Summary
The pathways by which oncogenes, such as MLL-AF9, initiate transformation and leukemia in humans and mice are incompletely defined. In a study of target cells and oncogene dosage, we found that Mll-AF9, when under endogenous regulatory control, efficiently transformed LSK (Lin− Sca1+ c-kit+) stem cells while committed granulocyte-monocyte progenitors (GMPs) were transformation-resistant and did not cause leukemia. Mll-AF9 was expressed at higher levels in hematopoietic stem (HSC) than GMP cells. Mll-AF9 gene dosage effects were directly shown in experiments where GMPs were efficiently transformed by the high dosage of Mll-AF9 resulting from retroviral transduction. Mll-AF9 up-regulated expression of 196 genes in both LSK and progenitor cells, but to higher levels in LSKs than in committed myeloid progenitors.
Significance
In a comparison of Mll-AF9 oncogene expression in retroviral and knockin models, we showed a direct relationship between transformation susceptibilty and oncogene dosage in committed progenitor cells. In the knockin model, where oncogene expression is under endogenous regulatory control, we found high Mll-AF9 gene expression levels and high expression levels of downstream target genes in stem compared to committed progenitor cells. These results encourage further analysis of physiologically-regulated oncogene dosage effects on genes that are critical to cell-specific transformation susceptibility. Studies of cell-specific effects are increasingly important with the recognition that certain oncogenes, such as the MLL fusion genes, are globally active.
Introduction
Cellular development proceeds in a hierarchical fashion from rare self-renewing stem cells to committed progenitor (transit-amplifying) cells to differentiated post-mitotic cells (Jordan et al., 2006). Currently, little is known as to whether naturally occurring cancers arise from normal stem cells or from committed progenitor cells, either of which could potentially acquire oncogenic mutations.
Research on translocations involving MLL fusion oncogenes has been extremely productive for more than 20 years and has revealed important information about the biology of leukemia including the role of HOX gene expression, histone modifications and leukemia stem cells (Krivtsov et al., 2007). The well studied MLL-AF9 oncogene initiates myeloid leukemia in both humans and mice (Dobson et al., 1999; Iida, Seto et al., 1993). An Mll-AF9 transgenic murine model that results in myeloid leukemia has been described and studied in some detail (Corral et al., 1996; Johnson et al., 2003; Kumar et al., 2004). In this model, the Mll-AF9 oncogene, introduced by homologous recombination, is under control of the endogenous Mll promoter, and thus expressed at physiologic levels. This model is potentially informative because it permits the study of well characterized mammalian hematopoietic stem and progenitor cells (Akashi et al., 2000; Spangrude et al., 1988).
A study of the MLL-ENL fusion gene introduced by retrovirus showed that both hematopoietic stem cells (HSCs) and committed myeloid progenitor cells were transformed by the fusion oncogene with highest efficiency in HSC population (Cozzio et al., 2003). More recent studies showed that MLL-AF9 introduced by retrovirus could transform both early hematopoietic progenitors (Somervaille et al., 2006) and committed myeloid progenitors (Krivtsov et al., 2006). A potential limiting factor in these previous studies comes from the utilization of retroviruses to introduce the oncogene. Retroviral introduction can result in non-controlled and potentially non-physiologic levels of oncogene expression, depending on the numbers of viral integrations and the type of promoters. The transforming effects of cellular oncogenes, including MLL fusions, MYC, BCR-ABL and CEBPA, may differ significantly depending on oncogene expression levels (Caslini et al., 2004; Chapiro et al., 2006; Ren, 2004). To circumvent these limitations, we studied the knockin Mll-AF9 murine model, which permits a direct comparison of the susceptibility to transformation of LSK (Lin−/c-kit+/Sca-1+, including hematopoietic stem cell HSC and common lymphoid progenitor CLP) stem and committed myeloid progenitor (common myeloid progenitor CMP and granulocyte-monocyte progenitor GMP) cells. The knockin model also permits expression of Mll-AF9 at physiologic gene dosages, which we postulate should differ across the hematopoietic stem and various progenitor cells populations based on studies of wild type Mll expression (Jude et al., 2007; McMahon et al., 2007).
We report differences in transformable cells (LSKs > CMPs > GMPs) when the MLL fusion oncogene is expressed at physiologic gene doses. We describe the importance of oncogene dosage which is suggested by 1) differences in Mll-AF9 expression in HSCs and GMPs, and 2) biologic differences between retrovirally introduced MLL-AF9 and endogenous Mll-AF9 expression.
Results
Mll-AF9 mice show increased HSCs, CLPs and CMPs
Bone marrow cells from 8 to 10 week old Mll-AF9 knockin mice or their wild type (WT) siblings were used in this study. At this age, Mll-AF9 mice show myeloid cell proliferation but do not develop leukemia until six months of age (Chen et al., 2006; Corral et al., 1996). We sorted bone marrow cells into previously well defined progenitor or stem cell populations. Lin−Sca1+c-kit+(LSK) (Ikuta et al., 1992; Spangrude et al., 1988) markers were used to sort the closely related self-renewal hematopoietic stem cells (HSCs) and common lymphoid progenitors (CLPs) (Kondoet al., 1997; So et al., 2003; Terskikh et al., 2003) The comparison groups of committed myeloid progenitors included common myeloid progenitors (CMPs) and granulocyte-monocyte progenitors (GMPs). Analysis of the sorting profiles of HSCs, CLPs, CMPs and GMPs revealed that Mll-AF9 resulted in increased percentages of c-kit+Sca1+cells in both the HSC and CLP populations (Figure 1A). We also saw an increased number of FcγRII/IIIlo (CMP) but not FcγRII/IIIhi (GMP) cells. In multiple experiments, sorted cell populations from Mll-AF9 mice showed a consistently higher percentage of HSCs, CLPs and CMPs, but not GMPs (Figure 1B) when compared to the ones from WT mice (Figure 1B).
Figure 1. Mll-AF9 results in the expansion of HSC, CLP and CMP populations.
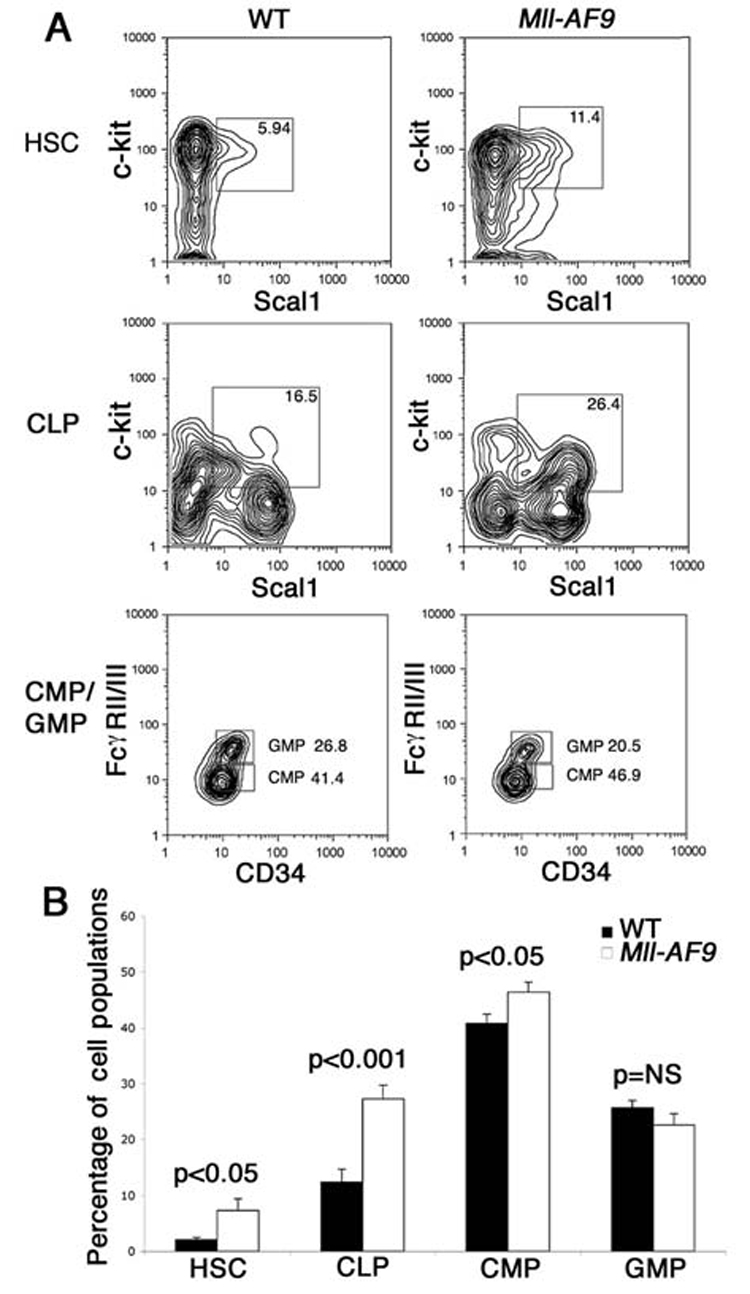
a, Sorting profiles of HSCs, CLPs, CMPs and GMPs showing the expansion of c-kit+Sca1+ HSCs (in Lin−IL-7R− population) and CLP (in Lin−IL-7R+ population) in Mll-AF9 mice. Expansion of the CMP population is also shown. b, Significantly higher percentages of HSCs, CLPs and CMPs were found in lineage negative marrow cells of Mll-AF9 than those of wild type mice. Error bars represent the standard error of the means.
Leukemia risk is dependent on the type of transplanted cells that express physiologic levels of Mll-AF9
MLL-ENL, when introduced into stem and progenitor cell populations (including HSCs, CMPs and GMPs) by retrovirus and under the control of the retroviral promoter, transformed those populations and produced leukemia in transplanted mice with additional events (Cozzio et al., 2003). Similarly, retrovirally transduced MLL-AF9 transformed GMP cells and produced leukemia (Krivtsov et al., 2006). In this study, we tested the ability of Mll-AF9, expressed under the control of the endogenous Mll promoter, to transform stem and progenitor cell populations and to produce leukemia in transplanted mice.
Lethally irradiated WT mice received 25–2500 sorted Mll-AF9 HSC, CLP, CMP or GMP cells. Results are shown in Figure 2A and with more details in Table 1. A hierarchy in the ability to produce leukemia was found: The progeny of only 100 HSCs were sufficient to produce fatal leukemia in 90% of animals. However, only the higher dose of 2500 (but not 250) CMPs caused disease, and with a longer latency than the recipients of LSKs (p<0.0001, Log-rank test). The relatively long latency to leukemia even with LSKs (HSCs/CLPs) suggests that additional events are required to develop fatal leukemia. In repeated experiments, none of the animals receiving GMPs developed leukemia, even at the maximum of 2500 GMP cells. To determine the minimum number of cells required to produce leukemia, five animals received only 25 HSCs each, two of which developed leukemia (Table 1). By limiting dilution analysis, the frequency of “transformable hematopoietic cells” (THCs) was 1:45 in HSCs and 1:57 in CLPs, which was significantly higher than the 1:1043 in CMPs (p<0.0001) (Table 1).
Figure 2. HSCs and CLPs are more efficient than CMPs and GMPs in producing leukemia in vivo.
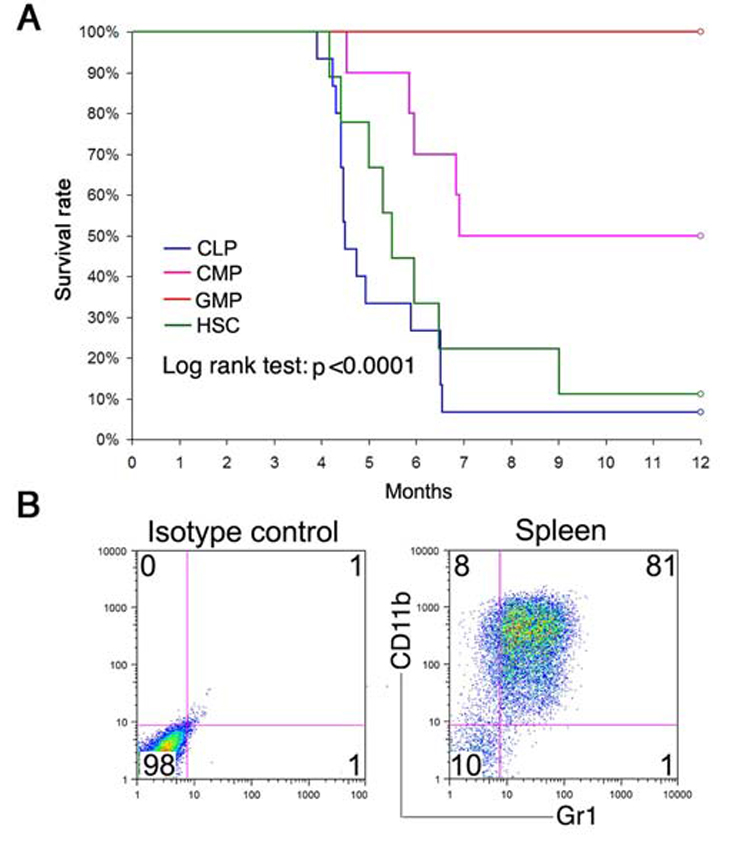
a,Survival of irradiated mice receiving 100–2500 cells. The survival rate was calculated using Kaplan-Meier analysis. The HSC and CLP groups had significantly poorer survival than the CMP and GMP groups (p<0.0001, log-rank test). b, Recipient mice developed myeloid leukemia after transplantation. All recipient mice showed the same high CD11b+Gr1+ profile in spleen previously described in Mll-AF9 leukemic mice. FACS on the spleen of a representative recipient is shown.
Table 1.
Summary data of transplantation experiments
| Starting Mll-AF9 Population | No. of Cells Transplanted | No. of Animals Transplanted | No. of Animals with AML (%) | Latency of AML (Median & 95% CI in Days) | Frequency of Transformable Hematopoietic Cells (THCs) |
|---|---|---|---|---|---|
| HSC | 25 | 5 | 2 (40%) | - *(167, -) | |
| 100 | 10 | 9 (90%) | 165 (152, 197) | 1: 45 | |
| CLP | 100 | 5 | 4 (80%) | 198 (134, NA) | |
| 250 | 5 | 5 (100%) | 136 (119, 179) | 1: 57 | |
| 2500 | 5 | 5 (100%) | 137 (131, 199) | ||
| CMP | 250 | 5 | 0 (0%) | - (-, -) | |
| 2500 | 5 | 5 (100%) | 181 (138, 210) | 1: 1043 | |
| GMP | 250 | 5 | 0 (0%) | - (-, -) | -- |
| 2500 | 6 | 0 (0%) | - (-, -) |
Five negative control mice injected with 2.5×105 WT bone marrow cells were all alive for the duration of the experiment.
inestimable
Although the different cell populations exhibited varying ability to cause disease, the type of leukemia caused by those cells was similar. Immunophenotyping revealed that the majority of cells from the enlarged spleens of recipient animals were myeloid (CD11b+Gr1+) in phenotype (Figure 2B). Leukemias could be transferred to secondary recipients and the immunophenotype remained the same as the transplanted cells from the primary recipients (data not shown). The demonstration that the relatively mature “downstream” myeloid leukemia cells are independent of the cell types transplanted is similar to that reported in the retroviral MLL-ENL and MLL-AF9 models (Cozzio et al., 2003; Somervaille et al., 2006).
Mll-AF9 induced transformation is highest in stem cells and lowest in GMPs in Mll-AF9 knockin mice
In a series of experiments we studied the mechanisms for the differences in leukemia outcomes based on the critical cell types. We first compared the self-renewal effects of Mll-AF9 knockin stem and progenitor cells using a myeloid colony forming assay (Johnson et al., 2003). Sorted cells were cultured in methylcellulose medium containing IL-3, IL-6, SCF and GM-CSF, replated every 7 days and colonies were studied at day 21. Figure 3A shows the significant increase in colony numbers from all Mll-AF9 stem/progenitor cells compared to wild type. Notably, LSKs formed the greatest number of colonies, with CMPs and GMPs forming significantly fewer colonies. We and others have previously shown that in addition to increased colony numbers, MLL fusion genes induce the formation of compact colonies which are composed predominantly of immature myeloid cells (Johnson et al., 2003; Somervaille et al., 2006). As shown in Figure 3B, significantly more compact colonies were found in Mll-AF9 LSK cultures than in those from CMP and GMP cultures. No compact colonies were found in any wild type cultures. Overall, colony assays showed that enhanced self-renewal induced by Mll-AF9 was greatest in LSKs (HSCs/CLPs) compared to the committed myeloid progenitor populations (CMPs and GMPs). Immunophenotyping revealed that cells from colonies in all Mll-AF9 groups were CD11b+Gr1+/− myeloid (Supplementary Figure 1).
Figure 3. In Mll-AF9 knockin mice, HSCs and CLPs produce more total and compact myeloid colonies with enhanced self-renewal in vitro than CMPs and GMPs. MLL-AF9 retrovirally transduced GMPs produce the most total and compact myeloid colonies.
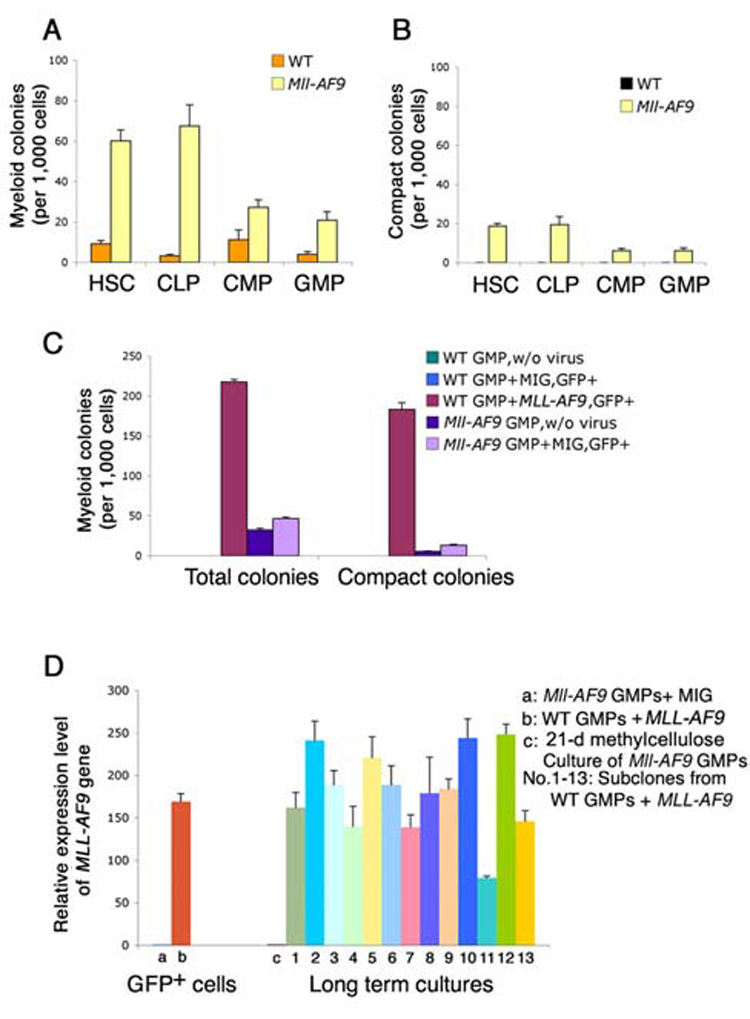
a, Total myeloid colony numbers were higher in HSCs and CLPs than in CMPs and GMPs in Mll-AF9 mice after 21-day culture under myeloid conditions. All the Mll-AF9 populations had significantly increased colonies compared to WT. b. Compact colony numbers were higher in HSCs/CLPs than in CMPs/GMPs in Mll-AF9 mice after 21-day culture. No compact colonies were detected in WT mice. Error bars represent standard error of the means. c, Total myeloid colony and compact colony numbers were the highest in MLL-AF9 retrovirally transduced GMP cells compared to the MIG (MSCV-IRES-GFP) vector transduced knockin Mll-AF9 GMPs and other knockin Mll-AF9 GMP controls as labeled. d, MLL-AF9 expression in retrovirally transduced GMPs and knockin Mll-AF9 GMPs. Error bars represent standard error of the means.
Retrovirus-induced expression of MLL-AF9 in GMPs results in increased myeloid colonies and long term self renewal in vitro; these changes are not found in Mll-AF9 knockin GMPs
In contrast to the GMPs transformed by knockin Mll-AF9 that did not produce leukemia, GMPs transformed by the MLL-AF9 retrovirus were capable of producing leukemia in transplanted animals (Krivtsov et al., 2006). Thus, we compared the effects of MLL-AF9 in GMPs transduced by retrovirus to those in GMPs from Mll-AF9 knockin mice. Wild type GMPs were transduced with MSCV-MLL-AF9-GFP retrovirus as previously described (Krivtsov et al., 2006), while Mll-AF9 knockin GMPs were transduced with the MSCV-GFP retrovirus as controls. The reagents and protocols for these studies were identical to those used by Krivtsov et al. In the first series of experiments, we compared myeloid colonies from both methods of fusion gene introduction. GFP+ cells were selected as previously described and myeloid colonies were counted after three sequential platings on day 21. Results in Figure 3C, Left show that total myeloid colonies were more than four times higher in the GMPs transduced by MLL-AF9 retrovirus than in Mll-AF9 knockin GMPs transduced with the MSCV-GFP control virus. Similarly, when colony types were examined, more compact immature colonies were found in the MLL-AF9 transduced cells than in the knockin cells that constitutively express Mll-AF9 (Figure 3C, Right). These data show enhanced self renewal of retroviral MLL-AF9 in vitro; Enhanced self renewal of retrovirally transformed cells was further shown in cytokine (IL3, IL6, SCF and GM-CSF) enriched liquid culture where Mll-AF9 knockin cells did not survive beyond 20 days while retroviral MLL-AF9 cells continued to grow in long term liquid culture (Supplementary Figure 2).
MLL-AF9 expression is significantly higher in retrovirally transduced GMPs than in Mll-AF9 knockin GMPs
The known strength of the retroviral promoter, combined with data from the colony assays and Southern blotting all suggested that expression of MLL-AF9 will be higher in retrovirally transduced GMPs than in Mll-AF9 knockin GMPs. Using primers that detected a sequence present in both retroviral MLL-AF9 and knockin Mll-AF9 constructs (but not in wild type mice), we compared the expression levels of the fusion gene in the MLL-AF9 transduced GMPs to those in MSCV-GFP transduced Mll-AF9 GMPs by real time quantitative RT-PCR. Figure 3D Left, showed that GFP+ MLL-AF9 retrovirally transduced cells had 170 fold higher expression of MLL-AF9 than knock-in GMPs with virus control. In long term culture, expression levels of MLL-AF9 in the subclones from MLL-AF9 retrovirally transduced GMPs remained very high (Figure 3D, Right). Results from southern blotting with GFP as a probe on the genomic DNA from these cultured cells showed more than one band; these results provide evidence that multiple MLL-AF9 integrations were likely (Supplementary Figure 3).
Gene expression profiles induced by Mll-AF9 expressed at physiologic levels
A goal of our study was to define the molecular pathways that would explain the differences between Mll-AF9 HSCs and GMPs. We compared the early (preleukemic) in vivo effects of the Mll-AF9 fusion gene on gene expression levels in the cells from Mll-AF9 knockin mice. RNA was extracted from sorted HSCs, CLPs, CMPs and GMPs and amplified for analysis by Affymetrix murine 430 2.0 microarrays. To identify genes differentially expressed as a result of Mll-AF9 expression, we performed a two-way ANOVA using a stratified permutation test (See Supplementary Methods). Allowing for a false discovery rate (FDR) of 10% (Benjamini et al., 1995), this analysis yielded 446 genes that were differentially expressed in Mll-AF9 compared to WT cells (Supplementary Figure 4). A clustering analysis was performed using this 446 gene set, with results shown in Figure 4A. The expected clustering of CMPs with GMPs, and HSCs with CLPs was found. Mll-AF9 HSCs and CLPs were clustered with each other instead of their wild type counterparts, suggesting very similar downstream effects of the fusion gene in the two related populations. Of the 446 genes selected by the two-way ANOVA, 192 were expressed at high levels in all four Mll-AF9 cell types compared to wild type while 179 genes displayed lower expression in the Mll-AF9 populations (Supplementary tables 1 and 2). The top 50 genes up-regulated in all four cell types are shown in the heat map inFigure 4B. These genes are ranked in decreasing order of fold changes in HSCs.
Figure 4. Hierarchical clustering of Mll-AF9 LSK (HSC/CLP), and CMP/GMP populations. Genes overexpressed in HSC, CLP, CMP and GMP populations.
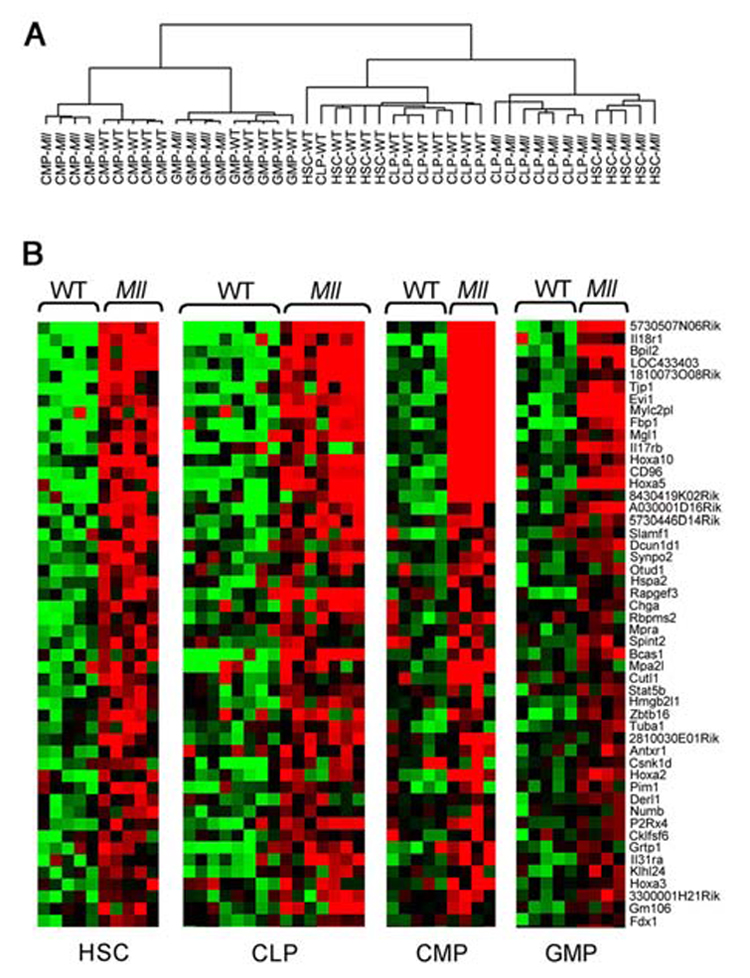
a, A two-way ANOVA with stratified permutation testing was performed to select genes differentially expressed in Mll-AF9 compared to wild type cells in each of the four populations. Hierarchical clustering performed with the 446 genes selected by the two-way ANOVA (FDR<0.1) separates the HSCs/CLPs from the CMPs/GMPs. WT = wild type, Mll=Mll-AF9. b, Mll-AF9 up-regulates expression of genes in multiple cell types (FDR<0.1, two-way ANOVA with permutation testing). Heat-maps showing the expression level of the top 50 genes up-regulated in the Mll-AF9 samples compared to WT, ranked in decreasing order of fold change up-regulation in HSCs. Expression levels are represented by colors: black=median red > median, green < median. Gene identifiers are at right.
Further analysis of the 192 up-regulated genes in the Mll-AF9 populations revealed 96 genes more highly expressed in transformation-sensitive LSKs compared to transformation-resistant CMPs/GMPs (FDR <0.1, Significance Analysis of Microarrays (Tusher et al., 2001) The top 50 of the 96 Mll-AF9 LSK overexpressed genes are shown in the heat map of Mll-AF9 transformed cells in Figure 5A. Representatives of the genes more highly expressed in LSK than in CMP/GMP group are well-known targets of Mll and Mll-fusion proteins - Hoxa5, Hoxa9, and Meis1. Also included is Evi1, not currently known to be a direct target of Mll or Mll fusion genes. Evi1 over-expression was confirmed by quantitative RT-PCR shown in Supplementary Figure 5. We analyzed in more detail the relative levels of known targets Hox5, Hoxa9, Meis1 and Evi1 in each population of cells with results shown in Figure 5B. Importantly, these four genes were most highly expressed in Mll-AF9 LSKs, expressed at intermediate levels in wild type LSKs and Mll-AF9 CMPs/GMPs and at the lowest levels in wild type CMP/GMPs. Several of the 192 genes that we found to be up-regulated by Mll-AF9 in all four cell populations, including Hoxa5, Hoxa9, Hoxa10, and Meis1 were previously found to be highly “immediately” expressed in GMP cells transformed by the MLL-AF9 retrovirus (Krivtsov et al., 2006).
Figure 5. A set of genes are up-regulated by Mll-AF9 to highest levels in LSKs (HSCs/CLPs) compared to both to Mll-AF9 CMPs/GMPs and to wild type HSCs/CLPs.
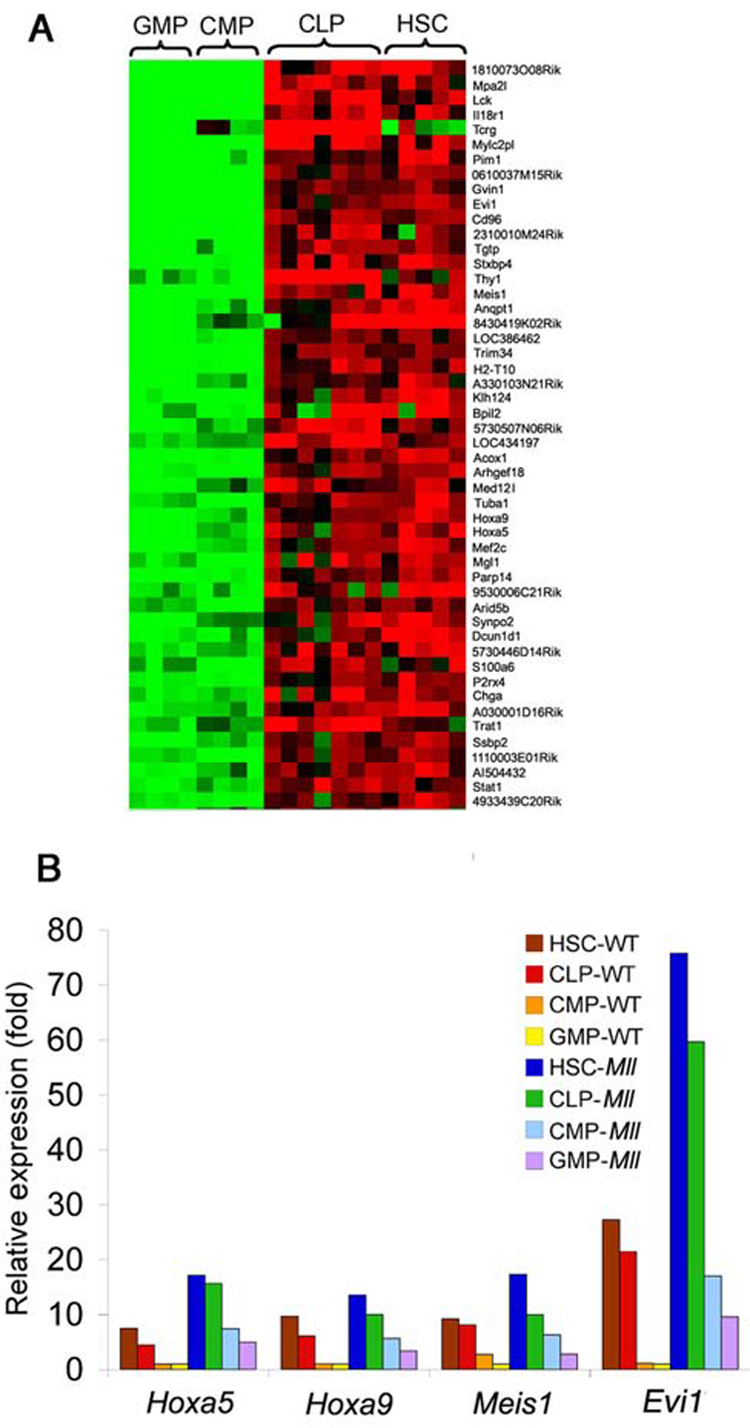
a, Mll-AF9 up-regulated genes are highly expressed in HSCs and CLPs compared to CMPs and GMPs. Of the 192 genes over-expressed in Mll-AF9 cells, 96 genes are expressed at higher levels in HSCs and CLPs compared to CMPs and GMPs (FDR<0.1, SAM). The top 50 genes in this subset are shown. Expression levels are represented by colors: black = median, red > median, green < median. b, Expression of Mll-AF9 up-regulated genes – Hoxa5, Hoxa9, Meis1 and Evi1 is highest in Mll-AF9 HSCs/CLPs. Data represent average expression relative to levels in wild type GMPs.
We also carried out a comparative analysis of the previously reported leukemias resulting from retrovirus transduced GMPs (Krivtsov et al., 2006) and our knockin preleukemia cells. This analysis showed 20 genes that were up-regulated in both groups (Supplementary Table 3). Included were the expected Hoxa5-9 and Meis1 genes plus novel genes, such as IL31 receptor A and Chemokine-like factor super family 6 genes. Since the previous study used only GMPs rather than the four cell types of this study and a different Affymetrix probe set, a complete comparison of the two is difficult and it is likely that the number of genes in common is actually greater than 20.
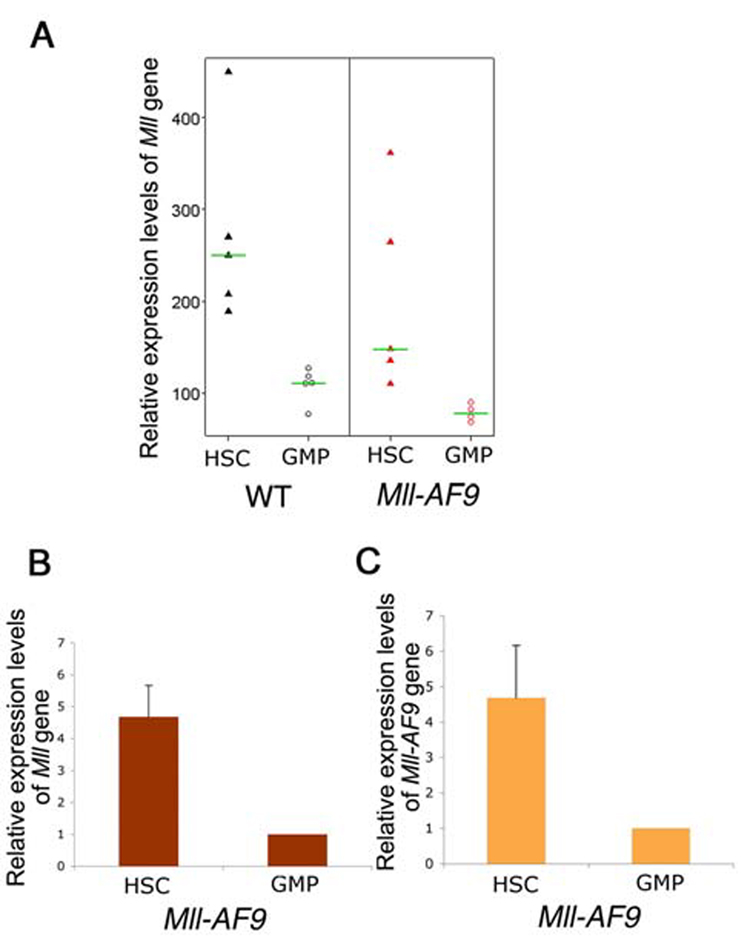
Mll and Mll-AF9 expression are higher in HSCs than in GMPs
We next evaluated the hypothesis that the up-regulated expression of genes known to be downstream of Mll, such as Hoxa genes in HSCs, could be a result of higher expression levels of the Mll-AF9 fusion gene in HSCs. The expression of wild type Mll has been shown to be highest in HSCs compared to more mature progenitors (Jude et al., 2007). Since the Mll-AF9 fusion gene is present in all cells of knockin mice, we studied expression levels of the oncogene in the various hematopoietic cells with the expectation that expression of Mll-AF9 would parallel that of Mll. The microarray results showed that the expression level of Mll was higher in WT than Mll-AF9 cells and was higher in HSCs than GMPs (Figure 6A). Quantitative real-time RT-PCR interrogating the 3′ end of Mll showed that Mll expression level in HSCs is 4–5 fold higher than in GMPs from Mll-AF9 mice (Figure 6B). Similarly, using a 5′ Mll primer/probe set, higher expression was found in HSCs compared to GMPs (data not shown). Quantitative real-time RT-PCR analysis was also performed to evaluate Mll-AF9 levels in HSCs and GMPs. Results in Figure 6C show 4–5 fold higher levels of Mll-AF9 in HSCs compared to GMPs.
Figure 6. Mll-AF9 expression is higher in HSC than GMP populations. Retrovirally transduced MLL-AF9 results in very high expression levels of the oncogene.
a, Mll expression in the HSCs and GMPs from WT and Mll-AF9 mice by microarray. b, Mll expression in the HSCs and GMPs from Mll-AF9 mice by real-time RT-PCR. c, Mll-AF9 expression in the HSCs and GMPs from Mll-AF9 mice by real-time RT-PCR. d, MLL-AF9 expression in retrovirally transduced GMPs and knockin Mll-AF9 GMPs. Error bars represent standard error of the means.
To summarize, the higher expression levels of Mll-AF9 in HSCs compared to GMPs in the physiologic model suggests the importance of Mll-AF9 gene dosage in producing downstream effects, in the Hoxa family and other genes, although other cell context-specific differences are also likely to be important and deserve further research.
Discussion
This study focused on the malignant transformation initiated by the fusion gene Mll-AF9 when expressed at physiologic levels in the knockin model or at supraphysiologic levels in the retroviral model. The data from the physiologic model showed highest levels of Mll and Mll-AF9 in the most transformable HSCs and lower levels in the more resistant committed myeloid progenitor GMPs. Complementary data showed high Mll-AF9 gene dosage in retrovirally transformed GMPs which were found to have enhanced self renewal and ability to grow in long term in vitro culture. While it is possible that the cell-type differences in transformation are unrelated to the expression levels of Mll-AF9 at physiologic dosages, we favor the hypothesis that the “superactivation” of target genes observed in HSCs/CLPs (LSKs) is likely to be oncogene dose related. In support of the role of Mll fusion gene expression in the different cell types are recent data with conditional knockout mice: Wild type Mll showed highest expression in HSCs compared to other cell types (Jude et al., 2007). Wild type MLL and likely MLL fusion proteins bind to the promoters of many genes and serve as a global regulator of gene transcription. As a result, a large number of downstream target genes are altered in regulation (Guenther et al., 2005; Milne et al., 2005; Scacheri et al., 2006). These cell type differences in expression of Mll target genes appear to be further enhanced by Mll fusion genes resulting in activation of cellular pathways, especially those that enhance self-renewal and block cellular differentiation. However, it is possible that additional molecular differences could result in increased sensitivity of HSCs to the fusion oncogene is possible. A combination of higher Mll-AF9 gene dosage and a more receptive cellular environment may be responsible for the superior transformation of LSKs.
Previously published studies with Mll fusion genes (MLL-ENL or MLL-AF9) have utilized retroviruses with strong promoters and multiple virus insertions resulting in non-controlled and potentially non-physiologic expression levels of oncogene. Non-physiologic expression could mask important cell type-specific effects on the promoters of target cells. One advantage of the Mll fusion gene knockin model over retroviral or physical methods is that the oncogene should be expressed at physiologic levels with cell-type specificity. In the current study, the lack of leukemia in lethally irradiated recipients of Mll-AF9 GMPs contrasts with the experiments in which the fusion gene is introduced into GMPs by retroviral transduction (Krivtsov et al., 2006). These results are supported by our in vitro data which showed significantly enhanced cell growth in retrovirally transformed MLL-AF9 cells but not in physiologically expressed Mll-AF9 cells. Enhanced self renewal of retrovirally transformed GMPs was shown in the ability of these cells to grow in long term culture in vitro. In previous results from a MLL-ENL model, leukemia developed in animals that received 800–2490 retrovirally transduced GMPs (Cozzio et al., 2003). Also, a shorter latency to leukemia development is found in the retroviral models compared to our knockin model (Cozzio et al., 2003; Krivtsov et al., 2006; Somervaille et al., 2006). We cannot rule out the possibility that the more rapid development of leukemia in retroviral models may in part or totally be due to retroviral enhancement of secondary cooperating events, but our short term myeloid colony data strongly suggest that the differences are immediate and very direct. Also, it is possible that the differences could result from the use of human MLL in the retroviral construct compared to the endogenous murine Mll in our studies. However, this is unlikely, as to date no differences in critical domains have been described for human and murine MLL, and the AF9 portion of both models is identical. With these caveats, it is likely that the differences between the retroviral MLL-AF9 and knockin Mll-AF9 experiments are due to gene dosage effects. This conclusion is also supported by 1) the presence of multiple integration sites in the retrovirally transduced cells shown by Southern blotting and 2) the strong MSCV-based retroviral promoter in this study and others (Krivtsov et al., 2006;Somervaille et al., 2006).
Our results showing that knockin Mll-AF9 HSCs and CLPs, representing the relatively undifferentiated LSK (Lin−c-kit+Sca-1+) hematopoietic cells, are most efficiently transformed are similar to those reported for retrovirally introduced MLL-ENL 15 (Cozzio et al., 2003). Also, similar to the retroviral MLL-ENL model and MLL-AF9 model (Somervaille et al., 2006), the bulk of cells of all the leukemias were relatively mature myeloid CD11b+Gr1+ in type, irrespective of the phenotype of the transplanted transformed cells. However, we did not determine the nature of the leukemia stem cells (LSCs) that initiate and maintain the leukemia in the animal. The long latency for development of the leukemias in animals suggests that there are important genetic and/or epigenetic events occurring during this latency period. These later events could also be important in determining the phenotype of the LSCs. The results presented have implications for therapy of both the early and later stages of leukemia.
In our knockin model, 192 genes were found to be up-regulated by Mll-AF9 in all four cell populations. Several, including Hoxa5, Hoxa9, Hoxa10, and Meis1, were previously found to be highly “immediately” expressed in GMP cells transformed by the MLL-AF9 retrovirus (Krivtsov et al., 2006). Also, as discussed in “Results”, we found 20 genes in common in our knockin preleukemia data set and the leukemia data set described earlier (Krivtsov et al., 2006) Another report showed that MLL-AF9 introduced by retrovirus resulted in up-regulation of several critical genes when leukemia stem cells (LSCs) were compared to the transformed preleukemic “initiating” cells (Somervaille et al., 2006). That study did not compare gene signatures in wild type compared to “initiating” cells.
We found very high expression of Evi1 in Mll-AF9 cells compared to the corresponding wild type cells. High levels of expression of Evi1, have been reported in human myeloid leukemias with MLL-rearrangements (Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2003; Valk, Verhaak et al., 2004). Also, Evi1 over-expression is sufficient to immortalize murine hematopoietic cells (Du et al., 2005), which suggest that this gene should be studied further for its role in the pathogenesis of MLL-fusion leukemias.
The knock in murine Mll-AF9 model is useful because the fusion gene is present and expressed in all progenitor and stem cells. The situation in humans is less clear, since the cell in which the human MLL-AF9-producing translocation develops is not defined. However, the human MLL-AF9 gene will be present both in the cells with the initial “hit” plus all progenitor cells and cells at later stages of differentiation. While it is possible that the transforming human MLL-AF9 translocation may take place at a maturation stage later than the HSC, murine studies suggest that this is much less likely to be functionally meaningful than a “hit” within the HSC population. Future studies will be necessary to further define this issue.
In conclusion, our results directly show that supraphysiologic oncogene doses of Mll-AF9 produced biologically different effects from physiologic doses in the same cell type. We also show an association between oncogene dosage and cell type-specific transformation susceptibility; however, the oncogene dosage differences are less in the physiologic model compared to the retroviral model. While we favor the hypothesis that both Mll-AF9 expression differences between cells types and other cell context differences are pathophysiologically important, direct evidence will need to be provided in future studies. Seminal earlier studies with myc and other oncogenes have shown that gene dosage effects are central to the pathophysiology of cancers that develop under natural conditions (Ren, 2004). Experimental studies that introduce oncogenes by viruses and other physical methods have been extremely important in cancer biology research. However, to the extent that they result in non-physiologic oncogene expression levels, experimental results may differ from those in naturally occurring cancers.
Experimental procedures
Mice
The Mll-AF9 mice were originally produced in the laboratory of Dr. Terence Rabbitts (Leeds, UK). Briefly, heterozygous mice were produced by fusing the human AF9 short form (breakpoint to 3′end) into exon 8 of the mouse Mll gene (Corral et al., 1996), and have been maintained on a C57BL/6 background. The wild type mice used in the experiments were the littermates of Mll-AF9 mice. All the mice were housed under specific pathogen-free conditions in an accredited facility at the University of Minnesota. All experiments were conducted after approval by the Institutional Animal Care and Use Committee (IACUC).
Cell sorting and FACS analysis
Single cell suspensions of bone marrow were obtained from 8 week old WT or Mll-AF9 mice. The purification of HSC population (Lin−Thy1.1lo Sca-1+ c-kit+) was similar to the method described before (Kondo et al., 1997; Terskikh, Miyamoto et al., 2003). Briefly, bone marrow cells were stained with biotin-conjugated lineage specific anti-IL-7R (Pharmingen, San Diego, CA) and cocktail antibodies from the Lineage Cell Depletion Kit (Miltenyi, Bergisch Gladback, Germany) according to manufacturer’s instructions. Lin+cells were partially removed by magnetic beads (MACS, Miltenyi, Bergisch Gladback, Germany). The remaining cells were stained with Streptavidin-PE-Cy5 conjugate, and further stained with APC-conjugated anti-c-kit, FITC-conjugated anti-Sca-1 and PE-conjugated anti-Thy1.1 antibodies (Pharmingen, San Diego, CA). The HSC population was sorted by FACSAriaTM (BD Biosciences Immunocytometry Systems, San Jose, CA).
The CLPs were sorted as Lin−IL-7R+Thy1.1− Sca-1lo c-kitlo(Kondo et al., 1997; So et al., 2003), using a similar method. CMPs (Lin−IL-7R− Sca-1−c-kit+CD34+ FcγRII/IIIlo) and GMPs (Lin− IL-7R−Sca-1−c-kit+CD34+FcγRII/IIIhi)were separated as described previously (Manz et al., 2001;Terskikh et al., 2003). The purity of sorted cell populations was > 95% by post-sort analysis.
Relative percentage of HSCs, CLPs, CMPs and GMPs from lineage negative marrow cells in wild type and Mll-AF9 marrow (Figure 1a, 1b) were calculated as follows: %HSCs = the percentage of Sca1+c-kit+Thy1lo cells in Lin−IL-7R−population; %CLPs = the percentage of Sca1+c-kit+cells in Lin−Thy1−IL-7R+population; %CMPs=the percentage of CD34+ FcγRII/IIIlo cells in Lin−Sca1−c-kit+population; %GMPs=the percentage of CD34+ FcγRII/IIIhi cells in Lin−Sca1−c-kit+ population. Statistical comparisons were performed using the two-tailed t-test.
For FACS analysis, single cell suspensions from either cultured cells or mouse hematopoietic organs (bone marrow or spleen) were stained with FITC or PE labeled anti-mouse antibodies: CD11b, Gr1 (Pharmingen or eBioscience, San Diego, CA) and acquired on a BD FACScalibur with Cell Quest software. Data were analyzed with FloJo software (Tree Star Inc, San Carlos, CA).
Methylcellulose culture
Sorted cells were cultured in methylcellulose medium under myeloid conditions, using methocult 3534 (StemCell Technologies, Vancouver, Canada), supplemented with 10ng/ml GM-CSF (R&D, Minneapolis, MN) (Chen et al., 2006). Cells were cultured in triplicate for 21 days transfers every 7 days. Colonies containing over 50 cells were counted and classified under the microscope as previously described (Jordan et al., 2006).
Mouse transplantation with sorted cell populations
Each sorted population was transplanted into mice at various doses. 25 and 100 sorted HSCs, 100, 250 and 2500 CLPs, 250 and 2500 CMPs or GMPs from Mll-AF9 were mixed with a radioprotective dose of 2.5×105 bone marrow cells from WT mice and injected into lethally irradiated (960 rad) recipients. Each group contained at least 5 mice. Five recipient mice injected with 2.5×105 WT bone marrow cells were used as negative controls. Mice were sacrificed when they became detectably ill. Necropsy, FACS, immunohistochemistry and histopathology evaluations were performed at the time of sacrifice. The survival rate was calculated using the Kaplan-Meier method. The frequency of transformable hematopoietic cells was calculated by limiting dilution analyses using L-calc software (StemCell technologies).
Retrovirus transduction
Retrovirus constructs MSCV-MLL-AF9-GFP, MSCV-GFP and package plasmid psi-Eco were used to produce retrovirus supernatant by co-transfection of 293T cells. Transduction of WT or Mll-AF9 GMPs was performed as previous described (Krivtsov et al., 2006) After transduction, GFP positive cells were sorted and put in methylcellulose culture for colony assays. RNA from these cells was extracted to detect MLL-AF9 expression by quantitative RT-PCR. DNA was purified and digested by EcoRI for Southern blotting.
Gene expression studies
For quantitative real time RT-PCR, reverse transcription was performed using the Supercript II reverse transcription kit (Invitrogen) and real-time PCR detection was performed using TaqMan primer/probe sets(Applied Biosystems Inc., Foster City, CA) and an ABI 7500 Real-Time PCR system. For MLL-AF9, real-time PCR detection was performed using SYBR Green. In all RT-PCR experiments, Gapdh was used as the housekeeping gene. Changes in expression calculations were performed by the 2▵▵CT method using the Relative Expression Software Tool (REST,www.gene-quantification.info).
Microarray Analysis
For gene expression profiling, total RNA was extracted from sorted cells and amplified (Affymetrix). Labeled cRNA was hybridized to Mouse 430 2.0 genomic arrays. Normalization and analysis of chip data were performed using the Expressionist package (GeneData Inc., Supplementary Method). Heat maps were generated using Cluster and Treeview http://rana.lbl.gov/EisenSoftware.htm. See Supplementary Data for detailed Microarray analysis. All microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10627.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (RO1-CA087053 to JHK, and K08-CA122191 to ARK), the Children’s Oncology Group and Minnesota Medical Foundation (ARK), and by the Children’s Cancer Research Fund (JHK and ARK). The authors thank Scott A. Armstrong and Andrei V. Krivtsov (Children’s Hospital/Harvard Medical School) for providing the reagents and protocols for the retroviral studies, discussions of the microarray studies and reading of the manuscript. Terry Rabbitts (Leeds, UK) originally produced the Mll-AF9 knockin mouse. The University of Minnesota Cancer Center Biostatistical Core (Robin Bliss and Yan Zhang), Flow Cytometry Core (Tucker LeBien), the Histopathology Core (Dr. Gerald O’Sullivan) and the Biomedical Genomics Facility were critical to this project. All the authors have no conflicts of interest to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;6774:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Lowenberg B, Delwel R. Low expression of MDS1-EVI1-like-1 (MEL1) and EVI1-like-1 (EL1) genes in favorable-risk acute myeloid leukemia. Exp. Hematol. 2003;11:1066–1072. doi: 10.1016/j.exphem.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statistical Society. 1995:289–300. [Google Scholar]
- Caslini C, Serna A, Rossi V, Introna M, Biondi A. Modulation of cell cycle by graded expression of MLL-AF4 fusion oncoprotein. Leukemia. 2004;6:1064–1071. doi: 10.1038/sj.leu.2403321. [DOI] [PubMed] [Google Scholar]
- Chapiro E, Russell L, Radford-Weiss I, Bastard C, Lessard M, Struski S, Cave H, Fert-Ferrer S, Barin C, Maarek O, et al. Overexpression of CEBPA resulting from the translocation t(14;19)(q32;q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;10:3560–3563. doi: 10.1182/blood-2006-03-010835. [DOI] [PubMed] [Google Scholar]
- Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 2006;2:669–677. doi: 10.1182/blood-2005-08-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, Rabbitts TH. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;6:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;24:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CL, Warren AJ, Pannell R, Forster A, Lavenir I, Corral J, Smith AJ, Rabbitts TH. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;13:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;12:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2005;24:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, Kamada N, Ariyoshi Y, Takahashi T, Ueda R. MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene. 1993;11:3085–3092. [PubMed] [Google Scholar]
- Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc. Natl. Acad. Sci. U. S. A. 1992;4:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JJ, Chen W, Hudson W, Yao Q, Taylor M, Rabbitts TH, Kersey JH. Prenatal and postnatal myeloid cells demonstrate stepwise progression in the pathogenesis of MLL fusion gene leukemia. Blood. 2003;8:3229–3235. doi: 10.1182/blood-2002-05-1515. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. N. Engl. J. Med. 2006;12:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and Independent Roles for MLL in Adult Hematopoietic Stem Cells and Progenitors. Cell Stem Cell. 2007;1:324. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;5:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;11:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;7104:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;5:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- Manz MG, Traver D, Miyamoto T, Weissman IL, Akashi K. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 2001;11:3333–3341. doi: 10.1182/blood.v97.11.3333. [DOI] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams' O, Brady HJM. Mll Has a Critical Role in Fetal and Adult Hematopoietic Stem Cell Self-Renewal. Cell Stem Cell. 2007;1:338. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;24:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Ren R. Modeling the dosage effect of oncogenes in leukemogenesis. Curr. Opin. Hematol. 2004;1:25–34. doi: 10.1097/00062752-200401000-00005. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006;4:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer. Cell. 2003;2:161–171. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer. Cell. 2006;4:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;4861:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;1:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;9:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Lowenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;16:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.