Abstract
The subcellular distribution of kinases and other signaling proteins is regulated in response to cellular cues; however, the extent of this regulation has not been investigated for any gene set in any organism. Here, we present a systematic analysis of protein kinases in the budding yeast, screening for differential localization during filamentous growth. Filamentous growth is an important stress response involving mitogen-activated protein kinase and cAMP-dependent protein kinase signaling modules, wherein yeast cells form interconnected and elongated chains. Because standard strains of yeast are nonfilamentous, we constructed a unique set of 125 kinase-yellow fluorescent protein chimeras in the filamentous Σ1278b strain for this study. In total, we identified six cytoplasmic kinases (Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p) that localize predominantly to the nucleus during filamentous growth. These kinases form part of an interdependent, localization-based regulatory network: deletion of each individual kinase, or loss of kinase activity, disrupts the nuclear translocation of at least two other kinases. In particular, this study highlights a previously unknown function for the kinase Ksp1p, indicating the essentiality of its nuclear translocation during yeast filamentous growth. Thus, the localization of Ksp1p and the other kinases identified here is tightly controlled during filamentous growth, representing an overlooked regulatory component of this stress response.
INTRODUCTION
In eukaryotes, protein function is regulated through mechanisms controlling transcription, translation, post-translational modification, protein degradation, and subcellular localization. In recent years, global studies, systematic studies, or both have been used to consider the majority of these regulatory mechanisms across a wide set of genes and proteins. DNA microarray technologies (DeRisi et al., 1997; Gasch et al., 2001) and mass spectrometry-based approaches (Gygi et al., 1999; Tang et al., 2005; Roth et al., 2006) have cataloged genome-wide changes in transcriptional levels, protein abundance, and posttranslational modifications; however, our understanding of regulated protein localization remains cursory, constructed piecemeal from individual reports of a given protein whose function is regulated by its localization.
Protein localization has been investigated most intensely in Saccharomyces. cerevisiae (Kumar et al., 2002; Huh et al., 2003), and reports of regulated protein localization have surfaced frequently in yeast-based studies. For example, several yeast proteins, such as the G1 cyclins Cln2p and Cln3p, are regulated by differential compartmentalization during cell cycle progression (Edgington and Futcher, 2001). The transcription factor Pho4p, involved in phosphate metabolism, is predominantly cytoplasmic under conditions of phosphate sufficiency, but it localizes to the nucleus during phosphate starvation (O'Neill et al., 1996). Components of the yeast Slt2p mitogen-activated protein kinase (MAPK) cell wall integrity pathway localize at sites of polarized growth in response to mating factor (Mazzoni et al., 1993; Buehrer and Errede, 1997). Collectively, from these and other studies (Shimada et al., 2000), we infer that a sizable protein complement may be regulated by differential localization in yeast, with protein kinases constituting one group particularly subject to such regulation.
Protein kinases play a prominent role in many developmental processes, and filamentous differentiation in yeast provides a strong example. In certain strains of yeast (e.g., Σ1278b), MAPK and cAMP-dependent protein kinase (PKA) pathways mediate a stress-induced transition to a multicellular, filamentous growth form (Gimeno and Fink, 1994; Kron, 1997; Cullen and Sprague, 2000; Erdman and Snyder, 2001). Specifically, nitrogen stress or growth in the presence of short chain alcohols initiates a developmental program characterized by the formation of filamentous chains of cells, called pseudohyphae (Lorenz et al., 2000). During filamentous growth, yeast cells delay in G2/M, exhibit an elongated morphology, display an altered budding pattern, remain physically attached after cytokinesis, and invade their growth substrate (Gimeno et al., 1992; Kron, 1997; Madhani and Fink, 1998). These morphological and genetic changes are brought about through signaling pathways encompassing the Kss1p MAPK cascade (Ste11p, Ste7p, and Kss1p) and PKA (Liu et al., 1993; Pan et al., 2000). In yeast, PKA consists of the regulatory subunit Bcy1p and the catalytic subunit isoforms Tpk1p, Tpk2p, and Tpk3p (Robertson and Fink, 1998). As yeast cells undergo filamentous growth, these kinase-based signaling modules function with additional genes and pathways governing cell polarity, bud site selection, and cell cycle progression (Chandarlapaty and Errede, 1998; Madhani et al., 1999; Miled et al., 2001). Extensive regulatory mechanisms are in place to coordinate signaling pathways during filamentous growth (Gimeno and Fink, 1994; Borneman et al., 2006), and the subcellular distribution of yeast kinases is likely controlled as part of this regulation.
To consider the degree to which protein localization is regulated during eukaryotic cell growth, and, specifically, the extent to which this contributes to the filamentous growth response, we screened all protein kinases in the budding yeast for differential localization during filamentous growth. This analysis revealed six kinases localized evenly across the cell during vegetative growth but localized predominantly in the nucleus under conditions of filamentous growth. Through localization-based epistasis studies, we found that the kinases form part of an interdependent network of regulated protein localization—the first such network identified in any eukaryote. Our results indicate a “regulatory/subordinate” relationship among kinases within this subnetwork; by using deletion mutants and kinase-dead alleles, we show that kinase translocation, in many cases, requires the presence/activity of another kinase. In addition, this study implicates the functionally uncharacterized Ser/Thr kinase Ksp1p in filamentous growth, while highlighting, in broader terms, the need to consider similar regulatory mechanisms in other eukaryotes.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The filamentous yeast strains Y825 and Y825/6 are derived from Σ1278b (Gimeno et al., 1992). The genotype of haploid Y825 is MATa ura3-52 leu2Δ0; the genotype of diploid Y825/6 is ura3-52/ura3-52 leu2Δ0/leu2Δ0. The nonfilamentous strain Y2269 is of the S288c genetic background, and it is a derivative of strain BY4743 (Giaever et al., 1999). Deletion mutants were constructed in strain Y825 by using a one-step polymerase chain reaction (PCR)-based gene disruption strategy (Baudin et al., 1993) with the G418 resistance cassette from plasmid pFA6a-KanMX6 (Longtine et al., 1998). The sks1-K39R kinase-dead allele was generated by site-directed mutagenesis by using oligonucleotides described in Yang and Bisson (1996). Kinase-dead alleles were obtained or generated on low-copy plasmids, except the ksp1-K47D allele, which is carried on pYEPs29K47D, described in Fleischmann et al. (1996).
Standard growth medium for microscopy was prepared using 0.17% yeast nitrogen base (YNB) without amino acids and ammonia, 2% glucose, and 5 mM ammonium sulfate (Guthrie and Fink, 1991). Haploid filamentous growth was induced in this standard medium supplemented with 1% (vol/vol) butanol (Lorenz et al., 2000) or on SLAD plates (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose, 50 μM ammonium sulfate, with essential amino acids) plus 1% (vol/vol) butanol (Gimeno et al., 1992; Lorenz et al., 2000). Diploid filamentous growth was induced in liquid low-nitrogen medium (0.17% YNB without amino acids and ammonia, 2% glucose, and 50 μM ammonium sulfate) (Gimeno et al., 1992). Invasive growth was assayed on YPD medium or synthetic complete (SC) −Ura as indicated previously (Guthrie and Fink, 1991; Lorenz et al., 2000).
Construction of Yeast Kinase-Fluorescent Protein Fusions
In this study, we constructed a Gateway-compatible (Invitrogen, Carlsbad, CA) yeast vector for the recombination-based cloning of promoter-gene cassettes as carboxy-terminal fusions to fluorescent protein (Walhout et al., 2000). This Gateway vector was constructed from the centromeric yeast shuttle vector YCp50 (Rose et al., 1987). To construct pDEST-vYFP, the Venus variant of yellow fluorescent protein (vYFP) (Nagai et al., 2002) was amplified by PCR from pBS7 (Yeast Resource Center, University of Washington, Seattle, WA) for introduction into the SphI-SalI site of YCp50. Subsequently, the YCp50 vector carrying vYFP was digested with SphI and made blunt with T4 DNA Polymerase (New England Biolabs, Ipswich, MA). Gateway cassette A (Invitrogen) was ligated with the blunt-ended vector, and EcoRI was used to identify the orientation of the cassette.
In total, we cloned 119 kinase genes into pDEST-vYFP. Briefly, we amplified by PCR each open reading frame along with 1 kb of upstream genomic DNA for introduction into pDEST-vYFP; PCR primers contained phage lambda att sites, allowing for recombination with the att-containing destination vector according to protocols described in Gelperin et al. (2005). On cloning, the PCR product creates a translational fusion between the 3′ end of the gene and vYFP, with a 10-codon linker encoding NPAFLYKVVI. The six remaining yeast kinase genes (MEC1, HRK1, RIM15, TEL1, TOR1, and TOR2) proved difficult to clone, and they were instead chromosomally tagged at their 3-ends with vYFP in the filamentous strain Y825/6; the genes HRK1, RIM15, TEL1, and TOR1 were also chromosomally tagged in Y825 as well. Integrated alleles were generated by standard protocols by using the vYFP-KanMX6 cassette from pBS7 (Yeast Resource Center, University of Washington).
Live Cell Microscopy
Cells were grown overnight, diluted to an OD600 of roughly 0.1 and grown in standard medium or inducing conditions as required. Filamentous growth in haploid strains was induced by inoculating diluted cultures into standard growth medium supplemented with 1% butanol for 4 h at 30°C. Diploid filamentous strains were induced as follows: overnight cultures were centrifuged, washed, and inoculated at an OD600 of 0.1 into low-nitrogen medium at 30°C for 4 h before observation. Osmotic stress and heat stress were induced by growing diluted overnight cultures in standard growth medium to mid-log phase, followed by either 20 min growth in medium containing 0.4 M NaCl (Ferrigno et al., 1998) or by 20 min growth at 37°C (Ferrigno et al., 1998). 4,6-Diamidino-2-phenylindole (DAPI) was added at a final concentration of 2 μg/ml for 30 min to stain DNA, marking the nucleus and mitochondria.
Phenotypic Assays for Filamentous Growth Defects
Colony morphology of deletion mutants was observed by streaking mid-log cultures grown in YPD onto SLAD plates supplemented with 1% (vol/vol) butanol (Lorenz et al., 2000) and incubating at 30°C for 10 d. Invasive growth was determined by the standard plate-washing assay of Gimeno et al. (1992). For this assay, mid-log phase cultures were spotted onto YPD plates and incubated for 5 d at 30°C; surface cells were then washed off under a gentle stream of water.
RESULTS
Localization of Yeast Kinases during Filamentous Growth
The yeast proteome encompasses 125 protein kinases, defined from data sets deposited in the Saccharomyces Genome Database as of August 2006 (www.yeastgenome.org). For purposes of this analysis, we include the regulatory subunit of protein kinase A, Bcy1p. The yeast kinase complement, or kinome, is listed in Supplemental Figure SF1, along with relevant functions and protein localization data describing the subcellular distribution of each protein in nonfilamentous lab strains (e.g., S288c). To investigate the subcellular dynamics of these kinases during filamentous growth, we constructed a unique plasmid-based collection of kinase-fluorescent protein fusions. This plasmid-based approach is well suited for localization studies in nonstandard genetic backgrounds, such as in the filamentous strain Σ1278b; the Σ1278b genetic background is preferred for studies of filamentous growth (Gimeno et al., 1992), and existing reagents for protein localization studies are not available in this strain. As part of this study, we designed a centromeric yeast shuttle vector, pDEST-vYFP, for the expression of cloned genes as chimeric proteins in which vYFP is fused to the carboxy terminus of the target protein (Nagai et al., 2002). The pDEST-vYFP vector carries phage lambda att sites for recombination-based cloning, accommodating gene coding sequences along with 1 kb of upstream promoter sequence. By virtue of this vector, genes are expressed under control of their native promoters at nearly endogenous levels.
In total, we cloned 119 kinase genes into pDEST-vYFP and subsequently introduced the kinase-vYFP fusion into the filamentous strain Σ1278b by the approach outlined in Figure 1. For six kinases (MEC1, HRK1, RIM15, TEL1, TOR1, and TOR2), C-terminal fusions to vYFP were generated as integrated alleles in Σ1278b (described in Materials and Methods). The kinase-vYFP chimeras were screened by fluorescence microscopy for differential localization under vegetative and filamentous growth conditions in both haploid and diploid strains of Σ1278b, because the filamentous growth response differs according to ploidy (Gancedo, 2001). A full listing of protein kinase localizations under these conditions is presented in Supplemental Table ST1.
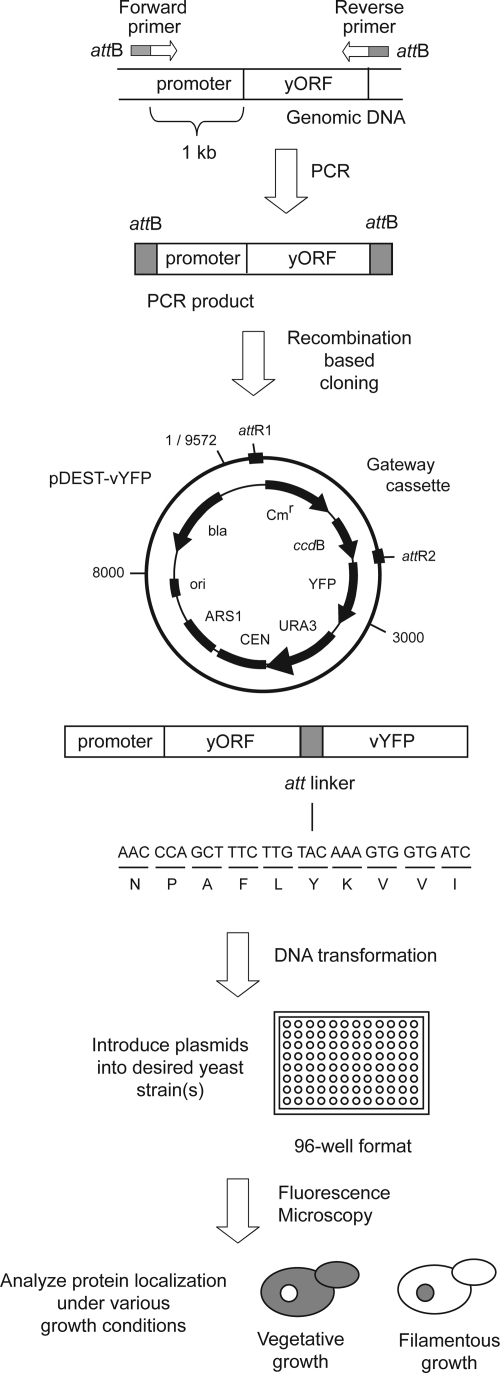
Figure 1.
Schematic overview of the construction and subsequent screening of carboxy-terminal YFP fusions. Primers for PCR amplification were designed with 5′-att sites such that resulting PCR products could be cloned into the pDEST-vYFP vector by phage λ-based recombination. The resulting gene fusion carries a 10-codon linker between the target open reading frame and YFP. Subsequent screening steps are as indicated, with a diagrammatic representation of a protein differentially localized to the nucleus under conditions of filamentous growth.
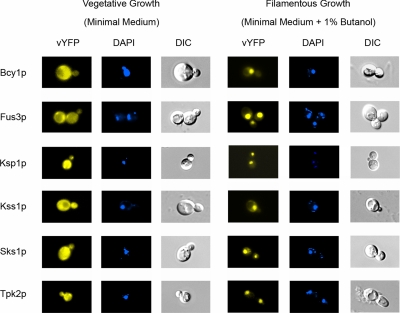
From this analysis, we found six kinases with altered subcellular distribution under filamentous growth conditions (Figure 2). The proteins Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p distributed evenly over the cell under conditions of vegetative growth, but they localized predominantly to the nucleus during filamentous growth. In each case, this nuclear shift was striking, with ∼80–95% of observed fluorescence concentrated within the nucleus. The differential localization of these kinases was evident in both haploid cells (shown in Figure 2) and diploid cells for Bcy1p, Fus3p, Kss1p, Sks1p, and Tpk2p; however, Ksp1p did not localize strongly to the nucleus during filamentous growth in diploid yeast.
Figure 2.
Differential localization of the protein kinases Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p. Kinase-vYFP fusions were visualized by fluorescence microscopy (left) under vegetative growth conditions and during filamentous growth. Yeast cells were stained with the DNA-binding dye DAPI (middle) to visualize the nucleus and mitochondria. The yeast cell shape and vacuoles were imaged by differential interference contrast (DIC) microscopy (right).
As mentioned previously, Bcy1p, Tpk2p, and Kss1p are known components of filamentous growth PKA and MAPK cascades, respectively. Fus3p is the MAPK mediating the yeast mating response (Elion et al., 1993; Choi et al., 1999), and Sks1p is a serine/threonine kinase involved in the cellular response to glucose limitation (Vagnoli and Bisson, 1998). The cellular function of Ksp1p is unknown, although its overexpression is known to suppress mutations in SRM1, a nucleotide exchange factor required for nucleocytoplasmic trafficking of macromolecules (Fleischmann et al., 1996).
To determine whether the observed localization shifts are specific to the yeast filamentous growth response, we further screened the six yeast kinases identified above for differential localization during osmotic stress (in 0.4 M NaCl) and heat stress in Σ1278b. Bcy1p is known to localize differentially during heat stress (Griffioen et al., 2001); however, the other kinase-vYFP fusions were distributed evenly over the cytoplasm and nucleus under the conditions tested. Thus, the observed nuclear shifts do not represent general stress responses, and they are likely specific to conditions inducing filamentous growth.
Phenotypic Analysis of Nuclear-localized Kinases
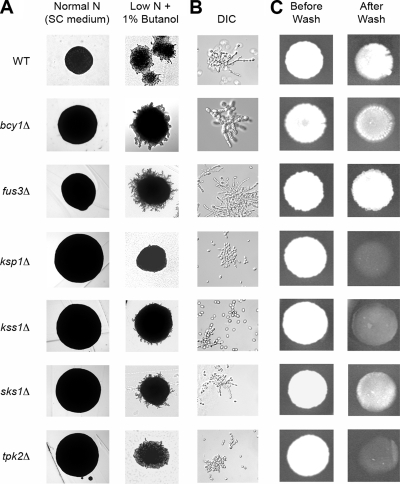
To clarify the potential roles of these six kinases in filamentous growth, we deleted each corresponding kinase gene in a haploid derivative of Σ1278b for phenotypic analysis of filamentation (Figure 3). Deletion mutants were screened for colony and cell morphologies, and invasive growth was assayed by the standard plate-washing assay of Gimeno et al. (1992). Filamentous growth phenotypes were consistent across all assays. Hyperfilamentous growth was evidenced by extended peripheral filamentation, elongated cell morphology, and increased invasive growth relative to wild type; the converse phenotypes were present in mutants defective in filamentous growth. In our assays, deletion of KSP1, KSS1, and TPK2 resulted in decreased filamentous growth, whereas the fus3Δ mutant was hyperfilamentous. Deletion of BCY1 and SKS1 did not affect filamentous growth. These phenotypic results are consistent with our understanding of filamentous growth functions associated with Bcy1p, Fus3p, Kss1p, and Tpk2p; however, Ksp1p had not been implicated previously in filamentous growth, and, in this context, the strong effect of Ksp1p on filamentous growth is particularly noteworthy.
Figure 3.
Phenotypic analysis of kinase deletion mutants in the filamentous Σ1278b genetic background. Each haploid deletion mutant was assayed for surface-spread filamentation (A), cell morphology (B), and invasive growth (C). Surface-spread filamentation was assayed on SLAD medium (see Materials and Methods) supplemented with 1% butanol. Cells from these colonies were inoculated into a small volume of water for DIC microscopy. Invasive growth was assayed on YPD medium as described in Materials and Methods. The Σ1278b background strain Y825 served as a wild-type control for these filamentous growth assays.
The kinase shifts reported here were identified using proteins tagged at their carboxy termini with vYFP; however, carboxy-terminal protein modifications can, in some cases, disrupt function. To consider the functionality of these kinase-vYFP chimeras, we assessed the ability of Fus3p-vYFP, Ksp1p-vYFP, Kss1p-vYFP, and Tpk2p-vYFP to complement corresponding deletion mutants for filamentous growth phenotypes. In each case, introduction of the kinase-vYFP fusion restored wild-type filamentous growth. The Bcy1p-vYFP chimera complemented the heat-sensitive phenotype of a bcy1Δ mutant, as assayed by the method described in Toda et al. (1987). The functionality of Sks1p-vYFP was not considered, because its null phenotype is not easily assayed.
Interdependent Kinase Translocation to the Nucleus during Filamentous Growth
The regulated localization of Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p during filamentous growth and the known interplay between kinases in signaling pathways led us to consider the possibility that the observed nuclear translocation of these kinases may be interdependent: the presence of one kinase may be required for the localization shift of another. To address this explicitly, we systematically examined the subcellular localization of these six kinases in mutant backgrounds individually deleted for one of the five other kinases. For example, the subcellular distribution of Bcy1p was examined under conditions of vegetative and filamentous growth in five haploid mutant backgrounds: fus3Δ, ksp1Δ, kss1Δ, sks1Δ, and tpk2Δ. The remaining five kinases were also examined accordingly, and the results from this study are presented in Figure 4.
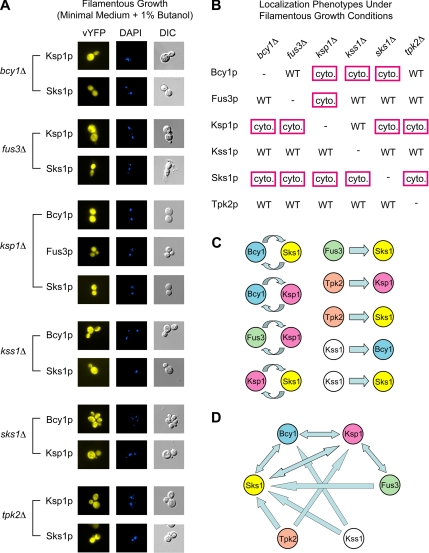
Figure 4.
Interdependent localization of yeast kinases during filamentous growth. (A) Each kinase-vYFP fusion was visualized by fluorescence microscopy under filamentous growth conditions in a haploid strain of Σ1278b deleted for the indicated kinase. DAPI-stained and DIC images are provided. Images of the kinase-vYFP fusions under vegetative growth conditions are presented in Supplemental Figure SF2. The mixed distribution of these kinases over the cytoplasm and nucleus signifies a loss of nuclear localization under filamentous growth conditions upon deletion of the indicated kinase gene. Only kinases exhibiting a loss of nuclear localization are shown here. (B) Matrix of kinase localizations in gene deletion backgrounds under conditions of filamentous growth. The results corresponding to the images shown in part A are boxed in red. Images of nuclear-localized kinase-vYFP fusions unaffected by the indicated gene deletions are presented in Supplemental Figure SF3. (C) Localization-based regulatory relationships among yeast kinases. Each forward pointed arrow indicates that the given kinase is required for the wild-type localization of the subordinate kinase; for example, Fus3p is required for the wild-type nuclear localization of Sks1p during filamentous growth. Reciprocal relationships between kinases are indicated by the circular arrows in the left column. (D) The network of regulated protein localization between Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p is illustrated here; arrows are drawn as described for C. Double-sided arrows indicate that the localization of the given kinase pair is reciprocally affected under filamentous growth conditions.
Interestingly, the nuclear shift observed for Bcy1p, Fus3p, Ksp1p, and Sks1p is dependent in each case upon the presence of at least one other kinase (Figure 4, A and B). Fus3p is distributed evenly across the cytoplasm and nucleus under conditions inducing filamentous growth in a haploid strain deleted for KSP1. The nuclear shift of Bcy1p requires the presence of Ksp1p, Kss1p, and Sks1p. The predominantly nuclear localization of Ksp1p during filamentous growth is lost in strains deleted for BCY1, FUS3, SKS1, or TPK2. The deletion of any kinase in this subset disrupts the nuclear shift of Sks1p under filamentous growth conditions. In contrast, the localization of Kss1p and Tpk2p during filamentous growth is unaffected by the deletion of these kinases.
Many kinases in this localized subgrouping exhibit reciprocal relationships (Figure 4C). The deletion of BCY1 affects the butanol-induced nuclear localization of Sks1p, and, reciprocally, deletion of SKS1 disrupts the nuclear shift of Bcy1p. We observed similar effects for the kinase pairs Bcy1p-Ksp1p, Fus3p-Ksp1p, and Ksp1p-Sks1p. Other kinase pairs, however, exhibit a unidirectional “regulatory/subordinate” relationship; for example, the presence of Fus3p is required for the nuclear shift of Sks1p, but SKS1 is not required for wild-type localization of Fus3p under conditions of filamentous growth. Similarly, Tpk2p contributes to the regulated localization of Ksp1p and Sks1p, but the reciprocal relationships are not evident. Kss1p also contributes to the butanol-induced nuclear shift of Bcy1p and Sks1p, although neither gene product affects the localization of Kss1p during filamentous growth.
Taken collectively, the six kinases identified here form part of an interdependent network of regulated protein localization during filamentous growth (Figure 4D). It is likely that additional proteins contribute to the regulated localization of these kinases; thus, the relationships indicated in Figure 4 represent a portion of a potentially larger network. In particular, our results highlight the key regulatory roles of Kss1p and Tpk2p; both proteins are required for the wild-type localization of members of this subnetwork (i.e., Bcy1p, Ksp1p, and Sks1p), but neither is affected by the presence of any other kinase tested here. Ksp1p plays a central role in this network, affecting the localization of Bcy1p, Fus3p, and Sks1p, while itself requiring the presence of Bcy1p, Fus3p, Sks1p, and Tpk2p for wild-type localization under conditions of filamentous growth. This is particularly interesting because Ksp1p had not been implicated previously in filamentous growth. Downstream or subordinate roles are also evident within this subnetwork; in particular, all five other kinases are required for the nuclear localization of Sks1p, whereas its presence is only required for the wild-type localization of Bcy1p and Ksp1p.
This localization-based interdependence suggests regulatory relationships that may be investigated through traditional epistasis studies with double-deletion mutants. For example, as outlined above, the localization of Ksp1p is affected upon deletion of BCY1, FUS3, SKS1, or TPK2; thus, we deleted each of these genes individually in a strain deleted for KSP1 to assess the impact of each mutation on the ksp1Δ filamentous growth phenotype. The results from this analysis are shown in Supplemental Figure SF4. The ksp1Δfus3Δ mutant exhibited exaggerated filamentous growth, as assessed by examination of colony morphology, cell morphology, and invasive growth; this mimics the phenotype of fus3Δ. As expected, the ksp1Δtpk2Δ double mutant shows no filamentous growth; however, deletion of BCY1 and SKS1, respectively, restores yeast filamentous growth in a ksp1Δ genetic background. At minimum, this indicates a filamentous growth effect associated with sks1Δ. It is interesting that the ksp1Δ phenotype is masked by the phenotype associated with the other gene deletion in each double mutant. From this, Ksp1p may serve an upstream role in filamentous growth pathways; however, it is very difficult to interpret these results, because we have no clear evidence that Ksp1p actually functions in a linear pathway with the PKA modules, MAPK modules, or both.
Kinase Activity Is Required for the Nuclear Translocation of Dependent Proteins
Kinases serve both structural and catalytic functions in signaling pathways, phosphorylating target proteins but also, in many cases, acting as scaffolds facilitating protein–protein interactions (Choi et al., 1994; Madhani and Fink, 1997). To specifically determine the role of kinase phosphorylation in controlling subordinate protein translocation to the nucleus, we examined the subcellular distribution of Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p in kinase-dead mutants of Fus3p (fus3-K42R), Ksp1p (ksp1-K47D), Kss1p (kss1-K42R), Sks1p (sks1-K39R), and Tpk2p (tpk2-K99R) (Fleischmann et al., 1996; Yang and Bisson, 1996; Madhani et al., 1997; Demlow and Fox, 2003; Zeitlinger et al., 2003); Bcy1p was omitted from this study because it does not possess a kinase domain. As indicated in Supplemental Figure SF5, each kinase-dead mutant exhibits filamentous growth phenotypes mirroring those observed upon gene deletion.
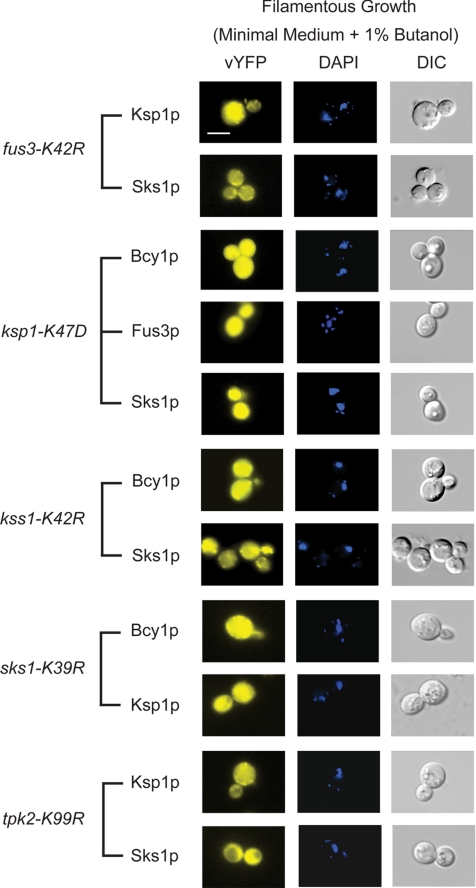
In this analysis, we individually introduced each kinase-dead allele into a haploid Σ1278b strain deleted for the corresponding kinase gene and analyzed the subcellular localization of kinases dependent upon this protein for nuclear translocation during filamentous growth (Figure 5). For example, the localization of Ksp1p and Sks1p was observed in a kinase-dead fus3-K42R mutant; the localization of Bcy1p, Fus3p, and Sks1p was assessed in the ksp1-K47D mutant, and so on. In each case, kinase activity was required for the observed localization shift. As shown in Figure 5, Ksp1p-vYFP and Sks1p-vYFP were evenly distributed across the nucleus and cytoplasm in a strain bearing the fus3-K42R allele under conditions of butanol-induced filamentous growth. Similar results were observed over the full panel of mutants tested. Thus, the kinase activity of the identified proteins is required for the nuclear translocation of dependent kinases. It should be noted that the required phosphorylation events may be indirect. Also, observed protein localization was not affected in any kinase-dead mutant under conditions of vegetative growth (Supplemental Figure SF6).
Figure 5.
The observed nuclear translocations are dependent upon the kinase activity of Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p, respectively. Each indicated kinase-vYFP fusion was imaged in a strain carrying the kinase-dead allele listed to the left. For example, the subcellular distribution of Ksp1p and Sks1p is shown in a haploid fus3Δ strain carrying the fus3-K42R kinase-dead allele on a low-copy plasmid. DAPI-stained and DIC images are as described previously. Note the mixed distribution of each kinase over the cytoplasm and nucleus upon loss of the indicated kinase activity. Images of these kinases in the kinase-dead background strains under vegetative growth conditions are presented in Supplemental Figure SF6. Bar, 3 μm.
Nuclear Translocation of Ksp1p Is Required for Wild-Type Filamentous Growth
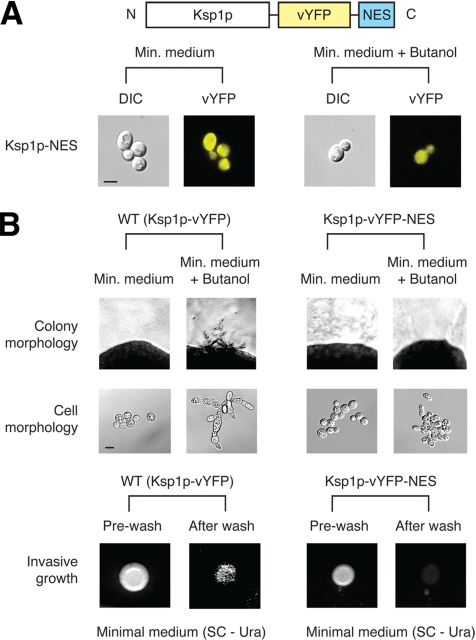
As evidenced by its null phenotype, Ksp1p is required for butanol-induced filamentous growth in haploid yeast; however, the importance of its observed localization shift during filamentous growth cannot be explicitly determined from this finding. To consider this point more directly, we generated a modified form of Ksp1p unable to translocate into the nucleus. Specifically, we fused a nuclear export sequence (NES) to the 3′ end of KSP1-vYFP, as illustrated in Figure 6A. The Ksp1p-vYFP-NES chimera was expressed from the native KSP1 promoter on the low-copy destination vector described in Figure 1. For this analysis, the NES was derived from the Rev protein of HIV-1 (Fritz et al., 1995), which has been used previously to direct nucleocytoplasmic trafficking in yeast (Murphy and Wente, 1996). Attachment of this NES prevented the nuclear shift of Ksp1p under conditions of butanol-induced filamentous growth (Figure 6A). Phenotypic analysis of this NES-tagged mutant revealed significantly reduced filamentous growth as compared with a haploid ksp1Δ strain expressing the Ksp1p-vYFP chimera: surface-spread filamentation and invasive growth was lost in the NES-tagged strain, and its cell morphology was rounded rather than elongated under conditions inducing filamentous growth (Figure 6B). The ksp1-K47D kinase-dead mutant is also defective in filamentous growth (Supplemental Figure SF5); therefore, the nuclear translocation of Ksp1p and its kinase activity are both required for wild-type filamentation, suggesting that Ksp1p likely phosphorylates one or more nuclear proteins during the filamentous growth response.
Figure 6.
Tagging Ksp1p with a NES abolishes filamentous growth. (A) Schematic diagram of the NES-tagged form of Ksp1p, where the NES (encoding LQLPPLERLTLD) is fused at the carboxy terminus of the Ksp1p-vYFP chimera. As visualized by fluorescence microscopy, addition of the NES prevents the butanol-induced nuclear translocation of Ksp1p. DIC and vYFP images are shown. Bar, 3 μm. (B) Phenotypic analysis of NES-tagged Ksp1p. For comparison, we have included a haploid ksp1Δ strain carrying a centromeric plasmid with the KSP1-vYFP fusion under transcriptional control of the native KSP1 promoter; this strain exhibits wild-type filamentous growth properties. Addition of the NES results in decreased filamentous growth, evidenced in a loss of surface-spread filamentation, rounded cell morphology, and decreased invasive growth. Bar, 3 μm.
DISCUSSION
From numerous individual examples, it is clear that the function of a protein can be controlled through mechanisms regulating its subcellular distribution; however, the extent to which this form of regulation occurs has not been investigated on a large scale. Accordingly, we undertook the first systematic analysis of differential protein localization for any protein set in any eukaryote, and by using the yeast kinome as the subject, we identified an interdependent subnetwork of proteins whose intracellular localization is tightly regulated during filamentous growth. This study, therefore, describes an underappreciated type of regulatory network, yields insight into the degree to which differential protein localization serves as a widespread regulatory mechanism, and also identifies the kinase Ksp1p as a new filamentous growth gene.
The plasmid collection described here constitutes a unique and versatile resource for the yeast scientific community, in complement to a very useful set of chromosomally integrated green fluorescent protein-fusions presented in Huh et al. (2003). Obviously, however, our plasmid collection is not without its limitations; carboxy-terminal fusions may perturb the functions of some proteins, and this is an important consideration in using these plasmids. For example, carboxy-terminal tagging is typically problematic in analyzing isoprenylated gene products and geranylgeranylated proteins (Bhattacharya et al., 1995), as well as proteins modified with palmitoyl and farnesyl groups (Sun et al., 2004; Roth et al., 2006). Although some proteins of the cell wall, endoplasmic reticulum, and peroxisomes may be adversely affected by carboxy-terminal modification, we do expect the majority of yeast proteins to function normally as C-terminal fluorescent protein fusions (Pelham et al., 1988). Plus, the alternative approach of amino-terminal tagging poses greater potential for protein mislocalization.
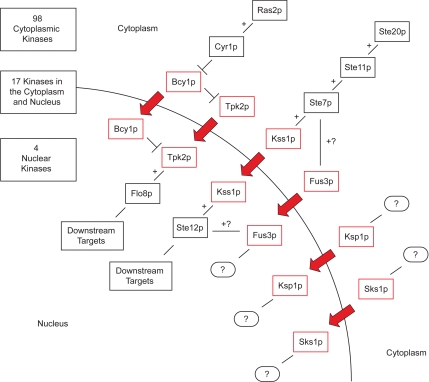
By live cell imaging of kinase-YFP chimeras, we found Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p differentially localized to the nucleus under filamentous growth conditions. These kinases have been characterized to varying degrees, and the pathway context of each is presented in Figure 7, along with a summary of our localization data. In many cases, the observed localization shifts can be reconciled easily with corresponding protein functions. Specifically, PKA phosphorylates nuclear proteins, such as the filamentous growth transcription factor Flo8p (Rupp et al., 1999), under filamentous growth conditions; the nuclear shift of Bcy1p and Tpk2p likely enables PKA to selectively phosphorylate nuclear-localized targets during filamentation. The filamentous growth MAPK Kss1p phosphorylates an incompletely defined set of nuclear proteins, including the transcriptional activator Ste12p. Kss1p does not shift its localization in response to mating factor (Ma et al., 1995), and this filamentous growth-induced shift may constitute one mechanism ensuring Kss1p signaling specificity.
Figure 7.
Subcellular localization of the yeast kinome during filamentous growth. This diagram summarizes the localization of 125 protein kinases, constituting the yeast kinome, under conditions of filamentous growth. In total, 119 kinases do not shift localization during the transition to filamentous growth, and the localization of these kinases is tallied in the top left. For simplicity, Yck3p, which localizes to the vacuole, is included in the cytoplasmic compartment. The six kinases that shift localization between vegetative and filamentous growth conditions are boxed in red, and the pathway context of each kinase is shown. Flo8p and Ste12p are included as representative targets of Tpk2p and Kss1p, respectively. Speculative and unknown interactions are indicated with question marks.
In regard to this study, it should be noted that filamentous growth can be induced by conditions of nitrogen deprivation as well as by growth in the presence of short-chain alcohols, such as butanol. Classically, pseudohyphal growth refers to a form of filamentous growth induced in diploid yeast by conditions of nitrogen deprivation on solid medium, wherein the yeast strain exhibits both surface-spread filamentation and invasive growth (Gimeno et al., 1992; Gancedo, 2001). Haploid strains of yeast undergo invasive growth on rich medium, but do not exhibit extensive surface-spread filamentation (Roberts and Fink, 1994). Growth in butanol can be used to induce filamentation in haploid and diploid yeast, yielding morphological properties resembling pseudohyphal growth, even in liquid medium (Lorenz et al., 2000). Compared with nitrogen deprivation, butanol induction involves several underlying genetic differences; for example, Lorenz et al. (2000) report that numerous upstream nutrient-sensing genes required for classic pseudohyphal growth are not required for butanol-induced filamentous growth. We, however, identified at least two of these genes (GPA2 and GPR1) in a disruption screen for genes essential in butanol-induced haploid filamentous growth (Jin et al., 2008); thus, further analysis will be required to understand the genetic basis of these induction mechanisms. To consider both induction schemes, in this study, we assayed kinase localizations in diploid yeast under conditions of nitrogen stress and in haploid yeast by growth in butanol. Furthermore, we have endeavored to make clear the growth conditions used in each study throughout this text.
As indicated from our data, Ksp1p represents an important new filamentous growth gene, because its deletion inhibits all characteristic filamentous growth landmarks in haploid yeast: cell elongation, surface-spread filamentation, and invasive growth. Furthermore, the localization shift of Ksp1p is required for filamentous growth, as is its kinase activity. This strongly suggests that Ksp1 phosphorylates one or more nuclear proteins as an essential step in the yeast filamentous growth response. At present, Ksp1p has no confirmed targets. In vitro phosphorylation studies using protein microarrays identify 187 putative substrates for Ksp1p (Ptacek et al., 2005); however, none of these putative substrates belong to known filamentous growth pathways. The nuclear shift of Ksp1p requires the presence of BCY1, TPK2, FUS3, and SKS1, and, at minimum, the kinase activity of Fus3p. Ksp1p is not an established target of these kinases, and the effect of Fus3p phosphorylation may be indirect. Interestingly, Ksp1p is a target for Hsf1p, Pho85p, and Pcl1p; hence, its predicted involvement in the cell cycle and in the yeast general stress response (Dephoure et al., 2005; Hashikawa et al., 2006). Because filamentous growth is coordinated with the cell cycle and general stress response machinery, Ksp1p may play an important role in the signaling link between these processes.
The nuclear shifts of Fus3p and Sks1p are surprising. Fus3p is phosphorylated by Ste7p and translocates to the nucleus in response to mating pheromone, where it phosphorylates Ste12p and the filamentous growth transcription factor Tec1p. In the latter case, phosphorylation of Tec1p targets it for degradation, thereby inhibiting the filamentous growth pathway during mating (Elion et al., 1993; Choi et al., 1999; Bao et al., 2004). During filamentous growth, the function of Fus3p in the nucleus is unclear, because it presumably cannot phosphorylate Tec1p under these conditions. Fus3p may still be phosphorylated by Ste7p, and it may still phosphorylate Ste12p during filamentous growth. Fus3p kinase activity is required for the nuclear translocation of Ksp1p and Sks1p during filamentation, suggesting that it possesses previously unappreciated roles in the yeast filamentous growth response. The role of Sks1p in filamentous growth is also unclear, because its known functions are apparently distinct from those mediating filamentous growth, and its deletion does not affect filamentation. In this context, kinase deletion phenotypes must be interpreted with caution, because kinases are known to engage in significant cross talk and compensatory activity (Madhani and Fink, 1997; McClean et al., 2007). Thus, the phenotype of a deleted kinase may be masked by activity from another kinase not normally functioning in a given process.
The observed localization shifts raise an interesting question regarding the mechanism by which these kinases translocate into the nucleus. Little has been reported describing nuclear localization signals (NLSs) in these proteins, and sequence analysis by the program PSORTII (Nakai and Horton, 1999) suggests the presence of a putative NLS in only Ksp1p. Thus, the remaining kinases are likely ferried into the nucleus through interaction with another protein. For example, under conditions of temperature stress in S288c, Bcy1p translocates from the nucleus to the cytoplasm through an interaction with the protein Zds1p (Griffioen et al., 2001). Similar interactions with other proteins may allow for the filamentous growth-induced translocation of the kinase subset reported here, although no obvious candidates are evident from interaction data sets.
It is tempting to extrapolate our findings over the yeast proteome as a whole; however, we expect that the kinome is particularly subject to this form of regulatory control and that the overall percentage of yeast proteins regulated by differential localization will be less than the 5% rate (6 of 125) observed here. We do expect the transcription factor complement in yeast to be similarly regulated by subcellular localization to a high degree, and, in complement to this study, it would be interesting to screen the full set of yeast transcription factors for differential localization during filamentous growth.
Collectively, this study presents the first large-scale analysis of differential protein localization in any eukaryote, and the kinase network identified here defines a previously overlooked mechanism of regulatory control during yeast cell growth and development. Conceptually, regulated protein localization provides a level of specificity to otherwise promiscuous kinase activities, and the network-based structure of this regulated localization confers greater specificity still. The implications of these findings extend beyond our understanding of yeast cell biology. Similar mechanisms are assuredly at play in higher eukaryotes as well, and subsequent investigations in higher organisms will clarify the degree to which these localization-based regulatory networks are evolutionarily conserved.
Supplementary Material
ACKNOWLEDGMENTS
We thank Damian Krysan (University of Rochester), Robert Fuller (University of Michigan), Anthony Borneman (Yale University), and Michael Snyder (Yale University) for providing filamentous yeast strains. We thank Hiten Madhani (University of California, San Francisco) for providing kinase-dead alleles of KSS1 and FUS3, Tom Fox (Cornell University) for providing the tpk2-K99R allele, and Markus Aebi (ETH Zurich) for providing the ksp1-K47D allele. This work was supported by grants RSG-06-179-01-MBC from the American Cancer Society, DBI 0543017 from the National Science Foundation, and Basil O'Connor Award 5-FY05-1224 from the March of Dimes (to A.K.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1199) on April 16, 2008.
REFERENCES
- Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R., Madhani H. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Chen L., Broach J. R., Powers S. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc. Natl. Acad. Sci. USA. 1995;92:2984–2988. doi: 10.1073/pnas.92.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer B. M., Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S., Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kranz J., Mahanty S., Park K., Elion E. Characterization of Fus3 localization: active Fus3 localizes in complexes of varying size and specific activity. Mol. Biol. Cell. 1999;5:1553–1568. doi: 10.1091/mbc.10.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Satterberg B., Lyons D. M., Elion E. Ste5 tethers multiple protein kinases in the MAPK cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Cullen P. J., Sprague G. F. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:13461–13463. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demlow C. M., Fox T. D. Activity of mitochondrially synthesized reporter proteins is lower than that of imported proteins and is increased by lowering cAMP in glucose-grown Saccharomyces cerevisiae cells. Genetics. 2003;165:961–974. doi: 10.1093/genetics/165.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Howson R. W., Blethrow J. D., Shokat K. M., O'Shea E. K. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc. Natl. Acad. Sci. USA. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Edgington N. P., Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- Elion E., Satterberg B., Kranz J. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;19:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann M., Stagljar I., Aebi M. Allele-specific suppression of a Saccharomyces cerevisiae prp20 mutation by overexpression of a nuclear serine/threonine protein kinase. Mol. Gen. Genet. 1996;250:614–625. doi: 10.1007/BF02174449. [DOI] [PubMed] [Google Scholar]
- Fritz C. C., Zapp M. L., Green M. R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Gancedo J. M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Huang M., Metzner S., Botstein D., Elledge S. J., Brown P. O. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D., et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Shoemaker D. D., Jones T. W., Liang H., Winzeler E. A., Astromoff A., Davis R. W. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Gimeno C. J., Fink G. R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Griffioen G., Branduardi P., Ballarini A., Anghileri P., Norbeck J., Baroni M. D., Ruis H. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 2001;21:511–523. doi: 10.1128/MCB.21.2.511-523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hashikawa N., Mizukami Y., Imazu H., Sakurai H. Mutated yeast heat shock transcription factor activates transcription independently of hyperphosphorylation. J. Biol. Chem. 2006;281:3936–3942. doi: 10.1074/jbc.M510827200. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jin R., Dobry C. J., McCown P. J., Kumar A. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell. 2008;19:284–296. doi: 10.1091/mbc.E07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron S. J. Filamentous growth in budding yeast. Trends Microbiol. 1997;5:450–454. doi: 10.1016/S0966-842X(97)01131-1. [DOI] [PubMed] [Google Scholar]
- Kumar A., et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Cutler N. S., Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Cook J. G., Thorner J. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol. Biol. Cell. 1995;6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Galitski T., Lander E. S., Fink G. R. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Styles C. A., Fink G. R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Mazzoni C., Zarov P., Rambourg A., Mann C. The SLT2 (MPK1) MAPK kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J. Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean M. N., Mody A., Broach J. R., Ramanathan S. Cross-talk and decision making in MAP kinase pathways. Nat. Genet. 2007;39:409–414. doi: 10.1038/ng1957. [DOI] [PubMed] [Google Scholar]
- Miled C., Mann C., Faye G. Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol. Cell. Biol. 2001;21:3714–3724. doi: 10.1128/MCB.21.11.3714-3724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R., Wente S. R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- O'Neill E. M., Kaffman A., Jolly E. R., O'Shea E. K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Pan X., Harashima T., Heitman J. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 2000;3:567–572. doi: 10.1016/s1369-5274(00)00142-9. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Hardwick K. G., Lewis M. J. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robertson L. S., Fink G. R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Roth A. F., Wan J., Green W. N., Yates J. R., Davis N. G. Proteomic identification of palmitoylated proteins. Methods. 2006;40:135–142. doi: 10.1016/j.ymeth.2006.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S., Summers E., Lo H. J., Madhani H., Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y., Gulli M.-P., Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- Sun B., Chen L., Cao W., Roth A. F., Davis N. G. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol. Biol. Cell. 2004;15:1397–1406. doi: 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Orlicky S., Liu Q., Willems A., Sicheri F., Tyers M. genome-wide surveys for phosphorylation-dependent substrates of SCF ubiquitin ligases. Methods Enzymol. 2005;399:433–458. doi: 10.1016/S0076-6879(05)99030-7. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Vagnoli P., Bisson L. F. The SKS1 gene of Saccharomyces cerevisiae is required for long-term adaptation of snf3 null strains to low glucose. Yeast. 1998;14:359–369. doi: 10.1002/(SICI)1097-0061(19980315)14:4<359::AID-YEA227>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Temple G. F., Brasch M. A., Hartley J. L., Lorson M. A., van den Heuvel S., Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Yang Z., Bisson L. F. The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast. 1996;12:1407–1419. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1407::AID-YEA36%3E3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., Young R. A. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.