Abstract
The geometry of the phosphodiester backbone was analyzed for 7739 dinucleotides from 447 selected crystal structures of naked and complexed DNA. Ten torsion angles of a near-dinucleotide unit have been studied by combining Fourier averaging and clustering. Besides the known variants of the A-, B- and Z-DNA forms, we have also identified combined A + B backbone-deformed conformers, e.g. with α/γ switches, and a few conformers with a syn orientation of bases occurring e.g. in G-quadruplex structures. A plethora of A- and B-like conformers show a close relationship between the A- and B-form double helices. A comparison of the populations of the conformers occurring in naked and complexed DNA has revealed a significant broadening of the DNA conformational space in the complexes, but the conformers still remain within the limits defined by the A- and B- forms. Possible sequence preferences, important for sequence-dependent recognition, have been assessed for the main A and B conformers by means of statistical goodness-of-fit tests. The structural properties of the backbone in quadruplexes, junctions and histone-core particles are discussed in further detail.
INTRODUCTION
The apparent simplicity of double-helical DNA, the icon of molecular biology, is deceiving. While the architecture of its antiparallel strands remains the same, subtle conformational variations suffice to guarantee its recognition by other molecules. The structural variations are critical especially for reliable recognition between DNA and proteins, which is the conditio sine qua non in the essential processes of replication, transcription and DNA chromatin compaction. Local conformational changes induced by interactions with other molecules can either leave the DNA structure unaltered (i.e. in the form of a straight double helix) or introduce bends and kinks within the double helix, as in sequence-dependent CAP/DNA complexes (1) or in DNA coiled around histone-core proteins (2).
The necessity of understanding DNA variability has become more urgent as the sequence-specific protein/DNA recognition required e.g. by transcription factors seems less likely to follow simple and generally applicable rules analogous to the rules governing DNA self-recognition by the complementarity of the Watson–Crick (W–C) paired bases (3). The idea of the general ‘code of recognition’ between amino acids and nucleotides (4) has not been confirmed despite extensive efforts. The lack of simple rules for general protein/DNA recognition has been explained by the existence of too many structural degrees of freedom at the protein/DNA interface (5), and so far only limited rules of recognition have been formulated within narrower groups of transcription factors with certain binding motifs, such as zinc fingers or helix–turn–helix (6–9).
Ultimately, the variability and plasticity of the local DNA structure, and thus its ability to recognize other molecules and be recognized by them, can be attributed to the properties of the bases and to their sequence-dependent arrangement. Base-pair and base-step morphology (10,11) has been widely analyzed to describe sequence-dependent deformability as observed in the crystal structures of DNA complexes with sequence-specific proteins (12,13) as well as in noncomplexed DNA (14). By combining descriptors of base morphology with constraints imposed by a simple model of the phosphodiester backbone, slide and shift have been suggested to describe the key sequence properties of dinucleotide steps (15). However, the backbone does not act as a passive link merely holding the bases at their positions, but its inherent flexibility contributes to, and limits, the base placement so that the local DNA structure results from the interplay between optimal base positions and preferred conformations of the sugar phosphate backbone. An analysis of the conformational space populated by the DNA backbone and the correlation between its conformation and the DNA sequence are therefore important for fully understanding DNA recognition.
The structural alphabet of the DNA double-helical A-, B- and Z-forms has been described in detail earlier (16,17). Nevertheless, DNA is known to adopt also other forms, such as triple (18) and quadruple helices (19), junction (cruciform) structures (20) and parallel helices (21). However unusual some of these DNA forms may be, their architecture is, in full analogy to the double helical DNA, almost completely based on the self-assembly of two or more DNA strands and does not form complicated folds analogous to RNA. The availability of some of these unusual DNA structures in well-refined crystal structures as well as the growing number and quality of more conventional DNA crystal structures present a challenge to undertake an analysis of the DNA conformational space in much greater detail than it was possible a few years ago (22).
This work presents a comprehensive analysis of the conformational space of the DNA backbone using a near-dinucleotide building block as a model. Dinucleotide conformations have been clustered as the local structural property without any consideration of the classification of the overall DNA architecture as, for instance, B- or A-type double helix. The study has been performed on almost 8000 dinucleotide units from 447 crystal structures of DNA, alone or in complexes with other molecules and has made use of a slightly modified procedure developed earlier for an analysis of RNA conformations (23). To assess the nature of the broadening of the DNA conformational space upon interacting with other molecules (mainly with proteins), the classified conformers of naked DNA have been compared to those of complexed DNA molecules. In addition, the structural properties of the backbone have been discussed in selected unusual structures like quadruplexes and histone-core particles. Because the possible sequence preferences of various conformers are important for the sequence-dependent recognition they have been assessed by means of rigorous nonparametric statistical testing within the group of naked B- and A-form double helices.
METHODS
The selection of structures used for the analysis was limited to nucleic acid (NA) crystal structures containing only DNA (thus excluding hybrids with RNA) present in the Nucleic Acid Database (24) on 19 July 2005. Four hundred and fifteen structures with crystallographic resolution better than or equal to 1.9 Å were selected; this resolution had previously been identified as limiting the ambiguity of the statistical treatment of torsional distributions (22). Four hexa- and one hepta-nucleotide sequences, CGATCG, CGTACG, CGCGCG, CGCGAA, GCGCGCG, were overrepresented in the original compilation of structures and 26 structures containing them were therefore removed from the analyzed set. Since the initial set of structures limited to 1.9 Å resolution does not contain some a priori important classes, it was further augmented by 58 structures with unusual topologies, such as G-quadruplexes, i-motif, four-way and three-way junctions, as well as by important types of protein/DNA complexes, such as DNA complexed with TATA-box binding proteins or histone-core proteins so that 447 structures were selected for the analysis. All modified and incomplete nucleotides were removed so that the complete data set (further referred to as Dataset 1) contains 7739 dinucleotides (Table 1); PDB codes of the analyzed structures are listed in Table 2, and all dinucleotides are fully characterized in Supplementary Table T1.
Table 1.
The datasets of the dinucleotides used in this study
| Dataset | Characterization | Number of structures | Number of dinucleotides |
|---|---|---|---|
| 1 | All dinucleotides analyzed by FT averaging and clustering | 447 | 7739 |
| 2 | Only noncomplexed DNA | 187 | 1861 |
| 3 | Dataset 2 without quadruplexes, Z-DNA, 1DC0 and all dinucleotides forming non-WC pairs | 46 of A-form 72 of B-form 118 in total | 391 in A-form structures 806 in B-form structures 1197 in total |
Table 2.
The PDB codes of the structures used in the analysis
| Structure Type | PDB Codes |
|---|---|
| Noncomplexed A-DNA (46) | 118d, 137d, 138d, 160d, 1d78, 1d79, 1dnz, 1kgk, 1m77, 1ma8, 1mlx, 1nzg, 1vj4, 1xjx, 1z7i, 1zex, 1zey, 1zf1, 1zf6, 1zf8, 1zf9, 1zfa, 213d, 243d, 260d, 295d, 2d94, 317d, 338d, 344d, 345d, 348d, 349d, 368d, 369d, 370d, 371d, 395d, 396d, 399d, 414d, 440d, 9dna, dh010, adh012, adh034 |
| Noncomplexed B-DNA (72) | 122d, 123d, 158d, 183d, 196d, 1bd1, 1bna, 1cw9, 1d23, 1d3r, 1d49, 1d56, 1d61, 1d8g, 1d8x, 1dou, 1dpn, 1edr, 1ehv, 1en3, 1en8, 1en9, 1ene, 1enn, 1fq2, 1g75, 1i3t, 1ikk, 1j8l, 1jgr, 1l4j, 1l6b, 1m6g, 1n1o, 1nvn, 1nvy, 1p4y, 1p54, 1s23, 1s2r, 1sgs, 1sk5, 1ub8, 1ve8, 1zf0, 1zf3, 1zf4, 1zf5, 1zf7, 1zfb, 1zff, 1zfg, 232d, 251d, 2d25, 307d, 355d, 3dnb, 403d, 423d, 428d, 431d, 436d, 454d, 455d, 456d, 460d, 463d, 476d, 477d, 5dnb, 9bna |
| DNA/drug and DNA/ protein complexes, Z-DNA, quadruplexes (329) | 110d, 115d, 131d, 145d, 151d, 152d, 159d, 181d, 182d, 184d, 190d, 191d, 1a1g, 1a1h, 1a1i, 1a1k, 1a2e, 1a73, 1aay, 1ais, 1azp, 1b94, 1b97, 1bf4, 1bqj, 1brn, 1c8c, 1cdw, 1ckq, 1cl8, 1cn0, 1d02, 1d11, 1d14, 1d15, 1d21, 1d22, 1d2i, 1d32, 1d37, 1d38, 1d40, 1d41, 1d45, 1d48, 1d53, 1d54, 1d58, 1d67, 1d76, 1d90, 1d9r, 1da0, 1da2, 1da9, 1dc0, 1dc1, 1dcg, 1dcr, 1dcw, 1dfm, 1dj6, 1dl8, 1dn4, 1dn5, 1dn8, 1dnf, 1dp7, 1dsz, 1e3o, 1egw, 1em0, 1emh, 1eo4, 1eon, 1esg, 1eyu, 1f0v, 1fd5, 1fdg, 1fhz, 1fiu, 1fms, 1fn1, 1fn2, 1g2f, 1g9z, 1gtw, 1gu4, 1h6f, 1hcr, 1hlv, 1hwt, 1hzs, 1i0t, 1i3w, 1ick, 1ign, 1ih4, 1ih6, 1imr, 1ims, 1j59, 1j75, 1jb7, 1jes, 1jft, 1jh9, 1jk1, 1jk2, 1jpq, 1jtl, 1juc, 1jux, 1jx4, 1k3w, 1k3x, 1k9g, 1kbu, 1kci, 1kx3, 1kx5, 1l1h, 1l1t, 1l1z, 1l3l, 1l3s, 1l3t, 1l3u, 1l3v, 1lat, 1lau, 1ljx, 1llm, 1lmb, 1m07, 1m19, 1m3q, 1m5r, 1m69, 1m6f, 1mf5, 1mj2, 1mjm, 1mjo, 1mjq, 1mnn, 1mus, 1mw8, 1nh2, 1njw, 1njx, 1nk0, 1nk4, 1nk7, 1nk8, 1nk9, 1nkc, 1nke, 1nkp, 1nnj, 1nqs, 1nr8, 1nt8, 1nvp, 1o0k, 1omk, 1orn, 1p20, 1p3i, 1p3l, 1p71, 1per, 1pfe, 1ph4, 1ph6, 1ph8, 1pji, 1pjj, 1puf, 1pup, 1puy, 1q3f, 1qda, 1qn3, 1qn4, 1qn5, 1qn6, 1qn8, 1qn9, 1qna, 1qnb, 1qne, 1qum, 1qyk, 1qyl, 1qzg, 1r2z, 1r3z, 1r41, 1r68, 1rff, 1rh6, 1rnb, 1rpe, 1rqy, 1run, 1s1k, 1s1l, 1s32, 1ssp, 1suz, 1sx5, 1sxq, 1t9i, 1tdz, 1tez, 1tro, 1u1p, 1u1q, 1u1r, 1u4b, 1ue2, 1ue4, 1uhy, 1v3n, 1v3o, 1v3p, 1vzk, 1w0u, 1wd0, 1wte, 1wto, 1wtp, 1wtq, 1wtr, 1wtv, 1xa2, 1xam, 1xc9, 1xjv, 1xo0, 1xuw, 1xux, 1xvn, 1xvr, 1xyi, 1ytb, 1ytf, 1zez, 1zf2, 1zna, 200d, 210d, 211d, 212d, 215d, 221d, 224d, 234d, 235d, 236d, 241d, 242d, 244d, 245d, 254d, 258d, 276d, 277d, 278d, 279d, 284d, 288d, 292d, 293d, 2bdp, 2bop, 2cgp, 2crx, 2dcg, 2des, 2hap, 2hdd, 2nll, 2or1, 2pvi, 304d, 306d, 308d, 313d, 314d, 331d, 334d, 336d, 351d, 352d, 360d, 362d, 366d, 367d, 383d, 385d, 386d, 3bam, 3bdp, 3cro, 3crx, 3hts, 3pvi, 400d, 417d, 427d, 432d, 441d, 442d, 443d, 452d, 453d, 465d, 467d, 473d, 481d, 482d, 4bdp, adh013, zdf013, zdfb03, zdfb06 |
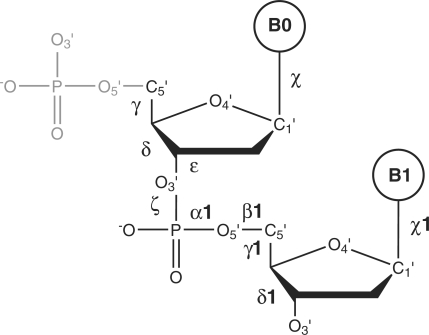
The DNA conformational space was investigated at the level of a dinucleotide unit with its 5′-end phosphate group removed; it was described by six backbone torsion angles between γ and δ + 1, plus two χ angles characterizing the glycosidic bond (Figure 1). This unit is identical to the 5′-end dinucleotide that naturally lacks the initial phosphate group, and similar to the ‘suite’ defined by Murray et al. (25), which covers the angles between δ and δ + 1. Two torsion angles at both 5′- and 3′-ends of the complete dinucleotide unit (α and β at the 5′-end and ɛ + 1 and ζ + 1 at the 3′-end) were not explicitly analyzed but they were monitored during the clustering process.
Figure 1.
The analyzed unit is defined by ten torsion angles from γ to δ + 1 along the backbone plus torsions χ and χ + 1 at the glycosidic bond. B0 and B1 symbolize the bases.
The presence of torsions α, β, and ɛ + 1, ζ + 1 in the analyzed nucleotides implies that neither 5′- nor 3′-end residues were among the 7739 analyzed dinucleotides of Dataset 1. All structural data are ‘crystallographically’ independent, i.e. all dinucleotide coordinates are gathered from the asymmetric units of the respective structures. However, information about symmetry-related strands was used when appropriate, e.g. when considering base-pairing patterns in double helical or quadruplex structures.
The analysis started by dividing the multidimensional torsional space into three-dimensional (3D) projections (maps). Based on a priori knowledge of the DNA (22) and RNA conformational spaces (23), the following nine 3D maps were selected: (ζ, α + 1, γ + 1), (ζ, α + 1, δ), (α + 1, γ + 1, δ + 1), (γ, δ, ζ), (γ, ζ, γ + 1), (ζ, γ + 1, δ + 1), (γ, ζ, β + 1), (α + 1, δ + 1, χ + 1) and (ζ, α + 1, χ). For each map, Fourier transform of the torsional values (τ1, τ2, τ3) was calculated as described earlier (23). The only methodological difference from the previous work on RNA conformations was the treatment of places with a high density of data points corresponding to the regions of prevalent double helical A, BI and BII conformations. The extreme density of the points in these areas strongly influenced the results of the Fourier transform in the whole 3D map. To eliminate this mathematical artifact, two 2D scattergrams were constructed for each map, the highest density region was manually selected in each scattergram, and the intersection of the selected points was randomly reduced by 95%. For example, the number of Fourier-transformed points in the (ζ, α + 1, γ + 1) map was reduced from 7739 to 1375. It should be emphasized that the reduced number of the data points was used only to improve the reliability of the Fourier averaging, whereas the full data set of 7739 points (dinucleotides) was utilized in all the subsequent analyses.
The distribution of the points (τ1, τ2, τ3)i was then transformed into pseudo-electron densities using standard crystallographic procedures implemented in the program XtalView (26) with the same set of parameters as had been used in ref. (23). The sites with a high density of points were transformed into peaks in the maps. The peaks thus correspond to areas with a high concentration of torsion angles and represent conformationally favored (and therefore interesting) regions. Eight to twelve peaks were identified within each of the nine analyzed maps, and each peak was assigned a symbolic name in the form of a letter or a letter and a number. The peak names are mere labels and carry no particular meaning. Each peak was then approximated by a sphere with a radius, typically of between 15° and 40°, estimated from the density contour. All data points lying inside the peak's sphere were labeled by the peak's name. If a data point lay within two or more peaks, it was assigned to the most intense one. The data points located outside the radii of all the peaks were not assigned to any peak. As nine maps were analyzed, each dinucleotide was characterized by a nine-letter string referred to as an imprint. The individual imprints were used to cluster dinucleotides with similar conformations by simple alphabetical sorting. The clusters were identified as a set of data points with (nearly) identical imprints. The sorting was based primarily on the imprints from the first four maps, (ζ, α + 1, γ + 1), (ζ + 1, α + 1, δ), (α + 1, γ + 1, δ + 1) and (γ, δ, ζ), whereas the other maps were used mainly to verify the quality of the sorting process. The final data matrix consisting of 7739 data points (dinucleotides) sorted into clusters is presented as Supplementary Table T1.
Within each cluster, the arithmetic means and the standard deviations were calculated for all 14 dinucleotide torsions using the formulas for the circular mean and circular standard deviation (27). The outliers leading to the degradation of the standard deviation were removed so that the final standard deviations of the torsional angles between γ and δ + 1 are typically better than 10°.
Dataset 1 was subdivided into two more data sets: Dataset 2 and Dataset 3 (Table 1). Dataset 2 was created by removing all structures of DNA/protein and DNA/drug complexes from Dataset 1 (Table 1) and was used to study the effect of complexation on dinucleotide conformation. To test the possible relationships between dinucleotide conformational classes and sequences in noncomplexed B- and A-form double helices, Dataset 2 was further modified by removing Z-DNA dinucleotides, quadruplexes (G-quadruplexes and i-motif structures), structure 1DC0 (BD0026) (28) and all the dinucleotides with non-W–C paired or non-paired bases, thus resulting in Dataset 3 (Table 1). The DNA dodecamer 1DC0 (28) was removed from Dataset 3, because of the uniqueness of its double helical architecture in combining features typical of the B- and A-forms. The 1DC0 structure will be discussed in the Results and discussion section below. The aim of classifying dinucleotide steps by means of combining Fourier averaging with clustering analysis was to define the conformational families with low variations of torsion angles unambiguously. A consequence of such a strict requirement was the relatively large number of dinucleotides not assigned to any cluster. Therefore, to improve the statistical significance of the sequence analyses, an additional round of conformational assignment of unclassified dinucleotides in Dataset 3 was performed. Further classification was accomplished by calculating both the Euclidean distance and Manhattan distance (known also as taxi-cab metric, L1 distance; the distance between two points measured along axes at right angles) distances between torsional angles δ, ɛ, ζ, α1, β1, γ1 and δ1 of the unassigned dinucleotides and the conformational families. A dinucleotide was assigned to the cluster with the lowest distance provided that both the Euclidean and Manhattan distances were smaller than 35°. Approximately one-half of the originally unassigned dinucleotides were classified in this procedure, leaving roughly 1/8 of the total number of dinucleotides unclassified.
The contingency tables of the counts of dinucleotide sequences (steps) were built for six broad conformational classes, BI, BII, AI, AII, B/A and A/B, whose detailed description can be found in the Results and discussion section. Those steps with unclassified conformations were attributed either to the RestB category if their parent structure was annotated as a B-type double helix by the NDB, or to the RestA category if they had originated from an A-type double helix. Only the assignment of these two categories, RestB and RestA, used an a priori classification of the double-helical architecture.
The sequence–conformation relationships of Dataset 3 were analyzed by means of statistical hypothesis testing. The contingency tables correspond to the product-multinomial model with fixed row margins. At the very beginning, the χ2 test of the homogeneity of the frequency distribution of the sequences of the individual conformational classes AI, AII, RestA, BI, BII, A/B, B/A and RestB (Table 5) was performed. This test compares the multinomial distributions between rows. Since this homogeneity was rejected (Pearson χ2-test statistic on 105° of freedom is 996.8 with a P-value <10−16), the sequences are not distributed homogeneously between conformational families and the sequence–conformation relationships were tested further.
Table 5.
Counts of 16 dinucleotide steps in conformational families of noncomplexed A- and B-form double helices (Dataset 3)
| Conformation | RR | RY | YR | YY | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GA | GG | AC | AT | GC | GT | CA | CG | TA | TG | CC | CT | TC | TT | |
| AI | 0 | 1 | 1 | 46 | 13 | 6 | 47 | 20 | 8 | 56 | 9 | 10 | 60 | 1 | 2 | 0 |
| AII | 0 | 0 | 1 | 16 | 1 | 0 | 0 | 0 | 0 | 27 | 8 | 0 | 1 | 0 | 0 | 0 |
| RestA | 0 | 2 | 0 | 13 | 4 | 1 | 7 | 0 | 1 | 9 | 0 | 1 | 14 | 2 | 3 | 0 |
| Total in A | 0 | 3 | 2 | 75 | 18 | 7 | 54 | 20 | 9 | 92 | 17 | 11 | 75 | 3 | 5 | 0 |
| BI | 53 | 16 | 39 | 4 | 5 | 29 | 12 | 9 | 1 | 48 | 12 | 8 | 11 | 12 | 31 | 50 |
| BII | 4 | 2 | 10 | 18 | 5 | 1 | 35 | 2 | 31 | 33 | 10 | 18 | 0 | 0 | 0 | 0 |
| A/B | 2 | 1 | 0 | 0 | 0 | 4 | 1 | 0 | 3 | 41 | 8 | 0 | 2 | 4 | 1 | 8 |
| B/A | 17 | 2 | 2 | 0 | 3 | 19 | 37 | 7 | 0 | 1 | 0 | 0 | 2 | 2 | 11 | 8 |
| RestB | 9 | 2 | 10 | 1 | 4 | 5 | 33 | 1 | 4 | 19 | 1 | 0 | 3 | 3 | 10 | 6 |
| Total in B | 85 | 23 | 61 | 23 | 17 | 58 | 118 | 19 | 39 | 142 | 31 | 26 | 18 | 21 | 53 | 72 |
Dinucleotides from RestA and RestB categories were not assigned to any of the conformations; R are purines, Y pyrimidines.
The first statistical experiment, further referred to as the test of the ‘uniformity of dinucleotide representation’, is a χ2 goodness-of-fit test of equality of the column margins. It compares the observed frequency of a given dinucleotide with a hypothetical frequency of 1/16 corresponding to the situation when all dinucleotides are distributed evenly. The uniformity of dinucleotide representation was measured for dinucleotides in A-like and B-like conformations from Dataset 3 including the unclassified ones (RestA was included in A-like, and RestB in B-like conformers). The test was performed for all 16 steps as well as for four pyrimidine/purine (Y/R) sequences. The actual frequencies of the palindromes (AT, GC, CG, TA) were counted twice. If the null hypothesis of the uniformity of dinucleotide representation in all sequences is rejected, the Pearson residuals provide evidence about a possible over- or underrepresentation of dinucleotide sequences with respect to the hypothetical equal frequencies. The critical values were calculated according to the rule of Bonferroni (29), which for the multiple-significance test ensures that the overall type I error is below 5%.
The second test further referred to as the test of ‘dinucleotide homogeneity’ examined whether a particular sequence was under- or over-represented within a particular conformational class. The count of the sequence in any conformation was then compared to the sum of the counts of this sequence in the remaining conformations considered. Like the test before, the test of dinucleotide homogeneity was performed for all 16 sequences as well as four pyrimidine/purine (Y/R) sequences, this time employing Standardized Pearson residuals (30), which are residuals adjusted to have asymptotic standard normal distribution. Too large a value of the standardized Pearson residual, exceeding the critical value, indicates a significant overrepresentation as compared with the null hypothesis in that cell, whereas a negative value below the negative of the critical value indicates a significant underrepresentation. The Bonferroni correction gives the conservative critical value to these tests, and values which are too large or too small are significant.
The described χ2-test of dinucleotide homogeneity was supported by an additional statistical analysis. A so-called ‘odds ratio’ is a measure indicating the violation of homogeneity in the individual cells of the contingency table. The odds ratio for a particular cell was computed by reducing the contingency table to a two-by-two table composed of the cell being investigated and merging the remaining rows together and the remaining columns together. The odds ratio then represents the ratio of the likelihood of the occurrence of an individual sequence in an individual conformation and the probability of the occurrence of this sequence in any other conformation. The odds ratio greater/smaller than one corresponds to over-/under-representation, respectively. Complete Tables of odds ratios are shown in Supplementary Tables S3–S6.
RESULTS AND DISCUSSION
This section characterizes DNA dinucleotide conformations (Table 4), compares them in structures of naked and of complexed oligonucleotides (Figure 2), and investigates sequence preferences in the naked A-DNA and B-DNA double helices (Tables 5–9). The differences between the dinucleotide conformers observed in DNA and RNA are also briefly discussed. Finally, the characteristic features of selected important classes of ‘untypical’ DNA structures, such as quadruplexes or histone-core particles, are annotated in the context of their dinucleotide conformations.
Table 4.
The main DNA conformational classes identified in the present work
| Description | N | Clustered torsions | Cluster number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| γ | δ | ɛ | ζ | α + 1 | β + 1 | γ + 1 | δ + 1 | χ | χ + 1 | |||
| ‘Canonical’ A-form, labeled AI | 192 | 54 | 82 | 205 | 285 | 294 | 172 | 55 | 83 | 201 | 202 | 8 |
| AII, A-form with an α + 1/γ + 1 switch | 44 | 52 | 82 | 195 | 291 | 149 | 194 | 182 | 87 | 204 | 188 | 19 |
| A with δ, δ + 1 close to O4′-endo | 9 | 44 | 101 | 192 | 281 | 297 | 182 | 44 | 99 | 210 | 211 | 25 |
| AI–BI, with δ C3′-, δ + 1 C2′-endo | 32 | 54 | 86 | 194 | 281 | 301 | 179 | 55 | 142 | 214 | 251 | 41 |
| AI–BI, with δ O4′-, δ + 1 C2′-endo | 34 | 54 | 99 | 186 | 274 | 297 | 178 | 51 | 141 | 235 | 264 | 47 |
| BI–AI, with δ1 O4′-endo | 100 | 51 | 130 | 183 | 267 | 297 | 171 | 51 | 106 | 250 | 239 | 32 |
| BII–AI, with an α + 1/γ + 1 switch, high β + 1 | 9 | 49 | 146 | 257 | 186 | 60 | 224 | 196 | 90 | 260 | 200 | 110 |
| BI variation in complexes | 412 | 45 | 137 | 178 | 255 | 304 | 187 | 45 | 139 | 252 | 256 | 58 |
| ‘Canonical’ B-form, labeled BI | 1,531 | 47 | 136 | 184 | 262 | 302 | 179 | 45 | 138 | 251 | 260 | 54 |
| BII variation in complexes | 269 | 43 | 140 | 201 | 216 | 314 | 156 | 46 | 140 | 261 | 253 | 86 |
| BII-form | 340 | 46 | 143 | 245 | 172 | 297 | 142 | 46 | 141 | 269 | 259 | 96 |
| BI, with an α + 1/γ + 1 switch | 109 | 46 | 139 | 195 | 245 | 32 | 196 | 296 | 150 | 252 | 253 | 116 |
| BI, 3′-mismatches with an χ + 1 syn, α + 1/γ + 1 switch | 8 | 50 | 137 | 196 | 225 | 33 | 187 | 295 | 145 | 257 | 70 | 122 |
| AI–BI, 3′-mismatches with χ + 1 syn | 14 | 58 | 91 | 214 | 280 | 295 | 176 | 56 | 139 | 238 | 67 | 121 |
| Z-form, Y–R step | 21 | 54 | 147 | 264 | 76 | 66 | 186 | 179 | 95 | 205 | 61 | 123 |
| ZI-form, R–Y step | 40 | 177 | 96 | 242 | 292 | 210 | 233 | 54 | 144 | 63 | 205 | 124 |
| ZII-form, R–Y step | 18 | 179 | 95 | 187 | 63 | 169 | 162 | 44 | 144 | 58 | 213 | 126 |
‘Description’ is a short annotation of the conformation, ‘N’ is the number of dinucleotides which define the conformation, ‘Clustered Torsions’ are the arithmetic means calculated for the torsions used in the clustering process, with the torsions being defined in Figure 1. Bold font is used merely to indicate the three most important DNA forms.
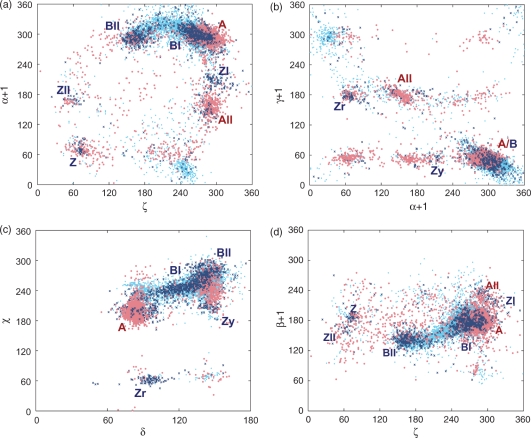
Figure 2.
Two-dimensional scattergrams of torsion angles in naked DNA (Dataset 2 from Table 1, dark blue) and in DNA from complexes (Dataset 1, cyan). A, B and Z are the respective double-helical forms, r stands for purines and y for pyrimidines. The conformations of almost 4000 RNA dinucleotides are plotted as pink dots for comparison.
Table 6.
The violation of the uniformity of dinucleotide representation for purines (R) and pyrimidines (Y) between A, B and combined conformational families as measured by the standardized Pearson residuals
 |
The underrepresented sequences are indicated by a gray background, the overrepresented are in bold, both exceeding the critical value of ± 2.50 (the 5% confidence level).
Table 7.
The violation of the homogeneity of purine (R) and pyrimidine (Y) dinucleotide steps between AI, AII and nonclassified (RestA) conformers in the A-form double helices as measured by the standardized Pearson residuals
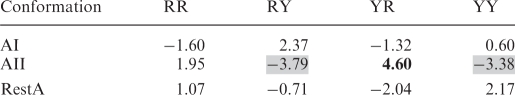 |
The underrepresented sequences are indicated by a gray background, the overrepresented are in bold, both exceeding the critical value of ±2.87 (the 5% level test).
Table 8.
The violation of the dinucleotide homogeneity for sequences between BI, BII, A/B, B/A and unclassified dinucleotides (RestB) in the B-form double helices measured by the standardized Pearson residuals
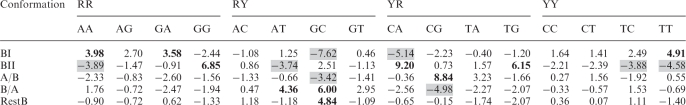 |
The underrepresented sequences are indicated by the gray background, the overrepresented are in bold, both exceeding the critical value of ±3.42 (the 5% level test).
Table 9.
A summary of the conformational preferences of dinucleotide steps in B-DNA helices
| Sequence | RR | RY | YR | YY | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GA | GG | AC | AT | GC | GT | CA | CG | TA | TG | CC | CT | TC | TT | |
| Conformation | BI | BI | BI | BII | – | B/A | B/A, RestB, BII | – | BII | A/B, (BII) | – | BII | (BI) | (BI) | BI | BI |
Some sequences were not assigned any conformational preference because of their low representation in the whole data set.
Overview of DNA conformations
Fourier averaging and clustering performed on the full data set of all available structures (Dataset 1 defined in Table 1) revealed a large number of conformational clusters (Supplementary Table T2), which may however be condensed into a much smaller number of families (Table 4). These main conformational families include all major well-characterized DNA double helical forms (BI, BII, AI, AII and Z) as well as less expected conformers, combining structural features typical of A- and B-forms. However, their structural diversity is far from matching that of RNA conformers (31).
A-DNA is a well-described conformation (32–34) characterized by the C3′-endo sugar pucker (δ ∼ 80°), with ζ and α + 1 ∼300° (gauche-), β + 1 ∼180° (trans-) and with its glycosidic torsion angle χ adopting a value near 200° (low anti). A-form conformers exhibit a relatively low dispersion of torsion angles and are sufficiently represented by two major conformations, the canonical A-form, represented by Cluster 8 and labeled also AI (Table 4) and the AII conformation (Cluster 19). AII is characterized by the α and γ torsions in the trans region. These values can be reached from the canonical α/γ values (300° and 60°, respectively) by the so-called ‘crankshaft motion’, which effectively compensates for the switch in torsion values in such a way that the overall course of the backbone does not alter dramatically. Both A-forms have torsion values close to those reported earlier (32,33) and are virtually the same as in their respective RNA conformations (31). What is important is the existence of other A-like conformers with a sugar pucker in the O4′-endo region (δ ∼ 100°, Cluster 25 in Table 4), observed both in noncomplexed and complexed DNA. Since the interconversion of C2′-endo to C3′-endo occurs preferentially via the O4′-endo state (35), dinucleotides forming Cluster 25 may be described as A-to-B transitional conformers; they are closer to the A-form, because their χ torsion is A-like.
Also both major B-form conformers, BI (Cluster 54 in Table 4) and BII (Cluster 96), have torsion values near those known from earlier studies (36,37). The canonical BI-conformation is by far the most frequent conformer both in naked and complexed DNA, respectively. It is characterized by the C2′-endo sugar pucker with δ ∼ 135°, ζ and α + 1 torsions in the gauche-range (however, its ζ-value near 260° is lower than in A-DNA) and by χ adopting much a higher value than in A-DNA; the χ values typical of the B-form close to 260° are called high anti. The variations within the BI conformers (Supplementary Table T2) result mainly from ɛ, ζ, α + 1 and β + 1 torsions, but the changes in these torsions mostly compensate each other. Only four or five BI conformers of the many which have been identified form larger clusters and only three (Clusters 50, 54 and 58) have been observed in structures of naked DNA.
In naked DNA structures, the BI- and BII-forms are separated by a gap between the ζ and ɛ torsions, and to a lesser extent also between β + 1 and χ torsions (37). However, such a distinction between these forms almost completely disappears in complexed DNA (Figure 2a and d). Despite the near-continuous BI-to-BII transition, the data from naked DNA (Dataset 3) clearly indicate that BII should be recognized as a distinct B-form characterized by ζ in the trans region, by a high value of ɛ and by a low β near 140°. Changes and mutual compensations of torsion angles in BI and BII are obvious from Table 4 by comparing the values for Clusters 54, 86 and 96 (all the clusters spanning the BI- and BII-forms are listed in the Supplementary Table T2): The BII-like conformers start at ɛ and ζ values close to the BI-form (illustrated by Cluster 86 with ɛ/ζ ∼ 200°/215°), gradually pass to ‘typical BII’ values of Cluster 96 (ɛ/ζ ∼ 245°/172°) and end with Cluster 105 having extreme values of ɛ and ζ (ɛ/ζ ∼ 264°/149°). The almost continuous transition from BI to BII is best described by the linear anticorrelation of ɛ and ζ values (ɛ = −0.73 ζ + 367°, R2 = 0.85, N = 2022, with the equation being valid within the limits of BI and BII conformations).
The occurrence of two consecutive BII conformers is infrequent, but it does occasionally occur both in naked and complexed structures, corroborating an earlier observation (22). In naked DNA, the BII–BII repetition has to be stabilized either by a crystal contact, or it is induced by specific structural features, such as the BII repetitions found in the four-way DNA junction 1L4J (38). The frequency of occurrence of BII–BII steps in protein complexes is similar to that in naked DNA, and BII–BII repetition is uncommon even in nucleosome or in strongly bent CAP/DNA structures. Three consecutive BII steps are rare but have been observed in four-way junctions (e.g. in the 1L4J structure) or in DNA/histone-core complexes. In summary, BII conformers tend to be isolated, with a typical pattern of BII-rich regions being the BI–BII–BI–BII repetition. Alternatively, BI may be repeated more than once while BII can be replaced by another deformed A–B conformer in this pattern.
The average values of the backbone torsions in a nucleotide (listed from α to ζ) were calculated for the most common A- (AI and AII) and B-forms (BI and BII) using only naked DNA structures (Dataset 3 in Table 1). The values listed in Table 3 allow for an easy comparison with other references (39,22). A careful selection of high-resolution DNA structures in Dataset 3, the numbers of observations for each structural class, and the intervals of reliability of all torsions imply that the torsion values in Table 3 are a source of reliable structural description of the sugar–phosphate backbone of the double helical forms.
Table 3.
Torsion angles [°] in nucleotides of the major A- and B-forms
| α | β | γ | δ | ɛ | ζ | χ | N | |
|---|---|---|---|---|---|---|---|---|
| Canonical A-form (AI) | 294.8 ± 0.9 | 172.7 ± 1.0 | 54.3 ± 0.9 | 82.1 ± 0.7 | 205.6 ± 1.0 | 285.4 ± 0.7 | 200.5 ± 1.0 | 180 |
| AII | 145.6 ± 2.3 | 191.9 ± 2.0 | 182.8 ± 1.7 | 85.0 ± 1.4 | 197.0 ± 2.0 | 289.2 ± 1.7 | 203.4 ± 1.1 | 49 |
| Canonical B-form (BI) | 299.0 ± 0.9 | 179.3 ± 1.0 | 48.4 ± 0.6 | 132.8 ± 1.0 | 181.7 ± 1.0 | 263.2 ± 0.8 | 250.3 ± 1.1 | 418 |
| BII | 292.6 ± 1.3 | 143.1 ± 1.3 | 46.0 ± 0.9 | 143.0 ± 0.9 | 251.1 ± 2.1 | 168.0 ± 1.4 | 277.8 ± 1.4 | 187 |
The data were obtained through the analysis of 118 naked (noncomplexed) DNA structures from Dataset 3 (Table 1). The confidence intervals for the mean values of the torsion angles were computed under the assumption that the angles at the 95% confidence level are distributed normally. N is the number of observations of each conformer, AI corresponds to Cluster 8, AII to Cluster 19, BI to Cluster 54 and BII to Cluster 98.
Several fairly populated conformers fall within neither the A- nor the B-form category. However, they can be characterized as conformations with one nucleotide of the B-type and the other of the A-type and having sugar in one or both nucleotides in the transitional O4′-endo pucker. The first such group of conformers, exemplified by Cluster 41 (Table 4), can be described as AI–BI. The nucleotide at the 5′-end of the analyzed dinucleotide unit is in an A-like conformation (C3′-endo sugar pucker, low χ), whereas the 3′-end nucleotide is of a BI-type (C2′-endo sugar pucker, χ higher than 200°). In another similar cluster (Cluster 47, Table 4), the sugar of the 5′-end nucleotide adopts the A-to-B transitional O4′-endo pucker, and this nucleotide should therefore be considered as an intermediate between the A- and B- forms. AI–BI conformers occurring both in naked and complexed B-DNA double helices are characterized by a strong sequence bias toward the Y–R sequences (see below for a detailed discussion on sequence preferences within the individual conformer families). The occurrence of the A-to-B conformations seems to reflect the inherent flexibility of certain, preferably Y–R, sequences, as they can be explained neither by the presence of interacting species (e.g. ions) nor by the packing effects.
In analogy with the A–B conformers, combined B–A clusters were also identified (Table 4). First such a group of conformers is exemplified by Cluster 32 (Table 4); it has the C5′-end nucleotide in the BI-form while the C3′-end adopts a transitional conformation between B- and A-forms (O4′-endo sugar pucker, χ ∼ 239°) and can be characterized as BI–AI. The BI–AI conformers occur both in naked and complexed DNA and the majority of their nucleotides are involved in W–C base pairing. Their sequence dependencies are more complex than those of the AI–BI conformations: The Y–R sequences are disfavored while the A–A and A–T sequences are preferred. A few small B–A clusters with high ɛ and low ζ can be characterized as BII–AI conformers (Supplementary Table T2) with the C5′-end residue in the BII-form (C2′-endo sugar pucker, high anti χ), and with the C3′-end residue in the A-form (C3′-endo, χ ∼ 200°). Other torsions in BII–AI conformers may also adopt unusual values, such as α + 1 at 60° and γ + 1 at 200°, in Cluster 110. Most of the BII–AI dinucleotides are found in the R–R sequences; some are involved in G–A or A–G mismatches adopting Hoogsteen base pairing (Cluster 107 in Supplementary Table T2). These clusters clearly show how localized deformation of the regular B-form is sufficient to accommodate the G/A mismatch into the double helix.
Both B- and A-forms accommodate the crankshaft motion compensation between α and γ (or α + 1 and γ + 1) torsions but differ in its realization. The A-form has its important substate, AII (Cluster 19, Table 4), with trans/trans α and γ torsions, observed in naked and complexed DNA, as well as in RNA. In contrast, trans/trans α/γ combination is never observed in the B-form where α and γ torsions may be flipped from their canonical g–/g+ values to the g+/g– combination (Cluster 116) virtually only by interactions with proteins.
The B–A and A–B clusters described so far combine features typical of the B- and A-forms in such a way that each nucleotide within the analyzed unit adopts predominantly one or the other form. However, there are three or four small clusters (Clusters 24–26 and to some extent also Cluster 3 in Supplementary Table T2, Cluster 25 is also in Table 4) which are examples of a true combination of the B- and A-forms. These dinucleotides are characterized by having both sugars at the O4′-endo sugar pucker, transitional form between the C2′- and C3′-endo puckers and the ζ value near 280°. These unusual conformers can be found not only in highly deformed regions of DNA complexed with the TATA-box-binding protein (40,41) but also in naked A-DNA structures (42).
The combining of B- and A-type conformers has been described using base morphology and helical parameters such as groove width and helical twist in DNA complexed with proteins (43,44) as well as in naked DNA (45). B- and A-forms have also been observed to coexist in several crystal structures. Entire B- and A-DNA double helices have been located in a single-crystal structure (46), demonstrating similarity in their thermodynamic stability. The scaffolding of this crystal lattice is built by A-DNA helices with B-DNA helices being interspersed in crystallographically disordered positions in the lattice interstices. In several structures, oligonucleotides (28,47,48) capture DNA in various phases of the B-to-A transition. Perhaps the most direct observation of the B-to-A transition itself has been achieved by solving a series of structures (1IH1–6) with sequences containing guanines and a varying number of methylated and brominated cytosines (48). In another structure, 1DC0 (28), the whole double helix has features of both B- and A-forms: The bases are perpendicular to the helical axis (have an almost zero inclination), which is typical of the B-form, but most of the other parameters, such as twist, sugar pucker, minor-groove width, slide and the distance of the P atom from the base plane Zp (49), adopt A-like values. The backbone torsions of this structure are strictly A-like, in fact, all its residues were classified as canonical A-DNA. This structure demonstrates the limits of any analysis using torsion angles only. To capture all the details of such a conformation, torsion angles should be complemented with other parameters, such as inclination, slide or Zp (49). However, it should be emphasized that the 1DC0 structure is an exception and that the distinction between the B- and A-type was possible in the other cases by analyzing the torsional space.
The above observations support the view describing the right-handed double-helical forms as one broad conformational family with a strong preference for the BI-form connected by a nearly continuous set of conformers to the AI- and AII-forms on the one hand and to the BII-form on the other.
The glycosidic angle in four clusters (Clusters 119–122, Table 4 and Supplementary. Table T2) characterizing the structure of non-W–C (mismatched) base pairs adopts a rare syn orientation (χ ∼ 70°). Most dinucleotides in these clusters are of a GG or GA sequence, but there are several GT and GC exceptions. Clusters 121 and 122 with the 3′-end base in syn orientation are listed in Table 4, whereas their 3′-end continuation, Clusters 119 and 120, are shown in Supplementary Table T2. All nucleotides forming Cluster 122 come from G-quadruplexes. Cluster 121 contains mainly unusual non-W–C pairs between the W–C edge of cytosine and the Hoogsteen edge of guanine. These nonplanar G–C pairs from the TATA box bound to the TATA-box-binding protein [e.g. the PDB entry 1QN3 (50)] correspond to the class ‘IV trans’ of the Leontis–Westhof classification (51).
While the single building unit both for A- and B-DNA is a nucleotide, the left-handed double-helical Z-DNA is constructed from dinucleotide steps with distinct conformations consisting of alternating pyrimidine–purine (Y–R) or purine–pyrimidine (R–Y) steps (52). The Y–R steps are implemented by one geometry described by Cluster 123 (Table 4), whereas the R–Y steps may adopt two distinct conformations characterized either as ZI (Cluster 124) or as ZII (Cluster 126) (53,54).
A comparison of conformations of naked and complexed DNA
A brief inspection of 2D scattergrams in Figure 2 shows that distributions of all torsions in naked (noncomplexed) DNA are significantly broadened upon complexation with proteins and small ligands (e.g. drugs). DNA molecules in the crystal phase are obviously not ‘naked’ but immersed in solvent, mainly water molecules and metal cations. These small solvent particles are indispensable for structural integrity of nucleic acids but their influence is not considered explicitly here. In our opinion, the fact that DNA crystallized from pure solvent is conformationally more compact than DNA co-crystallized with drug molecules and especially polymeric proteins indicates that (i) small solvent particles impose the smallest conformational constraints, and (ii) DNA–DNA crystal contacts are rare and/or relatively nonspecific.
The merging of the BI- and BII-forms upon complexation with proteins, perhaps the most significant case of the conformational broadening caused by complexation, was discussed in the previous section. Four distinct regions of the ζ/α + 1 scattergram (Figure 2a) induced by complexation with proteins are discussed below:
A fair number of conformations is present at very low α + 1 near 30° and ‘normal’ ζ at ∼240°. These well-defined conformers also appear in the upper left corner of the α + 1/γ + 1 scattergram near α + 1∼30° and γ + 1∼300° (Figure 2b). They correspond to B-like families with α + 1 and γ + 1 values flipped from their normal g–/g+ values to g+/g– conformation and are represented by Clusters 116 and 112 (Supplementary Table T2). These conformers occur at points of a substantial DNA bend like in complexes with DNA polymerase, histone-core proteins and transcription factors, or in ‘disordered’ regions of four-way junctions. However, not all conformations in this area originate from DNA complexes: The points pertaining to Cluster 122 come mostly from guanine quadruplexes (Cluster 122).
A small region of about 40 residues adopting the same α + 1 values (∼30°) but lower ζ (∼180°-230°) corresponds to nucleotides with higher β + 1 (∼190°–240°) and belongs to Cluster 110 (Supplementary Table T2). The rest of dinucleotides in this region of the ζ/α + 1 scattergram were not assigned to any cluster and correspond either to DNA in protein complexes, especially with endonucleases, or to DNA intercalated with drug molecules.
A rather diffuse region between ζ ∼ 170°–250° and α + 1 ∼90°–150° originates from nonclustered dinucleotides interacting strongly with histone-core proteins and with intercalated drugs. Interestingly, this conformation may be induced by the intercalation of a drug molecule to both the double helix and quadruplex and may thus reflect backbone adaptation to the intercalated drug.
The residues in the region with a rare combination of ζ/α + 1, ∼60°/∼200°, are not clustered; some are from structures of single-stranded DNA and likely to be a real DNA conformer. However, most of these residues are likely to result from an incorrectly fitted sugar pucker, which forces the backbone into extreme values of ɛ torsion (below 90°), subsequently implying the rare combination of ζ/α + 1.
The distribution of β torsion is also significantly broadened in complexes. The ζ/β + 1 scattergram (Figure 2d) shows how conformations separated into distinct regions in naked DNA broaden upon complexation. While the most prominent case of merged BI- and BII-forms was already discussed above, two other diffuse areas (not assigned to any of the identified clusters), occurring exclusively in complexes, are found near ζ/β + 1 ∼280°/80° and near ζ/β + 1 ∼170°-230/200–240°. In the former group, α + 1/γ + 1 torsions occupy the untypical g–/t region. Similar conformations exist also in RNA both for C3′/C3′ and C2′/C2′ sugar puckers as conformers 1e and 4s, respectively (31). In the latter group, approximately half of the residues can be mapped to the ζ/α + 1 group discussed above under Point (i), whereas the other half has no structural or functional characteristics in common.
Two groups of conformers can be observed in the α + 1/γ + 1 scattergram (Figure 2b) for complexed DNA. One large group around the α + 1/γ + 1 ∼30°/300° region was already discussed above as the ζ/α + 1 Group 1. Another is located in a small region of α + 1 ∼240°–270° and γ + 1 ∼170°. Although this area has not been identified as a distinct cluster, this conformation may be found either in the i-motif structures [e.g. 190D (55)] or in the noncanonical base pairs classified as WC/WC trans [Type 2 according to Leontis–Westhof classification (51)].
The two conformers described above (i-motif/noncanonical base pairs with α + 1 ∼240°–270° and γ + 1 ∼170°, and dinucleotides extended by intercalated drugs with ζ ∼ 170°–250° and α + 1 ∼90°-150°) represent, in our opinion, new unique DNA conformations. However, they were not clustered by the analysis, and their unequivocal identification as novel backbone conformers requires an analysis of new crystal structures.
The main change occurring in the δ/χ distribution (Figure 2c) upon the DNA complexation is the increase of the dispersion of χ values in the C3′-endo region, blurring the positive correlation between δ and χ torsions observed in noncomplexed structures. It should be emphasized that δ torsions near 100°, corresponding to the O4′-endo sugar pucker (as well as another C2′-to-C3′-endo transitional form with a sugar pucker in the C1′-exo region), are populated both in complexed and noncomplexed DNA. Apparently, the O4′-endo pucker is of a type of a distinct deoxyribose conformation and is highly unlikely to result from incorrect pucker assignment during the refinement process. Conformers with a sugar pucker between C3′-endo and O4′-endo (δ ∼ 90°), occurring both in naked and complexed DNA are mostly purine residues from Z-DNA and from guanine quadruplexes.
The syn orientation of the bases (χ ∼ 70°) is rare. Syn conformers with the C2′-endo (δ ∼ 140°) sugar pucker have been observed only in complexes with proteins; roughly 1/3 of them were classified as Clusters 121 and 122 (Table 4). This conformation is adopted by guanine in a syn orientation, either forming a Hoogsteen pair with cytosine in complexes with TATA-box-binding proteins, thus avoiding a possible sterical clash (50), or forming G–G pairs of guanine quadruplexes (56). However, the majority of the C2′-endo syn residues did not form any distinct conformation. These originated either from the same structures as the classified ones or are found in single-stranded DNA.
To summarize, complexation with proteins and small ligands (‘drugs’) induces a widening of torsional distributions of the DNA backbone. Some selected protein/DNA complexes have crystallographic resolution worse than the target value of 1.9 Å (22) and these structures are likely to blur the distributions by noise. Assuming that the refinement protocols do not systematically bias torsion distributions, at least in such a large statistical sample, the resolution-related broadening represents white noise. Nonrandom widening of torsion distributions caused by interactions between DNA and ligands should then be reflected by new conformers not seen in the naked DNA. This is indeed the case: While over 120 clusters were localized in all analyzed dinucleotides (Dataset 1), only 28 of them were found in naked DNA (Dataset 2).
One important reason for the conformational widening is the stabilization of A-like or combined B/A conformers induced by interacting molecules (33,43,57). Although the majority (70%) of dinucleotides from protein/DNA complexes adopt BI and BII conformations, their significant portion (30%) may be found in AI- and AII-forms. Such a plasticity of DNA, when the conformation is changed locally into an A-form, is one of the ways in which DNA achieves specificity in protein/DNA binding (58–60). Remodeling from the B- to A-form also provides a mechanism for smooth bending of the double helix and for the controlling of widths of major and minor grooves. By changing the accessibility of the edges of the individual base pairs (43), the narrowing and deepening of the major groove in A-DNA enables the appearance of sequence-specific contacts. Quite a large degree of distortion of the double helix required to achieve a specific protein binding may be attained by its local ‘deformation’ into an A-like conformation (61,62). The narrowing and deepening of the major and the widening of the minor grooves is also a reason for the increased population of BII conformers and for a smooth transition between the BI- and BII-forms in protein/DNA complexes (44).
Sequence preferences in double-helical B- and A-forms
The preferences of different sequences for different conformations were tested for dinucleotide steps in naked (noncomplexed) right-handed double-helical structures involved in W–C base pairs (Dataset 3). The conformational plasticity of a sequence probed by the crystal forces, which is statistically tested here, is de facto a consequence of the general structure-correlation principle formulated by Burgi and Dunitz (63,64).
In order to maximize the statistical significance of the sequence comparisons, the number of the conformational classes being analyzed was reduced, leaving eight statistical categories: AI, AII, BI, BII, A/B, B/A, RestA and RestB. The B-like clusters were labeled as BI (Clusters 48–85), BII (Clusters 86–106), A/B (clusters 22, 23, 38–47) or B/A (Clusters 27–37, 107–110). Similarly, the A-like clusters were assigned either to the AI (Clusters 1–21, 24–26) or AII (Cluster 19) category. For dinucleotides not assigned to any of these categories, an a priori NDB classification into the A- and B-form helices had to be used. If they appeared in A-form double helices, they were assigned to the RestA category, if they appeared in a B-form, they were assigned to the RestB category. The counts of all the 16 dinucleotide steps which were utilized in the statistical analyses are listed in Table 5.
It should be emphasized that the statistical tests performed are limited to sequences which were subjected to crystallization trials and succeeded in them. This fact must be borne in mind when interpreting the sequence preferences within our data sets. For instance, the underrepresentation of sequences with adenine and thymine in the A-form double helices may, and is likely to, reflect the thermodynamic preference of sequences containing these nucleotides. However, it may also reflect a lack of crystallization trials of such sequences after it was detected that they do not crystallize in A-DNA. Similarly, the preference of A-DNA for the GG sequences and of B-DNA for the AA sequences is likely to reflect real thermodynamic preferences, but we cannot completely exclude that certain sequences of a particular length were more popular in crystallization trials than others (here we allude to the known disposition of octameric sequences to crystallize in the A-form). A complicated interplay between sequences, their length, crystallization conditions and the resulting double-helical structure has been discussed since the early days of oligonucleotide crystallography (33,65,66). A seminal work by Dickerson et al. (67) shows that the crystal-packing forces probe the malleability of different sequences to adopt different conformations and that one sequence subjected to different environments may adopt several conformations.
Conformational space of dinucleotide sequences is mapped not only by crystal packing forces. Another force probing the polymorphism of individual sequences are the interactions with co-crystallized molecules where proteins, drugs and other small ligands impose different constraints on the DNA helices. However, complexed DNA structures have not been used to investigate sequence–structure correlations for two reasons: (i) their resolution is lower on average than that of naked DNA structures and (ii) the bias of the sequence space is likely to be even higher than in naked DNA, because sequences of DNA complexed with specific binders (e.g. transcription factors) need not be random in any way.
An analysis of the current data shows a strong general preference for the canonical BI conformer (Cluster 54) in all 16 dinucleotide steps. However, most steps are also capable of adopting a wide range of conformations, thus reflecting various local influences. Great differences have been found between counts in the A- and B-forms (Table 5, ‘Total in A’, ‘Total in B’ rows). The most apparent difference is the low number of adenosine and thymine residues in all A-like conformers (Table 5); while the AA step is highly populated in the B-form, it is completely missing in the A-form, and while GG is the most frequent step in the A-form, it is much less populated in the B-form. This observation was quantitatively confirmed by a test of uniformity of dinucleotide representation, performed for all 16 dinucleotide steps (Supplementary Table S2) between A-like (AI, AII, RestA) and B-like (BI, BII, A/B, B/A, RestB) dinucleotides. The qualitative difference between sequences of A-like and B-like conformers and the virtual lack of A/T nucleotides in the former one leads to the necessity of treating the A-like and B-like conformers separately in subsequent statistical tests (Table 6).
Another statistical test, the dinucleotide homogeneity test, allows for a more detailed analysis of sequence preferences within A- and B-forms. The sequences for this test were categorized either as purines/pyrimidines, or as actual nucleotides.
Well-founded conclusions for A-type conformers can only be drawn at the purine/pyrimidine level. Table 7 and Supplementary Table S3 show that the minor AII family prefers YR sequences while being under-represented for RY and YY sequences. The typical feature of the AII conformation, torsions α + 1 and γ + 1 near the trans region (Table 4), leading to an almost planar arrangement of six atoms O3′-P-O5′-C5′-C4′-C3′ at the 3′-end nucleotide, can be adopted by the purine nucleotide but only with difficulty by the pyrimidine nucleotide.
The dinucleotide homogeneity test performed for the B-like conformational families reveals their intrinsic sequence preferences. When tested for the Y/R sequences, the homogeneous steps (RR and YY) show significant preference within the BI family (Supplementary Table S5), which is usually not the case with combined steps (YR and RY). On the other hand, the combined steps are preferred in less populated conformational families, namely YR is abundant in the BII and A/B families, and RY in B/A families (nevertheless, many RY steps remain unclassified as RestB). Such a variability of conformations, especially of the YR steps, corresponds to their known role in bending and kinking (68).
The sufficient amount of data for the B-form DNA makes it possible to analyze all 16 dinucleotide steps separately. Both Pearson residuals (Table 8) and odds ratios (Supplementary Table S6) confirm the preference of YY or RR for adopting the BI conformation, with the only exception being the GG step (see below). The underrepresentation of combined Y/R sequences in BI is caused by the frequencies of the GC and CA steps being very low (Tables 5 and 8). The preference of the BII family for all four YR sequences can be inferred from the values of both Pearson residuals and odds ratios. The only two significantly populated YR sequences are TG and the complementary CA steps, which have high frequencies in the BII-form and low frequencies in other conformations. This indicates that the facing strands of the W–C paired tetranucleotide d(CA).d(TG) are likely to adopt the BII-form (69). Besides the CA step, also the CG step is often considered to be highly malleable to adopt the BII-form (70) but the current data do not support this view; the CG count in the BII-form is not statistically significant. Instead of preferring BII, the CG step may be considered plastic, it can adopt BI, A/B, and BII with comparable counts and was also found in a number of unclassified conformers (RestB). Structural variability of the CG step has been observed previously; CG conformation has been shown to depend not only on the immediately flanking nucleotides (37,71,72) but also on the more distant ones (73). The TA step has a similar count in the BI- and BII-forms, which contradicts the earlier observation that TA displays a low propensity to undergo BI-to-BII transition (70).
The YY steps disfavor the BII family to the extent that none of the CC, CT, TC, TT steps was identified as a BII conformer. The conformational preferences of the RR sequences are rather interesting: The three steps containing adenine (AA, AG, GA) are underrepresented while GG is significantly overrepresented in BII, which is the opposite in the case of the BI family. Although the lower number (23) of the GG steps in the B families calls for caution, their propensity to adopt the BII conformation (69) seems to be clearly pronounced.
The A/B and B/A families are less populated (Table 5), therefore any conclusion must be drawn carefully. Whereas the CG step can clearly adopt the A/B conformation, the GC step shows a high propensity for the B/A conformation. Some of these steps come from the CGC sequence, in which the central G nucleotide is responsible for the A-like features of the two consecutive B/A and A/B conformations. An analogous link can be made for T from the ATC or ATT sequences, where the AT step exists in the B/A conformation and TC or TT in the A/B one. On the other hand, several steps have seldom been observed in the combined B + A families. In particular, no YR step was classified as B/A, and only three RR and five RY steps adopted the A/B conformation, corroborating the general reluctance of purines to accept the C3′-endo sugar pucker in the B-like double helix.
Dinucleotide steps with an unclassified B-like conformation (RestB in Table 8) form about 14% of all steps in the B-form double helices, and the Pearson residuals of these unclassified dinucleotides are neutral for most sequences. An important exception is the significantly over-represented GC. The GC step, occurring with comparable counts in BII, B/A and RestB categories and underrepresented in the BI family, is the sequence with the most complicated conformational behavior. Multiple stable conformational states observed in this work for the GC step may be an indirect confirmation and generalization of its bistability in non-complexed DNA and ‘continuous flexibility’ in DNA/protein complexes (49).
The sequence preferences for the B-like conformation families are briefly summarized in Table 9. The BI conformation is numerically dominant in all the sequences (with the possible exception of CA) but it is significantly overrepresented in comparison with the other families only in some steps, notably in AA, TT and GA. Some steps, mainly GG, CA and TG show a propensity for the BII-form, whereas the CG step has a high propensity for the A/B conformation, and the AT and GC steps for the B/A conformation.
Annotation of selected DNA structures
Certain conformers occur mostly or exclusively in structurally and/or functionally distinct types of nondouble helical and deformed double-helical structures. The following paragraphs describe several such relationships in various DNA structures.
G-quadruplexes of the Oxytricha nova telomere are all conformationally similar structures which can be almost completely formed from clustered conformers. The central step of the quadruplex (Residues 2 and 3) in complexes with the telomere-end binding protein [structures 1JB7 (74), 1PH4, 1PH6 and 1PH8 (56)] as well as in the non-complexed quadruplex [1JPQ (75)] adopts the conformation of Cluster 122 (Supplementary Table T2), a B-like cluster with the canonical α + 1 and γ + 1 values flipped (‘α + 1/γ + 1 crank’) and with the syn orientation (χ ∼ 70°) of the second guanine base enabling non-W–Ck purine–purine base pair. The next step (Residues 3 and 4) adopt the conformation of Cluster 119, another B-like conformer with the first base in the syn orientation (χ ∼ 70°). The GT steps joining the G-quadruplex with TTTT loops have the conformation of Cluster 120; their G nucleotides are again characterized by the syn orientation (χ ∼ 70°) and by nontypical values of α and γ torsions, namely g+ (∼ 60°) and t (∼ 180°), respectively. The second thymine residue from the TTTT loop stacks on top of the 5′-terminal guanine from the second strand. This TT step, like the subsequent one, has its backbone deformed both at the 5′-end (α = 150°) and at the 3′-end (ζ + 1 = 60°). Its central sugar-to-sugar (δ-to-δ + 1) part is classified as the BI Cluster 85. The other residues in the Oxytricha nova G-quadruplex adopt the conformations of clusters in BI- and BII-forms.
i-motif or cytosine quadruplex
The i-motif or cytosine quadruplex (55) consists of two interlocked pairs of parallel strands of the CCCC sequence. Unlike in the case of the Oxytricha nova G-quadruplexes, nucleotide conformations in the i-motif do not cluster into distinct conformers and most dinucleotide steps were actually not classified. For instance, only three steps in the d(ACCCCT) structure [1BQJ, (76)] were classified as Clusters 11 and 15 (Supplementary Table T2), containing conformers with C3′-endo sugar puckers but more B-like ζ and χ torsion values. No steps were classified in the 1V3N and 1V3O (77) structures, and only one step was assigned to the BI-to-A Cluster 32 (Table 4) in 1V3P (77). The limited success of clustering the i-motif dinucleotides can be partially attributed to the small amount of data available and partially also to the extreme, and most likely incorrect, values of some torsional angles (most notably to the δ values near 170°) pushing other torsions to rarely populated regions during the refinement and preventing these residues from being identified by their clustering.
Four- and three-way junctions
Junctions between DNA helices are important as intermediates in DNA rearrangements and as components in the secondary structure of single-stranded DNA molecules, such as certain viral genomes. The most important of these is undoubtedly the four-way junction, the Holliday junction of genetic recombination (78). It is formed by an incomplete exchange of strands between two double-stranded helices. However, other junctions are also possible, namely three-way junctions, the simplest and most commonly occurring branched structures in biologically active, single-stranded nucleic acids.
The arms of the junctions are formed by B-type double helices, residues are classified either as BI, or as BII, the junction site itself is formed by a sharp turn in the phosphodiester backbone. This sharp turn is captured mainly by a change in the ɛ, ζ, α + 1, β + 1, and γ + 1 torsions, which adopt unusual values. Three conformationally distinct types of four-way junctions have been identified. However, the scarcity of structural data did not allow to classify the junction-site step as a distinct conformation in any of these structures.
Structures of Cre recombinase bound to a Holliday junction recombination intermediate [e.g. 2CRX (79), 4CRX (80)] contain DNA duplexes arranged in a nearly planar X-shaped structure. The junction is formed by a linkage between T and A nucleotides, which sharply bends DNA by an unusual combination of torsions ζ, α + 1, β + 1 and γ + 1. The values of these torsions vary, however, from one structure to another, thus preventing this step from clustering. For example, the ζ, α + 1, β + 1 and γ + 1 torsions of the junction in the 2CRX structure adopt a rare combination g+/g+/g+/t, which has not observed among stable conformers even in the more variable RNA.
The Holliday junction of the ‘inverted repeat sequence’ CCGGTACCGG [e.g. 1DCW (81), 1JUC (82)] is characterized by a high proportion of unclassified and BII-form residues, but only the residue joining two double-helical segments radically deviates from B-like torsion values, mainly in ζ and α + 1.
The third distinct architecture of the four-way DNA junction is exemplified by the decamer structures 467D (83) and 1ZF2 (84) with a sharp bend between Residues A6 and C7; the bend can be characterized by a combination of unusually high ɛ and ζ (∼290° and ∼260°, respectively).
Like four-way junctions, also a three-way junction in a complex with trimeric Cre recombinase [e.g. the 1F44 structure (85)] has only one phosphodiester linkage of the junction region in an unusual conformation while the arms retain a near-perfect B-form. In analogy to the four-way junctions listed under Point (iii) above, the only distinction between the junction site and the BI conformation is in the high values of both the ɛ and ζ torsions; ɛ adopts a value of 260°, typical of BII, and the value of ζ is higher (∼210°) than that expected for a BII conformation (∼150°).
DNA in the nucleosome-core particle (NCP)
The nucleosome-core particle consists of 146 or 147 bp of double-stranded DNA wrapped in 1.65 left-handed superhelical turns around four identical pairs of proteins individually known as histones and collectively known as the histone octamer. Nucleosomes, which are ubiquitous in eukaryotic DNA, have been shown to display preferred sequence positioning by bioinformatic analysis of a large volume of sequence data (86). Although these probabilistic relationships cannot yet be confirmed in crystal structures because of the limited volume and variety of crystallized DNA sequences, the atomic resolution of these structures allows for a detailed structural description of the DNA wrapped around the octamer of histone proteins. The bending of DNA around the histone core is achieved by an alteration of the twist angle (87,88) in all six NCPs analyzed in this work [1KX3 and 1KX5 (89), 1P3I and 1P3L (90), 1S32 (91) and 1M19 (92)].
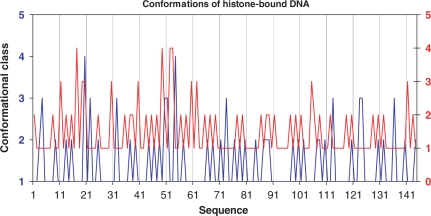
The assignment of individual conformational classes to the backbone of a histone-wrapped DNA makes it possible to describe how DNA bending changes the backbone conformation. The backbone in Structure 1KX5 (89) exhibits a fairly regular periodic alteration of BI and BII conformers, occasionally varied at points of direct protein/DNA contacts by more deformed B-type conformers, characterized by flipped α + 1 and γ + 1 torsions (‘switched BI’, Clusters 113–117, Table 4 and Supplementary Table T2) and by several residues with scattered values of ζ and α + 1. To visualize the structural periodicity, the conformations of the individual steps were plotted as a sequence of events along the polynucleotide chain (Figure 3) and nucleotides with non-BI backbone were plotted as Classes 2, 3, or 4 and appear as peaks in Figure 3. The plot reveals not only the periodic variations of different backbone conformations but also the existence of structural correlations between both chains, the deformed states either directly face each other or are shifted by up to three steps in Chain J with respect to their location in Chain I.
Figure 3.
Dinucleotide conformations in the crystal structure of the histone-core particle 1KX5 (89). Dinucleotides are classified into four conformational families and labeled as follows: BI–1, BII–2, BI conformers with a α + 1/γ + 1 switch (Clusters 113–117) – 3, unclassifiable conformers – 4. One DNA chain, labeled I in the PDB file and drawn in blue and marked in the left y axis in the Figure, is traced from the 5′-end to the 3′-end. The other chain, labeled J in the PDB file and drawn in red in the right y axis, is traced from the 3′-end to the 5′-end. Base paired nucleotides from chains I and J have therefore the same x coordinate.
A comparison of DNA and RNA conformations
The recent dramatic increase in the number of solved RNA structures and the apparent complexity of their folds have drawn great attention to the analysis of RNA conformational space (23,25,31,93–96). Although similar diversity of folds cannot be matched by known DNA structures, DNA also exhibits unusual architectures such as quadruplexes (97,98) or junctions (79,99). The most stable form in both DNA and RNA is the right-handed double-helical arrangement, namely the BI in DNA and AI in RNA (31). These forms are the majority (more than 2/3 in each case) of the analyzed dinucleotides. Despite this shared general feature, the apparent difference between the structural behavior of RNA with the abundance of its 3D folds and of DNA structures composed of self-assembled strands is reflected by a different conformational behavior at the local level. The vast majority of DNA dinucleotides form a bundle of similar conformers, which can transform to one another in an almost continuous fashion. Only specific sequences at high salt concentrations can form radically different Z-DNA. On the other hand, RNA dinucleotides (or ribose-to-ribose ‘suite’ units) form a set of conformers which differ from each other radically (31). The extra hydrogen-bond donor and acceptor, the hydroxyl -O2′H at the ribose ring, stabilizes nucleotide conformations leading to bulges, loops, and consequently to global folds of RNA. When RNA is disrupted from its most stable A-form, it flips to a conformer whose characteristics are very different from the rigid right-handed A helix. DNA, with its numerous, closely related conformers, is ‘soft’, whereas RNA, with fewer, but conformationally very different, conformers, is ‘rigid’ but ‘brittle’. The consequence of this qualitative observation is that RNA conformers’ nomenclature (31), which is based on discrimination between two distinct ribose puckers, C3′-endo and C2′-endo, cannot be used for the description of DNA. This molecule is rather plastic, undergoing very subtle conformational changes which lead to the attenuation of the bimodality of deoxyribose puckers by populating also O4′-endo and C1′-exo puckers.
CONCLUSIONS
The present work has used torsion angles to describe the structural variability of the sugar–phosphate backbone of DNA and to identify the main DNA conformers. The unit under scrutiny was slightly smaller than a dinucleotide (Figure 1). The principal disadvantage of analyses in torsional space, high dimensionality, was overcome by the application of the previously developed technique of Fourier averaging in combination with cluster analysis (23). The classification of the global DNA architecture was not taken into consideration in the clustering process, and all conformational clusters were determined as local conformers.
A detailed analysis of 7739 dinucleotide units from a large number of crystal structures of naked (noncomplexed) DNA structures (187) and complexed DNA structures (260) has confirmed that most conformational variation is covered by several major conformational families (BI, BII, AI, AII). However, the DNA duplex itself is far from being uniform, the dominant conformers have many variants, serving specific roles. Sequence preferences in double-helical A- and B-DNA forms were tested for dinucleotide steps in non-complexed structures while utilizing the nonparametric χ2 goodness-of-fit statistical tests supported by odds-ratio quantities.
- The analysis identified all the known major conformers (AI, AII, BI, BII and Z, Table 4), thereby confirming the validity of the procedure.
- The values of the torsion angles determined from high-resolution naked DNA structures (Table 3) are a reliable source for an accurate structural description of the double-helical forms.
- The BI-form is by far the most populated, and therefore stable, DNA form.
- The torsion values reported for B-DNA based on an analysis of fiber diffraction experiments [Models 4 and 5 in (39)] reflect a combination of the BI- and BII-forms and their values especially for ɛ, ζ and α + 1, β + 1 and γ + 1 (220°, 200°, 330°, 136° and 30°, for Model 4, respectively) significantly differ from the values derived from an analysis based on single crystal structures.
- Based on an analysis of naked DNA, BII should be recognized as a distinct B-form with ζ in the trans region, a high value of ε and a low value of β near 140°. In complexes, the distinction between these two forms almost disappears and the gradual transition from BI to BII is best characterized by a linear anticorrelation of the values of torsions ɛ and ζ: ɛ = −0.73 ζ + 367°.
- Two or more BII conformers only rarely follow each other in sequence; BII–BII steps need to be stabilized by external forces, by either crystal packing or interaction with a complexed molecule.
- Several fairly populated conformers which could not be classified as either A- or B-type conformers were identified. They could be characterized as conformations with one nucleotide of A-type (AI) and the other of B-type (both BI and BII were observed); some represent an A-to-B transitional geometry with O4′-endo or C1′-exo sugar pucker.
- The large total number of minor conformers identified by the analysis has revealed the existence of many energetically low-lying states, which is an important feature of the DNA conformational space.
- Conformers with O4′-endo and C1′-exo sugar puckers corresponding to the transition between the major pucker modes, the C2′-endo and C3′-endo, were identified both in noncomplexed and complexed DNA structures.
- Conformers assigned in naked B and A type double helical structures (Dataset 3) show some important sequence preferences (Tables 5 and 9):
- The BI conformation is numerically dominant in all the sequences (with the possible exception of CA) but it is significantly over-represented in homogenous RR and YY steps except for the GG, which is preferred by the AI-form.
- The observed general preference of YR steps for adopting the BII-form may be explained by the preference of the TG step and its W–C counterpart, the CA step, for this form. Also the GG step was found to prefer BII. On the other hand, the malleability of the CG step to adopt the BII-form, frequently mentioned in the literature, was not confirmed by our analysis.
- Whereas the CG step shows a high propensity for mixed A/B conformations, the GC sequence prefers mixed B/A conformations.
- No YR step was classified as a B/A conformer, and only a few RR and RY steps adopted an A/B conformer, confirming the general reluctance of purines to accept the C3′-endo sugar pucker in the B-like double helix.
- The GC step prefers to adopt either BII or B/A conformers, many GC steps are nonclassified. Of all dinucleotide sequences, this step has conformationally the most complicated behavior.
- The AII conformation is preferentially found in YR sequences but rarely in RY and YY sequences.
DNA crystallized only with solvent, water and metal cations, is conformationally most compact. Its torsion distributions are significantly broadened upon complexation with ligands, especially proteins. Many specific conformers are stabilized by interactions with ligands, they often mix features of the B and A forms enabling DNA to bend and form more specific interactions.
The wrapping of DNA around histone proteins in a nucleosome-core particle is attained by a fairly regular alteration of BI and BII conformers, occasionally substituted by deformed BI or combined B/A conformers (Figure 3).
Some conformers were allocated to structurally and/or functionally distinct types of different DNA forms. The best assignment was achieved in the case of G-quadruplexes from Oxytricha nova telomere, where all dinucleotides were successfully attributed to only a few distinct conformational classes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by Grants No. LC 512 and LC 06024 from the Ministry of Education, Youth and Sports of the Czech Republic, Grant No. A400550701 from the Grant Agency of the Academy of Sciences of the Czech Republic, and Grant No. AV0Z40550506 from the Academy of Sciences of the Czech Republic. Funding to pay the Open Access publication charges for this article was provided by Ministry of Education of the Czech Republic.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Matthews BW. No code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC, Rosenberg JM, Rich A. Sequence specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad.Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pabo CO, Nekludova L. Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki M. A framework for the DNA-protein recognition code of the probe helix in transcription factors: the chemical and stereochemical rules. Structure. 1994;2:317–326. doi: 10.1016/s0969-2126(00)00033-2. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki M, Brenner SE, Gerstein M, Yagi N. DNA recognition code of transcription factors. Protein Eng. 1995;8:319–328. doi: 10.1093/protein/8.4.319. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki M, Gerstein M. Binding geometry of α-helices that recognize DNA. Proteins. 1995;23:525–535. doi: 10.1002/prot.340230407. [DOI] [PubMed] [Google Scholar]
- 9.Mandel-Gutfreund Y, Schueler O, Margalit H. Comprehensive analysis of hydrogen bonds in regulatory protein–DNA complexes: in search of common principles. J. Mol. Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- 10.Dickerson RE, Bansal M, Calladine CR, Diekmann S, Hunter WN, Kennard O, von Kitzing E, Lavery R, Nelson H.CM, Olson W, et al. Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989;8:1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu X.-J, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 12.Olson WK, Gorin AA, Lu X.-J, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Hassan MA, Calladine CR. Two distinct modes of protein-induced bending in DNA. J. Mol. Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- 15.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol. 2000;295:71–83. doi: 10.1006/jmbi.1999.3236. [DOI] [PubMed] [Google Scholar]
- 16.Dickerson RE. DNA Structure from A to Z. how do you tell if a structure is right by reading the paper? Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- 17.Neidle S. Nucleic Acid Structure and Recognition. Oxford: Oxford University Press; 2002. [Google Scholar]
- 18.Felsenfeld G, Davies D, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 19.Morgan AR. Model for DNA replication by Kornberg's DNA polymerase. Nature. 1970;227:1310–1313. doi: 10.1038/2271310a0. [DOI] [PubMed] [Google Scholar]
- 20.Beerman TA, Lebowitz J. Further analysis of the altered secondary structure of superhelical DNA. Sensitivity to methylmercuric hydroxide a chemical probe for unpaired bases. J. Mol. Biol. 1973;79:451–470. doi: 10.1016/0022-2836(73)90398-7. [DOI] [PubMed] [Google Scholar]
- 21.van de Sande JH, Ramsing NB, Germann MW, Elhorst W, Kalisch BW, von Kitzing E, Pon RT, Clegg RC, Jovin TM. Parallel stranded DNA. Science. 1988;241:551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]
- 22.Schneider B, Neidle S, Berman HM. Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers. 1997;42:113–124. doi: 10.1002/(sici)1097-0282(199707)42:1<113::aid-bip10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 23.Schneider B, Moravek Z, Berman HM. RNA conformational classes. Nucleic Acids Res. 2004;32:1666–1677. doi: 10.1093/nar/gkh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh S.-H, Srinivasan AR, Schneider B. The Nucleic Acid Database - a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray LJ, Arendall W.B., III, Richardson DC, Richardson JS. RNA backbone is rotameric. Proc. Natl Acad. Sci. USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView/Xfit- -A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Reijmers TH, Wehrens R, Buydens LM. The influence of different structure representations on the clustering of an RNA nucleotides data set. J. Chem. Inf. Comput. Sci. 2001;41:1388–1394. doi: 10.1021/ci0103626. [DOI] [PubMed] [Google Scholar]
- 28.Ng HL, Kopka ML, Dickerson RE. The structure of a stable intermediate in the A <- -> B DNA helix transition. Proc. Natl Acad. Sci. USA. 2000;97:2035–2039. doi: 10.1073/pnas.040571197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonferroni CE. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. Istituto Superiore di Scienze Economiche e Commerciali di Firenze Firenze. 1936;Vol. 8:3–62. [Google Scholar]
- 30.Agresti A. Categorical data analysis. 2nd. New York: Wiley; 2002. [Google Scholar]
- 31.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Richardson DC, Ham D, Hershkowits E, Williams LD, Keating KS, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature. RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang AH-J, Fujii S, van Boom JH, Rich A. Molecular structure of the octamer d(G-G-C-C-G-G-C-C): modified A-DNA. Proc. Natl Acad.Sci. USA. 1982;79:3968–3972. doi: 10.1073/pnas.79.13.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shakked Z, Rabinovich D, Kennard O, Cruse W.BT, Salisbury SA, Viswamitra MA. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C) J. Mol. Biol. 1983;166:183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- 34.Jain SC, Zon G, Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC) Biochemistry. 1989;28:2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- 35.Olson WK, Sussman JL. How flexible is the furanose ring? I. A comparison of experimental and theoretical studies. J. Am. Chem. Soc. 1982;104:270–278. [Google Scholar]
- 36.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grzeskowiak K, Yanagi K, Privé GG, Dickerson RE. The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G: the effect of base pair reversal. J. Biol. Chem. 1991;266:8861–8883. doi: 10.2210/pdb1d23/pdb. [DOI] [PubMed] [Google Scholar]
- 38.Thorpe JH, Gale BC, Teixeira SC, Cardin CJ. Conformational and hydration effects of site-selective sodium, calcium and strontium ion binding to the DNA Holliday junction structure d(TCGGTACCGA)(4) J. Mol. Biol. 2003;327:97–109. doi: 10.1016/s0022-2836(03)00088-3. [DOI] [PubMed] [Google Scholar]
- 39.Chandrasekaran R, Arnott S. Springer, Berlin: 1989. Landolt-Börnstein Numerical Data and Functional Relationships in Science and Technology, Group VII/1b, Nucleic Acids. [Google Scholar]
- 40.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 41.Bleichenbacher M, Tan S, Richmond TJ. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 2003;332:783–793. doi: 10.1016/s0022-2836(03)00887-8. [DOI] [PubMed] [Google Scholar]
- 42.Savitha G, Viswamitra MA. An A-DNA structure with two independent duplexes in the asymmetric unit. Acta Cryst. D. 1999;55:1136–1143. doi: 10.1107/s0907444999003182. [DOI] [PubMed] [Google Scholar]
- 43.Lu X-J, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 44.Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: A structural analysis. J. Mol. Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 45.Ng H-L, Dickerson RE. Mediation of the A/B-DNA helix transition by G-tracts in the crystal structure of duplex CATGGGCCCATG. Nucleic Acids Res. 2002;30:4061–4067. doi: 10.1093/nar/gkf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doucet J, Benoit J.-P, Cruse W.BT, Prange T, Kennard O. Coexistence of A- and B-form DNA in a single crystal lattice. Nature. 1989;337:190–192. doi: 10.1038/337190a0. [DOI] [PubMed] [Google Scholar]
- 47.Malinina L, Fernandez LG, Huynh-Dinh T, Subirana JA. Structure of the d(CGCCCGCGGGCG) dodecamer: a kinked A-DNA molecule showing some B-DNA features. J. Mol. Biol. 1999;285:1679–1690. doi: 10.1006/jmbi.1998.2424. [DOI] [PubMed] [Google Scholar]
- 48.Vargason JM, Henderson K, Ho PS. A crystallographic map of the transition from B-DNA to A-DNA. Proc. Natl Acad. Sci. USA. 2001;98:7265–7270. doi: 10.1073/pnas.121176898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Hassan MA, Calladine CR. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Phil. Trans. R. Soc. Lond. A. 1997;355:43–100. [Google Scholar]
- 50.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang AH-J, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel GA, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 53.Wang AH-J, Quigley GJ, Kolpak FJ, van der Marel GA, van Boom JH, Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981;211:171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- 54.Gessner RV, Frederick CA, Quigley GJ, Rich A, Wang A.H.-J. The molecular structure of the left-handed Z-DNA double helix at 1.0-Å atomic resolution. J. Biol. Chem. 1989;264:7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, Cai L, Zhang X, Rich A. Crystal structure of a four-stranded intercalated DNA: d(C4) Biochemistry. 1994;33:13540–13546. doi: 10.1021/bi00250a005. [DOI] [PubMed] [Google Scholar]
- 56.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steffen NR, Murphy SD, Lathrop RH, Opel ML, Tolleri L, Hatfield GW. The role of DNA deformation energy at individual base steps for the identification of DNA-protein binding sites. Genome Inform. 2002;13:153–162. [PubMed] [Google Scholar]
- 58.Spolar RS, Record M.T., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 59.Dickerson RE, Chiu TK. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 60.Kono H, Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 61.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton NC, Perona JJ. Role of protein-induced bending in the specificity of DNA-recognition: Crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT) J. Mol. Biol. 1998;277:779–787. doi: 10.1006/jmbi.1998.1655. [DOI] [PubMed] [Google Scholar]
- 63.Burgi HB, Dunitz JD. From crystal statics to chemical-dynamics. Acc. Chem. Res. 1983;16:153–161. [Google Scholar]
- 64.Dunitz JD. From crystal statics to chemical dynamics. Acc. Chem. Res. 1983;16:153–161. [Google Scholar]
- 65.Dickerson RE, Grzeskowiak K, Grzeskowiak M, Kopka ML, Larsen T, Lipanov A, Prive GG, Quintana J, Schultz P, Yanagi K, et al. Polymorphism, packing, resolution, and reliability in single-crystal DNA oligomer analyses. Nucl. Nucl. 1991;10:1. [Google Scholar]
- 66.Berman HM. Crystal studies of B-DNA: the answers and the questions. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 67.Dickerson RE, Goodsell DS, Neidle S. … the tyranny of the lattice. Proc. Natl Acad. Sci. USA. 1994;91:3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki M, Yagi N. Stereochemical basis of DNA bending by transcription factors. Nucleic Acids Res. 1995;23:2083–2091. doi: 10.1093/nar/23.12.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Djuranovic D, Hartmann B. DNA fine structure and dynamics in crystals and in solution: the impact of BI/BII backbone conformations. Biopolymers. 2004;73:356–368. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 70.Bertrand H, Ha-Duong T, Fermandjian S, Hartmann B. Flexibility of the B-DNA backbone: effects of local and neighbouring sequences on pyrimidine-purine steps. Nucleic Acids Res. 1998;26:1261–1267. doi: 10.1093/nar/26.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lefebvre A, Mauffret O, Hartmann B, Lescot E, Fermandjian S. Structural behavior of the CpG step in two related oligonucleotides reflects its malleability in solution. Biochemistry. 1995;34:12019–12028. doi: 10.1021/bi00037a045. [DOI] [PubMed] [Google Scholar]
- 72.Baikalov I, Grzeskowiak K, Yanagi K, Quintana J, Dickerson RE. The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn. J. Mol. Biol. 1993;231:768–784. doi: 10.1006/jmbi.1993.1325. [DOI] [PubMed] [Google Scholar]
- 73.Lefebvre A, Mauffret O, Lescot E, Hartmann B, Fermandjian S. Solution structure of the CpG containing d(CTTCGAAG)2 oligonucleotide: NMR data and energy calculations are compatible with a BI/BII equilibrium at CpG. Biochemistry. 1996;35:12560–12569. doi: 10.1021/bi9606298. [DOI] [PubMed] [Google Scholar]
- 74.Zabin HB, Horvath MP, Terwilliger TC. Approaches to predicting effects of single amino acid substitutions on the function of a protein. Biochemistry. 1991;30:6230–6240. doi: 10.1021/bi00239a022. [DOI] [PubMed] [Google Scholar]
- 75.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 76.Weil J, Min TP, Yang C, Wang SR, Sutherland C, Sinha N, Kang CH. Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT) Acta Cryst. D. 1999;55:422–429. doi: 10.1107/s0907444998012529. [DOI] [PubMed] [Google Scholar]
- 77.Kondo J, Adachi W, Umeda S, Sunami T, Takenaka A. Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats. Nucleic Acids Res. 2004;32:2541–2549. doi: 10.1093/nar/gkh575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lilley DM. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 79.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eichman BF, Vargason JM, Mooers B.HM, Ho PS. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc. Natl Acad. Sci. USA. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thorpe JH, Teixeira SC, Gale BC, Cardin CJ. Structural characterization of a new crystal form of the four-way Holliday junction formed by the DNA sequence d(CCGGTACCGG)2: sequence versus lattice? Acta Cryst. D. 2002;58:567–569. doi: 10.1107/s0907444902001555. [DOI] [PubMed] [Google Scholar]
- 83.Ortiz-Lombardí M, González A, Eritja R, Aymamí J, Azorín F, Coll M. Crystal structure of a DNA holliday junction. Nat. Struct. Biol. 1999;6:913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- 84.Hays FA, Teegarden A, Jones Z.JR, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: A crystallographic map of DNA structure and conformation. Proc. Natl Acad. Sci. USA. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woods KC, Martin SS, Chu V, Baldwin EP. Quasiequivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a Lox three-way DNA junction. J. Mol. Biol. 2001;313:49–69. doi: 10.1006/jmbi.2001.5012. [DOI] [PubMed] [Google Scholar]
- 86.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 88.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 89.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 90.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Molecular recognition of the nucleosomal ‘supergroove’. Proc. Natl Acad. Sci. USA. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 93.Murthy VL, Srinivasan R, Draper DE, Rose GD. A complete conformational map for RNA. J. Mol. Biol. 1999;291:313–327. doi: 10.1006/jmbi.1999.2958. [DOI] [PubMed] [Google Scholar]
- 94.Hershkovitz E, Tannenbaum E, Howerton SB, Sheth A, Tannenbaum A, Williams LD. Automated identification of RNA conformational motifs: theory and application to the HM LSU 23S rRNA. Nucleic Acids Res. 2003;31:6249–6257. doi: 10.1093/nar/gkg835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sims GE, Kim S.-H. Global mapping of nucleic acid conformational space: dinucleoside monophosphate conformations and transition pathways among conformational classes. Nucleic Acids Res. 2003;31:5607–5616. doi: 10.1093/nar/gkg750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sykes MT, Levitt M. Describing RNA structure by libraries of clustered nucleotide doublets. J. Mol. Biol. 2005;351:26–38. doi: 10.1016/j.jmb.2005.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schultze P, Smith FW, Feigon J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG) Structure. 1994;2:221–233. doi: 10.1016/s0969-2126(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Patel DJ. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 99.Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution. Nat. Struct. Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.