the experimental conditions, cells were harvested for total RNA extraction as described below.
RNA extraction and microarray hybridization
The data and protocols described in this manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO, [20]) and are accessible through GEO Series accession number GSE9916.
Total RNA was isolated from THP-1 cells exposed to the above experimental conditions using the Trizol reagent (Invitrogen) according to the manufacturer's specifications. Microarray hybridization was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children's Research Foundation as previously described using the Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA) [21,22].
Analyses were performed using one experimental condition per chip, with each experimental condition carried out in triplicate (i.e. 3 biological replicates per experimental condition for a total of 9 individual chips). Image files were captured using an Affymetrix GeneChip Scanner 3000. CEL files were subsequently preprocessed using Robust Multiple-Array Average (RMA) normalization and GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA). All signal intensity-based data were used after RMA normalization, which specifically suppresses all but significant variation among lower intensity probe sets [23]. All chips were then normalized to the respective median values of the control condition.
Analysis of microarray data
Differences in mRNA abundance between LPS-stimulated cells and control cells, and between heat shock-treated cells and control cells were determined using GeneSpring GX 7.3 (Agilent Technologies). All statistical analyses used corrections for multiple comparisons. A list of genes differentially regulated between LPS-treated cells and control cells was generated using sequential expression and statistical filters as follows. Starting with all gene probes within the microarray (54,681 gene probes) we selected the genes having ≥ 2-fold expression difference (increased or decreased expression) between the respective median values of LPS-treated cells and control cells. The genes passing this expression filter were then subjected to a statistical test (ANOVA, Benjamini-Hochberg false discovery rate of 5%) using LPS-treated cells and control cells as the comparison groups. A list of genes differentially regulated between heat shock-treated cells and control cells was generated using identical sequential expression and statistical filters, but using heat shock-treated cells and control cells as the comparison groups.
Functional annotation analysis
Gene lists of differentially expressed genes were primarily analyzed for functional annotation enrichment using the PANTHER (Protein Analysis Through Evolutionary Relationships) Classification System [24,25]. The PANTHER Classification System is a publicly accessible relational database of functional gene annotations and is based on the existing biomedical literature. The application uses specific approaches to estimate significance (p values) based on non-redundant representations of the microarray chip and to convert the uploaded gene lists to gene lists containing a single value per gene. All PANTHER-derived data are corrected for multiple comparisons via Bonferroni.
Derivation of gene networks
Gene lists of differentially expressed genes were also analyzed for the presence of gene networks using the Ingenuity Pathways Analysis application (IPA, Ingenuity Systems, Redwood City, CA) [26]. The IPA application provides a tool for discovery of gene networks within the uploaded gene lists. The IPA application maps each gene identifier to its corresponding gene object in the IPA Knowledge Base. These focus genes are then overlaid onto a global molecular network developed from information contained in the Knowledge Base. Networks of these focus genes are then algorithmically generated based on their connectivity. Each individual IPA network has a maximum of 35 focus genes and is assigned a significance score (based on p value) representing the likelihood that the focus genes within the network are found therein by random chance. A high number of focus genes within a dataset leads to a higher network score (equal to the negative exponent of the respective p value such that a score of 3 corresponds to a p value of 10E-3). Each of the IPA-derived gene networks provided in the Results section has a significance score of ≥ 35 (i.e. p value ≥ 10E-35) and the gene networks are presented as merges of the top two scoring networks within each of the respective gene lists.
Derivation of gene network functional annotations
The functional annotations associated with the gene networks presented in Results section were generated using a relational database independent of the IPA application and the PANTHER database. The gene lists corresponding to the gene networks were analyzed using D.A.V.I.D. (Database for Annotation, Visualization and Integrated Discovery), which also allows public access to relational databases of functional gene annotations [27]. In the D.A.V.I.D. analytical output, "category" refers to the original database or resource from which the annotations are derived, and "term" refers to the enriched annotation terms associated with the given gene list. D.A.V.I.D is also based on the established biomedical literature and also uses specific approaches to estimate significance (p values) based on non-redundant representations of the microarray chip and to convert the uploaded gene lists to gene lists containing a single value per gene. The p values for a given category and term provide an estimate of the likelihood that a given annotation in enriched in a given gene list by chance alone. All of the p values presented in the D.A.V.I.D.-based analyses have been corrected for multiple comparisons using the Benjamini-Hochberg false discovery rate of 5%.
Results
Comparison of the genomic expression profiles of THP-1 cells exposed to either LPS or heat shock
Based on the filtering strategy described in the Methods section, there were 3,968 gene differentially expressed in THP-1 cells exposed to LPS. Of these 3,968 genes, 1,345 were upregulated and 2,623 were downregulated relative to control cells (see Additional file 1). In comparison, there were 2,921 genes differentially expressed in THP-1 cells exposed to heat shock. Of these 2,921 genes, 1,476 were upregulated and 1,445 were downregulated relative to control cells (see Additional file 2).
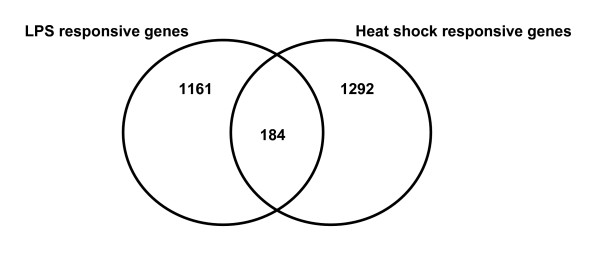
Figure 1 depicts a Venn analysis comparing the genes upregulated in response to LPS and heat shock, respectively. Eighty-six percent of the upregulated LPS-responsive genes were uniquely expressed in response to LPS, whereas 88% of the upregulated heat shock-responsive genes were uniquely expressed in response to heat shock. In contrast, 184 upregulated genes were common to both the LPS response and the heat shock response.
Figure 1.
Venn analysis comparing the 1,345 upregulated LPS-responsive genes and the 1,476 upregulated heat shock-responsive genes. See text for gene filtering strategy.
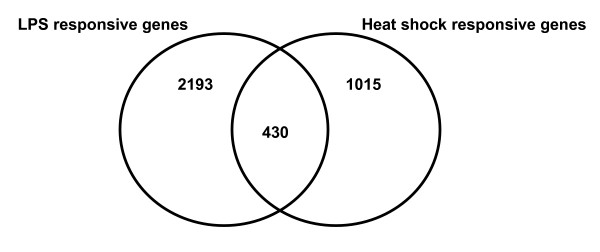
Figure 2 depicts a Venn analysis comparing the genes downregulated in response to LPS and heat shock, respectively. Eighty-four percent of the downregulated LPS-responsive genes were uniquely repressed in response to LPS, whereas 70% of the downregulated heat shock-responsive genes were uniquely repressed in response to heat shock. In contrast, 430 downregulated genes were common to both the LPS response and the heat shock response.
Figure 2.
Venn analysis comparing the 2,623 downregulated LPS-responsive genes and the 1,445 downregulated heat shock-responsive genes. See text for gene filtering strategy.
Collectively, these data demonstrate that a large proportion of genes differentially expressed in response to LPS or heat shock are unique relative to the specific stress signal. However, the two stress signals also share a relatively smaller proportion of commonly regulated genes.
Functional annotations corresponding to the upregulated LPS- and heat shock-responsive genes
To begin deriving biological meaning from the individual lists of upregulated LPS- and heat shock-responsive genes, we uploaded the respective upregulated gene lists to the PANTHER database and focused the analysis on enrichment for signaling pathways, biological processes, and molecular functions. As shown in Table 1, the upregulated LPS-responsive genes predominantly corresponded to several annotations related to inflammation, immunity, and signal transduction. In contrast, the upregulated heat shock-responsive genes predominantly corresponded to several annotations related to the stress response, molecular chaperones, and protein folding. Both upregulated gene lists corresponded to signaling pathways related to apoptosis. These data demonstrate that the individual upregulated gene lists of LPS- and heat shock-responsive genes are significantly enriched for unique functional annotations.
Table 1.
Functional annotations associated with the upregulated LPS- and heat shock-responsive genes, respectively.
| LPS | Heat Shock | |
| Pathway (p value) | • Apoptosis signaling (1.5E-10) | • Apoptosis signaling (1.8E-3) |
| • Inflammation mediated by chemokine and cytokine signaling (2.1E-7) | • Angiogenesis (3.2E-2) | |
| • Toll receptor signaling (9.6E-6) | ||
| • Integrin signaling (1.4E-2) | ||
| Biological process (p value) | • Immunity and defense (2.2E-25) | • Protein folding (2.1E-14) |
| • Signal transduction (5.8E-23) | • Protein metabolism and modification (1.7E-10) | |
| • Intracellular signaling cascade (3.0E-16) | • Stress response (1.8E-10) | |
| • Cell surface receptor mediated signaling (2.7E-10) | • Protein phophorylation (1.7E-6) | |
| • Cytokine and chemokine mediated signaling (3.1E-10) | • Immunity and defense (2.0E-5) | |
| Molecular function (p value) | • Signaling molecule (1.4E-10) | • Chaperone (1.2E-16) |
| • Other transcription factor (3.0E-7) | • Hsp 70 family chaperone (1.9E-7) | |
| • Chemokine (3.3E-6) | • Protein kinase (2.8E-6) | |
| • Cytokine receptor (1.2E-5) | • Transcription factor (2.0E-4) | |
| • Select regulatory molecule (8.9E-5) | • Chaperonin (1.2E-3) | |
Data are based on the PANTHER database using the default parameters in the database and Bonferroni correction for multiple comparisons.
Functional annotations corresponding to the downregulated LPS- and heat shock-responsive genes
To begin deriving biological meaning from the individual lists of downregulated LPS- and heat shock-responsive genes, we uploaded the respective downregulated gene lists to the PANTHER database and again focused the analysis on enrichment for signaling pathways, biological processes, and molecular functions. As shown in Table 2, the downregulated LPS-responsive genes predominantly corresponded to several annotations related to protein/carbohydrate metabolism, amino acid metabolism, and other biochemical processes (e.g. oxidoreductase, dehydrogenase, and lyase activity). In contrast, the downregulated heat shock-responsive genes predominantly corresponded to several annotations related to transcription and nucleotide processing. There were no common functional annotations between the downregulated LPS- and heat shock-responsive genes. These data demonstrate that the individual downregulated gene lists of LPS- and heat shock-responsive genes are significantly enriched for unique functional annotations.
Table 2.
Functional annotations associated with the downregulated LPS- and heat shock-responsive genes, respectively.
| LPS | Heat Shock | |
| Pathways | • None signficant | • None significant |
| Biological process (p value) | • Protein metabolism and modification (6.8E-7) | • Nucleoside, nucleotide and nucleic acid metabolism (1.3E-3) |
| • Amino acid metabolism (5.2E-6) | • mRNA transcription regulation | |
| • Other metabolism (1.4E-5) | • (2.7E-3) | |
| • Carbohydrate metabolism (1.3E-4) | • mRNA transcription (3.3E-2) | |
| • Protein modification (2.2E-4) | ||
| Molecular function (p value) | • Oxidoreductase (5.0E-7) | • Transferase (1.4E-3) |
| • Dehydrogenase (4.4E-6) | • Transcription factor (1.9E-3) | |
| • Synthase and synthetase (3.3E-4) | • Nucleic acid binding (9.5E-3) | |
| • Transferase (5.5E-4) | ||
| • Lyase (1.5E-3) |
Data are based on the PANTHER database using the default parameters in the database and Bonferroni correction for multiple comparisons.
Gene network expression corresponding to the upregulated LPS- and heat shock-responsive genes
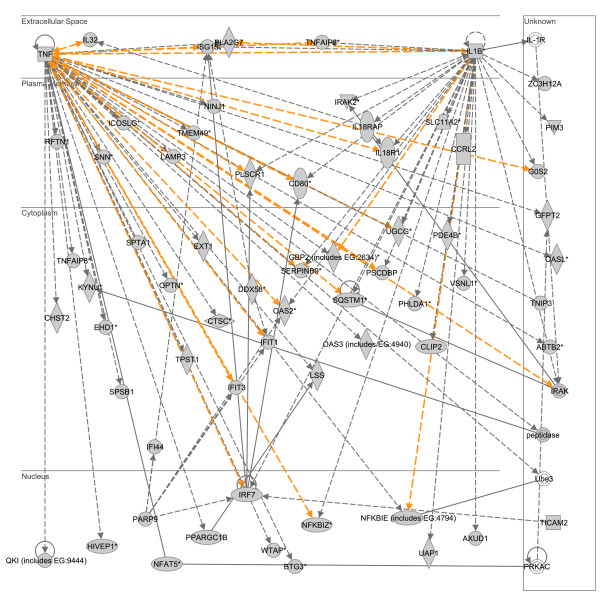
To derive further biological meaning from the individual lists of upregulated LPS- and heat shock-responsive genes, we uploaded the respective upregulated gene lists to the IPA application and focused the analysis on gene network expression. The two highest scoring, upregulated LPS-responsive gene networks were merged as depicted in Figure 3. This upregulated LPS-responsive gene network is dominated by two gene nodes, located in the extracellular compartment, and having the highest degree of connectivity relative to the other gene nodes in the network: interleukin-1β (IL1B) and tumor necrosis factor (TNF). The list of genes corresponding to this upregulated LPS-responsive gene network was subsequently uploaded to an independent database (D.A.V.I.D.) to derive the functional annotations corresponding to the gene network. The results of the D.A.V.I.D.-based analysis are provided in Table 3 and demonstrate that this upregulated LPS-responsive gene network is enriched for functional annotations corresponding to immunity and inflammation.
Figure 3.
Merge of two top scoring IPA networks derived from the list of 1,345 upregulated LPS-responsive genes. The merged network is depicted in the context of cellular compartments. This upregulated LPS-responsive network contains two nodes in the extracellular compartment having a high degree of connectivity to other network genes: tumor necrosis factor (TNF) and interleukin-1β (IL1B). Gold lines indicate connectivity between the two merged networks. See text for network derivation, and see Additional file 3 for network gene list and Additional file 4 for network legend.
Table 3.
Top 10 functional annotations for two merged networks upregulated in response to LPS (based on p value).
| Category | Term | # of genes | p-value |
| SP_PIR_KEYWORDS | interferon induction | 9 | 9.3E-11 |
| GOTERM_BP_ALL | response to biotic stimulus | 25 | 1.6E-8 |
| GOTERM_BP_ALL | immune response | 23 | 3.2E-8 |
| GOTERM_BP_ALL | defense response | 23 | 1.5E-7 |
| GOTERM_BP_ALL | response to stimulus | 28 | 8.4E-5 |
| GOTERM_BP_ALL | response to stress | 18 | 1.3E-4 |
| GOTERM_BP_ALL | response to external stimulus | 13 | 1.5E-4 |
| GOTERM_BP_ALL | organismal physiological process | 28 | 1.8E-4 |
| GOTERM_BP_ALL | response to pest/pathogen/parasite | 13 | 2.9E-4 |
| GOTERM_BP_ALL | response to other organism | 13 | 4.8E-4 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing.
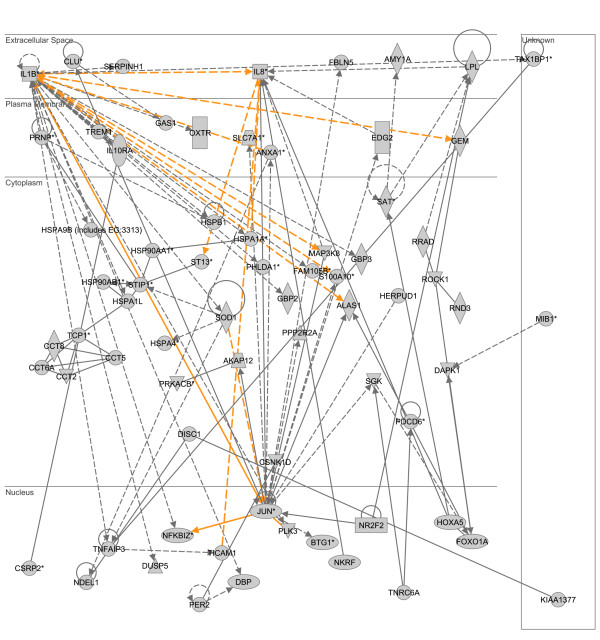
The two highest scoring, upregulated heat shock-responsive gene networks were merged and analyzed in a similar manner as described above. As shown in Figure 4, the upregulated heat shock-responsive gene network is dominated by two gene nodes having the highest degree of connectivity relative to the other gene nodes in the network: interleukin-1β (IL1B; extracellular compartment) and the jun oncogene (JUN; nuclear compartment). The results of the subsequent D.A.V.I.D.-based analysis are provided in Table 4 and demonstrate that this upregulated heat shock-responsive gene network is enriched for functional annotations corresponding to molecular chaperone and heat shock protein activity.
Figure 4.
Merge of two top scoring IPA networks derived from the list of 1,476 upregulated heat shock-responsive genes. The merged network is depicted in the context of cellular compartments. This upregulated heat shock-responsive network contains two nodes having a high degree of connectivity to other network genes: interleukin-1β (IL1B) located in the extracellular compartment and jun oncogene (JUN) located in the nuclear compartment. Gold lines indicate connectivity between the two merged networks. See text for network derivation, and see Additional file 5 for network gene list and Additional file 4 for network legend.
Table 4.
Top 10 functional annotations for two merged networks upregulated in response to heat shock (based on p value).
| Category | Term | # of genes | p-value |
| SP_PIR_KEYWORDS | chaperone | 11 | 1.4E-8 |
| GOTERM_BP_ALL | protein folding | 14 | 4.6E-7 |
| SP_PIR_KEYWORDS | heat shock | 8 | 1.2E-7 |
| GOTERM_BP_ALL | response to unfolded protein | 9 | 5.4E-7 |
| GOTERM_BP_ALL | response to protein stimulus | 9 | 5.4E-7 |
| SP_PIR_KEYWORDS | nucleotide-binding | 22 | 3.2E-7 |
| SP_PIR_KEYWORDS | molecular chaperone | 6 | 4.4E-7 |
| GOTERM_MF_ALL | unfolded protein binding | 11 | 4.5E-6 |
| SP_PIR_KEYWORDS | atp-binding | 18 | 6.6E-6 |
| INTERPRO_NAME | chaperonin TCP-1 | 5 | 3.8E-4 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing.
These data demonstrate that the individual upregulated gene lists of LPS- and heat shock-responsive genes correspond to distinct gene networks having unique functional annotations. Complete lists of the respective network genes and a network legend are provided in Additional files 3 to 5.
Gene network expression corresponding to the downregulated LPS- and heat shock-responsive genes
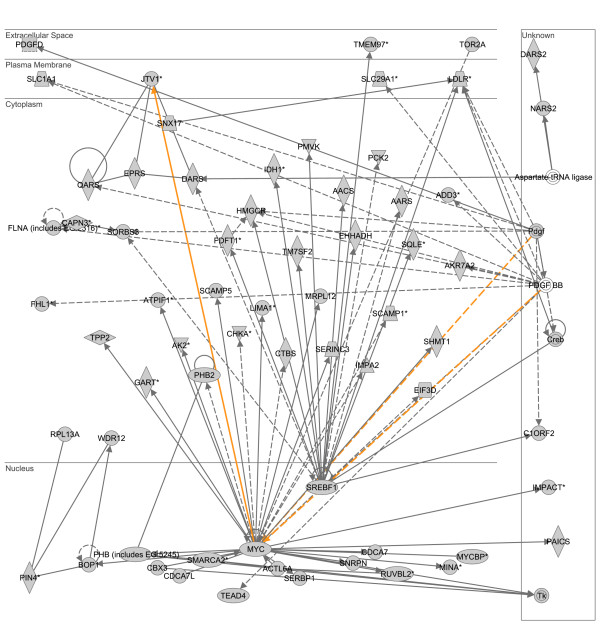
To derive further biological meaning from the individual lists of downregulated LPS- and heat shock-responsive genes, we conducted an identical analysis as described above based on the IPA application and the D.A.V.I.D. database. The two highest scoring, downregulated LPS-responsive gene networks were merged as depicted in Figure 5. This downregulated LPS-responsive gene network is dominated by two gene nodes, located in the nuclear compartment, and having the highest degree connectivity relative to the other gene nodes in the network: v-myc myelocytomatosis viral oncogene homolog (MYC) and sterol regulatory element binding transcription factor 1 (SREBF1). The results of the subsequent D.A.V.I.D.-based analysis are provided in Table 5 and demonstrate that this downregulated LPS-responsive gene network is enriched for functional annotations corresponding to protein metabolism/synthesis, tRNA-related biochemistry, and sterol metabolism.
Figure 5.
Merge of two top scoring IPA networks derived from the list of 2,623 downregulated LPS-responsive genes. The merged network is depicted in the context of cellular compartments. This downregulated LPS-responsive network contains two nodes in the nuclear compartment having a high degree of connectivity to other network genes: v-myc myelocytomatosis viral oncogene homolog (MYC) and sterol regulatory element binding transcription factor 1 (SREBF1). Gold lines indicate connectivity between the two merged networks. See text for network derivation and see Additional file 6 for network gene list and Additional file 4 for network legend.
Table 5.
Top 10 functional annotations for two merged networks downregulated in response to LPS (based on p value).
| Category | Term | # of genes | p-value |
| SP_PIR_KEYWORDS | aminoacyl-tRNA synthatase | 6 | 1.7E-4 |
| SP_PIR_KEYWORDS | protein biosynthesis | 8 | 5.2E-4 |
| GOTERM_BP_ALL | sterol metabolism | 7 | 6.0E-4 |
| GOTERM_BP_ALL | cellular physiological process | 57 | 7.0E-4 |
| GOTERM_BP_ALL | biosynthesis | 21 | 1.0E-3 |
| GOTERM_MF_ALL | aminoacyl-tRNA ligase activity | 6 | 1.3E-3 |
| GOTERM_MF_ALL | ligase activity, carbon-oxygen bonds | 6 | 1.3E-3 |
| GOTERM_BP_ALL | amino acid activation | 6 | 1.6E-3 |
| GOTERM_BP_ALL | tRNA aminoacylation for protein translation | 6 | 1.6E-3 |
| GOTERM_BP_ALL | tRNA amioacylation | 6 | 1.6E-3 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing.
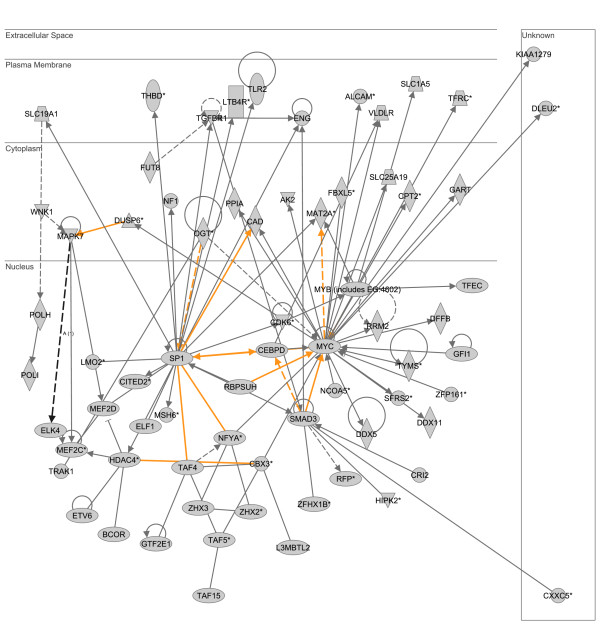
The two highest scoring, downregulated heat shock-responsive gene networks were merged as depicted in Figure 6. This downregulated heat shock-responsive gene network is dominated by two gene nodes, located in the nuclear compartment, and having the highest degree connectivity relative to the other gene nodes in the network: v-myc myelocytomatosis viral oncogene homolog (MYC) and Sp1 transcription factor (SP1). The results of the subsequent D.A.V.I.D.-based analysis are provided in Table 6 and demonstrate that this downregulated heat shock-responsive gene network is enriched for functional annotations corresponding to transcription and nucleic acid metabolism.
Figure 6.
Merge of two top scoring IPA networks derived from the list of 1,445 downregulated heat shock-responsive genes. The merged network is depicted in the context of cellular compartments. This downregulated heat shock-responsive network contains two nodes in the nuclear compartment having a high degree of connectivity to other network genes: v-myc myelocytomatosis viral oncogene homolog (MYC) and Sp1 transcription factor (SP1). Gold lines indicate connectivity between the two merged networks. See text for network derivation, and see Additional file 7 for network gene list and Additional file 4 for network legend.
Table 6.
Top 10 functional annotations for two merged networks downregulated in response to heat shock (based on p value).
| Category | Term | # of genes | p-value |
| SP_PIR_KEYWORDS | transcription | 28 | 1.7E-11 |
| SP_PIR_KEYWORDS | transcription regulation | 28 | 1.4E-11 |
| SP_PIR_KEYWORDS | nuclear protein | 37 | 4.0E-11 |
| GOTERM_BP_ALL | nucleic acid metabolism | 42 | 1.9E-7 |
| SP_PIR_KEYWORDS | repressor | 11 | 7.9E-7 |
| GOTERM_BP_ALL | regulation, nucleic acid metabolism | 32 | 4.7E-6 |
| GOTERM_BP_ALL | regulation of transcription | 31 | 1.1E-5 |
| GOTERM_BP_ALL | regulation of cellular metabolism | 32 | 9.6E-6 |
| GOTERM_BP_ALL | regulation of cellular process | 38 | 1.2E-5 |
| GOTERM_BP_ALL | regulation of metabolism | 32 | 1.2E-5 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing.
These data demonstrate that the individual downregulated gene lists of LPS- and heat shock-responsive genes correspond to distinct gene networks having unique functional annotations. Complete lists of the respective network genes and a network legend are provided in Additional files 4, 6, and 7.
Gene network expression corresponding to the common upregulated LPS- and heat shock-responsive genes
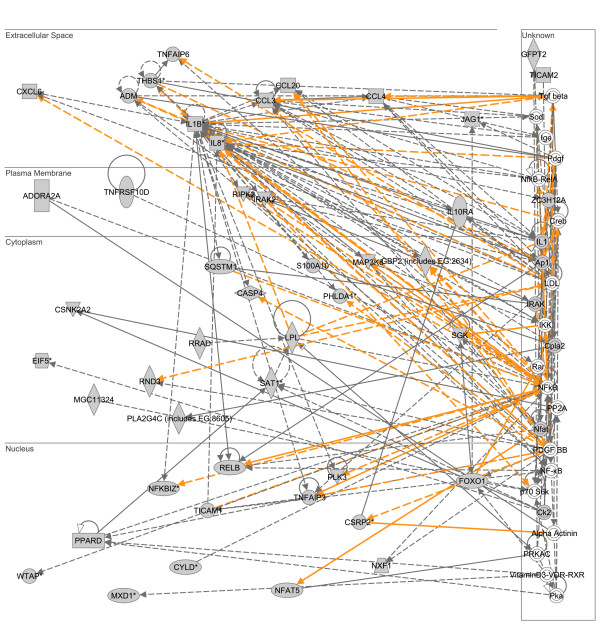
As previously described (Figure 1), there were 184 genes common to both the upregulated LPS-responsive gene list and the upregulated heat shock-responsive gene list. These 184 common genes were uploaded to the IPA application and the analytical focus was again placed on the presence of gene networks. The two highest scoring, upregulated gene networks corresponding to this 184 common gene list were merged as depicted in Figure 7. This upregulated gene network contains interleukin-1β (IL1B) and interleukin-8 (IL8) as two highly connected gene nodes, located in the extracellular compartment, and also having high level connectivity to the NF-κB pathway. The list of genes corresponding to this upregulated LPS/heat shock-responsive network was subsequently uploaded to an independent database (D.A.V.I.D.) to derive the functional annotations corresponding to the network. The results of the D.A.V.I.D.-based analysis are provided in Table 7 and demonstrate that this upregulated LPS/heat shock-responsive network is enriched for functional annotations corresponding to inflammation and chemokine-related biology. These data demonstrate that the common genes upregulated in response to LPS or heat shock correspond to a gene network associated with inflammation in general, and more specifically with chemokine-related biology. A complete list of the respective network genes and a network legend are provided in Additional files 4 and 8.
Figure 7.
Merge of two top scoring IPA networks derived from the list of 184 genes common to both the upregulated LPS-responsive gene list and the upregulated heat shock-responsive gene list. The merged network is depicted in the context of cellular compartments. This upregulated, common LPS/heat shock-responsive network contains two nodes in the extracellular compartment having a high degree of connectivity to other network genes: interleukin-1β (IL1B) and interleukin-8 (IL8). Both the IL1B and IL8 nodes also demonstrate high level connectivity to the NF-κB pathway. Gold lines indicate connectivity between the two merged networks. See text for network derivation, and see Additional file 8 for network gene list and Additional file 4 for network legend.
Table 7.
Top 10 functional annotations for two merged networks commonly upregulated in response to both LPS and heat shock (based on p value).
| Category | Term | # of genes | p-value |
| GOTERM_BP_ALL | inflammatory response | 12 | 2.9E-8 |
| GOTERM_BP_ALL | response to wounding | 14 | 4.6E-8 |
| GOTERM_BP_ALL | response to external stimulus | 14 | 8.2E-7 |
| INTERPRO_NAME | small chemokine, interleukin-8-like | 6 | 2.7E-4 |
| GOTERM_MF_ALL | protein binding | 29 | 1.5E-4 |
| GOTERM_MF_ALL | chemokine activity | 6 | 8.2E-5 |
| GOTERM_MF_ALL | chemokine receptor binding | 6 | 8.2E-5 |
| GOTERM_BP_ALL | response to stress | 16 | 9.3E-5 |
| SMART_NAME | SCY1 | 6 | 1.2E-4 |
| SP_PIR_KEYWORDS | chemotaxis | 6 | 3.2E-4 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing. 1Intercrine alpha family (small chemokine C-X-C) (chemokine CXC)
Gene network expression corresponding to the common downregulated LPS- and heat shock-responsive genes
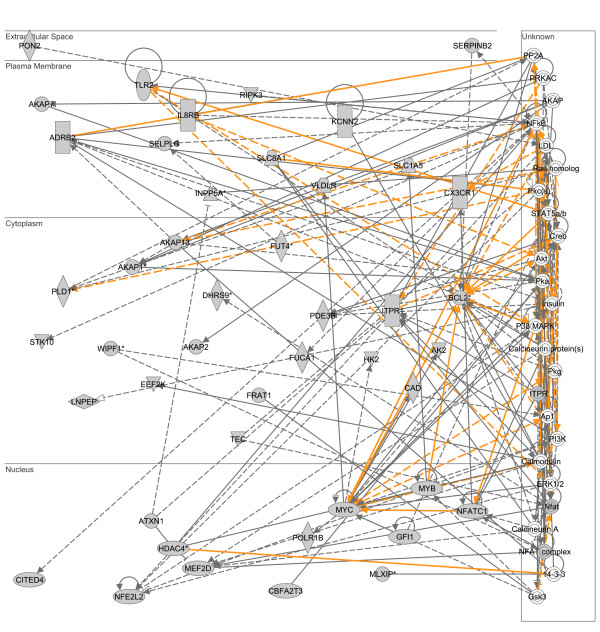
As previously described (Figure 2), there were 430 genes common to both the downregulated LPS-responsive gene list and the downregulated heat shock-responsive gene list. These 430 common genes were analyzed in a similar manner to that described above for the common upregulated genes. The two highest scoring, downregulated gene networks corresponding to this 430 common gene list were merged as depicted in Figure 8. This downregulated gene network contains B-cell CLL/lymphoma 2 (BCL2; cytoplasmic compartment) and v-myc myelocytomatosis viral oncogene homolog (MYC; nuclear compartment) as two highly connected gene nodes. The results of the D.A.V.I.D.-based analysis of this network are provided in Table 8 and demonstrate that this downregulated LPS/heat shock-responsive gene network is enriched for functional annotations corresponding to the cellular membrane and phosphorylation. These data demonstrate that the common genes downregulated in response to LPS or heat shock correspond to a gene network associated with the cellular membrane in general, and more specifically with phosphorylation events. A complete list of the respective network genes and a network legend are provided in Additional files 4 and 8.
Figure 8.
Merge of two top scoring IPA networks derived from the list of 430 genes common to both the downregulated LPS-responsive gene list and the downregulated heat shock-responsive gene list. The merged network is depicted in the context of cellular compartments. This downregulated, common LPS/heat shock-responsive network contains two nodes having a high degree of connectivity to other network genes: B-cell CLL/lymphoma 2 (BCL2) in the cytoplasmic compartment and v-myc myelocytomatosis viral oncogene homolog (MYC) in the nuclear compartment. Gold lines indicate connectivity between the two merged networks. See text for network derivation, and see Additional file 8 for network gene list and Additional file 4 for network legend.
Table 8.
Three significant functional annotations for two merged networks commonly downregulated in response to both LPS and heat shock (based on p value).
| Category | Term | # of genes | p-value |
| SP_PIR_KEYWORDS | membrane | 20 | 8.2E-3 |
| SP_PIR_KEYWORDS | phosphorylation | 14 | 1.0E-2 |
| SP_PIR_KEYWORDS | kinase | 9 | 3.8E-2 |
The analysis is based on the default parameters in D.A.V.I.D. and uses Benjamini-Hochberg correction for multiple testing. Unlike the other Tables, Table 8 lists only three functional annotations because all other related annotations did not have significant p values (< 0.05) when subjected to Benjamini-Hochberg correction.
Top genes expressed in response to LPS or heat shock stress
In order to assess the validity of the data presented above, we extracted the top ten genes expressed in response to LPS or heat shock stress (based on fold induction over control conditions). The majority of the top ten genes expressed in response to LPS stress (Table 9) are well established LPS-responsive genes. Similarly, the top ten genes expressed in response to heat shock stress (Table 10) are well established heat shock-responsive genes. These data demonstrate consistency with known LPS- and heat shock-related biology, thus adding an important level of validity to the data presented in the previous sections.
Table 9.
Top 10 genes (based on fold change versus control cells) expressed in response to LPS stress.
| Description | Gene Symbol | Fold Change |
| tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 1,696 |
| chemokine (C-C motif) ligand 20 Alias: MIP-3α | CCL20 | 1,036 |
| chemokine (C-C motif) ligand 4 Alias: MIP-1β | CCL4 | 749 |
| insulin-like growth factor binding protein 3 | IGFBP3 | 519 |
| interleukin 1, beta | IL1B | 446 |
| BCL2-related protein A1 | BCL2A1 | 399 |
| interleukin 8 | IL8 | 365 |
| lysosomal-associated membrane protein 3 Alias: CD208 | LAMP3 | 359 |
| Epstein-Barr virus induced gene 3 Alias: Interleukin-27 | EBI3 | 315 |
| interleukin receptor 7 | IL7R | 205 |
Table 10.
Top 10 genes (based on fold change versus control cells) expressed in response to heat shock stress.
| Description | Gene Symbol | Fold Change |
| heat shock 70 kDa protein 6 | HSPA6 | 3,580 |
| heat shock 70 kDa protein 1B | HSPA1B | 1,526 |
| immediate early response 5 | IER5 | 230 |
| collagen, type I, alpha 1 | COL1A1 | 183 |
| heat shock 70 kDa protein 1A | HSPA1A | 170 |
| Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II | P4HA2 | 118 |
| BCL2-associated athanogene 3 Alias: BAG-family molecular chaperone regulator-3 | BAG3 | 111 |
| DnaJ (Hsp40) homolog, subfamily B, member 4 Alias: DnaJ-like heat shock protein 40 | DNAJB4 | 106 |
| dual specificity phosphatase 1 Alias: MKP-1 | DUSP1 | 96 |
| v-jun sarcoma virus 17 oncogene homolog Alias: c-jun | JUN | 82 |
Discussion
We have characterized the genome-level responses of THP-1 cells (a model of peripheral blood mononuclear cells) following stimulation with one of two canonical stress/danger signals at a single time point. While previous high throughput-centered studies have focused on either LPS [7-12] or heat shock stress alone [28-34], to our knowledge the current work represents the first direct comparison of LPS and heat shock stress based on the same analytical approach. Importantly, the analytical approach employs three independent genomic expression discovery databases (i.e. PANTHER, IPA, and D.A.V.I.D.), thus adding to the trustworthiness of the derived functional annotations and networks.
The global view (Venn diagrams) of the genes differentially regulated in response to LPS or heat shock stress indicates that each stress signal regulates (increased or decreased expression) a relatively unique group of genes. Among the upregulated genes, > 85% of the genes were uniquely expressed in response to either LPS or heat shock stimulation. Among the downregulated genes, > 69% of the genes were uniquely repressed in response to either LPS or heat shock stimulation. Thus, the mononuclear cell response to LPS stress is generally divergent compared to that of heat shock stress.
The divergence between the LPS response and the heat shock response is evident by the functional annotations derived for the respective individual lists of differentially regulated genes. The functional annotations derived for the upregulated LPS-responsive genes were predominantly in the areas of inflammation and immunity. This observation is consistent with the well known role of LPS as the major pathogen associated molecular pattern of gram negative bacteria, and the well known role of peripheral blood mononuclear cells as sentinels of the innate immune system [35]. In contrast, the functional annotations derived for the upregulated heat shock-responsive genes were predominantly in the areas of heat shock proteins, protein unfolding, and molecular chaperones. This observation is also consistent with the existing literature surrounding the heat shock response and heat shock proteins [2,36].
The assertion of divergence is further supported by the functional annotations derived for the individual lists of differentially downregulated genes. The functional annotations derived for the downregulated LPS-responsive genes were predominantly in the areas of protein biosynthesis and sterol metabolism. These observations are indirectly supported by reports documenting LPS-mediated alterations of protein and lipid metabolism [37-42]. In contrast, the functional annotations derived for the downregulated heat shock-responsive genes were predominantly in the areas of nucleic acid metabolism and regulation of transcription. These observations are consistent with the aforementioned paradigm in which the heat shock response is characterized by a re-prioritization of gene expression toward heat shock protein transcription while non-heat shock protein gene transcription is globally repressed [4-6]. This paradigm is likely to be dependent on the time frame after heat shock. For example, global transcriptional repression may be most evident immediately following heat shock. However, the relatively large number of "non-heat shock protein" genes expressed four hours after THP-1 cells were subjected to heat shock indicates that the concept of global transcriptional repression following heat shock may be an oversimplification. The assertion of oversimplification is further supported by the work of previous investigators employing microarray technology to study the heat shock response of mammalian and non-mammalian systems [28-34]. These previous investigations were also notable for the expression of "non-heat shock protein" genes in response to heat shock.
In the context of our previous studies, several of the genes upregulated in response to heat shock or LPS warrant further discussion. Dual specificity phosphatase 1 (DUSP1, a.k.a. MKP-1) was substantially increased in THP-1 cells subjected to heat shock. MKP-1 is a dual specific phosphatase serving an important counter-regulatory function in the MAP kinase signaling pathway [15,43,44]. We previously demonstrated that MKP-1 is a heat shock-responsive gene in cultured murine macrophages and that heat shock-mediated regulation of the MKP-1 gene is dependent on both transcriptional and post-transcriptional mechanisms [14]. Sonna et al also reported MKP1/DUSP1 expression in primary human mononuclear cells exposed to heat shock [33].
Several laboratories, including our own, have previously reported that heat shock inhibits activation of the NF-κB signaling pathway [3,13,16-18], [45-47]. In this regard it was interesting to note that nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor zeta (NFKBIZ), NFKB inhibitor interacting Ras-like 2 (NKIRAS2), and NF-kappaB repressing factor (NKRF) were coordinately upregulated in THP-1 cells subjected to heat shock (see Supplementary Data). All three of these genes appear to play important negative regulatory roles in the NF-κB signaling pathway. NFKBIZ is a member of the ankryin-repeat family of the IκB group of proteins that bind and retain NF-κB in the cytoplasm in an inactive state [48]. NKIRAS2 is a Ras protein subclass reported to regulate the degradation of IκB [49]. Finally, NKRF has been reported to interact with specific negative regulatory elements that mediate transcriptional repression of NF-κB responsive genes [50] and has been previously reported to be expressed in primary human mononuclear cells exposed to heat shock [33].
LPS is a key pathogen associated molecular pattern that is well known to play an important role in the pathobiology of human septic shock [1,35]. In separate translational studies we have been conducting genome-wide expression profiling in children with septic shock [21,22,51]. One of the novel findings generated from this translational program is that pediatric septic shock is characterized by large scale downregulation of genes having zinc-related functional annotations. In keeping with this observation, we have also reported that nonsurvivors of pediatric septic shock have abnormally low serum zinc concentrations [21]. In light of these findings and the important role of LPS in septic shock, we searched the list of genes downregulated in THP-1 cells subjected to LPS stress for enrichment of zinc-related functional annotations. We found between 23 and 28 genes having functional annotations for the terms "zinc", "zinc ion binding", "zinc-finger", or "metal-binding" (data not shown, analysis based on the D.A.V.I.D. database). Thus, in keeping with our clinical, translational studies we have validated in vitro that LPS suppresses the expression of genes that either depend on zinc homeostasis or directly participate in zinc homeostasis.
The most novel information derived from the current data involves the list of genes commonly upregulated in response to LPS and heat shock stress. There were 184 genes common to both the upregulated LPS gene list and the upregulated heat shock-responsive gene list. This relatively limited number of genes corresponded to gene networks having functional annotations related to inflammation in general, and more specifically to chemokine-related biology. Thus, the response to heat shock stress is not completely divergent from that of the LPS response in that it shares a group of coordinately regulated genes related to chemokine signaling, a heretofore unreported observation.
Conclusion
We have directly compared the genomic response of THP-1 cells subjected to LPS stress or heat shock stress. The global gene expression patterns demonstrate that the two responses are predominantly divergent. The response to heat shock, however, is not necessarily characterized by global downregulation of non-heat shock protein genes. In fact, a large number of "non-heat shock protein" genes are expressed in response to heat shock stress and a number of these genes have overlap with inflammation-associated gene expression patterns classically attributed to LPS stress. Thus, the heat shock response and the LPS response are not necessarily mutually exclusive. Finally, the data generated from these experiments can serve as a valuable reference database for investigators in the field of stress/danger signaling.
Authors' contributions
HRW conceived and directed the overall study, conducted analyses, and drafted the initial manuscript. KO conducted all microarray experiments. BS assisted with data analysis and bioinformatics. All authors proof read the manuscript and agreed with its submission for review.
Supplementary Material
3968 genes regulate in lps treated thp1 cells.xls, list of genes regulated in response to LPS alone.
2921 genes regulate in heat shocked thp1 cells.xls, list of genes regulated in response to heat shock alone.
merged uregulated lps network.xls, list of genes corresponding to two merged, upregulated networks in response to LPS alone and shown in Figure 3.
merged upregulated heat shock network.xls, list of genes corresponding to two merged, upregulated networks in response to heat shock alone and shown in Figure 4.
merged downregulated LPS network.xls, list of genes corresponding to two merged, downregulated networks in response to LPS alone and shown in Figure 5.
merged downregulated heat shock network.xls, list of genes corresponding to two merged, downregulated networks in response to heat shock alone and shown in Figure 6.
genes common to both lps and heat shock.xls, list of genes commonly regulated in THP1 cells treated with either LPS or heat shock.
Acknowledgments
Acknowledgements
Supported by the National Institute of General Medical Sciences (R01GM061723 and RO1GM064619)
Contributor Information
Hector R Wong, Email: hector.wong@cchmc.org.
Kelli Odoms, Email: kelli.odoms@cchmc.org.
Bhuvaneswari Sakthivel, Email: bhuvana.sakthivel@cchmc.org.
References
- Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol. 2007;19:3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Wheeler DS, Wong HR. Heat shock response and acute lung injury. Free Radic Biol Med. 2007;42:1–14. doi: 10.1016/j.freeradbiomed.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Wong HR. Interactions between the heat shock response and the nuclear factor-kappa B signaling pathway. Crit Care Med. 2002;30:S89–95. doi: 10.1097/00003246-200201001-00012. [DOI] [PubMed] [Google Scholar]
- DeMeester SL, Buchman TG, Cobb JP. The heat shock paradox: does NF-kappaB determine cell fate? Faseb J. 2001;15:270–274. doi: 10.1096/fj.00-0170hyp. [DOI] [PubMed] [Google Scholar]
- Buchman TG, Abello PA, Smith EH, Bulkley GB. Induction of heat shock response leads to apoptosis in endothelial cells previously exposed to endotoxin. Am J Physiol. 1993;265:H165–70. doi: 10.1152/ajpheart.1993.265.1.H165. [DOI] [PubMed] [Google Scholar]
- Cabin DE, Buchman TG. Molecular biology of circulatory shock. Part III. Human hepatoblastoma (HepG2) cells demonstrate two patterns of shock-induced gene expression that are independent, exclusive, and prioritized. Surgery. 1990;108:902–912. [PubMed] [Google Scholar]
- Dos Santos S, Delattre AI, De Longueville F, Bult H, Raes M. Gene expression profiling of LPS-stimulated murine macrophages and role of the NF-kappaB and PI3K/mTOR signaling pathways. Ann N Y Acad Sci. 2007;1096:70–77. doi: 10.1196/annals.1397.071. [DOI] [PubMed] [Google Scholar]
- Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltathakis I, Alcantara O, Boldt DH. Expression of different NF-kappaB pathway genes in dendritic cells (DCs) or macrophages assessed by gene expression profiling. J Cell Biochem. 2001;83:281–290. doi: 10.1002/jcb.1231. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun. 2004;72:7247–7256. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos CC, Han B, Andrade CF, Bai X, Uhlig S, Hubmayr R, Tsang M, Lodyga M, Keshavjee S, Slutsky AS, Liu M. DNA microarray analysis of gene expression in alveolar epithelial cells in response to TNFalpha, LPS, and cyclic stretch. Physiol Genomics. 2004;19:331–342. doi: 10.1152/physiolgenomics.00153.2004. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Mages J, Heit A, Lang R, Wagner H. Transcriptional activation induced in macrophages by Toll-like receptor (TLR) ligands: from expression profiling to a model of TLR signaling. Eur J Immunol. 2004;34:2863–2873. doi: 10.1002/eji.200425228. [DOI] [PubMed] [Google Scholar]
- Dunsmore KE, Denenberg AG, Page K, Wong HR. Mechanism and function of heat shock-dependent IkappaBalpha expression. Inflamm Res. 2006;55:254–259. doi: 10.1007/s00011-006-0080-2. [DOI] [PubMed] [Google Scholar]
- Wong HR, Dunsmore KE, Page K, Shanley TP. Heat shock-mediated regulation of MKP-1. Am J Physiol Cell Physiol. 2005;289:C1152–8. doi: 10.1152/ajpcell.00138.2005. [DOI] [PubMed] [Google Scholar]
- Sanlorenzo L, Zhao B, Spight D, Denenberg AG, Page K, Wong HR, Shanley TP. Heat shock inhibition of lipopolysaccharide-mediated tumor necrosis factor expression is associated with nuclear induction of MKP-1 and inhibition of mitogen-activated protein kinase activation. Crit Care Med. 2004;32:2284–2292. doi: 10.1097/01.ccm.0000145580.96994.c9. [DOI] [PubMed] [Google Scholar]
- Wong HR, Ryan MA, Menendez IY, Wispe JR. Heat shock activates the I-kappaBalpha promoter and increases I-kappaBalpha mRNA expression. Cell Stress Chaperones. 1999;4:1–7. doi: 10.1006/csac.1998.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SC, Ryan MA, Shanley TP, Wong HR. Induction of the stress response with prostaglandin A1 increases I-kappaBalpha gene expression. Faseb J. 1998;12:1371–1378. doi: 10.1096/fasebj.12.13.1371. [DOI] [PubMed] [Google Scholar]
- Wong HR, Ryan M, Wispe JR. Stress response decreases NF-kappaB nuclear translocation and increases I-kappaBalpha expression in A549 cells. J Clin Invest. 1997;99:2423–2428. doi: 10.1172/JCI119425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja R, Odoms K, Dunsmore K, Shanley TP, Wong HR. Extracellular heat shock protein-70 induces endotoxin tolerance in THP-1 cells. J Immunol. 2006;177:7184–7192. doi: 10.4049/jimmunol.177.10.7184. [DOI] [PubMed] [Google Scholar]
- NCBI Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/
- Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–52. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Laramie JM, Chung TP, Brownstein B, Stormo GD, Cobb JP. Transcriptional Profiles of Human Epithelial Cells in Response to Heat: Computational Evidence for Novel Heat Shock Proteins. Shock. 2007. [DOI] [PubMed]
- Szustakowski JD, Kosinski PA, Marrese CA, Lee JH, Elliman SJ, Nirmala N, Kemp DM. Dynamic resolution of functionally related gene sets in response to acute heat stress. BMC Mol Biol. 2007;8:46. doi: 10.1186/1471-2199-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhang A, Lin B, Liu J, Xu LX. Study of heat shock response of human umbilical vein endothelial cells (HUVECs) using cDNA microarray. Int J Hyperthermia. 2007;23:225–258. doi: 10.1080/02656730701295441. [DOI] [PubMed] [Google Scholar]
- Koide T, Vencio RZ, Gomes SL. Global gene expression analysis of the heat shock response in the phytopathogen Xylella fastidiosa. J Bacteriol. 2006;188:5821–5830. doi: 10.1128/JB.00182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen ML, Nettleton D, Thacker EL, Edwards R, Minion FC. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect Immun. 2006;74:160–166. doi: 10.1128/IAI.74.1.160-166.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Gaffin SL, Pratt RE, Cullivan ML, Angel KC, Lilly CM. Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Visala Rao D, Boyle GM, Parsons PG, Watson K, Jones GL. Influence of ageing, heat shock treatment and in vivo total antioxidant status on gene-expression profile and protein synthesis in human peripheral lymphocytes. Mech Ageing Dev. 2003;124:55–69. doi: 10.1016/S0047-6374(02)00170-7. [DOI] [PubMed] [Google Scholar]
- Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C. Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. J Leukoc Biol. 2007;81:15–27. doi: 10.1189/jlb.0306167. [DOI] [PubMed] [Google Scholar]
- Oiknine J, Aviram M. Increased susceptibility to activation and increased uptake of low density lipoprotein by cholesterol-loaded macrophages. Arterioscler Thromb. 1992;12:745–753. doi: 10.1161/01.atv.12.6.745. [DOI] [PubMed] [Google Scholar]
- Funk JL, Feingold KR, Moser AH, Grunfeld C. Lipopolysaccharide stimulation of RAW 264.7 macrophages induces lipid accumulation and foam cell formation. Atherosclerosis. 1993;98:67–82. doi: 10.1016/0021-9150(93)90224-I. [DOI] [PubMed] [Google Scholar]
- Kazemi MR, McDonald CM, Shigenaga JK, Grunfeld C, Feingold KR. Adipocyte fatty acid-binding protein expression and lipid accumulation are increased during activation of murine macrophages by toll-like receptor agonists. Arterioscler Thromb Vasc Biol. 2005;25:1220–1224. doi: 10.1161/01.ATV.0000159163.52632.1b. [DOI] [PubMed] [Google Scholar]
- Jurasinski C, Gray K, Vary TC. Modulation of skeletal muscle protein synthesis by amino acids and insulin during sepsis. Metabolism. 1995;44:1130–1138. doi: 10.1016/0026-0495(95)90005-5. [DOI] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Jefferson LS, Kimball SR, Vary TC. Endotoxin-induced decrease in muscle protein synthesis is associated with changes in eIF2B, eIF4E, and IGF-I. Am J Physiol Endocrinol Metab. 2000;278:E1133–43. doi: 10.1152/ajpendo.2000.278.6.E1133. [DOI] [PubMed] [Google Scholar]
- Orellana RA, O'Connor PM, Bush JA, Suryawan A, Thivierge MC, Nguyen HV, Fiorotto ML, Davis TA. Modulation of muscle protein synthesis by insulin is maintained during neonatal endotoxemia. Am J Physiol Endocrinol Metab. 2006;291:E159–66. doi: 10.1152/ajpendo.00595.2005. [DOI] [PubMed] [Google Scholar]
- Shanley TP. Phosphatases: counterregulatory role in inflammatory cell signaling. Crit Care Med. 2002;30:S80–8. doi: 10.1097/00003246-200201001-00011. [DOI] [PubMed] [Google Scholar]
- Nimah M, Zhao B, Denenberg AG, Bueno O, Molkentin J, Wong HR, Shanley TP. Contribution of MKP-1 regulation of p38 to endotoxin tolerance. Shock. 2005;23:80–87. doi: 10.1097/01.shk.0000145206.28812.60. [DOI] [PubMed] [Google Scholar]
- Sun D, Chen D, Du B, Pan J. Heat shock response inhibits NF-kappaB activation and cytokine production in murine Kupffer cells. J Surg Res. 2005;129:114–121. doi: 10.1016/j.jss.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Pittet JF, Lee H, Pespeni M, O'Mahony A, Roux J, Welch WJ. Stress-induced inhibition of the NF-kappaB signaling pathway results from the insolubilization of the IkappaB kinase complex following its dissociation from heat shock protein 90. J Immunol. 2005;174:384–394. doi: 10.4049/jimmunol.174.1.384. [DOI] [PubMed] [Google Scholar]
- Yoo CG, Lee S, Lee CT, Kim YW, Han SK, Shim YS. Anti-inflammatory effect of heat shock protein induction is related to stabilization of I kappa B alpha through preventing I kappa B kinase activation in respiratory epithelial cells. J Immunol. 2000;164:5416–5423. doi: 10.4049/jimmunol.164.10.5416. [DOI] [PubMed] [Google Scholar]
- Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Janicke RU, Schulze-Osthoff K. A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem. 2006;281:12645–12654. doi: 10.1074/jbc.M511956200. [DOI] [PubMed] [Google Scholar]
- Fenwick C, Na SY, Voll RE, Zhong H, Im SY, Lee JW, Ghosh S. A subclass of Ras proteins that regulate the degradation of IkappaB. Science. 2000;287:869–873. doi: 10.1126/science.287.5454.869. [DOI] [PubMed] [Google Scholar]
- Feng X, Guo Z, Nourbakhsh M, Hauser H, Ganster R, Shao L, Geller DA. Identification of a negative response element in the human inducible nitric-oxide synthase (hiNOS) promoter: The role of NF-kappa B-repressing factor (NRF) in basal repression of the hiNOS gene. Proc Natl Acad Sci U S A. 2002;99:14212–14217. doi: 10.1073/pnas.212306199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HR. Pediatric septic shock treatment: new clues from genomic profiling. Pharmacogenomics. 2007;8:1287–1290. doi: 10.2217/14622416.8.10.1287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3968 genes regulate in lps treated thp1 cells.xls, list of genes regulated in response to LPS alone.
2921 genes regulate in heat shocked thp1 cells.xls, list of genes regulated in response to heat shock alone.
merged uregulated lps network.xls, list of genes corresponding to two merged, upregulated networks in response to LPS alone and shown in Figure 3.
merged upregulated heat shock network.xls, list of genes corresponding to two merged, upregulated networks in response to heat shock alone and shown in Figure 4.
merged downregulated LPS network.xls, list of genes corresponding to two merged, downregulated networks in response to LPS alone and shown in Figure 5.
merged downregulated heat shock network.xls, list of genes corresponding to two merged, downregulated networks in response to heat shock alone and shown in Figure 6.
genes common to both lps and heat shock.xls, list of genes commonly regulated in THP1 cells treated with either LPS or heat shock.