Abstract
Pathogenic mutations in presenilin 1 (PS1) are associated with ≈50% of early-onset familial Alzheimer disease. PS1 is endoproteolytically cleaved to yield a 30-kDa N-terminal fragment (NTF) and an 18-kDa C-terminal fragment (CTF). Using COS7 cells transfected with human PS1, we have found that phorbol 12,13-dibutyrate and forskolin increase the state of phosphorylation of serine residues of the human CTF. Phosphorylation of the human CTF resulted in a shift in electrophoretic mobility from a single major species of 18 kDa to a doublet of 20–23 kDa. This mobility shift was also observed with human PS1 that had been transfected into mouse neuroblastoma (N2a) cells. Treatment of the phosphorylated CTF doublet with phage λ protein phosphatase eliminated the 20- to 23-kDa doublet while enhancing the 18-kDa species, consistent with the interpretation that the electrophoretic mobility shift was due to the addition of phosphate to the 18-kDa species. The NTF and CTF eluted from a gel filtration column at an estimated mass of over 100 kDa, suggesting that these fragments exist as an oligomerized species. Upon phosphorylation of the PS1 CTF, the apparent mass of the NTF- or CTF-containing oligomers was unchanged. Thus, the association of PS1 fragments may be maintained during cycles of phosphorylation/dephosphorylation of the PS1 CTF.
Alzheimer disease (AD), the major cause of dementia in the elderly, is a progressive neurodegenerative disease. Both aging and genetic factors contribute to the predisposition to AD. The majority of early-onset AD, occurring at less than 60 years of age, is an inherited Mendelian disorder. Autosomal dominant familial AD (FAD) is associated with mutations in one of three different genes (1, 2). The first identified FAD gene (3–6), on chromosome 21, encodes the β-amyloid precursor protein (APP), from which is derived the Aβ peptide, a major component of the cerebral plaques that are characteristic of AD (7). The other two FAD genes are presenilin 1 (PS1) and presenilin 2 (PS2), located on chromosomes 14 and 1, respectively (8–11). Mutations in PS1 have been identified in about half of the early-onset FAD pedigrees studied (1). At least 30 different pathogenic mutations in PS1 have been identified, while only two pathogenic mutations in PS2 have been found despite exhaustive searches (2, 12).
PS1 and PS2 share significant identity over their amino acid sequences (8, 11, 13) and are predicted to have multiple transmembrane domains (14). The presenilins are homologous to the Caenorhabditis elegans sel-12 protein (15), which has a similar topology (16) and is involved in the Notch signaling pathway (17). The biological functions of the presenilins are as yet unknown.
Examination of presenilin metabolism both in vivo and in vitro has demonstrated that PS1 is proteolytically cleaved, yielding a 30-kDa N-terminal fragment (NTF) and an 18-kDa C-terminal fragment (CTF) (2, 18). Immunofluorescence studies, utilizing antibodies to either terminus of the protein, indicate that NTF and CTF are colocalized to the endoplasmic reticulum (ER) and Golgi apparatus (19), raising the possibility that the NTFs and CTFs remain associated with each other following cleavage. Neither the subcellular compartment where cleavage occurs nor the subcellular distribution of the fragments has been determined. The presenilins are neither glycosylated nor sulfated (20, 21), consistent with their localization to the ER and early Golgi (19, 20).
Several pathogenic mutations in PS1 occur in the major hydrophilic loop of the protein between amino acid residues 263 and 407 (2, 12). This region of the protein contains the site of endoproteolytic cleavage as well as several potential phosphorylation sites. One pathogenic mutation abolishes an mRNA splicing acceptor site (22), yielding a protein which fails to undergo proteolytic cleavage (18).
Since post-translational modifications may regulate either endoproteolytic cleavage of PS1 or subcellular distribution of the PS1 fragments, or both, we have examined the regulation of phosphorylation and the assembly of the proteolytic fragments of PS1. We demonstrate that the CTF of PS1 is phosphorylated in response to activation of either protein kinase C (PKC) or cAMP-dependent protein kinase (PKA) or to inhibition of protein phosphatase 1 (PP1) or protein phosphatase 2A (PP2A). In addition, we provide biochemical evidence suggesting that PS1 NTFs and CTFs assemble into multimeric complexes. As judged by size fractionation using gel filtration chromatography and discontinuous sucrose gradients, the apparent assembly of PS1 fragments was maintained after phosphorylation of the PS1 CTF.
MATERIALS AND METHODS
Materials.
Phorbol 12,13-dibutyrate (PDBu) (prepared as a 1 mM stock solution in dimethyl sulfoxide; LC Services, Woburn, MA), forskolin (2 mM in dimethyl sulfoxide), thapsigargin (1 mM in dimethyl sulfoxide), calyculin A (100 μM in dimethyl sulfoxide), and cyclosporin A (1 mM in ethanol; Calbiochem) were diluted to the indicated final concentrations immediately before use. All other chemicals were obtained from Sigma.
Ab 14 antiserum (anti-residues 1–25) and α PS1-loop antiserum (anti-residues 263–407) were prepared as described (18).
Cell Culture.
COS7 cells transfected with human PS1 (COS-huPS1; ref. 19), N2a cells transfected with human PS1 (23), and rat pheochromocytoma (PC12) cells were used. COS-huPS1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY)/10% fetal bovine serum/200 μg/ml G418 (Mediatech, Herndon, VA). Neuroblastoma (N2a) cells were cultured in 50% DMEM/45% Opti-MEM (Life Technologies)/5% fetal bovine serum/200 μg/ml G418. PC12 cells were cultured in DMEM/10% fetal bovine serum/5% horse serum. All cells were plated at equal densities and were maintained at 37°C in an atmosphere of 95% air/5% CO2 in 60- or 100-mm culture dishes.
Incubation and Lysis of Cells.
PDBu (1 μM), forskolin (5 μM), thapsigargin (100 nM), calyculin A (10 nM), or cyclosporin A (100 nM) (Calbiochem) was added as indicated, and incubation was for 45 min. Cells were washed once with PBS. Lysis buffer (0.5% Nonidet P-40/0.5% deoxycholate/PBS) was added to the dish, which was then incubated on ice for 5 min. Cells were harvested by scraping with a rubber policeman. Then, SDS was added (0.1% final concentration) and the cell lysate was sonicated for 10 sec in a Branson sonifier. Cells were then processed for immunoprecipitation.
Phosphate Labeling.
Cells were preincubated in phosphate-free medium (ICN) for 1 hr. Medium was then replaced with phosphate-free medium containing [32P]orthophosphate (NEN) at 500 μCi/ml (1 μCi = 37 kBq), and cells were incubated for an additional 3.5 hr. Then PDBu (1 μM), forskolin (5 μM), thapsigargin (100 nM), calyculin A (10 nM), or cyclosporin A (100 nM) (Calbiochem) was added and labeling was continued for 45 min. Cells were then processed for immunoprecipitation.
Immunoprecipitation.
Immunoprecipitation buffer was added to the sonicated cell lysate (final concentration 190 mM NaCl/50 mM Tris⋅HCl, pH 8.8/6 mM EDTA/2.5% Triton X-100). Primary antisera were added to samples as indicated and the mixtures were incubated at 4°C overnight. For collection of immune complexes, 25 μl of 50% (wt/vol) staphylococcal protein A Sepharose (Pharmacia) in PBS was added. After incubation for 2 hr at 4°C, samples were centrifuged at 15,000 × g for 30 sec and pellets were washed twice with buffer B (150 mM NaCl/10 mM Tris⋅HCl, pH 8.3/5 mM EDTA/0.1% Triton X-100) and once with PBS. Samples were resuspended in 30 μl of Laemmli sample buffer (24) and separated by SDS/12% PAGE. Either gels were dried and autoradiographed or separated proteins were transferred to nitrocellulose for immunoblot analysis.
Protein Phosphatase Treatment.
COS-huPS1 cells were incubated for 45 min in the presence of 1 μM PDBu or vehicle. Cells were lysed and immunoprecipitation was carried out as described above. The Sepharose–immune complex pellet was washed once with buffer B and twice with water and resuspended in 100 μl of λ protein phosphatase buffer (New England Biolabs). Then 400 units of phage λ protein phosphatase was added and the reaction mixture was incubated at 30°C for 2 hr. Following incubation, the immobilized immune complexes were washed with PBS. Laemmli sample buffer (50 μl) was added; samples were heated for 1 min at 80°C; and proteins were separated by SDS/12% PAGE and transferred to nitrocellulose for immunoblot analysis.
Immunoblot Analysis.
Following transfer of separated proteins to nitrocellulose, membranes were incubated in 5% (wt/vol) reconstituted powdered milk in 50 mM Tris-buffered saline (pH 7.5)/1% Tween 20 (TBST) for 1 hr. Antisera were diluted 1:1,000 in 5% milk/TBST and applied to blots for 3–4 hr, followed by three washes in TBST, 10 min each. For gel filtration column fraction analyses, blots were then incubated in a horseradish peroxidase-conjugated preparation of goat anti-rabbit IgG (Amersham). For all other experiments, blots were incubated in protein A-conjugated horseradish peroxidase (Sigma). After three washes in TBST, blots were incubated in chemiluminescence reagent (ICN) and exposed to x-ray film (Kodak).
Phosphoamino Acid Analysis.
COS-huPS1 cells, labeled with [32P]orthophosphate, were treated with PDBu and lysed as described above, immunoprecipitated with α PS1-loop antibody, and electrophoresed in an SDS/12% PAGE system. The gel was dried and the two major PS1 CTFs were excised, using the autoradiogram as a template. Gel fragments were reswollen in 25 mM ammonium bicarbonate, and proteins were digested in 50 μg/ml trypsin for 18 hr at 37°C. The digests were lyophilized and resuspended in 6 M HCl and heated at 110°C for 2 hr. Samples were then lyophilized and resuspended in 2% formic acid/8% acetic acid (pH 1.9), and aliquots and unlabeled phosphoamino acid standards were spotted on TLC plates and electrophoresed first at 500 V in pH 1.9 buffer, then at 400 V in pH 3.5 buffer, using a previously described method (25). Plates were dried and soaked with ninhydrin to stain phosphoamino acid standards. After air drying, plates were visualized using a Bio-Rad phosphorimager.
Separation of PS1 Fragments Under Nondenaturing Conditions.
COS-huPS1 proteins were extracted with 1% Nonidet P-40 in buffer A (20 mM Hepes, pH 7.4/2 mM EDTA/0.05% NaN3). Insoluble material was removed by centrifugation (15,000 × g, 5 min, 4°C). Two hundred microliters of the protein extracts was loaded on a Superose-12 column (Pharmacia) that had been equilibrated in buffer A with 0.15 M NaCl (0.3 ml/min). Proteins were eluted from the column with the same buffer, and fractions were subjected to immunoblot analysis.
In other experiments, COS-huPS1 proteins were extracted with 1% Nonidet P-40/20 mM Tris⋅HCl, pH 7.4/2 mM EDTA/0.5 mM EGTA. Protein extracts were loaded on a Mono-Q column that had been equilibrated in 20 mM Tris⋅HCl, pH 7.4/2 mM EDTA/0.5 mM EGTA. Proteins were eluted from the column by using a gradient from 0 to 1 M NaCl. Fractions were subjected to immunoblot analysis.
Sucrose Density Gradient Fractionation.
COS-huPS1 cells were incubated for 45 min with PDBu or vehicle. Cells were washed with PBS, collected in homogenization buffer (0.25 M sucrose/10 mM Tris⋅HCl, pH 7.4/1 mM MgCl2), centrifuged at 1,000 × g for 5 min, and resuspended in a final volume of 1.5 ml of homogenization buffer. Cells were homogenized in a steel-to-steel homogenizer, and 2.3 M sucrose was added to yield a final concentration of 1.4 M. Samples were then loaded on a discontinuous sucrose gradient (2.0 M/1.4 M/1.2 M/0.8 M/0.25 M) and centrifuged at 100,000 × g for 4.5 hr in a Beckman SW41 rotor. One-milliliter fractions were collected. Aliquots were applied to SDS/12% PAGE and subjected to immunoblot analysis.
RESULTS AND DISCUSSION
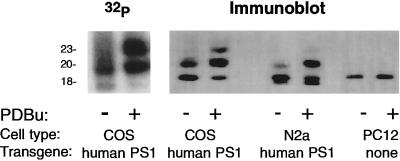
The incorporation of radioactive phosphate into the PS1 CTF of COS-huPS1 cells was dramatically increased in the presence of PDBu (which activates PKC; Fig. 1), forskolin (which indirectly activates protein kinase A; data not shown), or calyculin A (which inhibits protein phosphatases 1 and 2A; data not shown).
Figure 1.
Effect of PDBu on phosphorylation of PS1 CTF. (Left) COS-huPS1 cells were incubated for 3.5 hr in the presence of [32P]orthophosphate. Vehicle or 1 μM PDBu was added for an additional 45 min. Cells were lysed and immunoprecipitation was performed with α PS1-loop antiserum as described. (Right) COS-huPS1, human PS1-transfected N2a, and PC12 cells were treated with PDBu, lysed, and subjected to immunoprecipitation with α PS1-loop antiserum as described. Immunoblot analysis was performed using α PS1-loop antiserum.
This phosphorylation was associated with a shift in apparent molecular mass. Thus, in the absence of any addition, an 18-kDa fragment was observed. In the presence of PDBu, forskolin, or calyculin A two additional species appeared at approximately 20 kDa and 23 kDa, accompanied by a diminution in the 18-kDa species (Figs. 1 and 2). In a more limited series of studies, thapsigargin (which activates calcium-regulated protein kinases and protein phosphatases) and cyclosporin A (which inhibits protein phosphatase 2B) caused no increase in apparent molecular mass (Fig. 2). We tentatively assign identities of dephospho-, monophospho-, and diphospho- PS1 to the 18-, 20-, and 23-kDa species, respectively.
Figure 2.
Regulation of phosphorylation of PS1 CTF by various protein kinase activators and phosphoprotein phosphatase inhibitors. COS-huPS1 cells were incubated in the absence or presence of PDBu (PBu; 1 μM), calyculin A (Cal; 10 nM), cyclosporin A (Cyc; 100 nM), thapsigargin (THP; 100 nM), or forskolin (FSK; 5 μM). Cells were lysed and subjected to immunoprecipitation with α PS1-loop antiserum followed by immunoblot analysis using α PS1-loop antiserum.
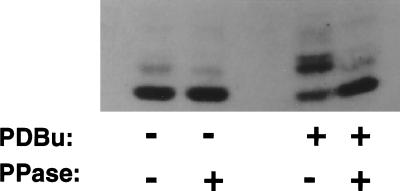
To further evaluate the possibility that the electrophoretic mobility shift was due to phosphorylation, we examined the effect of λ protein phosphatase on immunoprecipitates. Following such treatment, higher molecular mass proteins were eliminated and the 18-kDa species was enhanced (Fig. 3).
Figure 3.
Effect of phosphoprotein phosphatase on electrophoretic migration of PS1 CTF. COS-huPS1 cells were treated with vehicle or PDBu, lysed, and subjected to immunoprecipitation. Sepharose-bound immune complexes were treated with λ protein phosphatase. Proteins were eluted from the Sepharose beads with Laemmli sample buffer and applied to SDS/12% PAGE, and immunoblot analysis was performed using α PS1-loop antiserum.
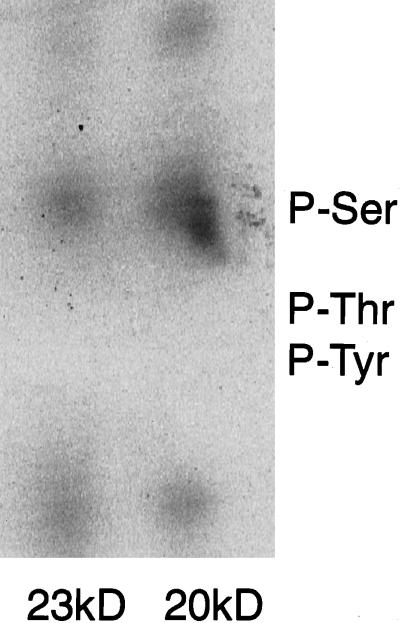
The 32P-phosphorylated 20-kDa and 23-kDa PS1 proteins resulting from incubation of cells with PDBu were subjected to phosphoamino acid analysis. Phosphorylation was found on serine but not on threonine or tyrosine (Fig. 4). PKC regulation of serine phosphorylation of the PS1 CTF, and a concomitant shift in electrophoretic mobility, has been found independently by other investigators (C. Haass, personal communication). In contrast to results obtained for the PS1 CTF, none of the compounds tested here affected phosphorylation or electrophoretic migration of the PS1 NTF (data not shown).
Figure 4.
Phosphoamino acid analysis of 20-kDa and 23-kDa PS1 CTFs. COS-huPS1 cells were labeled with [32P]orthophosphate, treated with PDBu, and subjected to immunoprecipitation. The two major phosphorylated CTF species were subjected to phosphoamino acid analysis and were visualized by phosphorimager.
We also examined the effects of PDBu on mouse neuroblastoma (N2a) cells transfected with human PS1 (Fig. 1). The doublet which appears at 17 and 18 kDa in N2a cells was shown previously to represent the endogenous mouse PS1 CTF and the transfected human PS1 CTF, respectively (18). Transfected human PS1 in N2a cells exhibited a shift in electrophoretic mobility upon exposure to phorbol ester similar to that seen in COS cells (Fig. 1). When rat pheochromocytoma (PC12) cells (Fig. 1), untransfected N2a, or rat primary neuronal cultures (data not shown) were treated with phorbol esters, no shift was observed in electrophoretic mobility of endogenous PS1. Thus, the human PS1 protein, but not the rat or mouse homologue, was readily phosphorylatable under the conditions tested here.
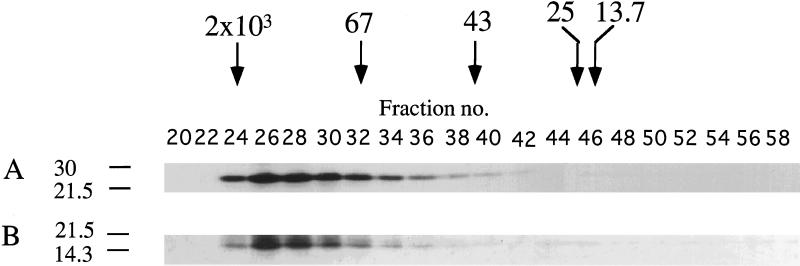
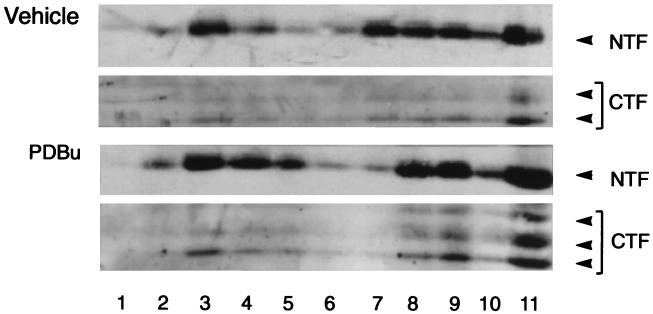
Extraction of COS-huPS1 cells with a mild detergent (Nonidet P-40), followed by gel filtration chromatography, indicated that the native PS1 NTF and the dephospho- and phospho-CTF coeluted with an apparent molecular mass in the range of 100 kDa (Fig. 5). This apparent mass is much higher than that observed on SDS/PAGE (i.e., 30 kDa, 18 kDa, and 20–23 kDa, respectively). Extraction of COS-huPS1 cells with Nonidet P-40, followed by ion-exchange chromatography using a Mono-Q column, indicated that the PS1 NTF and CTF also coeluted (data not shown). In another series of experiments, cells were incubated with PDBu or vehicle and then extracts were fractionated on discontinuous sucrose gradients (Fig. 6). Again, the NTF and phospho- and dephospho-CTF were found to comigrate. These various results support the concept that the fragments are associated in complexes.
Figure 5.
Gel filtration of proteins from COS-huPS1 cells. Nonidet P-40 extracts of cells were loaded onto a Superose 12 column and protein was eluted with buffer A containing 0.15 M NaCl. The indicated column fractions were analyzed by immunoblotting. Elution patterns of PS1 NTF (Ab 14) (A) and PS1 CTF (α PS1-loop) (B) are shown. The elution positions of molecular mass standards (kDa) are indicated by arrows.
Figure 6.
Sucrose gradient fractionation of COS-huPS1 cells. Cells were treated with vehicle or PDBu and lysed using a steel-to-steel homogenizer, and extracts were applied to discontinuous sucrose gradients. Fractions were collected, and proteins were separated by SDS/12% PAGE and immunoblotted with antisera against the PS1 NTF (Ab 14) and PS1 CTF (α PS1-loop). Fractions are numbered from top to bottom of gradient.
Protein phosphorylation has been demonstrated to regulate the processing of the APP (26–28). Stimulation of PKC (29) or protein kinase A (30) results in increased release of α-secretase-cleaved soluble APP with a concomitant decrease in Aβ production. This effect is due in part to regulation, by protein phosphorylation in the trans-Golgi network, of the biogenesis of APP-bearing transport vesicles (29, 30). The trans-Golgi network substrate which regulates vesicle budding is apparently integral (29, 30), prompting consideration of the presenilins as candidates for this integral phosphoprotein. However, stimulation of PKC results in a decrease in both Aβ-(1–42) and Aβ-(1–40) (31), while pathogenic mutations in PS1 lead to alterations in the ratio of Aβ-(1–42) to Aβ-(1–40) (23, 31–33), suggesting that PS1 is probably not the target of PKC that modulates APP processing. It will now be important to investigate the involvement of wild-type and mutant presenilins in the vesicle biology and protein processing events that constitute “γ-secretase” cleavage of APP (34).
Acknowledgments
This work was supported by U.S. Public Health Service Grants AG09464 (to P.G.), AG11508 and AG13780 (to S.G.), AG05689 (to M.S.), NS30428 (to R.E.T.) and grants from the Adler Foundation, the Develbiss Fund, and the Alzheimer’s Association (to S.S.S.). R.E.T. is a Pew Scholar. D.M.K. is a French Foundation Fellow. C.N. is supported by the Swedish Medical Research Council. P.StG.-H. is a Medical Research Council Scientist, and additional support was provided to P.StG.-H. and P.F. from the Medical Research Council and the Alzheimer’s Society of Ontario. S.S.S. is a recipient of an Alzheimer’s Association Zenith Award.
ABBREVIATIONS
- PS1
presenilin 1
- NTF
N-terminal fragment
- CTF
C-terminal fragment
- AD
Alzheimer disease
- APP
β-amyloid precursor protein
- PKC
protein kinase C
- PDBu
phorbol 12,13-dibutyrate
Note Added in Proof
The results cited as “C. Haass, personal communication” are described in detail in ref. 35.
References
- 1.Schellenberg G D. Proc Natl Acad Sci USA. 1995;92:8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanzi R E, Kovacs D M, Kim T-W, Moir R D, Guenette S Y, Wasco W. Neurobiol Dis. 1996;3:159–168. doi: 10.1006/nbdi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 3.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Nature (London) 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 4.Chartier-Harlin M-C, Crawford F, Houlden H, Warrem A, Hughes D, Fidani L, Goate A, Rossor M, Roques P, Hardy J, Mullan M. Nature (London) 1991;353:844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 5.Naruse S, Igarashi S, Kobayashi H, Aoki K, Inuzuka T, Kaneko K, Shimizu T, Ihara K, Kojima T, Miyatake T, Tsuji S. Lancet. 1991;337:978–979. doi: 10.1016/0140-6736(91)91612-x. [DOI] [PubMed] [Google Scholar]
- 6.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 7.Glenner G G, Wong C W. Biochem Biophys Res Commun. 1984;122:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 8.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. [Google Scholar]
- 9.Levy-Lahad E, Wijsman E M, Nemens E, Anderson L, Goddard K A B, Weber J L, Bird T D, Schellenberg G D. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 10.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, Mar L, Sorbi S, Macmias B, Piacentini S, Amaducci L, Chumakov I, Cohen D, Lannfelt L, Fraser P E, Rommens J M, St George-Hyslop P H. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 11.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C-E, Jondro P D, Schmidt S D, Wang K, Crowley A C, Fu Y-H, Guenette S Y, Galas D, Nemens E, Wijsman E M, Bird T D, Schellenberg G D, Tanzi R E. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 12.Van Broeckhoven C. Nat Genet. 1996;11:230–232. doi: 10.1038/ng1195-230. [DOI] [PubMed] [Google Scholar]
- 13.Slunt H H, Thinakaran G, Lee M K, Sisodia S S. J Amyloid: Int J Exp Clin Invest. 1995;2:188–190. [Google Scholar]
- 14.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 15.Levitan D, Greenwald I. Nature (London) 1996;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 17.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 18.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey A I, Gandy S E, Jenkins N A, Copeland N G, Price D L, Sisodia S S. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 19.Kovacs D M, Fausett H J, Page K J, Kim T-W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, Hyman B T, Tanzi R E, Wasco W. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 20.Cook D G, Sung J C, Golde T E, Felsenstein K M, Wojczyk B S, Tanzi R E, Trojanowski J Q, Lee V M-Y, Doms R W. Proc Natl Acad Sci USA. 1996;93:9223–9228. doi: 10.1073/pnas.93.17.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter J, Capell A, Grunberg J, Pesold B, Schindzielorz A, Prior R, Podlisny M B, Fraser P, St George-Hyslop P, Selkoe D J, Haass C. Mol Med (Oxford) 1996;2:673–691. [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Tur J, Froelich S, Prihar G, Crook R, Baker M, et al. Neuroreport. 1995;7:297–301. [PubMed] [Google Scholar]
- 23.Borchelt D R, Thinakaran G, Eckman C B, Lee M K, Davenport F, Ratovitsky T, Prada C-M, Kim G, Seekins S, Yager D, Slunt H, Wang R, Seeger M, Levey A I, Gandy S E, Copeland N, Jenkins N, Price D L, Younkin S G, Sisodia S S. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Nairn A C, Greengard P. J Biol Chem. 1987;262:7273–7281. [PubMed] [Google Scholar]
- 26.Buxbaum J D, Gandy S E, Cicchetti P, Ehrlich M E, Czernik A J, Fracasso R P, Ramabhadran T V, Unterbeck A J, Greengard P. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buxbaum J D, Koo E H, Greengard P. Proc Natl Acad Sci USA. 1993;90:9195–9198. doi: 10.1073/pnas.90.19.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung A Y, Haass C, Nitsch R M, Qiu W Q, Citron M, Wurtman R J, Growdon J H, Selkoe D J. J Biol Chem. 1993;268:22959–22962. [PubMed] [Google Scholar]
- 29.Xu H, Greengard P, Gandy S. J Biol Chem. 1995;270:23243–23245. doi: 10.1074/jbc.270.40.23243. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Sweeney D, Greengard P, Gandy S. Proc Natl Acad Sci USA. 1996;93:4081–4084. doi: 10.1073/pnas.93.9.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Citron M, Diehl T S, Gordon G, Biere A L, Seubert P, Selkoe D J. Proc Natl Acad Sci USA. 1996;93:13170–13175. doi: 10.1073/pnas.93.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird T D, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Nat Med. 1996;2:864–70. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 33.Duff K, Eckman C, Zehr C, Xin Y, Prada C-M, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgans D, Gordon M N, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu, H., Sweeney, D., Wang, R., Thinakaran, G., Lo, A. C. Y., Sisodia, S. S., Greengard, P. & Gandy, S. (1997) Proc. Natl. Acad. Sci USA 94, in press. [DOI] [PMC free article] [PubMed]
- 35.Walter, J., Grunberg, J., Capell, A., Pesold, B., Schindzielorz, A., Citron, M., Mendla, K., St George-Hyslop, P., Multhaup, G., Selkoe, D. & Haass, C. (1997) Proc. Natl. Acad. Sci. USA 94, in press. [DOI] [PMC free article] [PubMed]