Abstract
Biogenesis of the Golgi apparatus is likely mediated by the COPI vesicle coat complex, but the mechanism is poorly understood. Modeling of the COPI subunit βCOP based on the clathrin adaptor AP2 suggested that the βCOP C terminus forms an appendage domain with a conserved FW binding pocket motif. On gene replacement after knockdown, versions of βCOP with a mutated FW motif or flanking basic residues yielded a defect in Golgi organization reminiscent of that occurring in the absence of the vesicle tether p115. Indeed, βCOP bound p115, and this depended on the βCOP FW motif. Furthermore, the interaction depended on E19E21 in the p115 head domain and inverse charge substitution blocked Golgi biogenesis in intact cells. Finally, Golgi assembly in permeabilized cells was significantly reduced by inhibitors containing intact, but not mutated, βCOP FW or p115 EE motifs. Thus, Golgi organization depends on mutually interacting domains in βCOP and p115, suggesting that vesicle tethering at the Golgi involves p115 binding to the COPI coat.
INTRODUCTION
Golgi enzymes are present in stacked, biochemically distinct, membranous compartments or cisternae. Compartmentalization of the Golgi apparatus ensures optimal conditions for biosynthetic processing by promoting sequential substrate exposure, concentration of Golgi enzymes, and the creation of distinct reaction environments. Golgi compartmentalization is maintained despite a high flux of membrane and protein by the accurate transfer of specific components between cisternae. One view is that Golgi enzymes continuously move to antecedent cisternae in vesicles formed by the COPI coat complex, whereas the cargo being processed moves forward by simply remaining within the maturing cisternae (Bonifacino and Glick, 2004). Although not excluding concurrent mechanisms, this view emphasizes the importance of local, intracisternal recycling of Golgi enzymes by COPI vesicles in compartmentalization of the Golgi apparatus.
Newly synthesized Golgi proteins are initially concentrated by the COPII vesicle coat complex as they exit the endoplasmic reticulum (ER) with other cargo. Repeated rounds of COPII vesicle budding and fusion at distributed ER exit sites sustain recycling compartments adjacent to exit sites, termed intermediate compartments (ICs). COPI coats form on ICs and mediate retrieval of escaped ER proteins back to the ER, presumably further concentrating Golgi proteins remaining in the ICs (Martinez-Menarguez et al., 1999). The retrograde vesicles formed on ICs by COPI fuse with the ER by using the vesicle-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) Sec20, which binds the target membrane-associated (t)-SNARE complex of Ufe1, Sec22, and Slt1 (Burri et al., 2003; Dilcher et al., 2003). Efficiency and fidelity of the fusion may be promoted by the action of the putative Ds1 vesicle docking complex composed of ZW10 and RINT (Andag et al., 2001; Reilly et al., 2001; Vanrheenen et al., 2001; Hirose et al., 2004). Thus, in addition to escaped ER proteins, the COPI vesicle complex at ICs sorts Sec20 and the Ds1 complex to ensure correct targeting and fusion of the vesicles at the ER.
COPI-mediated sorting at ICs is apparently distinct from intracisternal COPI sorting. In mammalian cells, recycling from ICs is followed by inward movement of much of the remaining membrane to a pericentrosomal position where it becomes incorporated into the cis-aspect of the Golgi apparatus (Presley et al., 1997). Compartmentalization might start with sorting of cis-Golgi proteins away from the others by COPI vesicles. These vesicles fuse with newly generated ICs, thus enriching them in cis-elements. The medial-Golgi proteins will be sorted into COPI vesicles next, which fuse with the cis-cisternae, thus allowing it to mature. Such iterative local recycling of Golgi proteins, induced by a high affinity of the COPI coat for early Golgi proteins, with decreasing affinities toward later components, could mediate Golgi compartmentalization (Glick et al., 1997; Puthenveedu and Linstedt, 2005). Syntaxin5, when complexed with GOS28 and Ykt6 forms a t-SNARE that binds GS15 to mediate fusion of locally recycling COPI vesicles (Parlati et al., 2002; Xu et al., 2002). In the Golgi, local recycling may be enhanced by retention of vesicles in the local environment by protein interactions that tether these vesicles to the acceptor compartment. Because the proposed lengths of such vesicle “tethers” are more than the distance between the cisternae, it is possible that tethering is established even before vesicle budding is complete. Thus, Golgi-localized COPI coat complexes are expected to sort recycling Golgi proteins, Golgi-localized SNAREs, and Golgi-localized vesicle tethers, implying that the COPI complex might contain binding sites for these components.
COPI has few precisely mapped binding domains, but potential interaction sites have emerged from the recently reported γCOP structure (Hoffman et al., 2003; Watson et al., 2004). The COPI coat complex seems to contain an adaptor subcomplex that is structurally homologous to the AP2 adaptor of the clathrin coat, strongly suggesting conservation of the sorting functions known for the AP2 adaptor (Eugster et al., 2000). Similar to the α and β subunits of AP2, the γCOP subunit of COPI contains trunk, linker, and appendage domains (Hoffman et al., 2003; Watson et al., 2004), and βCOP might form an analogous structure, including a C-terminal appendage domain (Hoffman et al., 2003). For AP2, cargo interactions generally map to the trunk domains and transport or regulatory factor interactions map to the appendage domain (Owen et al., 2004). Interestingly, sequence alignment with αAP2 and βAP2 indicates that there is a conserved FW motif (FXXXW) in the γCOP and βCOP appendage domains (Hoffman et al., 2003; Watson et al., 2004). Mutation of the αAP2 and βAP2 FW motif in combination with flanking basic residues alters binding of regulatory factors (Owen et al., 1999; Traub et al., 1999; Owen et al., 2000), suggesting the potential importance of this region in γCOP and βCOP. Indeed, mutation in the FW motif of Sec26p, the yeast homologue of βCOP, causes temperature-sensitive growth defects (Hoffman et al., 2003; DeRegis et al., 2008). Thus, mutations in the FW motif of the γCOP or βCOP appendage domains might interfere with transport factor binding and possibly cause deficits in Golgi biogenesis.
In the present study, we analyzed the role of βCOP FW motif by using gene replacement after RNA interference (RNAi). Mutation of the βCOP FW motif or its flanking basic cluster yielded a defect in Golgi organization. The fragmented Golgi phenotype seemed similar to that observed after knockdown of the vesicle-tethering protein p115 (Puthenveedu and Linstedt, 2004), prompting a test for βCOP binding to p115. Indeed, the interaction occurred and depended on the βCOP FW motif and acidic residues, E19E21, in the p115 head domain. The interaction was functionally important because inverse charge substitution of p115 E19E21 also yielded the fragmented Golgi phenotype. Furthermore, the fragmented Golgi phenotype was observed in a permeabilized cell Golgi assembly assay carried out in the presence of peptide inhibitors directed against either the βCOP FW or the p115 E19E21 motifs. These results demonstrate a role for p115-βCOP binding in Golgi organization.
MATERIALS AND METHODS
Constructs and Reagents
Small interfering RNA (siRNA) against βCOP were either purchased (Integrated DNA Technologies, Coralville, IA) or synthesized using the Silencer siRNA kit (Ambion, Austin, TX). The siRNA target sequences in βCOP are as follows: AAAAGCCGTCTCCTTTGACTC (#2309) and AACTAGCTACACTAGGGGATC (#2276). Unless indicated, experiments used #2309. The rat βCOP cDNA was purchased fro American Type Culture Collection (Manassas, VA). Mutant versions of βCOP were generated either by restriction digests or by using the QuikChange site-directed mutagenesis kit (Stratagene, CA). Constructs were confirmed by restriction analysis and sequencing. Plasmid encoding RFP-KDEL was kindly provided by Dr. E. Snapp (Albert Einstein University).
Cell Culture and Immunofluorescence
Normal rat kidney (NRK) and HeLa cell lines were maintained, and immunofluorescence, including the ts045-VSVG trafficking assay, was performed as described previously (Puri and Linstedt, 2003; Puthenveedu and Linstedt, 2004). The antibodies and their dilutions were mouse anti-myc at 1:100 and mouse anti-βCOP at 1:500 (Sigma-Aldrich, St. Louis, MO), rabbit anti-giantin at 1:500 (Puthenveedu and Linstedt, 2001), sheep anti-TGN46 at 1:500 (Serotec, Raleigh, NC), mouse anti-VSVG (8G5; Lefrancois and Lyles, 1982; Sevier et al., 2000), mouse anti-golgin97 at 1:500 (Zymed Laboratories, South San Francisco, CA); mouse anti-ERGIC53 at 1:500 (Schweizer et al., 1990); rabbit anti-GRASP65 (Puthenveedu et al., 2006), rhodamine-labeled goat anti-mouse at 1:500 (Zymed Laboratories), Cy5-labeled goat anti-mouse at 1:500 (Zymed Laboratories), rhodamine-labeled goat anti-rabbit at 1:500 (Zymed Laboratories), and Cy5-labeled donkey anti-sheep at 1:500 (Zymed Laboratories).
RNA Interference and Replacement
For siRNA treatment, cells were plated in 35-mm dishes (containing glass coverslips for immunofluorescence analyses) and the day after plating, the cells were washed with PBS and incubated in 1.5 ml of minimal essential medium containing 10% fetal bovine serum. A mixture of 3 μl of 20 μM siRNA and 150 μl Opti-MEM (Invitrogen, Carlsbad, CA) was added to a mixture of 6 μl of Oligofectamine (Invitrogen) and 36 μl Opti-MEM. After 30 min at room temperature, the transfection mixture was added to the cells. After 48- or 72-h transfection, the cells were analyzed by immunofluorescence or immunoblotting. For gene replacement, 24 h after siRNA transfection, the cells were transfected using Transfectol (GeneChoice, San Diego, CA) with plasmids encoding Myc-tagged rat βCOP, Myc-tagged rat βCOP FW>AA, or Myc-tagged rat βCOP RK>AA. Cells were analyzed 24 or 48 h after transient transfection with the plasmid. Replacement of p115 was carried as described previously (Puthenveedu and Linstedt, 2004).
Image Analysis
Microscopy was performed as described previously (Guo and Linstedt, 2006). Optical sections at 0.3-μm spacing were acquired using the ImagingSuite software package (PerkinElmer Life and Analytical Sciences, Boston, MA). Individual experiments were performed with identical laser output levels, exposure times, and scaling. ImageJ (http://rsb.info.nih.gov/ij/) was used for colocalization analysis of two images. First, the average pixel intensity for the two images, in their entirety, was equalized using the divide function. Second, a threshold was chosen manually, based on comparison to the original gray-scale images. Third, the number of above-threshold pixels for each marker was determined per cell. Fourth, the number of above-threshold pixels showing colocalization was determined using the colocalization function with a fixed ratio of 0.75. Fifth, for each marker, the number of colocalized pixels was divided by the number of above-threshold pixels to yield the fraction of a given maker's area in a cell that colocalized with the other marker. Finally, the average of the two values, each representing the percentage of colocalization for each marker, was used as final colocalization value. To measure intensity of Golgi marker proteins, a fixed threshold was manually chosen and applied to all images. Individual cells were then selected with the free-hand tool, and the average intensity of the total above-threshold fluorescence was determined using the measure function of ImageJ. Values were normalized by dividing by the mean value of the entire data set for a given experiment. To measure Golgi size, three-dimensional (3D) rendering of confocal sections was generated though Volocity (PerkinElmer Life and Analytical Sciences). Then, the Golgi/remnant structures were selected using the magic wand function, and the size was determined using the analyze particle function.
Yeast Two-Hybrid Assay
The yeast dihybrid assay was carried out as described previously (James et al., 1996). Constructs in pOBD were transformed into PJ69-4A and interactions were tested by spotting the transformants on a minus-Leu Trp His 5 mM 3-aminotriazol plates. The p115 N terminus (1–655 amino acids) and βCOP were cloned in frame with the GAL4 activation and binding domains, respectively. The βCOP N terminus (1–666) and C terminus (333–953) were also cloned in frame with GAL4 binding domain.
Coimmunoprecipitation
HeLa cells from a 10-cm dish were lysed in 1 ml of HKT buffer (10 mM HEPES, pH 7.4, 100 mM KCl, 0.5% Triton (TX)-100, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml pepstatin A), allowed to sit on ice for 10 min, passed through a 25-gauge needle several times and centrifuged for 10 min at 15,000× g at 4°C. Aliquots of the supernatant (250 μl) were incubated 3 h at 4°C with antibody-bound beads. Beads were prepared by incubation of 10 μl of protein A-Sepharose (CL-4B; GE Healthcare, Chalfont St. Giles, United Kingdom) with 1 μl of either preimmune or rabbit anti-p115 antibody followed by washing. The immunobead-bound protein complexes were washed three times with 0.5 ml of HKT buffer, and they were analyzed by immunoblotting.
Protein Purification
Glutathione transferase (GST) fusion protein purification was carried out as described previously (Linstedt et al., 2000) for versions of βCOP-C (485–954 residues), βCOP-N (1–484 residues), and p115-N (1–655 residues). Individual colonies of DH5α Escherichia coli, transformed with plasmids encoding the GST fusion proteins, were grown to OD 0.6 in 500 ml of Luria broth (LB) at 37°C, and then expression was induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 5 h at 25°C. Cell pellets were collected, washed with 150 mM NaCl, and suspended in 4 ml of lysis buffer (50 mM Tris, pH 8.0, 5 mM EDTA, 150 mM NaCl, 10% glycerol, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml pepstatin A), incubated on ice for 30 min, adjusted to 0.5% Triton X-100, sonicated four times for 30 s each, and centrifuged at 50,000 rpm for 30 min in a TLA 100.3 rotor (Beckman Coutler, Fullerton, CA). The cleared lysate was collected and incubated with 0.5 ml of glutathione-agarose beads (Sigma-Aldrich) overnight at 4°C. The beads were washed with 40 ml of PD buffer (phosphate-buffered saline and 1 mM dithiothreitol) containing 0.1% Triton X-100, and then they were washed with 10 ml of PD buffer and eluted in PD buffer containing glutathione. After dialysis against HK buffer (10 mM HEPES, pH 7.4, and 100 mM KCl) at 4°C, the proteins were aliquoted and frozen. In some cases, the washed column beads were used directly in binding experiments. Hexahistidine-tagged protein purification was carried out according to the protocol provided by QIAGEN (Valencia, CA). Individual colonies of BL21 E. coli, transformed with plasmids encoding the hexahistidine-tagged fusion proteins, were grown to OD 0.6 in 500 ml of LB at 37°C, induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 5 h at 25°C. Cell pellets were collected, washed with phosphate-buffered saline, and frozen in liquid nitrogen. The pellets were thawed at 37°C and suspended in 4 ml of ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 14 mM β-mercaptoethanol, 1% TX-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml pepstatin A, pH 8.0), sonicated four times for 30 s each, and centrifuged at 50,000 rpm for 30 min in a TLA 100.3 rotor. The cleared lysate was collected and incubated with 0.25 ml of nickel-nitrilotriacetic acid agarose beads (Invitrogen) overnight at 4°C. The beads were washed with 30 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, and 14 mM β-mercaptoethanol, pH 8.0), then they were eluted in elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). After dialysis against HK buffer at 4°C, the proteins were aliquoted and frozen.
Binding Assays
Each assay was carried out with a 15-μl volume of packed glutathione-agarose beads coated with ∼5 μg of GST fusion protein. The beads were incubated with 1 ml of HK buffer containing 1 mg/ml bovine serum albumin (BSA) at 4°C for 1 h, collected, and then incubated with rotation with ∼1.5 μg of purified hexahistidine-tagged proteins in 30 μl of HK buffer for 90 min at 4°C. The beads were washed four times with 0.5 ml of HK buffer containing 0.5% TX-100 with a 1-min incubation on ice during each wash, and then the bound material was analyzed by immunoblotting. Antibodies and their dilutions used for immunoblotting were rabbit anti-His at 1:1000 (Bethyl Laboratories, Montgomery, TX), peroxidase-conjugated anti-rabbit at 1:5000 (Bio-Rad, Hercules, CA).
Permeabilized Cell Golgi Biogenesis Assay
NRK cells at 70% confluence on 12-mm coverslips were treated with 2.5 μg/ml brefeldin A (BFA) (Sigma-Aldrich) for 30 min followed by 50 μM H89 (Toronto Research Chemicals, North York, ON, Canada) for 10 min. The cells were then washed three times on ice with 0.5 ml of cold DMEM and two times with 0.5 ml of cold KOAc buffer (115 mM KOAc, 2.5 mM MgOAc, 25 mM HEPES, pH 7.2, and 1 mM dithiothreitol) containing 0.9 mM CaCl2 and 0.5 mM MgCl2. The washed cells were incubated in 50 μl of 0.4 μg/ml streptolycin O in KOAc buffer containing 0.1 mg/ml BSA on ice for 25 min followed by two washes with 0.5 ml of KTM buffer (KOAc buffer containing 2 mM K2CaEGTA, pH 7.4, 2 mM EGTA-KOH, pH 7.4, and 1 mM dithiothreitol). The cells were incubated with 0.5 ml of KTM buffer on ice for 5 min and permeabilized by incubation in KTM buffer containing 50 μM H89 at 37°C for 15 min. After permeabilization, the cells were washed two times with cold KTM buffer, and then they were incubated with KTM on ice for 10 min to extract the cytosol components followed by three washes with cold KTM buffer on ice. The coverslips were incubated at 37°C for 50 min in 50 μl of KTM buffer containing 10 mg/ml rabbit liver cytosol, an ATP regeneration system (0.5 mM ATP, 0.5 mM UTP, 50 μM GTP, 5 mM creatine phosphate, 25 μg/ml creatine phosphokinase, 0.05 mM EGTA, and 0.5 mM MgCl2), and 40 μg/ml green fluorescent protein. Where indicated, the incubation mix also contained 0.2 mg/ml βCOP-C, 0.2 mg/ml βCOP-C-FW>AA, 0.1 mg/ml p115-N, or 0.1 mg/ml p115-N-E>K. The coverslips were then washed two times with 0.5 ml of KTM buffer, fixed and stained as described above.
RESULTS
βCOP Knockdown Phenotype
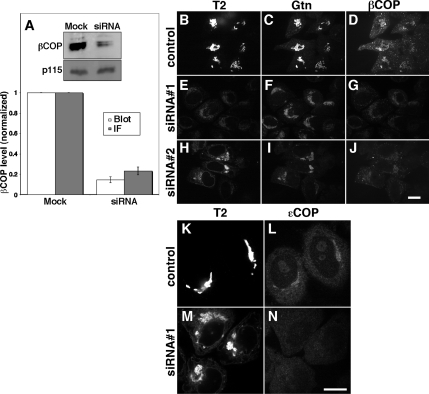
HeLa cells stably expressing N-acetylgalactosaminyl transferase-2 tagged with the green fluorescent protein (GalNAcT2-GFP) were treated with either of two βCOP siRNAs. Approximately 80% knockdown of βCOP expression was routinely achieved as indicated by quantification of immunoblots and by determining the βCOP fluorescence level across the cell population (Figure 1A). Although a complete knockdown of βCOP was not evident, the εCOP subunit of the COPI coat complex was dispersed in the βCOP knockdown cells, indicating that COPI assembly was significantly inhibited (Figure 1, K–N). Furthermore, and most critical to our analysis, the knockdown cells exhibited a striking Golgi phenotype. In contrast to the typical Golgi ribbon in control cells (Figure 1, B–D), cells with inhibited COPI assembly due to βCOP knockdown showed GalNAcT2-GFP redistributed to the ER and also to juxtanuclear-aggregated structures that were positive for the Golgi protein giantin (Figure 1, E–G). Two other golgins, GM130 (unpublished) and golgin-97 (see below), were also prominent in the juxtanuclear Golgi “remnants.” As evidence against nonspecific effects, each siRNA yielded the same phenotype (Figure 1, H–J), and the phenotype was rescued by expression of a siRNA-insensitive wild-type βCOP replacement construct (see below). Expression level of sec13, p115, giantin, and GalNAcT2-GFP were unaffected by βCOP knockdown (unpublished). Consistent with its partial accumulation in the ER in an immaturely glycosylated state, >50% of the GalNAcT2-GFP exhibited an increased mobility by SDS-polyacrylamide gel electrophoresis after βCOP knockdown. Thus, redistribution of GalNAcT2-GFP to the ER and to juxtanuclear Golgi remnants was a bone fide βCOP knockdown phenotype, and it was used to identify βCOP knockdown cells in subsequent morphological analyses.
Figure 1.
βCOP knockdown. (A) HeLa cells expressing GalNAcT2-GFP, grown in 35-mm dishes also containing a coverslip, were either mock transfected or transfected with βCOP siRNA. Total cell extracts (20 μg/lane) were analyzed by immunoblotting by using anti-βCOP, and, as a loading control, anti-p115 antibodies. The coverslips were analyzed by immunofluorescence by using anti-βCOP antibody. Quantified βCOP levels determined by immunoblot (Blot: mean βCOP/p115 ± SD; n = 2) and immunofluorescence (IF: mean ± SEM; >15 cells each) are presented after normalization to the mock-transfected cells. (B–J) HeLa cells expressing GalNAcT2-GFP were mock transfected (B–D) or transfected with either of two siRNAs against βCOP (E–J). The GalNAcT2-GFP, giantin (Gtn) and βCOP distributions were visualized 48 h later. Bar, 10 μm. (K–N) HeLa cells expressing GalNAcT2-GFP were mock transfected (K and L) or transfected with siRNA against βCOP (M and N) and GalNAcT2-GFP and eCOP distributions were visualized 48 h later. Bar, 10 μm.
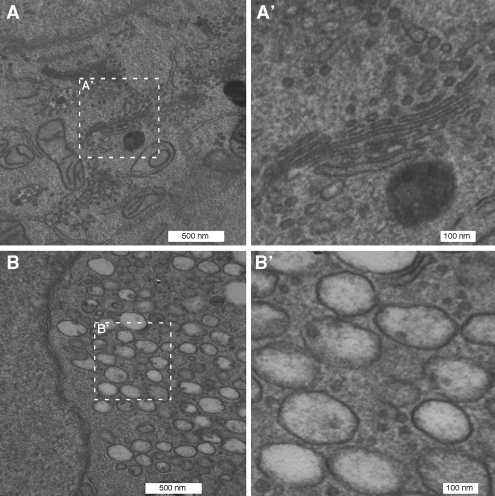
Ultrastructural analysis of the βCOP knockdown cells also revealed a clear and corresponding phenotype. In contrast to the stacked membranes comprising the Golgi ribbon in normal cells (Figure 2A), there were no detectable stacks in cells after βCOP knockdown. Instead, the juxtanuclear area of these cells exhibited a striking accumulation of aggregated membranes exhibiting diameters of ∼250 nm (Figure 2B).
Figure 2.
Absence of Golgi stacks. Mock (A) and siRNA (B) transfected GalNAcT2-GFP HeLa cells were processed for electron microscopy. Outlined regions are shown at higher magnifications in the adjacent panels as indicated (A′ and B′). Note the striking juxtanuclear accumulation of aggregated membranes exhibiting diameters of ∼250 nm in βCOP knockdown cells.
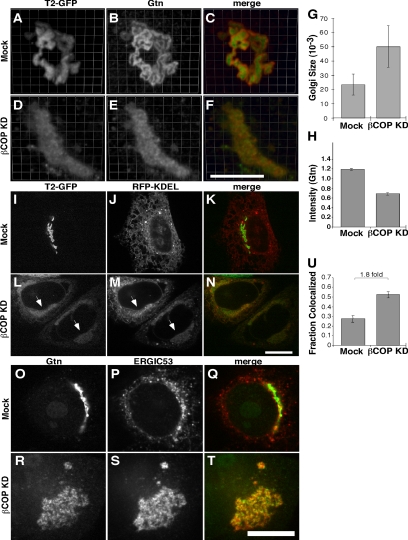
To characterize these remnant structures in terms of marker distribution, we analyzed apparent size, staining intensity and colocalization using immunofluorescence. Three-dimensional rendering of confocal sections highlighted the ribbon-like appearance of Golgi protein localization in control cells (Figure 3, A–C) and the collapsed, aggregated appearance in knockdown cells (Figure 3, D–F). The size of the remnants seemed larger and the intensity of the Golgi marker staining in these remnants seemed weaker. Indeed, this was supported by quantification of the apparent volume and staining intensity (Figure 3, G and H). Using giantin to mark the Golgi, the average volume of the fluorescent objects was double that of the controls and the mean pixel value was reduced by nearly half.
Figure 3.
Analysis of Golgi remnant structures after βCOP knockdown. (A–F) Mock- (A–C) or βCOP siRNA (D–F)-treated cells were processed to reveal GalNAcT2-GFP and giantin (Gtn) staining in juxtanuclear Golgi/remnants with 3D rendering. The merged images have GalNAcT2-GFP in green and giantin in red. Bar, 5 μm. (G) Golgi/remnant fluorescent size was determined in mock and βCOP knockdown cells (mean ± SD; >15 cells each). Size was total volume occupied by above-threshold fluorescence of giantin and showed a significant increase in βCOP knockdown cells. (H) Quantified giantin staining intensity for mock and βCOP knockdown cells is shown (n = 2, mean ± SD; >15 cells/condition/experiment). Intensity is the normalized mean pixel value in the Golgi/remnant. (I–N) HeLa cells stably expressing GalNAcT2-GFP were either mock (I–K) or βCOP siRNA (L–N) transfected, and, after 24 h, retransfected with a plasmid encoding KDEL-RFP. After another 24-h incubation, the cells were processed to reveal GalNAcT2-GFP and KDEL-RFP. The merged images have GalNAcT2-GFP in green and KDEL-RFP in red. Note the accumulation of KDEL-RFP in the juxtanuclear remnants induced by βCOP knockdown (arrows). (O–U) Mock- (O–Q) and βCOP (R–T) siRNA-transfected cells were costained using immunofluorescence to compare colocalization between giantin and ERGIC53. Colocalization was quantified (U) by determining the average of fraction of each marker's area that was also occupied by the other marker, where area is determined by above threshold fluorescence and corresponds to a Golgi/remnant structure (n = 2, mean ± SD; >15 cells each). Bar, 10 μm.
The apparent volume increase and reduction of staining intensity exhibited by Golgi markers in knockdown cells may reflect defects in membrane retrieval to the ER. Thus, we tested whether the remnants might contain ER proteins failing to undergo efficient retrieval. For this purpose, the cells were transfected with KDEL-tagged red fluorescent protein (Snapp et al., 2006). As expected, in control cells the construct was ER-localized and absent from post-ER structures (Figure 3, I–K). In contrast, after βCOP knockdown there was a clear accumulation of the construct in the juxtanuclear remnants (Figure 3, L–N). Although an effect on the ER cannot be excluded, it seemed relatively normal as indicated by the remaining amounts of KDEL-RFP present in the ER. Maintenance of normal ER structure was also supported by other markers showing ER staining in these cells, including GalNAcT2-GFP, which, outside of the remnants, colocalized with KDEL-RFP in a characteristic ER network pattern (Figure 3, L–N).
Costaining indicated an increase in Golgi marker colocalization after βCOP knockdown (Figure 3, A–F). Indeed, the cis-Golgi marker GPP130 (Linstedt et al., 1997) yielded a significant level of side-by-side staining with the predominately medial/trans-marker GalNAcT2-GFP (Rottger et al., 1998), and this separation collapsed to colocalized staining after βCOP knockdown (Supplemental Figure S1, A and B). This finding was supported by quantification in which the fraction of total marker area showing colocalization was determined (Supplemental Figure S1C). The same result was obtained when giantin, which marks the rim region of cis-cisternae, was compared with trans-Golgi markers golgin-97 or TGN46 (Supplemental Figure S1, D–I). In all cases examined, βCOP knockdown led to a 1.5- to 1.7-fold increase in colocalization. For comparison, we also analyzed Golgi marker colocalization in nocodazole and BFA-treated cells (Supplemental Figure S1, J–L). In BFA-treated cells, golgin and GRASP proteins are present in noncompartmentalized remnant structures, whereas in nocodazole-treated cells, they are in compartmentalized ministacks. As expected, colocalization between giantin and GRASP65 in nocodazole-treated cells was nearly identical to that in untreated control cells, whereas colocalization was significantly increased in BFA-treated cells. Indeed, the 1.6-fold increase in colocalization in the noncompartmentalized BFA remnants was equal in magnitude to the level of increase in βCOP knockdown cells. Interestingly, the IC marker ERGIC53, which is normally prominent in peripheral punctate structures as it cycles between the ER and Golgi, was also affected. After βCOP knockdown, ERGIC53 accumulated in the juxtanuclear Golgi remnants where it colocalized with giantin (Figure 3, O–U). This was not due to a collapse in the positioning of ER exit sites as staining for the COPII subunit sec13 yielded the normal peripheral distribution (unpublished). Furthermore, ER exit persisted because ts045-VSVG moved from the ER to the remnants upon 40 to 32°C temperature shift (unpublished). Thus, impaired COPI function causes accumulation in remnant structures of ER proteins targeted by retrieval, recycling IC markers, secretory cargo, and Golgi proteins, most strikingly the golgins.
The βCOP Appendage FW Motif Is Required for Golgi Biogenesis
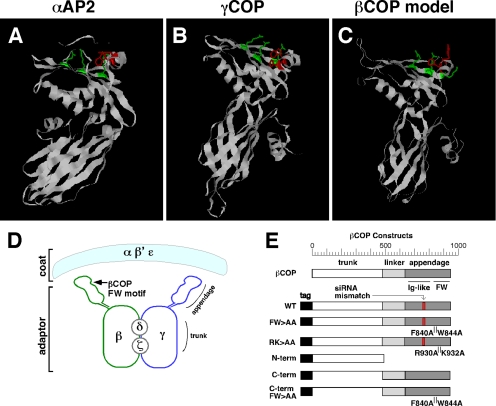
The structures of the AP2 α subunit and the COP1 γ subunit are strikingly similar (Figure 4, A and B; from Hoffman et al., 2003; Watson et al., 2004). Based on the sequence similarity of βCOP to γCOP, we used ModBase (Pieper et al., 2006) to generate a model of the βCOP C terminus (Figure 4C). This model suggests that the conserved FW residues (red) in βCOP lie on the surface exposed α helix of the appendage domain, and they are flanked by three basic amino acids (green) similar to αAP2 and γCOP (Figure 4, A and B). Therefore, the βCOP FW motif is likely to form an appendage domain binding pocket arranged on an adaptor complex analogous to AP2 (Figure 4D) and interfering with interactions at this site could dissect COPI function.
Figure 4.
Computational model of βCOP C terminus. (A–C) Shown are the determined structures for the C-termini of αAP2 and γCOP and the computed model for the analogous C terminus of βCOP. The βCOP model was generated using ModBase (Pieper et al., 2006) and based on βCOP sequence homology to γCOP. The position of the conserved FW residues is indicated in red. They lie on a surface exposed α-helix of the appendage domain, and they are flanked by three basic residues shown in green. (D) Schematic diagram of the COPI complex based on homology to AP2 (modified from Hoffman et al., 2003). (E) Schematic diagram of the βCOP constructs used in this study (numbering refers to amino acids).
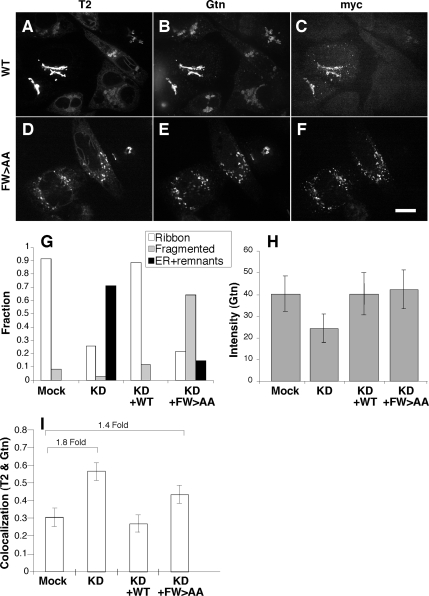
To assess the role of the FW motif, we generated myc-tagged βCOP replacement constructs corresponding to wild-type βCOP and a version with alanines substituted for F840 and W844 (Figure 4E). Mismatches were present at the siRNA target site to render the constructs insensitive to knockdown. Expression of the wild-type βCOP replacement construct was detected using myc staining, and it was found to restore normal Golgi morphology based on GalNAcT2-GFP and giantin staining patterns (Figure 5, A–C). Quantification indicated that >70% of the nonexpressing cells exhibited the ER/remnant phenotype, whereas ∼90% of the cells expressing the βCOP replacement construct showed normal Golgi ribbons (Figure 5G). This demonstrates that the knockdown phenotype is a specific consequence of βCOP inhibition and provides an assay for comparison with the FW mutant. Significantly, the FW>AA replacement construct yielded a phenotype distinct from both the knockdown and rescue phenotypes, despite its equivalent expression and its apparent proper targeting to Golgi membranes (Figure 5, D–F). In cells expressing FW>AA the Golgi proteins GalNAcT2-GFP and giantin were largely in peripheral punctate structures. The quantified results confirmed the conversion of the ER/remnant phenotype to the peripheral punctate pattern as being distinct from the complete rescue by the wild-type construct (Figure 5G). Based on these results, we determined Golgi protein intensity and colocalization in the peripheral structures after rescue with the FW>AA mutant. Giantin staining intensity, which is reduced by nearly half in remnant structures, was restored to control levels by both the FW>AA and wild-type βCOP constructs (Figure 5H). Furthermore, ER localization of RFP-KDEL was restored by expression of the FW>AA construct with no evidence of RFP-KDEL fluorescence in the peripheral punctate structures (unpublished). In contrast, analysis of marker colocalization indicated that, unlike the wild-type construct, the FW>AA construct failed to restore normal levels of marker separation between GalNAcT2-GFP and giantin (Figure 5I, 1.4-fold higher than controls) or GalNAcT2-GFP and GPP130 (Supplemental Figure S5, 1.4-fold higher than controls). Transport of ts045-VSVG was also tested in βCOP knockdown cells expressing the FW>AA replacement construct. In contrast to control cells, which exhibited clear surface staining 60 min after the 40 to 32°C temperature shift, cells expressing FW>AA failed to yield surface staining (Supplemental Figure S2). Thus, the βCOP FW binding pocket has a functional role in anterograde traffic and in biogenesis of the Golgi apparatus.
Figure 5.
βCOP appendage FW motif is required for Golgi biogenesis. (A–F) βCOP siRNA-transfected cells were retransfected after 24 h with plasmid encoding myc-tagged rat βCOP WT (A–C) or the FW>AA mutant (D–F). After an additional 24 h the cells were processed to assess Golgi integrity, by using GalNAcT2-GFP and giantin (Gtn) staining, and construct expression, by using myc-epitope staining. (G) Golgi integrity in mock, knockdown (KD) cells, and myc-positive knockdown cells (KD+WT or KD+FW>AA) was then scored as the fraction of cells showing normal Golgi ribbons (Ribbon), dispersed punctate structures (Fragmented), and juxtanuclear aggregated structure accompanied by partially ER-localized GalNAcT2-GFP (ER+remnants). Note that wild-type βCOP restored ribbon structure, whereas βCOP FW>AA yielded an IC pattern. (H–I) Quantification of giantin fluorescence intensity and giantin versus GalNAcT2-GFP colocalization was carried as described in the Figure 3 legend (mean ± SD; >15 cells each). Whereas wild-type βCOP restored marker intensity and separation, βCOP FW>AA only restored intensity.
The βCOP FW Binding Pocket Interacts with the N Terminus of the Vesicle Tether p115
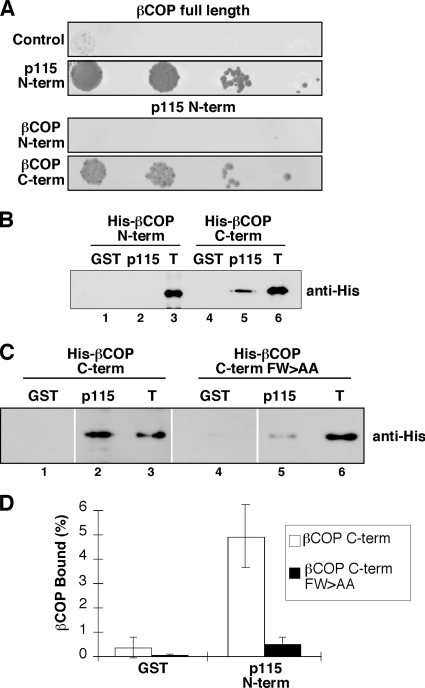
The fragmented Golgi structure, disrupted Golgi marker separation, and impaired anterograde trafficking observed in the βCOP knockdown cells expressing FW>AA are similar to the phenotype we observe in cells after p115 knockdown (Puthenveedu and Linstedt, 2004; Puthenveedu et al., 2006; see below), suggesting that p115 could be a regulatory factor that depends on an interaction with the βCOP FW motif. Indeed, evidence that p115 might interact with βCOP is present in the results of a proteomic human protein interaction screen (Rual et al., 2005). Furthermore, we observed that the p115 N terminus specifically interacted with βCOP in a yeast two-hybrid test and that this interaction involved the βCOP C terminus, which contains the appendage domain and not the βCOP N terminus (Figure 6A). Also, βCOP coimmunoprecipitated with p115 from HeLa cell lysates (Supplemental Figure S3). The interaction seemed direct because a purified, hexahistidine-tagged, C-terminal fragment of βCOP bound a purified N-terminal fragment of p115, whereas equal amounts of an N-terminal βCOP fragment did not (Figure 6B). Importantly, this interaction was significantly inhibited by introduction of the FW>AA substitution into the βCOP C-terminal fragment (Figure 6C). The 90% reduced binding level was comparable to background (Figure 6D).
Figure 6.
βCOP FW motif binds p115. (A) Yeast two-hybrid assay growth results for full-length βCOP (as bait) tested against a vector only control and the p115 N terminus. The p115 N terminus (as bait) was also tested against the βCOP N-terminal and C-terminal constructs diagrammed in Figure 4E. (B) Equivalent amounts (≈5 μg) of bead-bound GST and GST-p115-N were incubated with 1.5 μg of purified hexahistidine-tagged βCOP N-terminal and C-terminal constructs diagrammed in Figure 4E. Recovery of the βCOP constructs was determined by immunoblotting with an anti-histidine epitope antibody and compared with the 0.15 μg of signal representing 10% of total (T). (C) Equivalent amounts (≈5 μg) of bead-bound GST and GST-p115-N were incubated with 1.5 μg of purified hexahistidine-tagged βCOP C-terminal constructs with and without the FW>AA mutation as diagrammed in Figure 4E. Recovery of the βCOP constructs was determined by immunoblotting with an anti-histidine epitope antibody and compared with the 0.075-μg signal representing 5% of total (T). (D) The results showing a requirement for the FW motif in the βCOP–p115 interaction were quantified to yield the bound βCOP as a percentage of total (mean ± SD; n = 2).
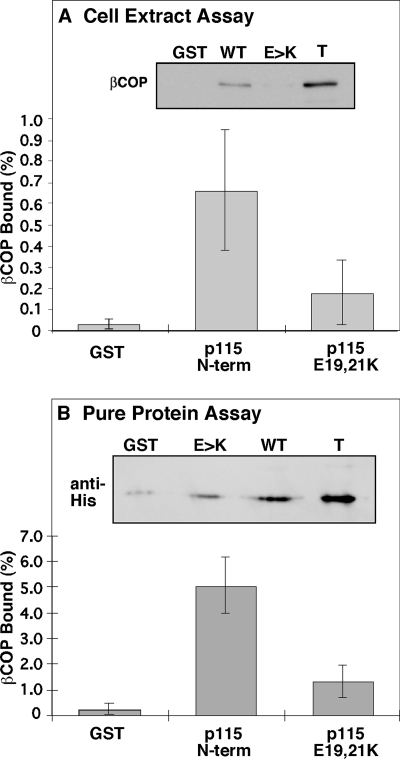
The p115 N terminus contains a conspicuous conserved region termed the homology region 1 (HR1) of unknown function (Sapperstein et al., 1995). HR1 contains a glutamic acid residue at position 21, and there is another at position 19. These were of interest because the βCOP FW motif is flanked by a cluster of positively charged residues that are not only also present in the AP2 α subunit (Figure 4, A–C) but also known to be functionally required for interactions within the AP2α FW binding pocket. Thus, we tested whether the p115 E19 and E21 residues were important for salt-bridge interactions with βCOP by changing the p115 residues to lysines. The GST-tagged p115 N terminus, with and without lysine substitutions, was purified and bound to glutathione agarose beads. The proteins were soluble and produced in high yields. After incubation with cytosolic extracts, recovery of βCOP was determined by immunoblot. Remarkably, βCOP was recovered with the wild-type p115 fragment but failed to interact with E19,21K, the lysine-substituted version (Figure 7A). Because βCOP is present assembled into the coatomer complex in cytosolic extracts (Waters et al., 1991), this result suggests that p115 interacted with assembled βCOP. To test the role of p115 E19 and E21 residues in direct binding to βCOP, the purified protein assay was also used. As expected, the lysine substitutions significantly interfered with the binding of p115 to the purified βCOP C terminus (Figure 7B). Thus, the interaction between p115 and βCOP involves E19 and E21 in p115 and F840 and W844 in the βCOP appendage domain. It remains to be tested whether this is actually an ionic interaction between the acidic p115 residues and the basic residues flanking the βCOP FW motif, because our focus here is the functional significance of the interaction.
Figure 7.
p115 E19E21 binds βCOP. (A) Equivalent amounts (≈5 μg) of bead-bound GST, GST-p115-N, and GST-p115-N with the E19,21K mutation were incubated with rabbit liver cytosol. Recovery of bound βCOP was determined by immunoblotting. Quantification presents the fraction bound as a percentage of total (mean ± SD; n = 3). (B) The purified hexahistidine-tagged βCOP C terminus (1.5 μg) was incubated with glutathione beads bearing ≈5 μg of GST, GST-p115-N, or GST-p115-N with the E19,21K mutation. Bound protein was determined by immunoblotting with the anti-histidine epitope antibody and compared with 0.075-μg signal representing 5% of total. The bound as a percentage of total is shown (mean ± SD; n = 3).
Golgi Biogenesis Requires Interacting Domains in βCOP and p115
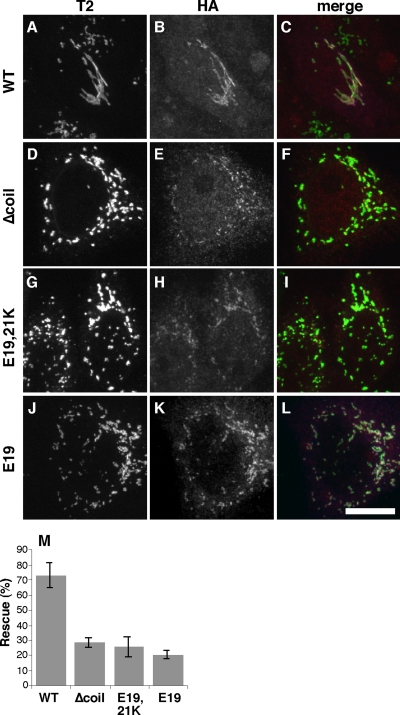
To test the functional significance of the lysine substitutions in p115 that disrupted binding to βCOP, we carried out gene replacement after p115 knockdown. As positive and negative controls, respectively, we used wild-type p115 and a version of p115 lacking the first coiled-coil domain, Δcoil1. Consistent with our previous work (Puthenveedu and Linstedt, 2004), expression of wild-type p115 rescued the knockdown phenotype and expression of Δcoil1, which lacks a mapped SNARE-binding domain, failed to rescue (Figure 8, A–F). Significantly, expression of the p115 E19,21K full-length replacement construct failed to rescue the phenotype (Figure 8, G–I). The p115 E19,21K expression level matched that of the wild-type protein, and p115 E19,21K was properly localized to the fragmented Golgi membranes, suggesting that the altered residues did not prevent p115 folding and targeting. Proper targeting yet failure of phenotype rescue was also the case for a full-length replacement construct bearing a single lysine substitution of E19 (Figure 8, J–L). Quantification of the results confirmed their statistical significance (Figure 8M).
Figure 8.
p115 E19E21 is required for Golgi biogenesis. (A–L) GalNAcT2-GFP expressing HeLa cells were transfected with p115 siRNA and after 72 h, the cells were microinjected with plasmids encoding hemagglutinin (HA)-tagged bovine p115 with sequences corresponding to wild-type (A–C), Δcoil1 (D–F), E19,21K (G–I), or E19K (J–L). After 5 h, the cells were fixed and stained with anti-HA antibody to detect microinjected cells, and GalNAcT2-GFP was visualized to determine Golgi integrity. (M) The results were quantified by determining percentage of HA-epitope–positive cells with an intact Golgi (n = 3, mean ± SD; >15 cells/experiment/condition).
Sequence alignment indicates that E21 is more highly conserved than E19, suggesting that E21 is actually more critical than E19. Further work is necessary to test this idea, but it is noteworthy that the E to K charge inversion at position 19 may have disrupted a salt-bridge interaction with βCOP involving p115 E21. Consistent with the involvement of an ionic interaction, there is a basic cluster in the βCOP FW platform (Figure 4C). To test its role, we generated a version of the βCOP replacement construct in which R930 and K932 within this cluster were mutated to alanines. Significantly, βCOP knockdown cells expressing the βCOP RK>AA replacement construct yielded a similar fragmented Golgi phenotype (Supplemental Figure S4).
Interestingly, analysis of Golgi marker staining intensity and colocalization in p115 knockdown cells indicated that both parameters matched those in βCOP knockdown cells expressing the FW>AA replacement construct. That is, GPP130 and GalNAcT2-GFP were strongly present and markedly colocalized in peripheral punctate structures (Supplemental Figure S5). For the p115 knockdown, this finding is consistent with previous work (Puthenveedu and Linstedt, 2001, 2004; Puthenveedu et al., 2006), and it is further supported by the fact that we did not detect stacked cisternae by EM after p115 knockdown (unpublished). Although some degree of membrane stacking could have been missed, indeed ministacks are evident in one report (Sohda et al., 2005), it is clear that Golgi biogenesis is disrupted by p115 knockdown to yield peripheral punctate structures that by objective, quantitative, fluorescent marker analysis bear remarkable similarity to those present after rescue of βCOP knockdown by FW>AA. Thus, mutations in βCOP that inhibit p115 binding block βCOP function in vivo and mutations in p115 that disrupt βCOP binding block p115 function in vivo, and, in both cases, the mutated proteins yield apparently identical phenotypes.
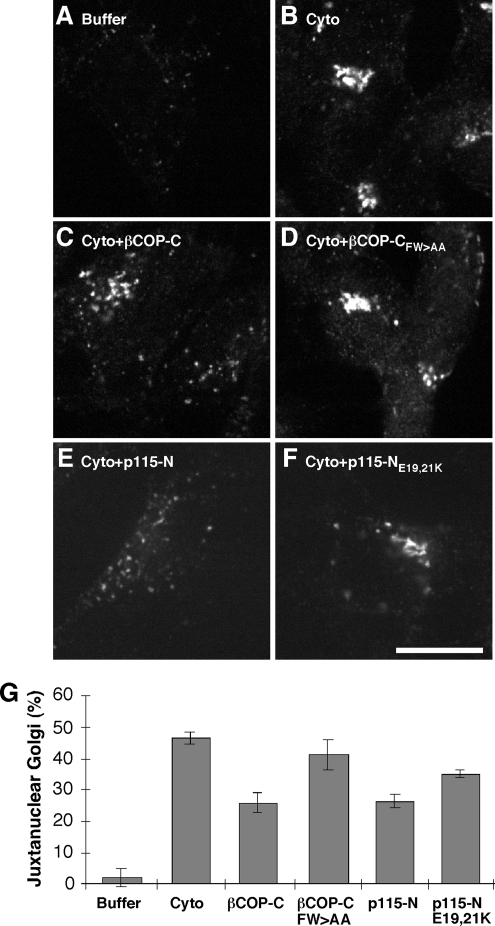
To extend these results, we used a permeabilized cell assay to measure Golgi biogenesis out of the ER. In intact cells, the Golgi apparatus efficiently and synchronously reassembles from the ER upon drug washout after sequential treatment with brefeldin A, to inhibit Arf1, and H89, to block COPII assembly (Puri and Linstedt, 2003). NRK cells were treated to collapse the Golgi into the ER, permeabilized with streptolysin O, salt-washed, and then incubated with buffer or cytosol. Incubations also contained purified GFP, to identify permeabilized cells, and an ATP-regenerating system. Whereas buffer alone yielded no Golgi assembly, cytosol addition promoted obvious Golgi assembly in 45% of the permeabilized cells (Figure 9, A, B, and G). Importantly, a significant inhibition of this assembly was observed when the cytosol incubation was carried out in the presence of 0.2 mg/ml (3.4 μM) purified C-terminal fragment of βCOP (Figure 9, C and G). In contrast, addition of βCOP FW>AA at the same concentration had no effect on the assay (Figure 9, D and G). Furthermore, addition of the purified N-terminal fragment of p115 also blocked Golgi assembly (Figure 9, E and G), and introduction of the E19,21K substitution, which blocks βCOP binding, reduced this inhibitory effect (Figure 9, F and G). The residual inhibition by p115-NE19,21K may be due to its ability to bind other factors, such as the COG complex (Sohda et al., 2007).
Figure 9.
In vitro Golgi assembly. (A–F) NRK cells were sequentially treated with BFA and H89 to collapse the Golgi into the ER, permeabilized with streptolysin-O, washed with salt solution, and incubated in reaction mix for 50 min at 37°C. The reaction mix contained buffer, 40 μg/ml GFP, and an ATP-regenerating system in the absence (A) or presence of cytosol (B) supplemented with either 0.2 mg/ml purified βCOP-C (C), 0.2 mg/ml βCOP-CFW>AA (D), 0.1 mg/ml p115-N (E), or 0.1 mg/ml p115-NE19,21K (F). After incubation, the cells were fixed and stained with anti-giantin antibody to assay Golgi assembly, and GFP was visualized to confirm permeabilization. (G) The results were quantified by determining percentage of GFP-positive cells with a juxtanuclear Golgi (n = 2; mean ± SD; >15 cells/experiment/condition).
Arrest of Golgi assembly in the presence of the βCOP-C and p115-N inhibitors resulted in accumulation of peripheral punctate structures reminiscent of the phenotype in intact cells in which the p115-βCOP interaction was disrupted by mutation of p115 or mutation of the βCOP FW binding pocket. Therefore, we tested whether inhibiting the βCOP–p115 interaction in the permeabilized assay also blocked marker separation. As expected, the juxtanuclear Golgi structures formed in the absence of added peptide (unpublished) or in the presence of either of the point mutated control peptides yielded side-by-side staining, rather than colocalization, of the Golgi markers giantin and GRASP65 (Supplemental Figure S6, A–C and G–I). Quantification indicated that the degree of colocalization was similar to that present in intact cells (Supplemental Figure S6M). In contrast, the peripheral punctuate structures in permeabilized cells treated with the inhibitory peptides showed significantly higher marker colocalization (Supplemental Figure S6, D–F and J–L). The overlap in staining of giantin and GRASP65 in the inhibited permeabilized cells, 1.4-fold higher than the control Golgi ribbon, was nearly identical to that evident in brefeldin A-treated intact cells and in striking contrast to the 0.9-fold level of colocalization in nocodazole-treated intact cells (compare Supplemental Figure S1L and S6M), supporting the conclusion that the βCOP-C and p115-N inhibitors blocked Golgi marker separation.
DISCUSSION
Analysis of divergent βCOP sequences in light of the solved crystal structures for αAP2, βAP2, and γCOP points to the presence of an appendage domain in βCOP and a conserved FW motif binding pocket in this domain. Our finding that the FW motif sequence is essential for Golgi biogenesis demonstrates a further degree of structural and functional conservation between AP2 and COPI. Furthermore, the AP2 FW binding pocket recruits transport factors, and we show that the βCOP FW binding pocket binds the vesicle tether p115. Significantly, interactions between coat complexes and docking factors can, in principle, increase specificity of trafficking by coupling sorting to fusion (Cai et al., 2007a). Thus our findings provide an important mammalian example of this emerging mechanism. Furthermore, our work identifies the functionally required E19E21 site in the p115 head domain, and it strongly suggests that COP1 is a key, although not necessarily exclusive, effector acting at this site because charge substitutions block both βCOP binding and Golgi biogenesis and because point mutations in βCOP that block p115 binding also block Golgi biogenesis yielding the same apparent phenotype. Evidence for the role of the interaction is not only provided through the in vivo phenocopy arising from the interaction-disrupting point mutations in βCOP and p115 but also by the inhibitors in the permeabilized assay that target the interaction and yield a similar block in Golgi biogenesis.
There is some controversy concerning whether p115 functions in the COPI or the COPII pathway. Compelling evidence indicates that Uso1p, the yeast homologue of p115, docks COPII vesicles (Cao et al., 1998) and also that abrogated Uso1p function has effects on COPII-mediated sorting (Morsomme and Riezman, 2002). However, the identity of the Uso1p interactors and its mechanism of docking remain unknown. In mammalian cells, p115 is recruited by rab1 during COPII assembly (Allan et al., 2000) and inhibition of p115 interferes with ER-to-Golgi transport (Alvarez et al., 1999; Allan et al., 2000; Puthenveedu and Linstedt, 2004). Also, p115 is required in vitro for IC formation (Bentley et al., 2006). In contrast, in the absence of p115, IC-like and/or ministack structures form and trafficking out of the ER into these structures (and even beyond, but with dramatically reduced efficiency) is observed (Alvarez et al., 1999; Kondylis and Rabouille, 2003; Puthenveedu and Linstedt, 2004; Sohda et al., 2005). Furthermore, p115 is required for in vitro intra-Golgi traffic of ts045-VSVG mediated by COPI vesicles (Waters et al., 1992) and p115 stimulates binding of COPI-coated vesicles to Golgi membranes (Sonnichsen et al., 1998). Our finding that p115 interacts with COPI in a manner functionally critical for Golgi biogenesis provides further strong support for the role of p115 in the COPI pathway.
The mechanism of p115 action remains unclear. It has been implicated in docking vesicles to acceptor compartments and in regulating SNARE function. p115 has a multitude of known interactions. Its overall shape is similar to myosin II in that it forms a homodimer with two large N-terminal head domains linked by a long coiled-coil tail. The head domain has two required interactions: E19E21 binding to COPI as reported here and HR2 binding to the COG docking complex (Sohda et al., 2007). The coiled-coil I domain, which most strongly interacts with a set of ER–Golgi SNARE proteins (Shorter et al., 2002), is also required for Golgi biogenesis (Puthenveedu and Linstedt, 2004). In contrast, the extreme C terminus, which is highly acidic and binds both giantin and GM130 (Linstedt et al., 2000), is dispensable (Puthenveedu and Linstedt, 2004). As suggested above, the p115–βCOP interaction likely serves to recruit p115 to COPI vesicles as they form. This may be the sole function of the interaction, but, interestingly, binding of the p115 head domain to the appendage of the COPI adaptor complex could position the coiled-coil domain so that it projects out from the vesicle and remains available for additional interactions. Thus, recruitment to the vesicle may be an initial step followed by subsequent interactions that still depend on binding to COPI and thus must occur before vesicle uncoating. Recent work revealed an interaction between the COPII coat and the TRAPP docking complex that underlies COPII vesicle docking, suggesting that coats are involved in docking and therefore, that docking can precede, rather than always follow, vesicle uncoating (Cai et al., 2007b). It follows that the p115–βCOP interaction may be at the core of a tethering complex in which p115 links vesicles to their target by simultaneously binding the COPI coat on the vesicle and a syntaxin5-based t-SNARE complex on the target membrane (Figure 10). Besides p115, other tethers have been shown to interact with COPI coats such as the aforementioned, ER-localized Dsl1 complex, the cis-Golgi–localized COG complex, and the trans-Golgi/early endosome-localized TRAPPII complex (Cai et al., 2007a). Structural and functional analysis of these interactions could reveal the mechanism governing specificity of targeting of COPI vesicles in the various COPI transport pathways.
Figure 10.
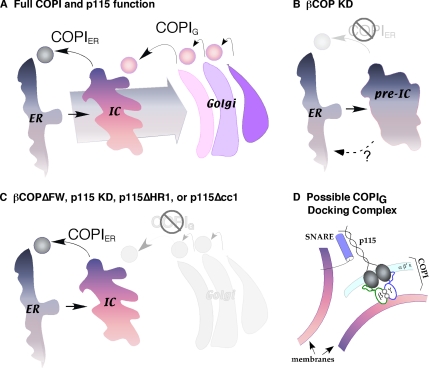
Model depicting COPIER and COPIG pathways. (A) Two COPI retrieval pathways are diagrammed. The COPIER pathway, which is p115-independent, functions to concentrate Golgi proteins via ER retrieval of non-Golgi proteins. The COPIG pathway, which is dependent on p115 binding to βCOP, functions to compartmentalize Golgi proteins via local retrieval between Golgi cisternae and from the Golgi to the IC. (B) In βCOP knockdown cells, both COPIER and COPIG are blocked, but COPII vesicle formation and fusion continues. This induces accumulation of pre-ICs contaminated with ER resident proteins and other proteins that fail to recycle. In the absence of recycling, Golgi proteins fail to concentrate. (C) Inhibition of the p115-βCOP interaction leaves the COPIG pathway blocked, whereas COPIER functions. Retrieval by COPIER allows concentration of Golgi proteins in ICs, but the membranes fail to compartmentalize without cycles of COPIG retrieval. (D) A hypothetical COPIG sorting and tethering complex is diagramed. Recruitment of p115 into COPI vesicles occurs when the E19E21 motif in p115 head domain binds the βCOP appendage FW domain. Before vesicle uncoating, this interaction could also dock the vesicles when the p115 coiled-coil domain simultaneously binds a SNARE complex on the target membrane.
In our present and past work, we used fluorescent image analysis to distinguish between Golgi phenotypes after knockdown and rescue. Interestingly, there were obvious shared features exhibited by βCOP knockdown cells rescued with βCOP FW>AA and p115 knockdown cells and in permeabilized cells treated with p115-N-term or βCOP-C-term inhibitors. Specifically, Golgi markers were in distributed punctate structures that exhibited signal intensities similar to control cells but increased Golgi marker colocalization. These phenotypes differed from that for βCOP knockdown in which Golgi markers were in juxtanuclear remnants and lacked normal signal intensity. A straightforward interpretation is that these results reveal the presence of two COPI-dependent steps: COPIER and COPIG (Figure 10). COPIER is p115-independent and retrieves cycling proteins and escaped ER residents, thereby concentrating Golgi proteins in post-ER membranes. COPIG requires p115 and mediates intra-Golgi retrieval thereby compartmentalizing Golgi proteins. In the absence of COPIER function, Golgi proteins leave the ER and accumulate in structures we term “pre-IC” because they likely represent the products of COPII vesicle budding and fusion that have not yet undergone retrieval of cycling and contaminating ER proteins. In the absence of COPIG function, retrieval to the ER by COPIER allows formation of an IC containing concentrated Golgi enzymes, but these membranes fail to compartmentalize without iterative cycles of COPIG budding and fusion.
The juxtanuclear positioning of the remnants formed when COPI function is impaired by βCOP knockdown is interesting because brefeldin A-induced inactivation of Arf1, which is required for COPI assembly (Donaldson et al., 1992; Helms and Rothman, 1992), leads to accumulation of Golgi remnants adjacent to peripheral ER exit sites (Ward et al., 2001). Arf1 has multiple effectors (Godi et al., 1998; Fucini et al., 2000, 2002), so this difference is likely the consequence of brefeldin A inhibiting a larger set of reactions than βCOP knockdown. Indeed, brefeldin A caused redistribution of the Golgi components present in the pre-ICs of βCOP knockdown cells to peripheral locations just as it does to Golgi components in control cells (unpublished). Thus, Arf1 may play a role in Golgi positioning that is independent of its role in COPI assembly. Note that ER exit sites remained in a peripheral location after βCOP knockdown, so positioning of the remnants is not simply a function of ER exit site collapse. Nevertheless, the juxtanuclear positioning of the remnants may not be an entirely active process because the remnants were largely insensitive to microtubule depolymerization by nocodazole (unpublished). Perhaps it is the positioning of remnants at peripheral exit sites that is active (Appenzeller-Herzog and Hauri, 2006) and, in the absence of this activity, the large size of the remnants leads to their juxtanuclear accumulation simply due to space considerations. Regardless, the insensitivity to nocodazole, which was applied for 3 h, is consistent with impaired recycling of Golgi components to the ER in the absence of COPI function because nocodazole is thought to induce Golgi-to-ER trafficking of Golgi components followed by their reemergence at peripheral ER exit sites (Cole et al., 1996; Storrie et al., 1998; but see Pecot and Malhotra, 2006). It is also interesting that partial restoration of COPI, i.e., when βCOP FW>AA is expressed, yields positioning of Golgi components at peripheral ER exit sites. We speculate that maturation of the IC membranes by COPIG is required to fully recruit, activate, or both the microtubule motor cytoplasmic dynein and hence the ICs fail to move inward.
Given that COPI mediates multiple membrane trafficking steps and that p115 has been implicated in vesicle docking, our identification of a p115 binding site in the COPI coat suggests that p115 might confer specificity in targeting for a particular COPI trafficking pathway (Malsam et al., 2005). This view is supported by the difference between the βCOP knockdown phenotype and the phenotype resulting from blocking the p115/βCOP interaction. p115 recruitment to COPI vesicles might be spatially restricted by a dependence on interaction of p115 with one of its multiple Golgi-localized interacting partners (Nakamura et al., 1997; Nelson et al., 1998; Lesa et al., 2000; Linstedt et al., 2000; Shorter et al., 2002). If so, this reaction could be favored at the Golgi, as opposed to the IC, and conditions at the IC would favor recruitment of a distinct set of targeting factors acting to target COPIER vesicles to the ER. Indeed, the Dsl1 complex, which is implicated in docking COPIER vesicles at the ER, binds COPI although not through βCOP (Andag et al., 2001; Reilly et al., 2001; Andag and Schmitt, 2003).
In summary, these experiments describe the βCOP knockdown phenotype, reveal a role for the βCOP FW motif, identify an interaction at this motif with p115, map the binding site in p115, and show a role for this p115 site in Golgi biogenesis. The disparity in COPI phenotypes indicates FW-dependent and -independent roles for COPI and the βCOP–p115 interaction suggest a mechanism for p115 sorting into COPI and possibly docking.
Supplementary Material
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1236) on April 23, 2008.
REFERENCES
- Allan B. B., Moyer B. D., Balch W. E. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Alvarez C., Fujita H., Hubbard A., Sztul E. ER to Golgi transport: requirement for p115 at a pre-Golgi VTC stage. J. Cell Biol. 1999;147:1205–1222. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andag U., Neumann T., Schmitt H. D. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- Andag U., Schmitt H. D. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Hauri H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Bentley M., Liang Y., Mullen K., Xu D., Sztul E., Hay J. C. SNARE status regulates tether recruitment and function in homotypic COPII vesicle fusion. J. Biol. Chem. 2006;281:38825–38833. doi: 10.1074/jbc.M606044200. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Sollner T. H., Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007a;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cai H., Yu S., Menon S., Cai Y., Lazarova D., Fu C., Reinisch K., Hay J. C., Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007b;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- Cao X., Ballew N., Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N. B., Sciaky N., Marotta A., Song J., Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRegis C. J., Rahl P. B., Hoffman G. R., Cerione R. A., Collins R. N. Mutational analysis of betaCOP (Sec26p) identifies an appendage domain critical for function. BMC Cell Biol. 2008;9:3. doi: 10.1186/1471-2121-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Finazzi D., Klausner R. D. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Eugster A., Frigerio G., Dale M., Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini R. V., Chen J. L., Sharma C., Kessels M. M., Stamnes M. Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol. Biol. Cell. 2002;13:621–631. doi: 10.1091/mbc.01-11-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini R. V., Navarrete A., Vadakkan C., Lacomis L., Erdjument-Bromage H., Tempst P., Stamnes M. Activated ADP-ribosylation factor assembles distinct pools of actin on Golgi membranes. J. Biol. Chem. 2000;275:18824–18829. doi: 10.1074/jbc.M000024200. [DOI] [PubMed] [Google Scholar]
- Glick B. S., Elston T., Oster G. A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 1997;414:177–181. doi: 10.1016/s0014-5793(97)00984-8. [DOI] [PubMed] [Google Scholar]
- Godi A., et al. ADP ribosylation factor regulates spectrin binding to the Golgi complex. Proc. Natl. Acad. Sci. USA. 1998;95:8607–8612. doi: 10.1073/pnas.95.15.8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Linstedt A. D. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J. Cell Biol. 2006;174:53–63. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J. B., Rothman J. E. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., Tagaya M. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G. R., Rahl P. B., Collins R. N., Cerione R. A. Conserved structural motifs in intracellular trafficking pathways: structure of the gammaCOP appendage domain. Mol. Cell. 2003;12:615–625. doi: 10.1016/j.molcel.2003.08.002. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois L., Lyles D. S. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology. 1982;121:168–174. doi: 10.1016/0042-6822(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Lesa G. M., Seemann J., Shorter J., Vandekerckhove J., Warren G. The amino-terminal domain of the golgi protein giantin interacts directly with the vesicle-tethering protein p115. J. Biol. Chem. 2000;275:2831–2836. doi: 10.1074/jbc.275.4.2831. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D., Jesch S. A., Mehta A., Lee T. H., Garcia-Mata R., Nelson D. S., Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J. Biol. Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D., Mehta A., Suhan J., Reggio H., Hauri H. P. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell. 1997;8:1073–1087. doi: 10.1091/mbc.8.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J., Satoh A., Pelletier L., Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- Martinez-Menarguez J. A., Geuze H. J., Slot J. W., Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- Morsomme P., Riezman H. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell. 2002;2:307–317. doi: 10.1016/s1534-5807(02)00133-8. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nelson D. S., Alvarez C., Gao Y. S., Garcia-Mata R., Fialkowski E., Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Vallis Y., Noble M. E., Hunter J. B., Dafforn T. R., Evans P. R., McMahon H. T. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Vallis Y., Pearse B. M., McMahon H. T., Evans P. R. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., Varlamov O., Paz K., McNew J. A., Hurtado D., Sollner T. H., Rothman J. E. Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl. Acad. Sci. USA. 2002;99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot M. Y., Malhotra V. The Golgi apparatus maintains its organization independent of the endoplasmic reticulum. Mol. Biol. Cell. 2006;17:5372–5380. doi: 10.1091/mbc.E06-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U., et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Puri S., Linstedt A. D. Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1253–1256. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Subcompartmentalizing the Golgi apparatus. Curr. Opin. Cell Biol. 2005;17:369–375. doi: 10.1016/j.ceb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Reilly B. A., Kraynack B. A., VanRheenen S. M., Waters M. G. Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol. Biol. Cell. 2001;12:3783–3796. doi: 10.1091/mbc.12.12.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S., White J., Wandall H. H., Olivo J. C., Stark A., Bennett E. P., Whitehouse C., Berger E. G., Clausen H., Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J. Cell Sci. 1998;111:45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- Rual J. F., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc. Natl. Acad. Sci. USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A., Fransen J. A., Matter K., Kreis T. E., Ginsel L., Hauri H. P. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur. J. Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- Sevier C. S., Weisz O. A., Davis M., Machamer C. E. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell. 2000;11:13–22. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. L., Sharma A., Lippincott-Schwartz J., Hegde R. S. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Ogata S., Sakisaka S., Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8:270–284. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Sakisaka S., Ogata S., Fujimoto J., Kiyokawa N., Ikehara Y. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem. Biophys. Res. Commun. 2005;338:1268–1274. doi: 10.1016/j.bbrc.2005.10.084. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., Lowe M., Levine T., Jamsa E., Dirac-Svejstrup B., Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., White J., Rottger S., Stelzer E. H., Suganuma T., Nilsson T. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M., Downs M. A., Westrich J. L., Fremont D. H. Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl. Acad. Sci. USA. 1999;96:8907–8912. doi: 10.1073/pnas.96.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrheenen S. M., Reilly B. A., Chamberlain S. J., Waters M. G. Dsl1p, an essential protein required for membrane traffic at the endoplasmic reticulum/Golgi interface in yeast. Traffic. 2001;2:212–231. doi: 10.1034/j.1600-0854.2001.020307.x. [DOI] [PubMed] [Google Scholar]
- Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K., Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Clary D. O., Rothman J. E. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Serafini T., Rothman J. E. ‘Coatomer’: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- Watson P. J., Frigerio G., Collins B. M., Duden R., Owen D. J. Gamma-COP appendage domain–structure and function. Traffic. 2004;5:79–88. doi: 10.1111/j.1600-0854.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- Xu Y., Martin S., James D. E., Hong W. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol. Biol. Cell. 2002;13:3493–3507. doi: 10.1091/mbc.E02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.