Abstract
We report that branched polyamines, including polyamidoamide dendimers, polypropyleneimine, and polyethyleneimine, are able to purge PrPSc, the protease-resistant isoform of the prion protein, from scrapie-infected neuroblastoma (ScN2a) cells in culture. The removal of PrPSc by these compounds depends on both the concentration of branched polymer and the duration of exposure. Chronic exposure of ScN2a cells to low noncytotoxic concentrations of branched polyamines for 1 wk reduced PrPSc to an undetectable level, a condition that persisted at least 3 wk after removal of the compound. Structure–activity analysis revealed that a high surface density of primary amino groups is required for polyamines to eliminate PrPSc effectively from cells. The removal of PrPSc by branched polyamines is attenuated by chloroquine in living cells, and exposure of scrapie-infected brain extracts with branched polyamines at acidic pH rendered the PrPSc susceptible to protease in vitro, suggesting that endosomes or lysozomes may be the site of action. Our studies suggest that branched polyamines might be useful therapeutic agents for treatment of prion diseases and perhaps a variety of other degenerative disorders.
Keywords: neurodegeneration, protein conformation
Prion diseases are a group of fatal neurodegenerative disorders that can occur in hereditary, sporadic, and infectious forms (1). These illnesses occur in humans and a variety of other animals (2). Prions are infectious proteins. The normal cellular form of the prion protein (PrP), designated PrPC, contains three α-helices and has little β-sheet; in contrast, the protein of the prions, denoted the scrapie form of PrP (PrPSc), is rich in β-sheet structure. The accumulation of PrPSc in the central nervous system precedes neurologic dysfunction accompanied by neuronal vacuolation and astrocytic gliosis.
The spectrum of human prion diseases includes kuru (3), Creutzfeldt–Jakob disease (CJD) (4), Gerstmann—Sträussler–Scheinker disease, fatal familial insomnia (5, 6), and a new form of human prion disease, new variant CJD (nvCJD), which has emerged in Great Britain and France (7–9). Several lines of evidence have suggested a link between the nvCJD outbreak and a preceding epidemic of bovine spongiform encephalopathy (7, 10–12). Although it is too early to predict the number of nvCJD cases that might eventually arise in Great Britain and elsewhere (8), it is clear that effective therapeutics for prion diseases are urgently needed. Unfortunately, although a number of compounds including amphotericins, sulfated polyanions, Congo red dye, and anthracycline antibiotics, have been reported as prospective therapeutic agents (13–16), all have demonstrated only modest potential to impede prion propagation, and none have been shown to effect the removal of preexisting prions from an infected host.
Here we report that noncytotoxic concentrations of branched polyamines can rapidly eliminate PrPSc from chronically infected ScN2a cells. These compounds appear to act by stimulating normal cellular mechanisms to destroy PrPSc. Once purged of prions, the treated cells remain free from evidence of scrapie infection during repeated serial passage in polyamine-free media. It remains to be determined whether branched polyamines will find use as therapeutic agents in prion diseases and other degenerative central nervous system disorders characterized by deposits of abnormal proteins such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and frontotemporal dementia (17–23).
Materials and Methods
Chemicals.
High molecular-weight (MW) polyethyleneimine (PEI) was purchased from Fluka. N-(1-[2,3-dioleoyloxy]propyl)-N,N,N-trimethylammonium methylsulfate (DOTAP), cationic lipid was purchased from Boehringer Mannheim, and SuperFect transfection reagent was purchased from QIAGEN (Valencia, CA). All other compounds were purchased from Sigma–Aldrich. All test compounds were dissolved in water at stock concentration of 3 mg/ml and filtered through a Millipore 0.22-μm filter.
Cultured Cells.
Stock cultures of ScN2a cells were maintained in MEM with 10% FBS/10% Glutamax (GIBCO BRL)/100 units/ ml penicillin/100 mg/ml streptomycin (supplemented MEM). Immediately before addition of test compounds, the dishes were washed twice with fresh supplemented MEM. After exposure to test compounds, dishes were drained of media, and cells were harvested by lysis in 0.25–1 ml 20 mM Tris, pH 8.0, containing 100 mM NaCl, 0.5% Nonidet P-40, and 0.5% sodium deoxycholate to obtain a total protein concentration of 1 mg/ml measured by the bicinchoninic acid assay (Pierce). Nuclei were removed from the lysate by centrifugation at 2,000 × g for 5 min. For samples not treated with proteinase K, 40 μl of whole lysate (representing 40 μg total protein) was mixed with an equal volume of 2× SDS reducing sample buffer. For proteinase K digestion, 20 mg/ml proteinase K (Boehringer Mannheim) (total protein/enzyme ratio, 50:1) was added, and the sample was incubated for 1 h at 37°C. Proteolytic digestion was terminated by the addition of Pefabloc (Boehringer Mannheim) to a final concentration of 5 μM. One-milliliter samples were centrifuged at 100,000 × g for 1 h at 4°C, the supernatants were discarded, and the pellets were resuspended in 80 μl of reducing SDS sample buffer for SDS/PAGE.
Brain Homogenates.
Brain homogenates from RML scrapie-affected CD-1 mice [10% (wt/vol) in sterile water] were prepared by repeated extrusion through syringe needles of successively smaller size, from 18 to 22 gauge. Nuclei and debris were removed by centrifugation at 1,000 × g for 5 min. The bicinchnoninic acid protein assay was used to determine protein concentration. Homogenates were adjusted to 1 mg/ml protein in 1% Nonidet P-40. For reactions, 0.5 ml homogenate was incubated with 25 μl 1.0 M buffer (sodium acetate for pH 3–6 and Tris acetate for pH 7–10) plus or minus 10 μl of polyamine stock solution (3 mg/ml) for 2 h at 37°C with constant shaking. The final pH value of each sample was measured directly with a calibrated pH electrode (Radiometer, Copenhagen). After incubation, each sample was neutralized with an equal volume 0.2 M Hepes, pH 7.5, containing 0.3 M NaCl and 4% Sarkosyl. Proteinase K was added to achieve a final concentration of 20 μg/ml, and samples were incubated for 1 h at 37°C. Proteolytic digestion was terminated by the addition of Pefabloc to a final concentration of 5 μM. Ten microliters of digested brain homogenate was mixed with equal volume 2 × SDS sample buffer and analyzed by SDS/PAGE followed by Western blotting.
Western Blotting.
After electrophoresis, Western blotting was performed as previously described (24). Samples were boiled for 5 min and cleared by centrifugation for 1 min at 14,000 × g in a Beckman 5415-C ultrafuge. SDS/PAGE was carried out in 1.5-mm 12% polyacrylamide gels (25). Membranes were blocked with 5% nonfat milk protein in calcium- and magnesium-free PBS plus 0.1% Tween 20 (PBST) for 1 h at room temperature. Blocked membranes were incubated with primary RO73 polyclonal antibody [to detect mouse PrP (MoPrP)] (26) or 3F4 monoclonal antibody (to detect MHM2 PrP) (27) at 1:5,000 dilution in PBST overnight at 4°C. After incubation with primary antibody, membranes were washed 3 × 10 min in PBST, incubated with horseradish peroxidase-labeled secondary antibody (Amersham Life Sciences), diluted 1:5,000 in PBST for 30 to 60 min at 4°C, and washed again for 3 × 10 min in PBST. After chemiluminescent development with enhanced chemiluminescence (ECL) reagent (Amersham) for 1 min, blots were sealed in plastic covers and exposed to ECL Hypermax film (Amersham). Films were processed automatically in a Konica (Japan) film processor.
Results
Elimination of PrPSc from ScN2a Cells Is Dose and Time Dependent.
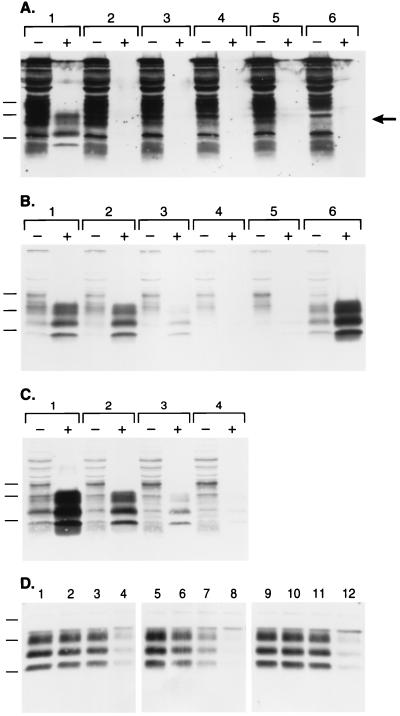
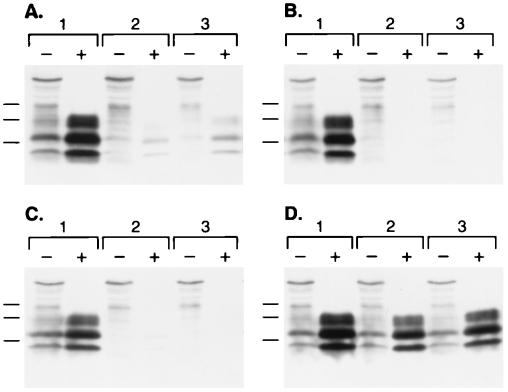
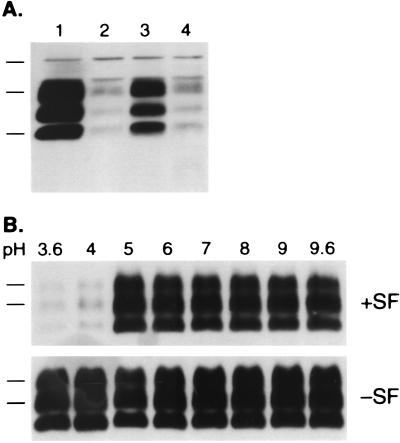
During an effort to optimize the transfection of ScN2a cells with pSPOX expression plasmids (28), we evaluated a transfection protocol that used SuperFect reagent (QIAGEN). Initially, we found that epitope-tagged (MHM2) PrPSc (28) could not be detected in ScN2a cells after SuperFect-mediated transfection, whereas MHM2 PrPSc was efficiently formed when a cationic liposome method for DNA delivery was used (Fig. 1A, lane 1). Close scrutiny revealed that, before protease digestion, SuperFect-transfected samples expressed MHM2 bands (lanes 2–5), which are not seen in the background pattern of an untransfected sample (lane 6). The 3F4 monoclonal antibody does not react with MoPrP but does exhibit high background staining on Western blots of mouse ScN2a cells as shown in lane 6. In lanes 1–5, increased immunostaining in the 20- to 30-kDa region (indicated by the arrow) was observed compared with the nontransfected sample loaded in lane 6. These observations led us to conclude that MHM2 PrP was successfully expressed by using SuperFect transfection reagent, but that conversion of MHM2 PrPC to protease-resistant MHM2 PrPSc was inhibited by SuperFect.
Figure 1.
Branched polyamines inhibit formation of nascent PrPSc and induce clearance of preexisting PrPSc. (A) Western blot probed with 3F4 monoclonal antibody recognizes newly expressed MHM2 PrP (region indicated by arrow). ScN2a cells were exposed to SuperFect for 3 h and harvested 3 d after removal of SuperFect. Minus (−) symbol denotes undigested control sample, and plus (+) symbol designates sample subjected to limited proteolysis. Lane 1: N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP)-mediated transfection. Lane 2: 30 μg/ml SuperFect, 5 μg pSPOX MHM2. Lane 3: 75 μg/ml SuperFect, 5 μg pSPOX MHM2. Lane 4: 150 μg/ml SuperFect, 5 μg pSOX MHM2. Lane 5: 150 μg/ml SuperFect, 10 μg pSPOX MHM2. Lane 6: No addition of either transfection reagent or DNA. Forty microliters of undigested cell lysate was used, whereas those samples subjected to limited digestion with proteinase K were concentrated 25-fold before SDS/PAGE. One milliliter of the digest was centrifuged at 100,000 × g for 1 h at 4°C and the pellets suspended in 80 μl of SDS sample buffer before SDS/PAGE followed by Western blotting. Apparent molecular masses based on migration of protein standards are 34.2, 28.3, and 19.9 kDa. (B) The blot in A was stripped of the mAb and redeveloped with the RO73 polyclonal antibody, which identifies all rodent PrPs including endogenous MoPrP. (C) Dose-dependent removal of PrPSc. Minus (−) symbol denotes undigested control sample, and plus (+) symbol designates sample subjected to limited proteolysis. Lane 1: No SuperFect. Lane 2: 30 μg/ml SuperFect. Lane 3: 75 μg/ml SuperFect. Lane 4: 150 μg/ml SuperFect. ScN2a cells were exposed to SuperFect for 3 h and harvested 3 d after removal of SuperFect. Apparent molecular masses based on migration of protein standards are 33.9, 28.8, and 20.5 kDa. (D) Time-dependent removal of PrPSc. ScN2a cells were exposed to 7.5 μg/ml: SuperFect (lanes 1–4), PEI (average-MW ≈60,000) (lanes 5–8), or PAMAM, generation 4.0 (lanes 9–12). Duration of exposure times for each polyamine: 0 h (lanes 1, 5, and 9), 4 h (lanes 2, 6, and 10), 8 h (lanes 3, 7, and 11), 16 h (lanes 4, 8, and 12). All samples shown were subjected to limited proteolysis to measure PrPSc. Apparent molecular masses based on migration of protein standards are 38, 26, and 15 kDa.
To investigate this apparent inhibition, we reprobed the Western blot with RO73 polyclonal antiserum to detect endogenous MoPrPSc, the presence of which is diagnostic for prion infection in ScN2a cells (29). We were surprised to find that the SuperFect-treated ScN2a cells no longer contained detectable quantities of MoPrPSc (Fig. 1B, lanes 4 and 5). To investigate the mechanism by which SuperFect reduced the level of preexisting PrPSc in chronically infected ScN2a cells, we measured endogenous PrPSc in ScN2a cells exposed to various concentrations of SuperFect in the absence of plasmid DNA. Indeed, treatment with SuperFect caused the disappearance of PrPSc from ScN2a cells in a dose-dependent manner (Fig. 1C). The concentration of SuperFect required to eliminate >95% of preexisting PrPSc with a 3-h exposure was 150 μg/ml (lane 4). Duration of treatment also influenced the ability of SuperFect to remove PrPSc from ScN2a cells: exposure to 150 μg/ml SuperFect for 10 min did not affect PrPSc levels (data not shown), whereas 7.5 μg/ml SuperFect eliminated all detectable PrPSc with a t1/2= 8 h (Fig. 1D).
Polyamines with a High Surface Amino Group Density Are the Most Effective at Eliminating PrPSc.
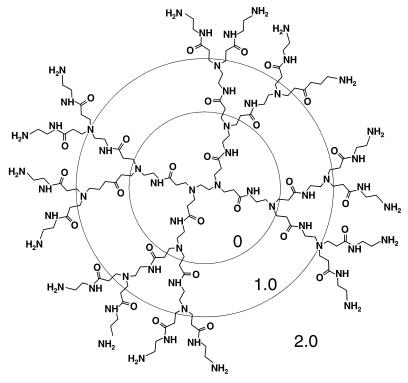
SuperFect is a mixture of branched polyamines derived from heat-induced degradation of a polyamidoamide (PAMAM) dendrimer (30). Therefore, we investigated the ability of several other branched and unbranched polymers to eliminate PrPSc from ScN2a cells (Table 1). The branched polymers investigated include various preparations of PEI as well as intact PAMAM and polypropyleneimine (PPI) dendrimers. Dendrimers are manufactured by a repetitive divergent growth technique, allowing the synthesis of successive well-defined “generations” of homodisperse structures (Fig. 2). The potency of both PAMAM and PPI dendrimers in eliminating PrPSc from ScN2a cells increased as the generation level increased: the most potent compounds with respect to eliminating PrPSc were PAMAM generation 4.0 and PPI generation 4.0, whereas PAMAM generation 1.0 showed very little ability to eliminate PrPSc (Table 1). Similarly, a high-MW fraction of PEI was more potent than low-MW PEI.
Table 1.
Removal of PrPSc by polymer compounds
| Compound | Molecular weight | Primary NH2 groups | IC50, ng/ml |
|---|---|---|---|
| PAMAM generation 0.0 | 517 | 4 | >10,000 |
| PAMAM generation 1.0 | 1,430 | 8 | >10,000 |
| PAMAM generation 2.0 | 3,256 | 16 | 2,000 |
| PAMAM generation 3.0 | 6,909 | 32 | 400 |
| PAMAM generation 4.0 | 14,215 | 64 | 80 |
| PAMAM-OH generation 4.0 | 14,279 | 0 | >10,000 |
| PPI generation 2.0 | 773 | 8 | 2,000 |
| PPI generation 4.0 | 3,514 | 32 | 80 |
| Low-MW PEI | ∼25,000 | 2,000 | |
| Average-MW PEI | ∼60,000 | 400 | |
| High-MW PEI | ∼800,000 | 80 | |
| Linear PEI | ∼60,000 | 2,000 | |
| poly(l-lysine) | ∼60,000 | >500 | 10,000 |
| SuperFect | 400 |
All compounds were tested at five different concentrations. PrPSc levels were measured by densitometry of Western blot signals.
IC50, approximate concentration of polymer required to reduce PrPSc to 50% of control levels in ScN2a cells after exposure for 16 h.
Figure 2.
Schematic diagram of PAMAM, generation 2.0 (ethylene diamine core). Successive full generations are indicated by concentric circles.
From the foregoing data, it is clear that for all three branched polyamines tested, increasing molecular size corresponded to an increased potency for eliminating PrPSc. To determine whether this trend was directly attributable to increased surface density of amino groups on the larger molecules, we also tested PAMAM-OH generation 4.0, a dendrimer that resembles PAMAM generation 4.0 except that hydroxyls replace amino groups on its surface. Unlike PAMAM generation 4.0, PAMAM-OH generation 4.0 did not cause a reduction of PrPSc levels even at the highest concentration tested (10 μg/ml), establishing that the amino groups are required for the elimination of PrPSc by PAMAM (Table 1). In an effort to assess the contribution of the branched architecture to the clearing ability of polyamines for PrPSc, we also tested the linear molecules poly(l-lysine) and linear PEI. Both of these linear compounds were less potent than a preparation of branched PEI with similar average-MW (Table 1), establishing that a branched molecular architecture optimizes the ability of polyamines to eliminate PrPSc, presumably because the branched structures achieve a higher density of surface amino groups.
Kinetics of PrPSc Elimination by Polyamines.
The preceding results demonstrate the potent ability of branched polyamines to clear PrPSc from ScN2a cells within a few hours of treatment. To explore whether these compounds could be used as a potential therapeutic for treatment of prion disease, we tested whether they were cytotoxic for ScN2a cells, using as criteria cell growth, morphology, and viability as measured by trypan blue staining. None of the compounds was cytotoxic to ScN2a cells after exposure for 1 wk at concentrations up to 7.5 μg/ml (data not shown). To determine whether branched polyamines can cure ScN2a cells of scrapie infection without affecting cell viability, we next examined the kinetics of prion clearance in the presence of a noncytotoxic concentration (7.5 μg/ml) of three different branched polyamines. ScN2a cells were exposed to SuperFect, PEI, or PAMAM generation 4.0 (Fig. 1D) for varying periods of time, and the kinetics of PrPSc elimination were assessed by Western blotting. All three compounds caused a substantial reduction in PrPSc levels after 8–16 h of treatment, and of the three compounds, PEI appeared to remove PrPSc most quickly, with a t1/2= 4 h (Fig. 1D).
Curing Neuroblastoma Cells of Scrapie Infection.
Encouraged by our success in reversing the accumulation of PrPSc in ScN2a cells under noncytotoxic conditions, we established that an extended exposure to even lower levels of the branched polyamines (1.5 μg/ml) was sufficient to eliminate PrPSc (data not shown). On the basis of these findings, we used this protocol to determine whether the severe reduction in PrPSc levels after exposure to branched polyamines would persist after removal of the compounds. After the exposure of ScN2a cells to a 1.5 μg/ml SuperFect for 1 wk, PrPSc was reduced to <1% of the baseline level but then increased back to ≈5% of the baseline level after 3 additional wk in culture in the absence of polyamine (Fig. 3A). In contrast, after exposure to 1.5 μg/ml of either PEI or PAMAM generation 4.0 for 1 wk, PrPSc was completely eliminated and did not return even after 3 wk of culture in control media without polyamines (Fig. 3 B and C). A more intensive course of treatment with 1.8 μg/ml SuperFect for 9 d also cured ScN2a cells of scrapie infection fully, manifested by the absence of PrPSc 1 mo after removal of SuperFect (data not shown).
Figure 3.
Persistent reduction in PrPSc levels. ScN2a cells were exposed to 1.5 μg/ml: (A) SuperFect, (B) PEI (average-MW ≈60,000), (C) PAMAM, generation 4.0, or (D) no addition. Cells were harvested: lane 1, before addition; lane 2, immediately after 1 wk continuous exposure to test compounds; and lane 3, 3 wk in polyamine-free medium after removal of test compounds. Minus (−) symbol denotes undigested control sample, and plus (+) symbol designates sample subjected to limited proteolysis. Apparent molecular masses based on migration of protein standards are 33.9, 28.8, and 20.5 kDa.
Evidence for Polyamines Acting Within an Acidic Compartment.
The potent activity of branched polyamines in rapidly clearing scrapie prions from cultured ScN2a cells led us to explore the mechanism by which these drugs might act. All of the compounds that effect removal of PrPSc from ScN2a cells are known to traffic through endosomes (31, 32). Because PrPC is converted into PrPSc in caveolae-like domains or rafts (33–36) and is then internalized through the endocytic pathway (37, 38), we reasoned that polyamines might act on PrPSc in endosomes or lysosomes. To test this hypothesis, we first investigated the effect of pretreatment with the lysosomotropic agents chloroquine and NH4Cl on the ability of polyamines to eliminate PrPSc. These lysosomotropic agents alkalinize endosomes and have no effect on PrPSc levels when administered to ScN2a cells (39). In our experiments, 100 μM chloroquine, but not 30 mM NH4Cl, blocked the ability of PEI to eliminate PrPSc (Fig. 4A). Similar results were obtained with SuperFect and PAMAM, generation 4.0 (data not shown). The ability of chloroquine to attenuate the ability of branched polyamines to remove PrPSc is consistent with the notion that these agents act in either endosomes or lysosomes. More work is required to determine the precise mechanism by which chloroquine attenuates the clearance of PrPSc by branched polyamines.
Figure 4.
Acidic conditions favor polyamine removal of PrPSc. (A) ScN2a cells were treated, lane 1: Control media. Lane 2: 7.5 μg/ml PEI (average-MW ≈60,000). Lane 3: PEI plus 100 μM chloroquine. Lane 4: PEI plus 30 mM NH4Cl. Chloroquine and NH4Cl were added 1 h before addition of PEI. Cells were harvested 16 h after addition of PEI. All samples shown were subjected to limited proteolysis to measure PrPSc. Apparent molecular masses based on migration of protein standards are 38, 26, and 15 kDa. (B) In vitro mixture of crude mouse brain homogenates with SuperFect under a range of pH conditions was performed as described in Methods (measured pH of each sample before neutralization denoted above the lanes). Addition of 60 μg/ml SuperFect denoted as “+SF” and control with no addition as “-SF”. All samples shown were adjusted to pH 7.5 and then subjected to limited proteolysis to measure PrPSc. Apparent molecular masses based on migration of protein standards are 30 and 27 kDa.
Nonetheless we considered the possibility that in an acidic environment, branched polyamines, by indirectly interacting either with PrPSc or with another cellular component, could cause PrPSc to become susceptible to hydrolases present in the endosome/lysozome. In an effort to test this hypothesis, we developed an in vitro degradation assay to evaluate the effect of pH on the ability of polyamines to render PrPSc sensitive to protease. Crude homogenates of scrapie-infected mouse brain were exposed to a broad range of pH values in the presence or absence of SuperFect and then treated with proteinase K before Western blotting. Whereas PrPSc remained resistant to protease hydrolysis throughout the pH range (3.6–9.6) in the absence of SuperFect, addition of the branched polyamine at pH 4.0 or below caused PrPSc to become almost completely degraded by protease (Fig. 4B). The conformational change of PrP induced by polyamines required only transient exposure to acidic pH and could not be reversed by neutralization, even after 16 hr.
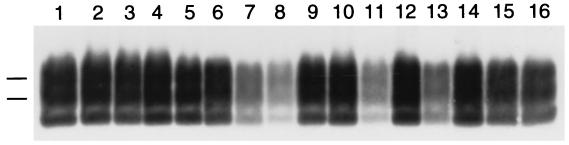
The dramatic effect of polyamine addition observed showing that clearance in vitro is optimized below pH 4 (Fig. 4B) is consistent with our hypothesis that polyamines act on PrPSc in an acidic compartment. To establish that our in vitro degradation assay (Fig. 4B) is a valid approximation of the mechanism by which branched polyamines enhance the clearance of PrPSc from cultured cells, we performed a structure activity analysis with several of the compounds tested in cultured cells (Table 1). We found an excellent correlation between the clearance of PrPSc in cultured ScN2a cells (Table 1) and the ability to render PrPSc susceptible to protease at acidic pH in vitro (Fig. 5). Notably, PAMAM-OH generation 4.0 failed to render PrPSc susceptible to protease, whereas PAMAM generation 4.0 and PPI, generation 4.0 exhibited an even stronger activity than SuperFect in vitro (Fig. 5), as expected from their observed potency in cultured ScN2a cells (Table 1).
Figure 5.
Removal of PrPSc from brain homogenate in vitro by various polyamines. Samples were incubated with polyamines at pH 3.6 and processed as described in Methods. Each polyamine was tested at 60 μg/ml concentration. Lanes 1 and 2: control. Lane 3: poly(l-lysine). Lane 4: PAMAM, generation 0.0. Lane 5: PAMAM, generation 1.0. Lane 6: PAMAM, generation 2.0. Lane 7: PAMAM, generation 3.0. Lane 8: PAMAM, generation 4.0. Lane 9: PAMAM-OH, generation 4.0. Lane 10: PPI, generation 2.0. Lane 11: PPI, generation 4.0. Lane 12: linear PEI. Lane 13: high-MW PEI. Lane 14: low-MW PEI. Lane 15: average-MW PEI. Lane 16: SuperFect. All samples shown were subjected to limited proteolysis to measure PrPSc. Apparent molecular masses based on migration of protein standards are 30 and 27 kDa.
Discussion
We described certain branched polyamines that cause the rapid elimination of PrPSc from ScN2a cells in a dose- and time-dependent manner. These compounds demonstrate a potent ability to remove prions from cultured cells at concentrations that are completely noncytotoxic, and the cells may be maintained indefinitely in culture in the presence of therapeutic levels of branched polyamines. Furthermore, when ScN2a cells were exposed to these compounds for ≈1 wk, PrPSc was reduced to undetectable levels and remained so for at least 1 mo after removal of the polyamine. Although it remains to be established whether these compounds may eventually be useful in treatment of humans or domestic animals with prion disease, our results clearly establish the potential of branched polyamines to promote the elimination of detectable PrPSc. Additional bioassay studies in transgenic mice are in progress to confirm that prion infectivity is also eliminated by these branched polyamines.
Clarification of the exact mechanism of PrPSc elimination by branched polyamines is an important objective. Although a number of possible scenarios exist, several possibilities may be excluded already. One possibility that we considered, that polyamines act by induction of chaperones such as heat-shock proteins that mediate PrP refolding, can be eliminated because of our apparent success in reproducing the phenomenon in vitro. Furthermore, polyamines seem to offer advantages over other putative therapeutics that would seek to promote refolding: at very high concentrations, DMSO and glycerol act as direct “chemical chaperones” and inhibit the formation of new PrPSc (40), but these compounds cannot reduce preexisting PrPSc levels. Furthermore, polyamines inhibit PrPSc formation at much lower concentrations than these agents. The ability of polyamines to effect the rapid clearance of PrPSc also contrasts with the activity of other potential prion therapeutics. Sulfated polyanions may inhibit PrPSc accumulation in ScN2a cells by directly binding to PrPC (41, 42), but because branched polyamines are able to clear preexisting PrPSc, their mechanism of action cannot simply involve binding to PrPC and inhibiting de novo synthesis.
Another possible mechanism that can be excluded is endosomal rupture. The branched polyamines that were effective in clearing PrPSc from ScN2a cells in our experiments, PEI, SuperFect, and PAMAM, are also potent lysosomotropic osmotic agents that can swell in acidic environments and rupture endosomes (31, 32). This might suggest that branched polyamines clear PrPSc from ScN2a cells by rupturing endosomes and exposing PrPSc to cytosolic degradation processes. However, it is known that the lysosomotropic endosome-rupturing agents NH4Cl, chloroquine, and monensin do not interfere with the formation of PrPSc in ScN2a cells (39). Furthermore, we found that chloroquine interferes with the ability of branched polyamines to clear PrPSc, and that polyamines can clear PrPSc in vitro at acidic pH in the absence of cell membranes. Together, these observations rule out endosome rupture as the mechanism by which branched polyamines remove PrPSc.
From our preliminary data, it seems likely that branched polyamines require the acidic environment of intact endosomes or lysosomes to destroy PrPSc. The structure–activity profile of polymers we tested reveals that the most active compounds possess densely packed regularly spaced amino groups, suggesting that these compounds may bind to a ligand that has periodically spaced negative charges. Several scenarios remain possible. (i) Branched polyamines may bind directly to PrPSc arranged as an amyloid with exposed negatively charged moieties and induce a conformational change under acidic conditions. (ii) Treatment of PrP 27–30 with acid decreases turbidity and increases α-helical content, suggesting that such conditions might dissociate PrPSc into monomers (43). It is therefore possible that polyamines bind to an equilibrium unfolding intermediate of PrPSc present under acidic conditions. (iii) Alternatively, polyamines might sequester a cryptic negatively charged component bound to PrPSc that is essential for protease resistance, but which is released only when PrPSc undergoes an acid-induced conformational change. Such a component might act as a chaperone for PrPSc inside endosomes or lysosomes. (iv) Finally, another possibility is that polyamines activate an endosomal or lysosomal factor that can induce a conformational change in PrPSc. Clearly, more work will be required to determine the precise mechanism by which branched polyamines destroy PrPSc.
The in vitro assay described here may find more general application in the search for drugs that effectively treat as well as prevent a number of degenerative and inherited diseases, where the accumulation of proteins seems to mediate the pathogenesis of these illnesses. By simulating lysosomes, where proteases hydrolyze proteins under acidic conditions, we were able to evaluate rapidly the efficacy of a variety of polyamines to induce degradation of PrPSc (Table 1). It may be possible to develop similar screening assays with wild-type and mutant proteins like the amyloid precursor protein, α-synuclein, superoxide dismutase, tau, amylin, and transthyretin, found to accumulate in some of the disorders mentioned below.
Besides using the in vitro assay to screen for potential drugs, branched polyamines might provide a new tool for exploring the conversion of PrPC into PrPSc. The mechanism by which branched polyamines render PrPSc susceptible to proteolysis remains to be established. Whether the interaction of branched polyamines with PrPSc is reversible is unknown. In addition, we do not know whether branched polyamines are able to solubilize PrPSc without irreversibly denaturing the protein. Whatever the mechanism by which branched polyamines interact with PrPSc, it is likely to be different from that found with chaotropes as well as denaturing detergents and solvents (44).
In conclusion, certain specific branched polyamines appear to mediate the clearance of PrPSc from cultured cells under noncytotoxic conditions. These compounds offer the intriguing possibility of therapeutic reagents for prion diseases. Because the drugs appear to act by stimulating normal cellular pathways of protein degradation to destroy PrPSc, it is tempting to speculate that this class of compounds might be of value in the treatment of other degenerative and hereditary disorders where abnormally folded, wild-type, or mutant proteins accumulate. Such an approach may find merit in developing effective therapeutics for one or more of the common degenerative illnesses, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, frontotemporal dementia, adult onset diabetes mellitus, and the amyloidoses (17–23, 45). Whether branched polyamines might also prove efficacious in a variety of inherited disorders where the accumulation of abnormal proteins is a hallmark of the illness remains to be established; these genetic maladies include heritable forms of prion disease, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, frontotemporal dementia, Pick’s disease, and amyloidosis, as well as the triplet repeat diseases including Huntington’s disease, spinal cerebellar ataxias, and myotonic dystrophy (46, 47). Perhaps branched polyamines will find utility in preventing or delaying the onset of these genetic diseases, where carriers can often be identified decades in advance of detectable neurologic or systemic dysfunction.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) (NS14069, AG08967, AG02132, and AG10770) and the American Health Assistance Foundation, as well as a gift from the Leila and Harold Mathers Foundation. Surachai Supattapone is supported by the Burroughs Wellcome Fund Career Development Award and an NIH Clinical Investigator Development Award (K08 NS02048–02).
Abbreviations
- PAMAM
polyamidoamide
- PPI
polypropyleneimine
- PEI
polyethyleneimine
- ScN2a
scrapie-infected neuroblastoma
- PrP
prion protein
- PrPC
cellular form of PrP
- PrPSc
scrapie isoform of PrP
- MW
molecular weight
- MoPrP
mouse PrP
Note Added in Proof
We have recently observed that PPI generation 4.0 does not clear PrPSc from Sc237-infected Syrian hamster brain homogenates using the procedure described here, suggesting that prions may display strain- or sequence-dependent differences in their susceptibility to branched polyamines.
References
- 1.Prusiner S B. Annu Rev Microbiol. 1989;43:345–374. doi: 10.1146/annurev.mi.43.100189.002021. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajdusek D C, Gibbs C J, Jr, Alpers M. Nature (London) 1966;209:794–796. doi: 10.1038/209794a0. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs C J, Jr, Gajdusek D C, Asher D M, Alpers M P, Beck E, Daniel P M, Matthews W B. Science. 1968;161:388–389. doi: 10.1126/science.161.3839.388. [DOI] [PubMed] [Google Scholar]
- 5.Goldfarb L G, Petersen R B, Tabaton M, Brown P, LeBlanc A C, Montagna P, Cortelli P, Julien J, Vital C, Pendelbury W W. Science. 1992;258:806–808. doi: 10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 6.Medori R, Montagna P, Tritschler H J, LeBlanc A, Cortelli P, Tinuper P, Lugaresi E, Gambetti P. Neurology. 1992;42:669–670. doi: 10.1212/wnl.42.3.669. [DOI] [PubMed] [Google Scholar]
- 7.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 8.Cousens S N, Vynnycky E, Zeidler M, Will R G, Smith P G. Nature (London) 1997;385:197–198. doi: 10.1038/385197a0. [DOI] [PubMed] [Google Scholar]
- 9.Will R G, Cousens S N, Farrington C P, Smith P G, Knight R S G, Ironside J W. Lancet. 1999;353:979. doi: 10.1016/s0140-6736(99)01160-5. [DOI] [PubMed] [Google Scholar]
- 10.Bruce M E, Will R G, Ironside J W, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, et al. Nature (London) 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 11.Hill A F, Desbruslais M, Joiner S, Sidle K C L, Gowland I, Collinge J, Doey L J, Lantos P. Nature (London) 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 12.Lasmézas C I, Deslys J-P, Demaimay R, Adjou K T, Lamoury F, Dormont D, Robain O, Ironside J, Hauw J-J. Nature (London) 1996;381:743–744. doi: 10.1038/381743a0. [DOI] [PubMed] [Google Scholar]
- 13.Ingrosso L, Ladogana A, Pocchiari M. J Virol. 1995;69:506–508. doi: 10.1128/jvi.69.1.506-508.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagliavini F, McArthur R A, Canciani B, Giaccone G, Porro M, Bugiani M, Lievens P M-J, Bugiani O, Peri E, Dall’Ara P, et al. Science. 1997;276:1119–1122. doi: 10.1126/science.276.5315.1119. [DOI] [PubMed] [Google Scholar]
- 15.Masullo C, Macchi G, Xi Y G, Pocchiari M. J Infect Dis. 1992;165:784–785. doi: 10.1093/infdis/165.4.784. [DOI] [PubMed] [Google Scholar]
- 16.Ladogana A, Casaccia P, Ingrosso L, Cibati M, Salvatore M, Xi Y-G, Masullo C, Pocchiari M. J Gen Virol. 1992;73:661–665. doi: 10.1099/0022-1317-73-3-661. [DOI] [PubMed] [Google Scholar]
- 17.Beyreuther K, Masters C L. Nat Med. 1997;3:723–725. doi: 10.1038/nm0797-723. [DOI] [PubMed] [Google Scholar]
- 18.Masters C L, Beyreuther K. Br Med J. 1998;316:446–448. doi: 10.1136/bmj.316.7129.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selkoe D J. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 20.Selkoe D J. Nature (London) 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 21.Wong P C, Pardo C A, Borchelt D R, Lee M K, Copeland N G, Jenkins N A, Sisodia S S, Cleveland D W, Price D L. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 22.Spillantini M G, Crowther R A, Jakes R, Hasegawa M, Goedert M. Proc Natl Acad Sci USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutton M, Lendon C L, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Nature (London) 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 24.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, et al. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 27.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler D A, Scott M R D, Bockman J M, Borchelt D R, Taraboulos A, Hsiao K K, Kingsbury D T, Prusiner S B. J Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M X, Redemann C T, Szoka F C J. Bioconjugate Chem. 1996;7:703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 31.Boussif O, Lezoualc’h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haensler J, Szoka F C J. Bioconjugate Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 33.Gorodinsky A, Harris D A. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner S B. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond S J, Smart E J, Anderson R G, Taraboulos A, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caughey B, Raymond G J, Ernst D, Race R E. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borchelt D R, Taraboulos A, Prusiner S B. J Biol Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 39.Taraboulos A, Raeber A J, Borchelt D R, Serban D, Prusiner S B. Mol Biol Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatzelt J, Prusiner S B, Welch W J. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 41.Gabizon R, Meiner Z, Halimi M, Ben-Sasson S A. J Cell Physiol. 1993;157:319–325. doi: 10.1002/jcp.1041570215. [DOI] [PubMed] [Google Scholar]
- 42.Caughey B, Brown K, Raymond G J, Katzenstein G E, Thresher W. J Virol. 1994;68:2135–2141. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Biochemistry. 1994;33:8375–8383. doi: 10.1021/bi00193a027. [DOI] [PubMed] [Google Scholar]
- 44.Prusiner S B, Groth D, Serban A, Stahl N, Gabizon R. Proc Natl Acad Sci USA. 1993;90:2793–2797. doi: 10.1073/pnas.90.7.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone M J. Blood. 1990;75:531–545. [PubMed] [Google Scholar]
- 46.Fu Y-H, Kuhl D P A, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J M H, Holden J J A, Fenwick R G, Jr, Warren S T, et al. Science. 1992;255:1256–1259. [Google Scholar]
- 47.The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]