Abstract
Reversible topoisomerase I (Top1)-DNA cleavage complexes are the key DNA lesion induced by anticancer camptothecins (e.g. topotecan and irinotecan) as well as structurally perturbed DNAs (e.g. oxidatively damaged DNA, UV-irradiated DNA, alkylated DNA, uracil-substituted DNA, mismatched DNA, gapped and nicked DNA, and DNA with abasic sites). Top1 cleavage complexes arrest transcription and trigger transcription-dependent degradation of Top1, a phenomenon termed Top1 down-regulation. In the current study, we have investigated the role of Top1 down-regulation in the repair of Top1 cleavage complexes. Using quiescent (serum-starved) human WI-38 cells, camptothecin (CPT) was shown to induce Top1 down-regulation, which paralleled the induction of DNA single-strand breaks (SSBs) (assayed by comet assays) and ATM autophosphorylation (at Ser-1981). Interestingly, Top1 down-regulation, induction of DNA SSBs and ATM autophosphorylation were all abolished by the proteasome inhibitor MG132. Furthermore, studies using immunoprecipitation and dominant-negative ubiquitin mutants have suggested a specific requirement for the assembly of Lys-48-linked polyubiquitin chains for CPT-induced Top1 down-regulation. In contrast to the effect of proteasome inhibition, inactivation of PARP1 was shown to increase the amount of CPT-induced SSBs and the level of ATM autophosphorylation. Together, these results support a model in which Top1 cleavage complexes arrest transcription and activate a ubiquitin-proteasome pathway leading to the degradation of Top1 cleavage complexes. Degradation of Top1 cleavage complexes results in the exposure of Top1-concealed SSBs for repair through a PARP1-dependent process.
Eukaryotic DNA topoisomerase I (Top1)3 catalyzes the breakage/reunion of DNA by transiently nicking one strand of the DNA duplex, through the formation of a reversible Top1-DNA covalent complex (reviewed in Refs. 1-4). It has been well established that anticancer camptothecins (e.g. irinotecan and topotecan) as well as various structurally perturbed DNAs (e.g. UV adducts, abasic sites, base mismatches, uracil incorporation, nicks and gaps, and oxidized DNA lesions) stabilize reversible Top1-DNA covalent complexes, often referred to as Top1 cleavable or cleavage complexes (reviewed in Refs. 3 and 5).
Studies using camptothecins (CPTs) have generated a wealth of information on the structure and biology of Top1 cleavage complexes (reviewed in Refs. 3 and 5). It is well established that CPTs exert their antitumor activity through their specific stabilization of Top1 cleavage complexes (6). Top1 cleavage complexes are unique DNA lesions, characterized by their reversibility and Top1-concealed single-strand breaks (SSBs) (6). It has been hypothesized that reversible Top1 cleavage complexes are processed into DNA damage through their interactions with cellular machineries associated with active DNA replication and transcription (7-11).
The role of active DNA replication in the processing of Top1 cleavage complexes into DNA damage was initially suggested from the observations that CPTs are exquisitely cytotoxic to S phase cells, and the arrest of active DNA replication with aphidicolin (APH) or other DNA synthesis inhibitors abolishes CPT cytotoxicity without any effect on the amount of Top1 cleavage complexes (12, 13). Analysis of the aberrant replication intermediates generated in the SV40 cell-free replication system has led to the suggestion of a replication fork collision model for the S phase-specific cytotoxicity of CPT-induced Top1 cleavage complexes (7, 10). In this model, a polarity-specific collision occurs between the advancing replication fork and the reversible Top1 cleavage complex, resulting in the arrest of the replication fork and the concomitant formation of a DNA double-strand break (DSB) and a Top1-DNA cross-link at the site of collision (7, 10, 14). Indeed, many DNA damage signals, such as phosphorylated RPA, ATM, Chk1, Chk2, p53, NF-κB, and H2AX (γ-H2AX) are detected upon CPT treatment, and most of them have been shown to be replication-dependent (reviewed in Ref. 5). As a consequence of the collision, the cells are arrested at the G2/M phase of the cell cycle (9) and cell death ensues (12, 13).
In addition to the replication-dependent processing, accumulating evidence has also pointed to a second mechanism involving transcription in the processing of Top1 cleavage complexes into DNA damage (8, 15). Treatment of cells with CPT results in rapid transcription arrest, followed by recovery of transcription (8). Recovery of transcription depends on proteasome activity and is correlated with transcription-dependent proteasomal degradation of Top1 (termed Top1 down-regulation) and the largest subunit of RNA polymerase II (8). It has been suggested that Top1 cleavage complexes arrest transcription elongation, triggering proteasomal degradation of Top1 and the largest subunit of RNA polymerase II (8). Proteasomal degradation of Top1 cleavage complexes at the collision sites has been speculated to expose the otherwise Top1-concealed SSBs for repair (8, 16).
In the current studies, we seek to provide evidence supporting the role of a ubiquitin-proteasome pathway in the transcription-dependent processing Top1 cleavage complexes into DNA SSBs. To eliminate the complication arising from DNA replication, quiescent (serum-starved) WI-38 cells were employed for analysis of the role of proteasome in the processing of Top1 cleavage complexes. Like Top1 down-regulation, CPT-induced SSBs and ATM autophosphorylation were shown to be transcription- and proteasome-dependent, consistent with the notion that transcription and proteasome are required for the processing of Top1 cleavage complexes into DNA damage. The involvement of a ubiquitin-proteasome pathway was further supported by the demonstration of the formation of Top1-ubiquitin conjugates and a specific requirement for the assembly of Lys-48-linked polyubiquitin chains for CPT-induced down-regulation. By contrast, the amount of CPT-induced SSBs and the level of ATM autophosphorylation were found to be increased upon PARP1 inactivation. Together, these results support the model in which a ubiquitin-proteasome pathway is involved in the processing of Top1 cleavage complexes into SSBs, and PARP1-mediated repair occurs downstream of the formation of SSBs.
EXPERIMENTAL PROCEDURES
Immunoblotting—Immunoblotting was performed as described previously (8). Antibodies against p-Ser-1981-ATM (p denotes phosphorylation) (Upstate Immunochemicals), γ-H2AX (Upstate), and p-Tyr15-cdc2 (Cell Signaling) were purchased from commercial sources. Anti-hTop2α antibody was kindly provided by Dr. Jaulang Hwang (Institute of Molecular Biology, Academia Sinica, Taiwan). Anti-Top1 antibodies were obtained from sera of Scleroderma 70 patients as described before (25). The hybridoma cell line that produces monoclonal antibody 12G10 (α-tubulin) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA).
Cell Culture—The human fibroblast cell line WI-38 was obtained from American Type Culture Collection (Manassas, VA). WI-38 cells (at passages 18-23) and HeLa TetOn cells (Clontech) were cultured in DMEM supplemented with 10% FetalPlex animal serum complex (Gemini Bio-Products, West Sacramento, CA), l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 37 °C incubator with 5% CO2. For serum starvation, then cells were washed with serum-free medium and cultured in DMEM containing 0.2% serum for 72 h. For neuronal culture, mixed cortical neurons were isolated from embryonic day 17.5 C57BL/6 mouse embryos as described (23). HeLa and V79 cells were cultured in DMEM supplemented with 10% FetalPlex animal serum complex.
Isolation of parp1+/- and parp1-/- Mouse Embryonic Fibroblasts (MEFs)—Embryonic day 12.5 mouse embryos were dissected free of brains and livers, finely minced, and then suspended in a trypsin-EDTA solution. The cell suspensions were incubated at 37 °C for 5 min, followed by termination with DMEM supplemented with 10% FetalPlex animal serum complex and glutamine/penicillin/streptomycin. The cells were pelleted and resuspended in fresh medium, followed by culturing in plastic plates in a 5% CO2 incubator at 37 °C.
Neutral and Alkaline Comet Assays—The comet assays were performed according to the Trevigen CometAssay™ kit protocol with slight modifications. WI-38 cells were pretreated with various inhibitors for 30 min, followed by co-treatment with 25 μm CPT for 1 h. Treated cells were trypsinized with 0.005% trypsin (50× lower than the normal concentration) at 37 °C for 5 min. Equal amount of drug-free medium (with 10% serum) was then added to quench the trypsin activity. The final cell density was approximately 10,000 cells/ml. 50 μl of the cell suspension was then mixed with 500 μl of 0.5% low melting point agarose (Invitrogen) (in phosphate-buffered saline) at 37 °C. 50 μl of the cell/agarose mixture was transferred onto glass slides. The slides were then immersed in prechilled lysis buffer (2.5 m NaCl, 100 mm EDTA, 10 mm Tris, pH 10.0, 1% Triton X-100, and 10% Me2SO) for 1 h. For neutral comet assay, the slides were immersed in 1× TBE (Tris/Borate/EDTA) for 30 min at room temperature, followed by electrophoresis in 1× TBE buffer at 0.8 V/cm for 8-10 min at room temperature. For alkaline comet assay, the slides were immersed in alkaline buffer (300 mm NaOH, 1 mm EDTA) for 5 min, followed by electrophoresis in alkaline buffer at 0.5 volt/cm for 20 min. After electrophoresis, the slides were immersed in neutralization buffer (0.4 m Tris, pH 7.5) for 5 min, followed by dehydration in 70% alcohol for 30 min and air drying overnight. The cells were then rehydrated in H2O and stained with 0.2 μg/ml ethidium bromide for 1 h. The images were visualized under a fluorescence microscope and captured with a CCD camera. The comet tail moment was defined in Ref. 48. The mean and the S.E. were obtained from at least 100 cells for each treatment group. Statistical analysis was performed using two-tailed unpaired Student's t test.
Immunocytochemistry—Immunocytochemistry was performed following the standard protocol as described previously (23). Briefly, the cells cultured on coverslips were fixed with 3.7% paraformaldehyde followed by incubation with anti-p-Ser-1981-ATM antibody (Cell Signaling) (1:500 dilution) and then with an appropriate Cy3-conjugated secondary antibody (Jackson Immunoresearch). The cells were then washed with phosphate-buffered saline followed by counterstaining with 4′,6′-diamino-2-phenylindole. The fluorescence images were captured using a CCD camera mounted on a Zeiss fluorescence microscope.
Top1 Down-regulation—CPT-induced Top1 degradation was monitored by immunoblotting of the alkaline lysates as described previously (8) with slight modification. Briefly, for each 35-mm dish, 100 μl of an alkaline lysis buffer (200 mm NaOH, 2 mm EDTA) was added, and the cells were then scraped by a rubber policeman. Alkaline lysates were neutralized by addition of 16 μl of 1 m HCl, 600 mm Tris, pH 8.0, followed by mixing with 13 μl of 10× S7 nuclease buffer (50 mm MgCl2, 50 mm CaCl2, 5 mm dithiothreitol, 1 mm EDTA, and a protease inhibitor mixture) and 60 units of staphylococcal S7 nuclease (30). The digestion was performed on ice for 20 min for releasing Top1 from covalent Top1-DNA complexes. After nuclease digestion, 100 μl of 6× SDS gel sample buffer was added, followed by boiling for 10 min. The samples were then analyzed by SDS gel electrophoresis and immunoblotted with anti-hTop1 antibodies.
Transfection of HA-tagged Mutant Ubiquitin cDNAs—HA-Ub, K48R-Ub, K29R-Ub, and K63R-Ub plasmids were obtained from Dr. Cam Patterson (University of North Carolina at Chapel Hill, NC). HA-K48-only-Ub, and HA-K63-only-Ub plasmids were obtained from Dr. Vishva M. Dixit (Genentech Inc., CA). The cells were transfected with 1.5-3 μg of DNA using the PolyFect transfection reagent (Qiagen). 20 h post-transfection, the cells were treated with CPT for various time, followed by immunoprecipitation (see below) or Top1 down-regulation analysis (see above).
Immunoprecipitation—For immunoprecipitation, cells growing on 60-mm dishes were lysed with 400 μl of an alkaline lysis buffer, followed by neutralization with 75 μl of a neutralization buffer (described in Top1 down-regulation assay) with brief sonication. 60 μl of 10× S7 buffer was then added to the cell lysates, followed by S7 nuclease treatment (180 units). The lysates were cleared by brief centrifugation in an Eppendorf centrifuge. Supernatants were mixed with 550 μl of 2× radioimmune precipitation assay buffer, protease inhibitor mixture, and phenylmethylsulfonyl fluoride, followed by clarifying with 30 μl of protein L-conjugated agarose beads (Santa Cruz). Top1 were immunoprecipitated by Top1 antiserum and protein L-conjugated agarose beads. For immunoblotting, 50 μl of the 6× SDS gel sample buffer was added to 30 μl of radioimmune precipitation assay buffer-washed beads, followed by boiling for 10 min. The samples were then analyzed by SDS gel electrophoresis and immunoblotted with anti-hTop1 or anti-HA antibodies.
RESULTS
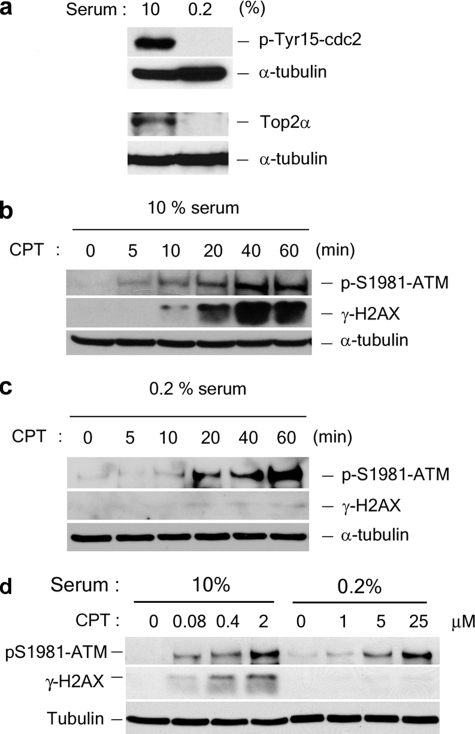
CPT Induces Distinct DNA Damage Signals Depending on the Growth State of WI-38 Cells—To study the roles of DNA replication and transcription in processing Top1-DNA cleavage complexes, CPT-induced DNA damage signals were measured in both proliferating (cultured in 10% serum) and nonproliferating (cultured in 0.2% serum) primary human lung fibroblasts, WI-38 cells. WI-38 cells are known to cease proliferation and enter a quiescent G0/G1 state upon serum starvation (17). As shown in Fig. 1a, WI-38 cells ceased proliferation after 72 h of being cultured in 0.2% serum as evidenced by the lack of p-Tyr-15 cdc2 (a cell cycle marker known to be elevated at G1/S and S/G2) (18) and a much reduced level of Top2α (a cell proliferation marker) (19, 20). Previous studies have demonstrated that CPT induces both H2AX phosphorylation at Ser-139 (γ-H2AX) and ATM autophosphorylation at Ser-1981 in tumor cells (21). Consistent with these observations, CPT was shown to induce γ-H2AX and ATM autophosphorylation at Ser-1981 in proliferating WI-38 cells (Fig. 1b). However, CPT was shown to induce little γ-H2AX, but relatively robust ATM autophosphorylation, in quiescent (0.2% serum) WI-38 cells (Fig. 1c). Indeed, a dose dependence study (Fig. 1d) also showed that CPT-induced γ-H2AX was greatly (over 300-fold) reduced in nonproliferating than in proliferating WI-38 cells, whereas CPT-induced ATM autophosphorylation was only approximately 10-fold reduced. These results suggest that ATM autophosphorylation and γ-H2AX may represent distinct DNA damage responses induced by CPT.
FIGURE 1.
CPT induces DNA damage signals in both proliferating and quiescent WI-38 cells. a, WI-38 cells were cultured in DMEM supplemented with either 10% or 0.2% serum as described under “Experimental Procedures.” The lysates were immunoblotted with antibodies as indicated. b, WI-38 cells were cultured in 10% serum and treated with 25 μm CPT for various time, followed by immunoblotting using antibodies as indicated. c, the same as in b except that cells were cultured in 0.2% serum. d, WI-38 cells cultured in 10 or 0.2% serum were treated with indicated concentrations of CPT for 1 h, followed by immunoblotting using antibodies as indicated.
CPT-induced γ-H2AX Is Replication-dependent—Because γ-H2AX is a well characterized marker for DSBs (22), CPT-induced γ-H2AX formation in proliferating, but not quiescent, cells suggests the involvement of DNA replication in the processing of Top1 cleavage complexes into DSBs, as predicted by the replication fork collision model (7, 10). Studies using the DNA replication inhibitor, APH, further confirmed the involvement of DNA replication in CPT-induced γ-H2AX in proliferating WI-38 cells (10% serum) (supplemental Fig. S1a). The replication inhibitor, APH (10 μm), was shown to largely abolish the CPT-induced γ-H2AX signal (supplemental Fig. S1). By contrast, the transcription inhibitor, DRB had essentially no effect on the CPT-induced γ-H2AX signal (supplemental Fig. S1a). These results suggest that CPT-induced DNA DSBs (as evidenced by γ-H2AX) are replication-dependent but transcription-independent in proliferating WI-38 cells. Indeed, direct measurement of DSBs by the neutral comet assay also confirmed the inhibitory effect of APH, but not DRB, on CPT-induced DNA DSBs (supplemental Fig. S1b). By contrast, both APH and DRB partially inhibited CPT-induced ATM autophosphorylation (approximately 2-5-fold reduction for each inhibitor) in proliferating WI-38 cells (supplemental Fig. S1a), suggesting the involvement of both DNA replication and transcription in CPT-induced ATM autophosphorylation. These results again suggest that CPT-induced γ-H2AX and ATM autophosphorylation may represent two separate, albeit overlapping, repair responses to CPT-induced DNA lesions in proliferating WI-38 cells, the former being linked to DNA replication and the latter to both replication and transcription.
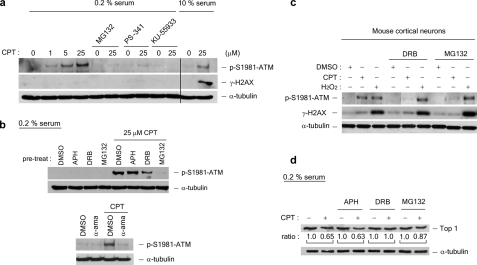
CPT-induced ATM Autophosphorylation Is Transcription- and Proteasome-dependent in Nonproliferating Cells—The differential roles of DNA replication and transcription in CPT-induced DNA damage responses were also investigated in quiescent WI-38 cells. As shown in Fig. 2a (middle row), CPT did not induce any detectable γ-H2AX signal, consistent with the notion that DNA replication is needed to process Top1 cleavage complexes into DNA DSBs. By contrast, CPT was shown to induce robust ATM autophosphorylation in quiescent WI-38 cells (Fig. 2a, top row). In addition, CPT-induced ATM autophosphorylation was unaffected by the replication inhibitor, APH (Fig. 2b, upper panels), but greatly reduced by the transcription inhibitors, DRB (150 μm) and α-amanitin (5 μg/ml; Fig. 2b, lower panels). The fact that α-amanitin fails to induce ATM autophosphorylation by itself (Fig. 2b) also excludes the possibility that CPT-induced ATM autophosphorylation is due to the arrest of transcription elongation complexes. These results suggest that transcription, but not DNA replication, is primarily involved in CPT-induced ATM activation in quiescent WI-38 cells. We have also shown that the proteasome inhibitors, MG132 and PS-341 (bortezomib), largely abolished CPT-induced ATM autophosphorylation in nonproliferating WI-38 cells (Fig. 2a), suggesting a requirement of proteasome activity for CPT-induced ATM autophosphorylation. CPT-induced ATM autophosphorylation was shown to depend on ATM kinase activity because the ATM kinase-specific inhibitor KU-55933 completely abolished CPT-induced ATM autophosphorylation (Fig. 2a).
FIGURE 2.
CPT-induced ATM autophosphorylation is transcription- and proteasome-dependent in quiescent WI-38 cells. WI-38 cells cultured in DMEM supplemented with 0.2% serum were treated with CPT (1, 5, or 25 μm) for 1 h in the presence and absence of various inhibitors. A proteasome inhibitor (2 μm MG132 or 1 μm PS-341) was added 30 min prior to CPT addition. The ATM inhibitor KU-55933 (10 μg/ml) was added 60 min prior to CPT addition. The cell lysates were immunoblotted with antibodies as indicated. As controls (the two right-most lanes in a), lysates from proliferating WI-38 cells (cultured in 10% serum) were immunoblotted. b, upper panels, WI-38 cells cultured in DMEM supplemented with 0.2% serum were preincubated with the replication inhibitor APH (10 μm) or the transcription inhibitor DRB (50 μm) for 30 min prior to CPT addition. Co-treatment with CPT was for 1 h. The cell lysates were immunoblotted with antibodies as indicated. b, lower panels, the same as in b (upper panels), except that the RNA polymerase inhibitorα-amanitin (5 μg/ml) was used during co-treatment with CPT. c, post-mitotic mouse cortical neurons were treated with CPT (20 μm) or H2O2 (100 μm) in the presence of MG132 (4 μm) or DRB (150 μm) for 1 h. The cell lysates were immunoblotted. d, Top1 down-regulation in WI-38 cells cultured in DMEM supplemented with 0.2% serum. The cells were pretreated with different inhibitors (APH, DRB, or MG132) for 30 min, followed by co-treatment with CPT (25 μm) for 3 h. Following a 30-min incubation in fresh medium (without inhibitors or CPT), the cells were lysed by an alkaline lysis procedure, and the cell lysates were subject to staphylococcal nuclease S7 treatment (see “Experimental Procedures”). The cell lysates were then analyzed by immunoblotting using hTop1 antisera. The relative band intensities (expressed in ratios; the relative band intensities of samples without CPT treatment were taken as 1.0) were quantified by KODAK one-dimensional image analysis software. DMSO, dimethyl sulfoxide.
The requirement of transcription and proteasome activity in CPT-induced DNA ATM autophosphorylation is reminiscent of CPT-induced Top1 down-regulation, which is also transcription- and proteasome-dependent (Fig. 2d). Previous studies have shown that CPT-induced Top1 down-regulation is dependent on transcription, but not new protein synthesis (8). Here, we showed that CPT-induced ATM autophosphorylation in quiescent WI-38 cells was unaffected by the protein synthesis inhibitor, cycloheximide (1 and 10 μm) (supplemental Fig. S2). The similar requirement for transcription and proteasome, but not new protein synthesis, for CPT-induced Top1 down-regulation and ATM autophosphorylation suggests the possibility that they may be related phenomena.
To further confirm that CPT induces transcription- and proteasome-dependent ATM autophosphorylation in nonproliferating cells, post-mitotic mouse cortical neurons were isolated and treated with CPT. As shown in Fig. 2c, CPT induced robust ATM autophosphorylation in mouse cortical neurons, but minimal γ-H2AX. CPT-induced ATM autophosphorylation was abolished by the transcription inhibitor, DRB, and the proteasome inhibitor, MG132. By contrast, hydrogen peroxide induced remarkable γ-H2AX in mouse cortical neurons, indicating the formation of DSBs (23). Different from CPT-induced ATM autophosphorylation, hydrogen peroxide-induced ATM autophosphorylation was inhibited by neither DRB nor MG132, suggesting that the hydrogen peroxide-induced DNA damage is independent of transcription and proteasome (Fig. 2c). Together, these results demonstrate that CPT is able to induce ATM autophosphorylation in both post-mitotic neurons and quiescent WI-38 cells, suggesting the presence of a replication-independent mechanism for CPT-induced DNA damage. Furthermore, ATM autophosphorylation induced by CPT, but not hydrogen peroxide, is transcription- and proteasome-dependent, further demonstrating the specific requirement for transcription and proteasome in CPT-induced DNA damage.
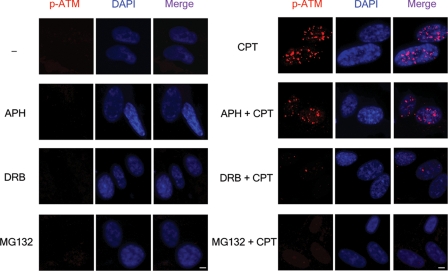
Immunocytochemistry was also performed to examine the possible formation of Ser-1981 autophosphorylated ATM (labeled p-ATM in Fig. 3) foci in CPT-treated quiescent WI-38 cells. As shown in Fig. 3, CPT induced a large number of p-ATM foci. Inhibition of DNA synthesis with APH (10 μm) had no significant effect on the number of p-ATM foci. However, inhibition of transcription with DRB (150 μm) or inhibition of proteasome with MG132 (2 μm) caused a dramatic reduction in the number of p-ATM foci. These results are consistent with the immunoblotting results (Fig. 2) and suggest the involvement of transcription and proteasome in CPT-induced ATM activation in quiescent cells.
FIGURE 3.
In situ visualization of autophosphorylated ATM foci in CPT-treated quiescent WI-38 cells. WI-38 cells cultured in 0.2% serum were pretreated with DMSO (0.1%), APH (10 μm), DRB (150 μm), or MG132 (2 μm) for 30 min, followed by 1-h co-treatment with CPT (25 μm). The cells were fixed by 3.7% paraformaldehyde and stained with both anti-p-Ser-1981-ATM antibody and 4′,6′-diamino-2-phenylindole (DAPI). Scale bar, 5 μm.
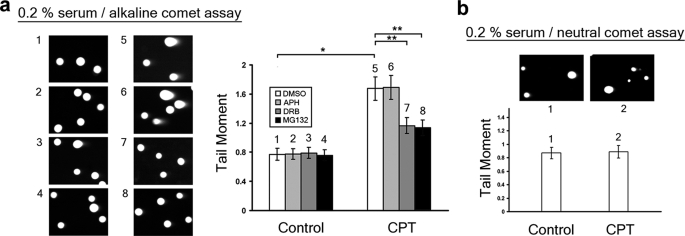
CPT Induces DNA Single-strand Breaks in a Transcription- and Proteasome-dependent Manner in Quiescent WI-38 Cells—The above studies have suggested that CPT-induced DNA damage is transcription- and proteasome-dependent in nonproliferative cells. To investigate the nature of CPT-induced DNA damage in nonproliferative cells, DNA strand breaks were measured by both alkaline and neutral comet assays in quiescent WI-38 cells treated with CPT. As shown in Fig. 4a, CPT induced a large increase in the tail moment of quiescent WI-38 cells (p = 2.7 × 10-7, determined by two-tailed unpaired Student's t test) as measured by the alkaline comet assay, suggesting the formation of strand breaks. The CPT-induced increase in the tail moment of quiescent WI-38 cells was significantly abolished by co-treatment with either the transcription inhibitor DRB or the proteasome inhibitor MG132, but not the replication inhibitor APH (Fig. 4a) (p = 0.002, 0.0008, and 0.9 for DRB, MG132, and APH treatments, respectively) (8, 16, 24). By contrast, CPT treatment had no effect on the tail moment of quiescent WI-38 cells as measured by neutral comet assay (p = 0.88) (Fig. 4b), suggesting that CPT did not induce any detectable DSBs in quiescent WI-38 cells, consistent with the lack of γ-H2AX induction under the same conditions (Fig. 1c). Consequently, our results suggest that CPT induces primarily SSBs in a transcription- and proteasome-dependent manner in quiescent WI-38 cells.
FIGURE 4.
CPT induces single-strand breaks in a transcription- and proteasome-dependent manner in quiescent WI-38 cells. a, WI-38 cells cultured in DMEM supplemented with 0.2% serum were pretreated with APH (10 μm), DRB (150 μm), or MG132 (2 μm) for 30 min, followed by co-treatment with CPT (25 μm) for 1 h. SSBs were analyzed by alkaline comet assay as described under “Experimental Procedures.” Left panel, representative comet images. Right panel, the histogram of the tail moment plotted against each treatment condition. b, WI-38 cells cultured in DMEM supplemented with 0.2% serum were treated with Me2SO or CPT (25 μm) for 1 h. DSBs were analyzed by neutral comet assay as described under “Experimental Procedures.” Upper panel, representative comet images. Lower panel, the histogram of the tail moment.
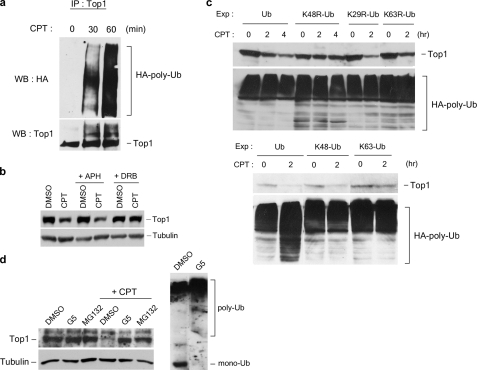
Assembly of Lys-48-linked Polyubiquitin Chains on Top1 Is Required for CPT-induced Top1 Down-regulation—The possible involvement of a ubiquitin-proteasome pathway in CPT-induced Top1 down-regulation is suggested from results based on the use of the proteasome inhibitors and the temperature-sensitive E1 mutant cell line, ts85 (25). To further characterize the ubiquitin-proteasome pathway involved in CPT-induced Top1 down-regulation, three different sets of experiments were performed. First, CPT was shown to induce the formation of Top1-ubiquitin conjugates in HA-Ub-transfected HeLa cells (Fig. 5a) and V79 cells (supplemental Fig. S3) based on immunoblotting of Top1 immunoprecipitates using anti-HA antibody. In addition, CPT-induced formation of Top1-ubiquitin conjugates was inhibited by DRB (supplemental Fig. S3), suggesting the involvement of transcription in the CPT-induced formation of Top1-ubiquitin conjugates. The requirement for transcription was also shown for CPT-induced Top1 down-regulation in HeLa cells (Fig. 5b), suggesting that the formation of Top1-ubiquitin conjugates is likely to be linked to Top1 down-regulation. Second, the involvement of ubiquitination in CPT-induced Top1 down-regulation was also studied in HeLa cells using dominant-negative Ub mutants. As shown in Fig. 5c, CPT-induced Top1 down-regulation was abolished in HeLa cells transfected with the dominant-negative Ub mutant, K48R-Ub (Lys-48 is mutated to Arg on HA-Ub). By contrast, CPT-induced Top1 down-regulation was unaffected in HeLa cells transfected with other dominant-negative Ub mutants (i.e. K29R-Ub and K63R-Ub) (Fig. 5c, upper panels), suggesting that the formation of Lys-48-linked polyubiquitin chains is specifically required for CPT-induced Top1 down-regulation. Consistent with this idea, CPT-induced Top1 down-regulation was unaffected in HeLa cells transfected with K48-Ub (all lysine residues on Ub were mutated to arginine residues except Lys-48), but abolished in cells transfected with K63-Ub (all lysine residues on Ub were mutated to arginine residues except Lys-63) (Fig. 5c, lower panels). Third, CPT-induced Top1 down-regulation was largely abolished in HeLa cells pretreated with G5 (Fig. 5d), a ubiquitin isopeptidase inhibitor, which depletes the free Ub pool (26). Indeed, the free Ub (labeled mono-Ub) pool was depleted in HeLa cells pretreated with G5 (Fig. 5d, right panel). Together, these results strongly suggest that CPT-induced down-regulation requires the assembly of Lys-48-linked polyubiquitin chains.
FIGURE 5.
CPT-induced Top1 down-regulation is dependent on the assembly of K48-linked polyubiquitin chains. a, HeLa cells were transfected with the HA-Ub expression plasmid. Transfection, CPT treatment, and immunoprecipitation (IP) were performed as described under “Experimental Procedures.” b, HeLa cells were pretreated with 10 μm APH or 150 μm DRB for 30 min, followed by co-treatment with 25 μm CPT for 3 h. Top1 down-regulation was analyzed as described under “Experimental Procedures.” c, HeLa cells were transfected with various plasmids to overexpress HA-tagged wild type and mutant ubiquitin proteins. Transfection, CPT treatment (25 μm), and Top1 down-regulation assay were performed as described under “Experimental Procedures.” Ub, wild type ubiquitin; K48R-Ub, K48R mutant ubiquitin; K29R-Ub, K29R mutant ubiquitin; K63R-Ub, K63R mutant ubiquitin; K48-Ub, K48-only ubiquitin (all lysine residues were mutated to arginine residues except Lys-48); K63-only Ub (all lysine residues were mutated to arginine residues except Lys-63). The amount of protein-HA-Ub conjugates in cells transfected with each construct was also determined by immunoblotting using anti-HA antibody (lower panels). d, Top1 degradation requires ubiquitin and proteasome. HeLa cells were pretreated with 5 μm MG132 or 5 μm G5 for 30 min, followed by co-treatment with 25 μm CPT for 3 h. Top1 down-regulation was determined by immunoblotting of cell lysates with anti-hTop1 antibody. Cell lysates (from control cells and G5-treated cells) were also immunoblotted with anti-ubiquitin antibody to assess the levels of free ubiquitin (mono-Ub) and ubiquitin-protein conjugates (poly-Ub). WB, Western blot; DMSO, dimethyl sulfoxide.
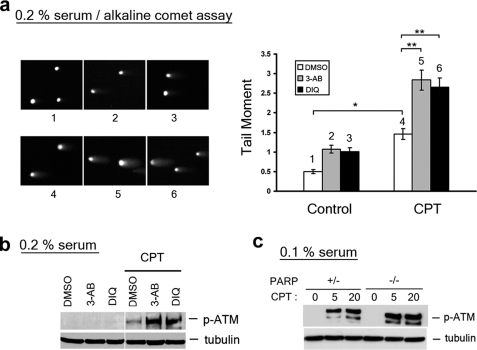
PARP1 Inactivation Leads to Elevated SSBs and ATM Autophosphorylation in Nonprolierating Cells Treated with CPT—PARP1, known to be involved in base excision repair, has also been shown to participate in the repair of CPT-induced DNA damage (27, 28). To test whether CPT-induced SSBs are the substrates for PARP1-mediated repair, PARP1 inhibitors 3-aminobenzamide (3-AB) and 1,5-dihydroxyisoquinoline (DIQ) were employed in the study. DIQ is known to be a more potent and specific inhibitor of PARP1 than 3-AB (29). Both 3-AB (4 mm) and DIQ (100 μm) were shown to increase the tail moment (measured by alkaline comet assay) of CPT-treated quiescent WI-38 cells (Fig. 6a) (p = 3.2 × 10-10 and 4.9 × 10-8, respectively), suggesting that CPT-induced SSBs are likely to be the substrates for PARP1-mediated repair. As shown in Fig. 6b, CPT-induced ATM autophosphorylation was also greatly elevated in both proliferating and quiescent WI-38 cells treated with either 3-AB or DIQ, suggesting that ATM may be activated by CPT-induced SSBs that are repaired by a PARP1-dependent pathway.
FIGURE 6.
Inactivation of PARP1 elevates the amount of CPT-induced SSBs and the level of ATM autophosphorylation. WI-38 cells cultured in DMEM supplemented with 0.2% serum were pretreated with 4 mm 3-AB or 100 μm DIQ for 30 min, followed by co-treatment with CPT (25 μm) for 1 h. The cells were subjected to either alkaline comet assay (see representative comet images to the left and the histogram of the tail moment to the right; p values for comparisons marked * and ** were < 0.005 as determined by two-tailed Student's t test) (a) or immunoblotting with indicated antibodies (b). c, parp+/- and parp-/- primary MEFs cultured in 0.1% serum (for 3 days) were treated with CPT for 1 h, followed by immunoblotting with antibodies as indicated. DMSO, dimethyl sulfoxide.
To further confirm that PARP1 is involved in the repair of CPT-induced SSBs, CPT-induced ATM autophosphorylation was also measured in wild type and PARP1 knock-out MEFs. As shown in Fig. 6c, CPT-induced ATM autophosphorylation was significantly increased in parp1-/- MEFs compared with parp1+/- MEFs cultured in 0.1% serum (nonproliferating condition), further suggesting that PARP1 acts downstream of CPT-induced SSBs.
DISCUSSION
Our current studies have demonstrated that Top1 cleavage complexes induced by CPT can generate distinct DNA damage signals and strand breaks depending on the proliferation state of cells. In proliferating WI-38 cells, CPT induces DNA DSBs (measured by neutral comet assay) and γ-H2AX, both of which can be completely abolished by the replication inhibitor APH but not by the transcription inhibitor DRB. These results are consistent with the proposed replication fork collision model for the replication fork-dependent processing of Top1 cleavage complexes into DNA damage (e.g. DNA DSBs) (7, 10). CPT also induces ATM autophosphorylation at Ser-1981 in proliferating WI-38 cells. However, CPT-induced ATM autophosphorylation is partially inhibited by either the replication inhibitor APH or the transcription inhibitor DRB. Inhibition of CPT-induced ATM autophosphorylation by APH is expected because DSBs, known to activate ATM (21), are generated at the collision sites according to the replication fork collision model (7, 10). However, it is unexpected that CPT-induced ATM autophosphorylation is also partially inhibited by the transcription inhibitor DRB, which could suggest a possible involvement of transcription in CPT-induced DNA damage, which is distinct from replication-dependent formation of DSBs.
Using quiescent WI-38 cells, we have investigated CPT-induced DNA damage in the absence of DNA replication. CPT was shown to induce robust ATM autophosphorylation at Ser-1981 but little γ-H2AX in quiescent WI-38 cells. In addition, CPT-induced ATM autophosphorylation is inhibited by the transcription inhibitors such as DRB and α-amanitin, suggesting the involvement of transcription, but not replication, in CPT-induced DNA damage in quiescent WI-38 cells. Similar results were obtained in post-mitotic mouse cortical neurons, which further confirms the involvement of transcription in CPT-induced DNA damage in nonproliferating cells. It is noted that hydrogen peroxide-induced ATM autophosphorylation in post-mitotic cortical neurons is neither transcription- nor proteasome-dependent, suggesting a specific requirement for transcription and proteasome in CPT-induced ATM autophosphorylation. The requirement for transcription in CPT-induced ATM autophosphorylation is not due to a requirement for new protein synthesis because the protein synthesis inhibitor cycloheximide has no effect on CPT-induced ATM autophosphorylation. The requirement for transcription but not protein synthesis in CPT-induced ATM autophosphorylation is reminiscent of CPT-induced down-regulation of Top1, which is also transcription-dependent but protein synthesis-independent. It has been suggested that CPT-induced down-regulation of Top1 depends on the process of active transcription but not newly synthesized protein products (8).
Analysis of CPT-induced ATM autophosphorylation in quiescent WI-38 cells has also revealed a dependence on proteasome activity because the proteasome inhibitors, MG132 and PS-341 (bortezomib), abolished CPT-induced ATM autophosphorylation. Interestingly, using comet assays, CPT was shown to induce primarily SSBs in a transcription- and proteasome-dependent manner. The correlation between ATM autophosphorylation (determined by the immunoblotting assay) and the formation of SSBs (determined by the alkaline comet assay) suggests that ATM autophosphorylation could be the DNA damage signal resulting from CPT-induced SSBs. The robust induction of ATM by CPT-induced SSBs is unexpected because ionization radiation-induced ATM activation is closely correlated with the number of DSBs but not SSBs (31). Our results could suggest that Top1 cleavage complexes induced by CPT may be processed through a transcription- and proteasome-dependent manner into a unique type of DNA lesion, which resembles but is not identical to normal SSBs induced by other agents such as hydrogen peroxide and IR. These unconventional SSBs (see Fig. 7 for discussion on SSB*) could lead to ATM activation by an unknown mechanism. Alternatively, CPT-induced ATM activation could involve changes of the local chromatin structure at the collision sites. It has been suggested that altered chromatin structure may be sufficient to activate ATM in the absence of DSBs (32). Further studies are necessary to reveal this potential novel mechanism for ATM activation by Top1-associated SSBs.
FIGURE 7.
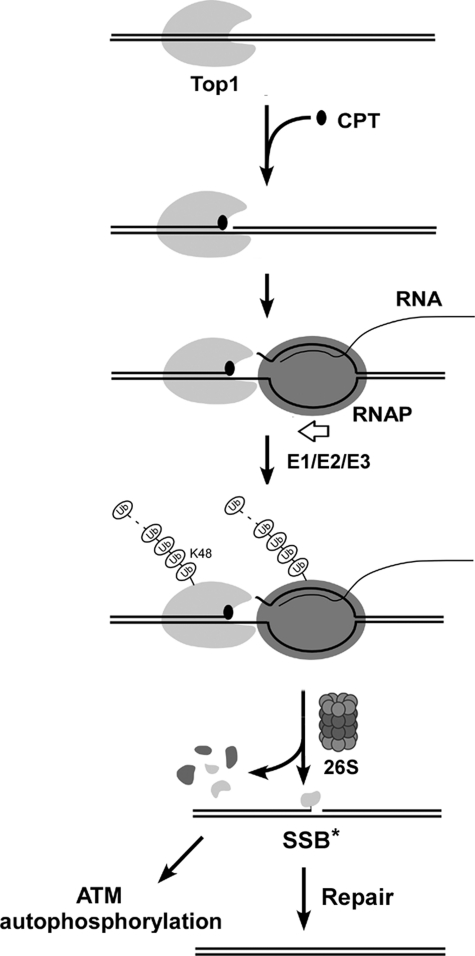
A proposed model for transcription-dependent processing of Top1-DNA covalent complexes into strand breaks. CPT stabilizes reversible Top1 cleavage complexes on chromosomal DNA within the actively transcribed regions. Upon collision with the elongating RNA polymerase complex, transcription is arrested, and a ubiquitin-proteasome pathway is activated, leading to the assembly of a Lys-48-linked polyubiquitin chain on Top1 and subsequent degradation by the 26 S proteasome. The fate of the RNA polymerase is not known. However, the largest subunit of RNA pol II has been shown to be extensively degraded (8). The proteasomal degradation of Top1 cleavage complexes presumably exposes the otherwise Top1-concealed SSB into SSB*. The SSB* is likely to be a SSB with 3′-phosphoryl end covalently linked to a Top1 active site tyrosine-containing peptide. The formation of SSB* (or its subsequent processing) can be lead to ATM autophosphorylation. Repair of SSB* occurs through a PARP1-dependent pathway.
The similar requirements (i.e. transcription and proteasome dependence) suggest that CPT-induced Top1 down-regulation is tightly linked to CPT-induced SSBs and ATM autophosphorylation. These results can best be explained by a model presented in Fig. 7, in which CPT-induced Top1 down-regulation is the result of a collision between the Top1 cleavage complex and the elongating RNA polymerase. Such a collision results in transient arrest of the elongating RNA polymerase complex and the activation of a proteasome pathway, leading to the assembly of Lys-48-linked polyubiquitin chain on Top1 and the subsequent degradation of the Top1 cleavage complex at the collision site and the concomitant exposure of the otherwise Top1-concealed SSB. The formation of the SSB* (an unconventional SSB that is covalently associated with the active site tyrosine-containing Top1 peptide) could lead to ATM activation and allow subsequent repair through a PARP1-dependent process (see discussion in later paragraphs). Following Top1 down-regulation and repair of the SSB*, transcription can then restart.
Recent studies have demonstrated that proteasome-mediated degradation can occur through both ubiquitin-dependent and ubiquitin-independent pathways (reviewed in Ref. 33). An increasing number of proteins have been shown to undergo degradation through ubiquitin-independent proteasome pathways (e.g. p21/Cip1 and TCRα) (34, 35). Using the ubiquitin isopeptidase inhibitor, G5, we have demonstrated the involvement of the free ubiquitin pool in CPT-induced Top1 down-regulation. The involvement of ubiquitin was further supported by the demonstration of transcription-dependent formation of Top1-ubiquitin conjugates in CPT-treated cells. Most importantly, using dominant-negative ubiquitin mutants, we have shown that the assembly of Lys-48-linked polyubiquitin chains on Top1 is required for CPT-induced Top1 down-regulation. Previous studies have demonstrated that a Cullin 3-based E3 ligase (Cul3) is involved in CPT-induced Top1 down-regulation (36). It seems likely that the Cul3 may be involved in the assembly of the Lys-48-linked polyubiquitin chains on Top1. The requirement for transcription for the assembly of the polyubiquitin chains on Top1 is interesting. Previous studies have suggested that proteasome components are associated with RNA polymerase II and are required for efficient transcription elongation (37, 38). It is possible that the collision between the Top1 cleavage complex and the elongating RNA polymerase may bring the proteasome components into close proximity of the colliding complexes and facilitate proteasomal degradation of Top1.
Our current studies have also established a role for PARP1 in CPT-induced DNA damage. We showed that upon PARP1 inactivation, CPT-induced SSBs and ATM autophosphorylation were significantly increased (Fig. 6). These results can best be explained by a model showing that PARP1 is involved in the repair of CPT-induced SSBs (a step downstream of processed Top1-DNA covalent complexes as shown in Fig. 7). Our results are consistent with previous results showing the involvement of PARP1 in CPT-induced DNA damage (27). PARP1 is known to be involved in base excision repair and form a base excision repair complex with XRCC1 and DNA Ligase IIIα, which is involved in the repair of SSBs (39). Interestingly, TDP1 has also been identified as a component of the same complex (40, 41). TDP1 has been shown to be involved in the removal of the residual Top1 peptides prior to SSB repair based on studies using TDP1-mutated SCAN1 (spinocerebellar ataxia with axonal neuropathy 1) lymphoblastoid cells and TDP1 knockout mice (41, 42). It is plausible that proteasome-processed Top1-DNA covalent complexes may generate unconventional SSBs (e.g. Top1 peptide-linked SSBs) that are further processed by TDP1 into conventional SSBs for repair.
In addition to S phase cytotoxicity, which is primarily the result of CPT-induced mitotic catastrophe (43), CPT is also known to induce cell death in post-mitotic neurons (44). It seems possible that the transcription-dependent processing of the Top1-DNA covalent complex may lead to the generation of an intermediate that, if not repaired, is lethal to cells (e.g. post-mitotic neurons). The precise structure of this lethal intermediate is not known. However, it is possible that this lethal intermediate is the Top1 peptide-linked SSB that is generated from the proteasome-degraded Top1-DNA covalent complex. It is interesting to note that this lethal intermediate (i.e. Top1 peptide-linked SSB) is accumulated in tdp1-/- cells, leading to topotecan (a CPT derivative) hypersensitivity (42).
Our results may have relevance to neurodegenerative diseases. It has been shown that oxidatively damaged DNA can trap Top1-DNA covalent complexes (45, 46). These Top1-DNA covalent complexes could be processed in a transcription- and proteasome-dependent manner into the proposed lethal intermediate that may contribute to neurodegeneration. It has been proposed that this lethal intermediate is normally processed by PARP1, TDP1, or polynucleotide kinase 3′-phosphatase into normal SSBs and subsequently repaired by other SSB repair components (47). In the absence of TDP1, this lethal intermediate would accumulate, leading to cell death (e.g. neuronal cell death). The elevated levels of autophosphorylated ATM in the presence of PARP1 inhibitors reported here suggest that ATM could be activated by these Top1-associated lethal intermediates, effecting a repair pathway for cell survival. Consequently, results reported here may reveal a significant pathway, relating ATM-dependent repair of Top1-DNA covalent complexes to human neurodegenerative diseases such as the spinocerebellar ataxia with axonal neuropathy disorder.
Supplementary Material
Acknowledgments
We thank Betty Zheng and Dr. Changshun Shao (Rutgers University) and Ren-Kuo Lin for technical assistance on the isolation and characterization of parp1+/- and parp1-/- MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA39662. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: Top1, topoisomerase I; DRB, 5,6-dichloro-1-β-d-ribobenzimidazole; APH, aphidicolin; MG132, carbobenzoxy-l-leucyl-l-leucyl-l-leucinal; Ub, ubiquitin; ATM, ataxia telangiectasia mutated; PARP1, poly(ADP-ribose) polymerase family, member 1; TDP1, tyrosyl-DNA phosphodiesterase 1; MEF, mouse embryonic fibroblast; CPT, camptothecin; SSB, single-strand break; DSB, double-strand break; DMEM, Dulbecco's modified Eagle's medium; E1, ubiquitin-activating enzyme; E3, ubiquitin-protein isopeptide ligase; HA, hemagglutinin; 3-AB, 3-aminobenzamide; DIQ, 1,5-dihydroxyisoquinoline.
References
- 1.Wang, J. C. (1996) Annu. Rev. Biochem. 65635. [DOI] [PubMed] [Google Scholar]
- 2.Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3430 -440 [DOI] [PubMed] [Google Scholar]
- 3.Li, T. K., and Liu, L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 4153 -77 [DOI] [PubMed] [Google Scholar]
- 4.Champoux, J. J. (2001) Annu. Rev. Biochem. 70369 -413 [DOI] [PubMed] [Google Scholar]
- 5.Pommier, Y. (2006) Nat. Rev. Cancer 6789 -802 [DOI] [PubMed] [Google Scholar]
- 6.Hsiang, Y. H., Hertzberg, R., Hecht, S., and Liu, L. F. (1985) J. Biol. Chem. 26014873 -14878 [PubMed] [Google Scholar]
- 7.Hsiang, Y. H., Lihou, M. G., and Liu, L. F. (1989) Cancer Res. 495077 -5082 [PubMed] [Google Scholar]
- 8.Desai, S. D., Zhang, H., Rodriguez-Bauman, A., Yang, J. M., Wu, X., Gounder, M. K., Rubin, E. H., and Liu, L. F. (2003) Mol. Cell. Biol. 232341 -2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao, Y. P., D'Arpa, P., and Liu, L. F. (1992) Cancer Res. 521823 -1829 [PubMed] [Google Scholar]
- 10.Tsao, Y. P., Russo, A., Nyamuswa, G., Silber, R., and Liu, L. F. (1993) Cancer Res. 53 5908. [PubMed] [Google Scholar]
- 11.Shao, R. G. (1999) EMBO J. 181397 -1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm, C., Covey, J. M., Kerrigan, D., and Pommier, Y. (1989) Cancer Res. 496365 -6368 [PubMed] [Google Scholar]
- 13.D'Arpa, P., Beardmore, C., and Liu, L. F. (1990) Cancer Res. 506919. [PubMed] [Google Scholar]
- 14.Zhang, H., D'Arpa, P., and Liu, L. F. (1990) Cancer Cells 223 -27 [PubMed] [Google Scholar]
- 15.Wu, J., and Liu, L. F. (1997) Nucleic Acids Res. 254181 -4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai, S. D., Li, T. K., Rodriguez-Bauman, A., Rubin, E. H., and Liu, L. F. (2001) Cancer Res. 615926 -5932 [PubMed] [Google Scholar]
- 17.Dimri, G. P., Hara, E., and Campisi, J. (1994) J. Biol. Chem. 26916180 -16186 [PubMed] [Google Scholar]
- 18.Norbury, C., Blow, J., and Nurse, P. (1991) EMBO J. 103321 -3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiang, Y. H., Wu, H. Y., and Liu, L. F. (1988) Cancer Res. 483230 -3235 [PubMed] [Google Scholar]
- 20.Earnshaw, W. C., Halligan, B., Cooke, C. A., Heck, M. M., and Liu, L. F. (1985) J. Cell Biol. 100 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuta, T., Takemura, H., Liao, Z.-Y., Aune, G. J., Redon, C., Sedelnikova, O. A., Pilch, D. R., Rogakou, E. P., Celeste, A., Chen, H. T., Nussenzweig, A., Aladjem, M. I., Bonner, W. M., and Pommier, Y. (2003) J. Biol. Chem. 27820303 -20312 [DOI] [PubMed] [Google Scholar]
- 22.Thiriet, C., and Hayes, J. J. (2005) Mol Cell 18617 -622 [DOI] [PubMed] [Google Scholar]
- 23.Zhang, A., Lyu, Y. L., Lin, C. P., Zhou, N., Azarova, A. M., Wood, L. M., and Liu, L. F. (2006) J. Biol. Chem. 28135997 -36003 [DOI] [PubMed] [Google Scholar]
- 24.El-Khamisy, S. F., Hartsuiker, E., and Caldecott, K. W. (2007) DNA Repair (Amst) 61485 -1495 [DOI] [PubMed] [Google Scholar]
- 25.Desai, S. D., Liu, L. F., Vazquez-Abad, D., and D'Arpa, P. (1997) J. Biol. Chem. 272 24159. [DOI] [PubMed] [Google Scholar]
- 26.Aleo, E., Henderson, C. J., Fontanini, A., Solazzo, B., and Brancolini, C. (2006) Cancer Res. 669235 -9244 [DOI] [PubMed] [Google Scholar]
- 27.Malanga, M., and Althaus, F. R. (2004) J. Biol. Chem. 2795244 -5248 [DOI] [PubMed] [Google Scholar]
- 28.Smith, L. M., Willmore, E., Austin, C. A., and Curtin, N. J. (2005) Clin. Cancer Res. 118449 -8457 [DOI] [PubMed] [Google Scholar]
- 29.Virag, L., and Szabo, C. (2002) Pharmacol Rev 54375 -429 [DOI] [PubMed] [Google Scholar]
- 30.Mao, Y., Desai, S. D., Ting, C. Y., Hwang, J., and Liu, L. F. (2001) J. Biol. Chem. 27640652 -40658 [DOI] [PubMed] [Google Scholar]
- 31.Ismail, I. H., Nystrom, S., Nygren, J., and Hammarsten, O. (2005) J. Biol. Chem. 2804649 -4655 [DOI] [PubMed] [Google Scholar]
- 32.Bakkenist, C. J., and Kastan, M. B. (2003) Nature 421499 -506 [DOI] [PubMed] [Google Scholar]
- 33.Hoyt, M. A., and Coffino, P. (2004) Cell Mol. Life Sci. 611596 -1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5403 -410 [DOI] [PubMed] [Google Scholar]
- 35.Yu, H., and Kopito, R. R. (1999) J. Biol. Chem. 27436852 -36858 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H. F., Tomida, A., Koshimizu, R., Ogiso, Y., Lei, S., and Tsuruo, T. (2004) Cancer Res. 641114 -1121 [DOI] [PubMed] [Google Scholar]
- 37.Gillette, T. G., Gonzalez, F., Delahodde, A., Johnston, S. A., and Kodadek, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 1015904 -5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinyamu, H. K., and Archer, T. K. (2007) Mol. Cell. Biol. 274891 -4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almeida, K. H., and Sobol, R. W. (2007) DNA Repair (Amst) 6695 -711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plo, I. (2003) DNA Repair 21087 -1100 [DOI] [PubMed] [Google Scholar]
- 41.El-Khamisy, S. F. (2005) Nature 434108 -113 [DOI] [PubMed] [Google Scholar]
- 42.Katyal, S., El-Khamisy, S. F., Russell, H. R., Li, Y., Ju, L., Caldecott, K. W., and McKinnon, P. J. (2007) EMBO J. [DOI] [PMC free article] [PubMed]
- 43.Roninson, I. B., Broude, E. V., and Chang, B. D. (2001) Drug Resist. Updat 4 303-313 [DOI] [PubMed] [Google Scholar]
- 44.Morris, E. J., and Geller, H. M. (1996) J. Cell Biol. 134757 -770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daroui, P., Desai, S. D., Li, T. K., Liu, A. A., and Liu, L. F. (2004) J. Biol. Chem. 27914587 -14594 [DOI] [PubMed] [Google Scholar]
- 46.Pourquier, P., Ueng, L. M., Fertala, J., Wang, D., Park, H. J., Essigmann, J. M., Bjornsti, M. A., and Pommier, Y. (1999) J. Biol. Chem. 2748516 -8523 [DOI] [PubMed] [Google Scholar]
- 47.Rass, U., Ahel, I., and West, S. C. (2007) Cell 130991 -1004 [DOI] [PubMed] [Google Scholar]
- 48.Lyu, Y. L., Kerrigan, J. E., Lin, C. P., Azarova, A. M., Tsai, Y. C., Ban, Y., and Liu, L. F. (2007) Cancer Res. 678839 -8846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.