Abstract
Maintenance of genomic integrity relies on a proper response to DNA injuries integrated by the DNA damage checkpoint; histone modifications play an important role in this response. Dot1 methylates lysine 79 of histone H3. In Saccharomyces cerevisiae, Dot1 is required for the meiotic recombination checkpoint as well as for chromatin silencing and the G1/S and intra-S DNA damage checkpoints in vegetative cells. Here, we report the analysis of the function of Dot1 in the response to alkylating damage. Unexpectedly, deletion of DOT1 results in increased resistance to the alkylating agent methyl methanesulfonate (MMS). This phenotype is independent of the dot1 silencing defect and does not result from reduced levels of DNA damage. Deletion of DOT1 partially or totally suppresses the MMS sensitivity of various DNA repair mutants (rad52, rad54, yku80, rad1, rad14, apn1, rad5, rad30). However, the rev1 dot1 and rev3 dot1 mutants show enhanced MMS sensitivity and dot1 does not attenuate the MMS sensitivity of rad52 rev3 or rad52 rev1. In addition, Rev3-dependent MMS-induced mutagenesis is increased in dot1 cells. We propose that Dot1 inhibits translesion synthesis (TLS) by Polζ/Rev1 and that the MMS resistance observed in the dot1 mutant results from the enhanced TLS activity.
GENOME integrity is constantly threatened by DNA damage. This damage can arise from endogenous sources, as a consequence, for example, of free radicals resulting from oxidative metabolism or of collapsed replication forks. Other sources of damage include effects of exogenous agents, such as ionizing radiation (IR), UV radiation, or certain chemotherapeutic drugs. There is also programmed DNA damage that must occur during certain biological processes, such as the DNA double-strand breaks (DSBs) that initiate recombination during meiosis (Roeder 1997), as well as V(D)J and class-switch recombination in mammals (Gellert 1996) or mating-type switching in yeast (Haber 1998).
Therefore, to maintain genomic integrity, cells must be able to properly react to DNA damage, and eukaryotic cells have evolved a complex surveillance mechanism, the so-called DNA damage checkpoint, which integrates a series of cellular responses to the presence of genome injuries (Nyberg et al. 2002). Essentially, the DNA damage checkpoint is composed of sensors that detect the damage and generate a signal that is transmitted to effectors, which eventually act on targets responsible for the different cellular responses to DNA damage. In Saccharomyces cerevisiae, the Mec1-Ddc2 kinase complex and the “9-1-1” (Ddc1-Rad17-Mec3) clamp, together with the Rad24 clamp loader, are independently recruited to the sites of lesions and induce the activation of the Rad53 and Chk1 effector kinases by Mec1-dependent phosphorylation in a process mediated by the Rad9 adaptor (Harrison and Haber 2006). The main responses resulting from activation of the checkpoint kinases include an arrest or delay in cell cycle progression and the activation of DNA repair at several levels, such as transcriptional induction of repair genes, direct activation of repair proteins, or relocalization of repair factors to the sites of damage (Zhou and Elledge 2000).
Among the different types of DNA damage, DSBs are perhaps the most dangerous because, if they are not properly repaired, they can lead to chromosome rearrangements, aneuploidy, loss of genetic information, and cell death. Repair of DSBs occurs by two processes: homologous recombination (HR) and nonhomologous end joining (NHEJ). Basically, during HR the DSB ends are resected to generate single-strand DNA, which invades an intact template to copy information. In mitotic cells, the preferred template for HR is the sister chromatid. On the other hand, during NHEJ direct religation of the DSB ends takes place, sometimes after limited processing; therefore, while HR maintains fidelity, NHEJ can be error prone (Paques and Haber 1999). In S. cerevisiae, the Rad52 protein is essential for all types of HR (Symington 2002), while NHEJ is abolished in the absence of a functional Ku complex (Boulton and Jackson 1996a,b).
In addition to HR and NHEJ, there are other DNA repair mechanisms that significantly contribute to genomic stability and, although they are not strictly DSB repair pathways, they can contribute to preventing DSB formation because they eliminate lesions that could potentially lead to replication blocks (Kupiec 2000). These mechanisms are the nucleotide excision repair (NER) and the base excision repair (BER) pathways. Whereas NER is specialized for the removal of bulky lesions that disrupt the DNA helix, such as pyrimidine dimers and photoproducts of UV light, BER removes nonbulky lesions, such as those resulting from alkylation damage (Sancar 1996).
In addition to these DNA repair pathways, there are also damage tolerance mechanisms, including that of translesion synthesis (TLS). These mechanisms are also critical for survival in the presence of genotoxic agents, allowing replication forks to pass through lesions and complete DNA replication although damage has not been removed from the template strand (Kunz et al. 2000; Prakash et al. 2005; Ulrich 2005; Klein 2007). Replicative polymerases are not able to carry out TLS because their catalytic centers are optimized for a perfect match between primer and template. However, damage-tolerant polymerases, specialized for TLS, contain more relaxed catalytic centers, which can accommodate lesions in the template, although fidelity is compromised (Kunkel 2004). In S. cerevisiae, TLS is carried out by the Polη, Polζ, and Rev1 polymerases, all of which have human orthologs (Kunz et al. 2000). Polη is encoded by the RAD30 gene and is dedicated to the repair of UV-induced TT dimers in an error-free manner. In contrast, Polζ, formed by the Rev3 and Rev7 subunits, in cooperation with Rev1, participates in error-prone TLS.
In eukaryotes, detection, signaling, and repair of the DNA lesions do not take place on naked DNA, but rather in the context of highly structured chromatin, which poses a barrier for the access of the cellular machinery involved in these processes. It is therefore not surprising that histone modifications and chromatin remodeling play an important role in the cellular response to DNA damage (Peterson and Cote 2004; Lydall and Whitehall 2005; van Attikum and Gasser 2005; Downs et al. 2007). One of the first chromatin modifications that occurs in response to DSBs is the Mec1- and Tel1-dependent phosphorylation of serine 129 of the yeast histone H2A in the chromosomal region flanking the break. Mutants lacking this phosphorylation site are defective in NHEJ and are sensitive to methyl methanesulfonate (MMS) and phleomycin (Downs et al. 2000).
Another histone modification involved in the DNA damage response is the methylation of lysine 79 of histone H3 (H3K79) mediated by Dot1 (Feng et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002). The first indication of the participation of Dot1 in a DNA-related checkpoint mechanism came from studies in meiotic cells, where Dot1 is required for the so-called pachytene checkpoint or meiotic recombination checkpoint, which blocks meiotic cell cycle progression until recombination has been completed (San-Segundo and Roeder 2000). In addition, Dot1 prevents the repair of meiotic DSBs using the sister chromatid as a template in the absence of Dmc1 (San-Segundo and Roeder 2000). More recently, it has been shown that Dot1 also participates in the DNA damage response in vegetative cells, being required for the Rad9-mediated activation of Rad53 in the G1/S and intra-S DNA damage checkpoints (Giannattasio et al. 2005; Wysocki et al. 2005). The dot1 mutant shows mild sensitivity to UV light and IR, and it has been proposed that Dot1-mediated H3K79 methylation is required for the repair of UV- and IR-induced lesions (Game et al. 2005, 2006; Toh et al. 2006; Bostelman et al. 2007).
Here, we investigate the role of Dot1 in the response to the alkylating agent MMS. We find that, surprisingly, the dot1 mutant is more resistant than the wild type to high MMS doses, but this increased resistance is not a consequence of fewer MMS-promoted lesions. On the contrary, the dot1 mutant displays a higher number of MMS-induced Rad52 repair centers and higher levels of histone H2A phosphorylation. Moreover, deletion of DOT1 also totally or partially suppresses the MMS sensitivity of mutants in HR, NHEJ, BER, and NER pathways. However, inactivation of the error-prone Rev3- and Rev1-dependent TLS pathway abolishes the increased MMS resistance conferred by the absence of Dot1, and the dot1 mutant shows augmented MMS-induced mutagenesis dependent on Rev3, indicating that Dot1 negatively regulates this TLS pathway.
MATERIALS AND METHODS
Strains and plasmids:
Yeast strain genotypes are listed in Table 1. Strains are in the BR1919, W303, BY4741/BY4742, or JKM179 backgrounds, as indicated. All strains used and compared in each particular experiment were isogenic, i.e., of the same genetic background. Gene disruptions were introduced either by direct transformation or by genetic crosses in an isogenic background. Plasmids pSS30 and pSS44 were used to generate dot1∷URA3 and dot1∷TRP1, respectively (San-Segundo and Roeder 2000). pSM20 and pSM31 (provided by D. Schild, Berkeley) were used to generate rad52∷LEU2 and rad54∷LEU2, respectively. Plasmid pES28 was used for sir2∷URA3 (Chien et al. 1993) and pKL12 was used for sir3∷TRP1 (Stone et al. 1991). Other gene deletions were made by a PCR-based approach (Longtine et al. 1998; Goldstein and McCusker 1999).
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| BR1919αa | MATα leu2-3,112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 |
| BR1919a | MATaleu2-3,112 his4-260 ura3-1 ade2-1 thr1-4 trp1-289 |
| YP163 | BR1919α dot1∷TRP1 |
| YP210 | BR1919α PCH2-HA |
| YP345 | BR1919α dot1∷URA3 PCH2-HA |
| YP370 | BR1919α rad52∷LEU2 PCH2-HA |
| YP371 | BR1919α rad52∷LEU2 dot1∷TRP1 PCH2-HA |
| YP372 | BR1919α rad52∷LEU2 sir2∷URA3 PCH2-HA |
| YP513 | BR1919a yku80∷kanMX2 PCH2-HA |
| YP514 | BR1919a yku80∷kanMX2 dot1∷URA3 PCH2-HA |
| YP515 | BR1919a yku80∷kanMX2 rad52∷LEU2 PCH2-HA |
| YP516 | BR1919a yku80∷kanMX2 rad52∷LEU2 dot1∷URA3 PCH2-HA |
| YP544 | BR1919(a or α) sir3∷TRP1 |
| YP545 | BR1919a yku80∷kanMX2 sir3∷TRP1 |
| YP546 | BR1919a yku80∷kanMX2 sir3∷TRP1 dot1∷URA3 |
| YP547 | BR1919a sir3∷TRP1 dot1∷URA3 |
| YP548 | BR1919α rad52∷LEU2 sir3∷TRP1 |
| YP549 | BR1919α rad52∷LEU2 sir3∷TRP1 dot1∷URA3 |
| YP550 | BR1919a rad52∷LEU2 yku80∷kanMX2 sir3∷TRP1 |
| YP551 | BR1919α rad52∷LEU2 yku80∷kanMX2 sir3∷TRP1 dot1∷URA3 |
| YP574 | BR1919α rad54∷LEU2 PCH2-HA |
| YP576 | BR1919a dot1∷TRP1 |
| YP587 | BR1919α rad54∷LEU2 dot1∷URA3 PCH2-HA |
| YP588 | BR1919a rad54∷LEU2 yku80∷kanMX2 PCH2-HA |
| YP589 | BR1919a rad54∷LEU2 yku80∷kanMX2 dot1∷URA3 PCH2-HA |
| YP978 | BR1919a rad1∷kanMX4 |
| YP979 | BR1919a rad14∷hphMX4 |
| YP980 | BR1919a apn1∷kanMX4 |
| YP981 | BR1919a rad1∷kanMX4 dot1∷URA3 |
| YP983 | BR1919a rad1∷kanMX4 rad52∷LEU2 |
| YP985 | BR1919a rad1∷kanMX4 rad52∷LEU2 dot1∷URA3 |
| YP987 | BR1919a rad14∷hphMX4 dot1∷URA3 |
| YP989 | BR1919a rad14∷hphMX4 rad52∷LEU2 |
| YP990 | BR1919a rad14∷hphMX4 rad52∷LEU2 dot1∷URA3 |
| YP992 | BR1919a apn1∷kanMX4 dot1∷URA3 |
| YP994 | BR1919a apn1∷kanMX4 rad52∷LEU2 |
| YP995 | BR1919a apn1∷kanMX4 rad52∷LEU2 dot1∷URA3 |
| W303-1A | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 rad5-G535R |
| W303-1B | MATα leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 rad5-G535R |
| YP435 | W303-1A zip1∷LEU2 PCH2-HA |
| YP436 | W303-1A PCH2-HA |
| YP439 | W303-1A yku80∷kanMX2 zip1∷LEU2 PCH2-HA |
| YP452 | W303-1A dot1∷URA3 zip1∷LEU2 PCH2-HA |
| YP454 | W303-1A yku80∷kanMX2 dot1∷URA3 zip1∷LEU2 PCH2-HA |
| YP506 | W303-1A dot1∷URA3 PCH2-HA |
| YP520b | W303-1A rad9∷HIS3 |
| YP521b | W303-1A rad24∷TRP1 |
| YP522b | W303-1A rad9∷HIS3 rad24∷TRP1 |
| YP523b | W303-1A rad9∷HIS3 dot1∷URA3 |
| YP524b | W303-1B rad24∷TRP1 dot1∷URA3 |
| YP525b | W303-1A rad9∷HIS3 rad24∷TRP1 dot1∷URA3 |
| W3749-14Cc | W303-1A RAD5 ADE2 bar1∷LEU2 RAD52∷YFP |
| W3483-10Ac | W303-1A RAD5 ADE2 bar1∷LEU2 MRE11∷YFP |
| YP741 | W303-1A RAD5 ADE2 bar1∷LEU2 RAD52∷YFP dot1∷TRP1 |
| YP756 | W303-1A RAD5 ADE2 bar1∷LEU2 MRE11∷YFP dot1∷kanMX6 |
| YP943 | W303-1A leu2∷SFA1 ade3∷GAL∷HO |
| YP944 | W303-1A leu2∷SFA1 ade3∷GAL∷HO dot1∷TRP1 |
| YP945 | W303-1A ade3∷GAL∷HO rad52∷LEU2 |
| YP946 | W303-1A ade3∷GAL∷HO rad52∷LEU2 dot1∷TRP1 |
| BY4741 | MATahis3Δ0 leu2Δ0 ura3Δ0 met15Δ0 |
| BY4742 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 |
| BY4741-dot1Δ | BY4741 dot1∷kanMX4 |
| BY4741-rad30Δ | BY4741 rad30∷kanMX4 |
| BY4741-rev1Δ | BY4741 rev1∷kanMX4 |
| BY4741-rev3Δ | BY4741 rev3∷kanMX4 |
| YP811 | BY4741 rad52∷LEU2 |
| YP1080 | BY4741 rad30∷kanMX4 dot1∷URA3 |
| YP1081 | BY4741 rev1∷kanMX4 dot1∷URA3 |
| YP1082 | BY4741 rev3∷kanMX4 dot1∷URA3 |
| YP1125 | BY4741 rev3∷kanMX4 rad52∷LEU2 |
| YP1126 | BY4741 rev3∷kanMX4 rad52∷LEU2 dot1∷URA3 |
| YP1196 | BY4741 rev1∷kanMX4 rad52∷LEU2 |
| YP1197 | BY4741 rev1∷kanMX4 rad52∷LEU2 dot1∷URA3 |
| JKM179d | MATα hml∷ADE1 hmr∷ADE1 ade1-110 leu2, 3-112 lys5 trp1∷hisG ura3-52 ade3∷GAL10∷HO |
| YP815 | JKM179 dot1∷TRP1 |
| YP1167 | JKM179 yku80∷kanMX2 |
| YP1168 | JKM179 yku80∷kanMX2 dot1∷TRP1 |
BR1919α was provided by Shirleen Roeder (Yale University) and corresponds to BR1919-8B (Rockmill and Roeder 1990). BR1919a was generated by switching the mating type of BR1919α.
These strains are segregants from the cross YP506 × DLY262. The strain DLY262 (W303-1B rad9∷HIS3 rad24∷TRP1) was provided by Ted Weinert (University of Arizona). The presence of PCH2-HA was not followed in these segregants.
These strains, provided by Rodney Rothstein, have been described (Lisby et al. 2003, 2004).
Provided by Jim Haber (Lee et al. 1998).
Sensitivity to MMS and HO endonuclease:
Logarithmically growing cells were serially diluted and spotted onto YPDA plates (YPD supplemented with 50 μg/ml adenine) or YPDA plates containing MMS (Sigma) at various concentrations. MMS plates were freshly made and used within 12–24 hr, except those of Figure 8C, which were used after 2 days to further decrease the effective concentration of MMS, allowing the growth of the extremely sensitive rad52 rev3 and rad52 rev1 cells. Continuous expression of HO was induced on plates containing 2% galactose. To analyze viability after transient HO expression, cells were grown in liquid YP–galactose (2%) for 4 hr and serial dilutions were spotted onto YPDA plates. In all cases, plates were incubated at 30° and the growth of colonies was monitored and recorded over time.
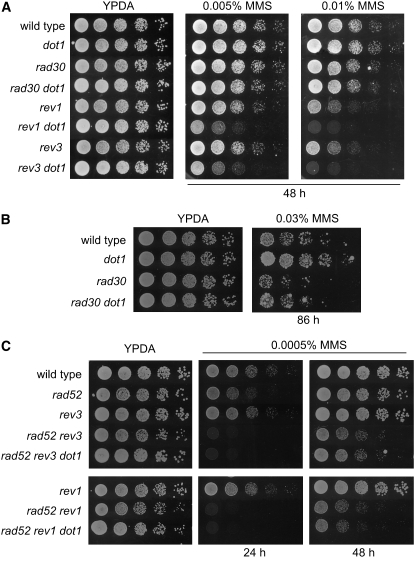
Figure 8.—
Genetic interaction of dot1 with TLS mutants. (A) Deletion of DOT1 enhances the MMS sensitivity of the rev1 and rev3 mutants. Fivefold serial dilutions of exponentially growing cells were spotted onto YPDA and 0.005 or 0.01% MMS plates. Strains are BY4741 (wild type), BY4741-dot1Δ (dot1), BY4741-rad30Δ (rad30), YP1080 (rad30 dot1), BY4741-rev1Δ (rev1), YP1081 (rev1 dot1), BY4741-rev3Δ (rev3), and YP1082 (rev3 dot1). (B) Deletion of DOT1 partially suppresses the MMS sensitivity of the rad30 mutant. Fivefold dilutions of exponentially growing cells were spotted onto YPDA and 0.03% MMS plates. Strains are BY4741 (wild type), BY4741-dot1Δ (dot1), BY4741-rad30Δ (rad30), and YP1080 (rad30 dot1). (C) Suppression of the rad52 MMS sensitivity by dot1 requires Rev3 and Rev1 function. Fivefold dilutions of exponentially growing cells were spotted onto YPDA and 0.0005% MMS plates. Strains are BY4741 (wild type), YP811 (rad52), BY4741-rev3Δ (rev3), YP1125 (rad52 rev3), YP1126 (rad52 rev3 dot1), BY4741-rev1Δ (rev1), YP1196 (rad52 rev1), and YP1197 (rad52 rev1 dot1).
NHEJ assays:
To analyze the repair by NHEJ of the HO-induced DSB at the MAT locus, JKM179-derived strains, lacking HMRa and HMLα and grown in YPDA, were diluted to 0.2 OD600 in YP–raffinose (2%) and incubated for ∼6 hr to log phase, and then galactose (2%) was added to half of the culture. After incubation for 3 hr, cells were plated on YPDA. Colonies were counted after 3 days of incubation at 30°. The efficiency of NHEJ is expressed as the viability of the cells incubated in galactose relative to the cells grown in raffinose.
For the plasmid religation assay, cells were transformed with 240 ng of BamHI-digested pRS314 or 60 ng of uncut plasmid and selected on SC–Trp plates. Repair is expressed as the ratio of colony formation from linear relative to circular plasmid transformations.
Western blot analysis:
To monitor phosphorylation of histone H2A at serine 129, trichloroacetic acid cell extracts were prepared as described (Longhese et al. 1997), separated by SDS–PAGE in 15% gels, and transferred to Immobilon-P (Millipore) membranes. The rabbit polyclonal anti-phospho-H2AS129 ab15083 antibody (Abcam) was used at 1:2000 dilution. Anti-PGK monoclonal antibody 22C5 (A-6457, Molecular Probes) was used at 1:5000 dilution as a loading control. Anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase (GE Healthcare) were used at 1:5000 dilution. Signal was detected with the ECL kit (GE Healthcare).
Microscopy:
To analyze the formation of Rad52-YFP foci, cells were grown on synthetic complete (SC) medium at 25°, treated with 0.02% MMS, and observed by fluorescence microscopy. Images were captured using a Leica DMRXA microscope equipped with an Orca-AG (Hamamatsu) CCD camera, a ×63 1.4NA objective, and a band-pass YFP filter set (excitation 500/20 nm, dichroic 515 nm, emission 535/30 nm). For each field, eight Z-positions at 0.4-μm intervals were captured and processed with Image J software (http://rsb.info.nih.gov/ij/). The experiment was repeated two times, and 200–600 cells were scored for each time point in every experiment.
Pulsed-field gel electrophoresis:
Exponentially growing cells were treated with 0.05% MMS. Samples were taken at 15 and 30 min and washed with 1 ml of 10 mm Tris, 50 mm EDTA, and 0.1% sodium azide, pH 8. Genomic DNA samples from 6 × 107 cells were prepared in agarose plugs essentially as described (Lengronne et al. 2001). Chromosomes were separated by pulsed-field gel electrophoresis (PFGE) in a Bio-Rad CHEF DRII system. Electrophoresis was performed for 24 hr at 6 V/cm with a switch time of 60–120 sec in 0.5× TBE at 14°. The gel was stained with 0.5 μg/ml ethidium bromide and photographed. For Southern blot analysis, chromosomes were transferred to Hybond N+ membrane (GE Healthcare) and hybridized with a P32-labeled URA3 probe (1.1-kb HindIII fragment) following standard procedures. The radioactive signal was captured on a Fuji imaging plate (BAS-MS 2040), scanned in a Fuji BAS1500 phosphorimager, and quantified using Image Gauge v4.2 software. The percentage of chromosome breakage is represented as the signal present below the intact chromosome V band relative to the total signal on each lane.
Mutagenesis assay:
To measure the frequency of spontaneous and MMS-induced forward mutagenesis at the CAN1 locus, cells were grown in YPDA to log phase, MMS (0.005%) was added to half of each culture, and cells were further incubated for ∼20 hr. Then, appropriate dilutions from each culture were made and plated onto YPDA and SC lacking arginine and containing 60 μg/ml canavanine. Plates were incubated at 30° for 3–4 days and colonies were counted. Mutagenesis frequency (measured by the appearance of Canr colonies) was obtained by a fluctuation test as the median value of seven independent cultures, with and without added MMS, for each strain. The given mutagenesis frequency is the mean and standard deviation of the median values from four independent experiments.
RESULTS
Genetic interaction of dot1 with DNA damage checkpoint mutants:
To better understand the role of Dot1 in the DNA damage response, we analyzed the sensitivity of the dot1 mutant to the DNA alkylating agent MMS, which, among other lesions, is thought to generate DNA double-strand breaks resulting from the processing of alkylated damage during DNA replication (Chlebowicz and Jachymczyk 1979; Wyatt and Pittman 2006). Although Dot1 is required to slow down cell cycle progression in response to the presence of MMS during the G1 and S phases (Giannattasio et al. 2005; Wysocki et al. 2005), we found that the dot1 mutant was not sensitive to MMS (Figure 1). However, when the deletion of DOT1 was combined with the absence of the Rad24 checkpoint protein, the double mutant was significantly more sensitive to MMS than the rad24 single mutant (Figure 1). In contrast, the rad9 dot1 double mutant was not more sensitive than the rad9 single mutant, and the MMS sensitivity of the rad9 rad24 dot1 triple mutant was similar to that of the rad9 rad24 double mutant (Figure 1). Genetic studies of DNA damage checkpoint mutants have placed RAD9 and RAD24 in different epistasis groups (Lydall and Weinert 1995; de la Torre-Ruiz et al. 1998). Our results therefore suggest that DOT1 belongs to the RAD9 group, consistent with the recent findings indicating the involvement of Dot1 in Rad9-dependent activation of Rad53 (Giannattasio et al. 2005; Wysocki et al. 2005).
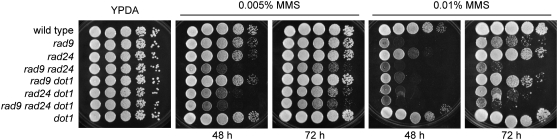
Figure 1.—
Deletion of DOT1 increases the MMS sensitivity of the rad24, but not the rad9, checkpoint mutant. Tenfold serial dilutions of exponentially growing cells were spotted onto YPDA and 0.005% or 0.01% MMS plates. Strains are YP436 (wild type), YP520 (rad9), YP521 (rad24), YP522 (rad9 rad24), YP523 (rad9 dot1), YP524 (rad24 dot1), YP525 (rad9 rad24 dot1), and YP506 (dot1).
Deletion of DOT1 suppresses the MMS sensitivity of HR and NHEJ mutants:
The increased MMS sensitivity of the rad24 dot1 double mutant might be explained by the impaired response in arresting or delaying cell cycle progression upon DNA damage, but it might also be a consequence of the participation of Dot1 in DNA repair. To investigate this possibility, we combined the deletion of DOT1 with mutations conferring defects in DSB repair, either by HR, such as rad52, or by NHEJ, such as yku80. The yku80 mutant is only slightly MMS sensitive, but deletion of DOT1 completely suppressed the MMS sensitivity of yku80 (Figure 2A). Interestingly, in contrast to MMS, mutation of DOT1 did not suppress the yku80 thermosensitivity derived from the altered telomere metabolism in the absence of a functional Ku complex (Figure 2A, middle) (Boulton and Jackson 1998). Moreover, deletion of DOT1 also partially suppressed the MMS hypersensitivity of rad52 (Figure 2B; compare second and third rows) and even that of the rad52 yku80 double mutant, which lacks both pathways of DSB repair (Figure 2B; compare fourth and fifth rows). This suppression is not exclusive of rad52; deletion of DOT1 also attenuated the MMS sensitivity of other HR mutants, such as rad54 (supplemental Figure S1). Strikingly, the absence of Dot1 suppressed the sensitivity of several DSB repair mutants to different degrees and also, notably, at high MMS doses, the dot1 single mutant was even more resistant to MMS than the wild type in all the strain backgrounds tested (Figure 2C; see also Figures 7A and 8B and supplemental Figure S3).
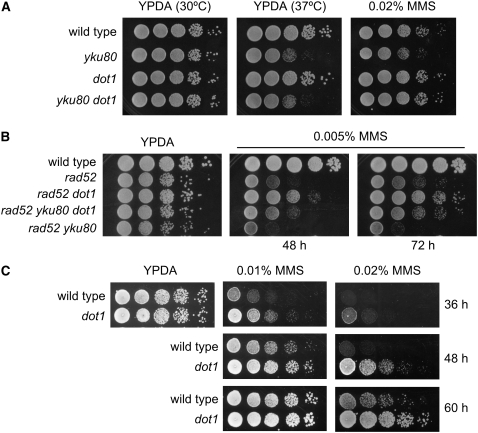
Figure 2.—
Deletion of DOT1 alleviates the MMS sensitivity of the yku80 and rad52 mutants. (A) Tenfold serial dilutions of exponentially growing cells were spotted onto YPDA plates incubated at 30° or 37° and a 0.02% MMS plate incubated at 30°. (B) Tenfold serial dilutions of exponentially growing cells were spotted onto YPDA and 0.005% MMS plates and incubated at 30° for 48 and 72 hr. Strains are BR1919α (wild type), YP513 (yku80), YP345 (dot1), YP514 (yku80 dot1), YP370 (rad52), YP371 (rad52 dot1), YP516 (rad52 yku80 dot1), and YP515 (rad52 yku80). (C) The dot1 mutant shows increased MMS resistance. Fivefold serial dilutions of exponentially growing cells were spotted onto YPDA and 0.01 or 0.02% MMS plates and incubated at 30° for 36, 48, and 60 hr. Strains are YP210 (wild type) and YP345 (dot1).
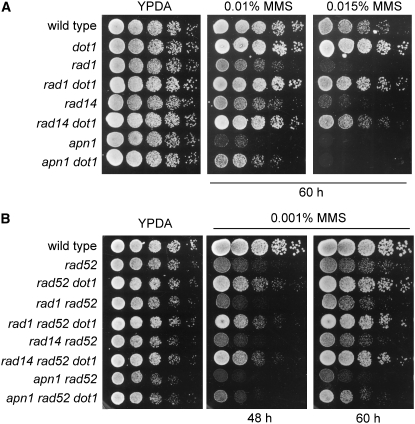
Figure 7.—
Genetic interaction of dot1 with BER and NER mutants. (A) Deletion of DOT1 suppresses the MMS sensitivity of BER and NER mutants. Fivefold serial dilutions of exponentially growing cells were spotted onto YPDA and 0.01 or 0.015% MMS plates. Strains are BR1919α (wild type), YP163 (dot1), YP978 (rad1), YP981 (rad1 dot1), YP979 (rad14), YP987 (rad14 dot1), YP980 (apn1), and YP992 (apn1 dot1). (B) Suppression of the rad52 MMS sensitivity by dot1 does not depend on NER or BER function. Fivefold dilutions of exponentially growing cells were spotted onto YPDA and 0.001% MMS plates. Strains are BR1919α (wild type), YP370 (rad52), YP371 (rad52 dot1), YP983 (rad1 rad52), YP985 (rad1 rad52 dot1), YP989 (rad14 rad52), YP990 (rad14 rad52 dot1), YP994 (apn1 rad52), and YP995 (apn1 rad52 dot1).
MMS resistance conferred by the absence of Dot1 does not depend on Sir function:
In the dot1 mutant, the Sir proteins are redistributed from the normally silenced loci, resulting in defective silencing (Singer et al. 1998; San-Segundo and Roeder 2000; van Leeuwen et al. 2002). The Sir complex is involved in DSB repair, at least through its function in silencing mating-type genes (Lee et al. 1999), and is also relocated in the genome when DSBs occur (Martin et al. 1999); therefore, it is possible that the increased MMS resistance observed in the absence of Dot1 could be derived from the delocalization of Sir proteins to other chromosomal locations, where they might either facilitate DNA repair or somehow prevent the formation of DNA lesions upon MMS treatment.
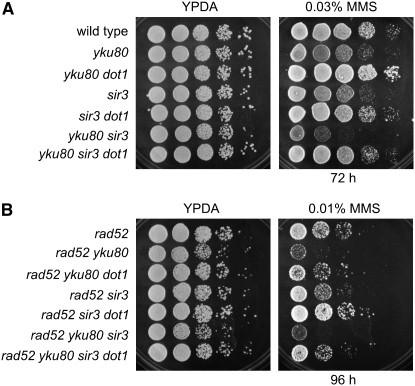
To explore this possibility, we combined the deletion of SIR3 with those of YKU80, RAD52, and DOT1 and analyzed the MMS sensitivity of all mutant combinations. As previously described, the sir3 mutant was slightly MMS sensitive (Martin et al. 1999), but this sensitivity was suppressed in sir3 dot1 (Figure 3A, rows 4 and 5). The yku80 sir3 double mutant was more sensitive to MMS than the single mutants were, but deletion of dot1 also alleviated this sensitivity (Figure 3A, rows 6 and 7). In addition, the absence of Dot1 conferred increased MMS resistance to rad52 sir3 (Figure 3B, rows 4 and 5) and to rad52 yku80 sir3 (Figure 3B, rows 6 and 7). Thus, the effect of DOT1 deletion on MMS resistance is not exerted through the redistribution of the Sir complex throughout the genome (Rudner et al. 2005).
Figure 3.—
Suppression of MMS sensitivity of yku80 and rad52 by dot1 does not depend on Sir3. Tenfold serial dilutions of exponentially growing cells were spotted onto YPDA and a 0.03% MMS plate (A) or a 0.01% MMS plate (B). Strains are BR1919α (wild type), YP513 (yku80), YP514 (yku80 dot1), YP544 (sir3), YP547 (sir3 dot1), YP545 (yku80 sir3), YP546 (yku80 sir3 dot1), YP370 (rad52), YP515 (rad52 yku80), YP516 (rad52 yku80 dot1), YP548 (rad52 sir3), YP549 (rad52 sir3 dot1), YP550 (rad52 yku80 sir3), and YP551 (rad52 yku80 sir3 dot1).
Additional relationships between Dot1 and Sir proteins are known to exist; for example, Dot1 is required for proper Sir2 meiotic localization and, like dot1, the sir2 mutant (but not sir3 or sir4) is defective in the meiotic recombination checkpoint (San-Segundo and Roeder 1999, 2000). However, in contrast to dot1, the absence of Sir2 did not suppress the MMS sensitivity of rad52; indeed, the sir2 rad52 double mutant was slightly more MMS sensitive than rad52 (supplemental Figure S2).
Mutation of DOT1 does not suppress the sensitivity of rad52 or yku80 to an HO-induced DSB:
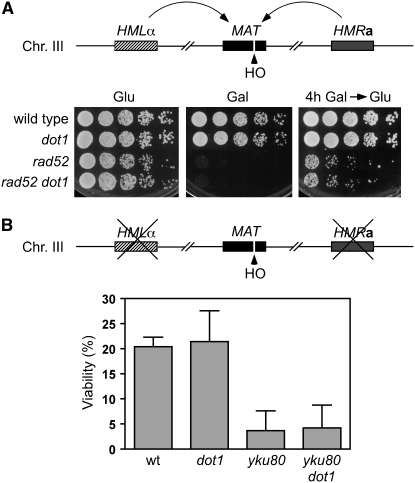
To determine whether the suppression of damage sensitivity of DSB repair mutants by deletion of DOT1 was specific to MMS lesions, we analyzed the sensitivity to a single DSB generated at the MAT locus by the HO endonuclease expressed under control of the GAL1-10 promoter. In strains where the break can be repaired by homologous recombination (gene conversion) between MAT and the HMRa or HMLα loci, deletion of DOT1 did not alter the sensitivity of the rad52 mutant to either sustained or transient HO induction (Figure 4A). In JKM179 strains carrying a deletion of HMLα and HMRa (Lee et al. 1998), where the HO-induced break can be repaired only by NHEJ, the viability of the dot1 mutant was similar to that of the wild type, and the yku80 dot1 double mutant was as sensitive as the yku80 strain, indicating that Dot1 most likely does not participate in NHEJ (Figure 4B). Also, using a plasmid religation assay, the dot1 mutant did not show any defect in NHEJ (supplemental Table S1). Thus, these results collectively suggest that the partial or total suppression of the DNA damage sensitivity of HR and NHEJ mutants observed in the absence of Dot1 is unique to the DNA damage generated by agents, like MMS, that may cause breaks indirectly by interfering with replication fork progression, but do not occur for DSBs that are generated directly by an endonuclease.
Figure 4.—
Deletion of DOT1 does not suppress the sensitivity of the rad52 and yku80 mutants to a DSB generated by the HO endonuclease at the MAT locus (A) Fivefold serial dilutions of log-phase cells growing in YPDA were spotted onto YPDA or YP–galactose plates (bottom, left and middle, respectively). Fivefold serial dilutions of log-phase cells incubated in liquid YP–galactose for 4 hr were spotted onto a YPDA plate (bottom right). Strains are YP943 (wild type), YP944 (dot1), YP945 (rad52), and YP946 (rad52 dot1). (B) Viability of JKM179-derived strains lacking HMLα and HMRa after incubation in YP–galactose for 3 hr to induce HO expression. Strains are JKM179 (wild type), YP815 (dot1), YP1167 (yku80), and YP1168 (yku80 dot1). Average and standard deviation values of three independent experiments are shown.
Chromosome fragmentation is not reduced in the dot1 mutant upon MMS treatment:
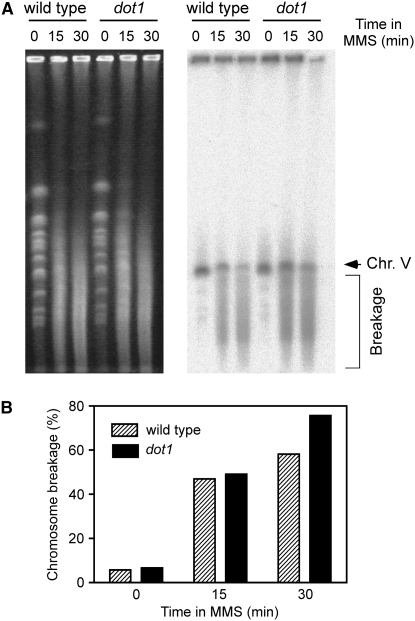
One attractive possibility for explaining the increased dot1 MMS resistance is that, in the absence of Dot1, there could be less DNA damage generated by MMS, either because the chromatin in such cells is more refractory to MMS in the absence of H3K79 methylation or because fewer MMS-methylated bases are converted into lethal lesions in the dot1 mutant. We used PFGE to detect chromosome fragmentation after MMS treatment as a way of monitoring the extent of primary damage generated by MMS. It has been reported that MMS produces heat-labile DNA that leads to chromosome fragmentation arising during the PFGE procedure, but the breakage does not result from DSBs formed in vivo (Lundin et al. 2005); thus, we reasoned that the extent of DNA fragmentation would reflect the number of labile sites created by MMS. We found no strong difference between wild type and dot1 in the amount of broken DNA generated after short exposure to MMS before repair has taken place (Figure 5), suggesting that DNA methylation by MMS is not reduced in the dot1 mutant.
Figure 5.—
Analysis of MMS-induced chromosome fragmentation by PFGE. (A) Exponentially growing wild-type (BR1919a) and dot1 (YP576) cells were treated with 0.05% MMS. After 15 and 30 min, agarose plugs were prepared and chromosomes were separated by PFGE. Chromosomes were visualized by ethidium bromide staining (left), transferred to a membrane, and hybridized with a probe specific for chromosome V (right). Chromosome V fragmentation results in a smear (bracket) below the band representing the whole chromosome (arrow). (B) Quantification of the radioactive signal present in A. Chromosome breakage is expressed as the ratio between the signal detected below the intact chromosome V band and the total signal detected on each lane.
The number of Rad52 foci and levels of histone H2A phosphorylation in response to MMS are increased in the dot1 mutant:
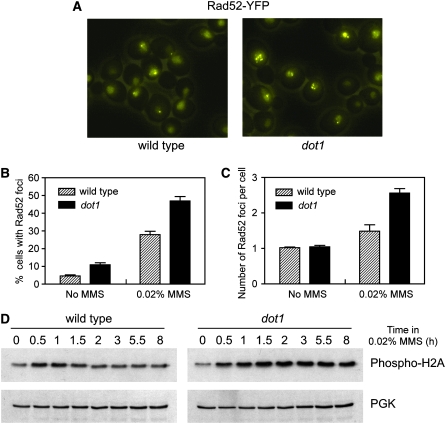
To monitor the formation and repair of MMS-induced recombinogenic damage in dot1 living cells, we first examined the formation of Rad52-YFP foci. Rad52-YFP accumulates in bright foci representing DNA repair centers in which multiple lesions are processed (Lisby et al. 2001, 2003). We treated wild-type and dot1 mutant cells with MMS for 60 min and analyzed by fluorescence microscopy the formation of Rad52-YFP foci (Figure 6A). We found that the number of cells displaying Rad52 foci was higher in the dot1 mutant compared to the wild type (Figure 6B), suggesting either that there are more damaged cells in dot1 or that Rad52 foci last longer in the absence of Dot1. Interestingly, whereas most wild-type cells displayed a single Rad52 focus, in the dot1 mutant we frequently found cells with two or more foci (Figure 6, A and C). The percentage of cells containing more than one focus after MMS treatment was 10.5% in the wild type and 33.2% in the dot1 mutant.
Figure 6.—
MMS-induced Rad52 foci formation and histone H2AS129 phosphorylation are increased in the dot1 mutant. (A) Rad52-YFP foci in wild-type and dot1 cells treated with 0.02% MMS for 1 hr. Representative fields are shown. Images are the maximum intensity projection from z-stacks of eight sections separated by 0.4 μm. The percentage of cells containing Rad52 foci (B) and the number of Rad52 foci per cell (C) are represented. (D) Western blot analysis of histone H2A phosphorylation at serine 129 in wild-type and dot1 cells treated with 0.02% MMS. Phosphoglycerate kinase (PGK) was used as a loading control. Strains are W3749-14C (wild type) and YP741 (dot1).
We also monitored phosphorylation of histone H2A in response to MMS, as a marker for the formation of DSBs, using a phospho-specific antibody (Downs et al. 2000; Fernandez-Capetillo et al. 2004). In wild-type cells treated with MMS, phosphorylation of histone H2A increased above basal levels shortly after addition of MMS and declined progressively throughout the experiment. In contrast, in the dot1 mutant, high levels of phospho-H2A persisted even at late time points (Figure 6D), consistent with either the existence of more breaks or a lower efficiency of DSB repair in the absence of Dot1. Together, these observations argue strongly against the hypothesis that there are fewer MMS-induced lesions in the dot1 mutant and suggest that other explanations must exist to account for MMS resistance conferred by dot1.
Genetic interaction of dot1 with BER and NER mutants:
An alternative possibility for explaining the enhanced MMS resistance of the dot1 mutant is that Dot1 could somehow limit the activity of repair pathways involved in the elimination of MMS-induced lesions. If that were the case, in the absence of Dot1, a higher fraction of the MMS primary genotoxic damage would be repaired by these pathways before being converted into lesions to be repaired by HR or NHEJ. The primary MMS-induced lesions are alkylated bases, which can be processed by the BER and NER systems. If the increased MMS resistance of dot1 were caused by an enhanced activity of BER or NER, then inactivation of these pathways would result in the suppression of the MMS resistance conferred by the absence of Dot1. To test this possibility, we combined the deletion of dot1 with that of genes involved in BER or NER.
Apn1 is the major endonuclease involved in BER (Boiteux and Guillet 2004). As expected, the apn1 mutant was sensitive to MMS, but the dot1 apn1 double mutant was more resistant to MMS than the apn1 single mutant (Figure 7A; 0.01% MMS) and the absence of Dot1 also partially suppressed the strong MMS sensitivity of the HR- and BER-deficient rad52 apn1 double mutant (Figure 7B).
We also deleted DOT1 in the NER-deficient rad1 mutant. During NER, the Rad1-Rad10 nuclease cleaves the 5′ side of the lesion, but it also has a role in recombination (Schiestl and Prakash 1988; Bardwell et al. 1994); therefore, we also analyzed the rad14 mutant, which is exclusively defective in NER (Prakash and Prakash 2000). The dot1 rad1 and dot1 rad14 double mutants were more resistant to MMS than the corresponding NER-deficient single mutants (Figure 7A). Furthermore, deletion of DOT1 also partially alleviated the MMS sensitivity of the HR- and NER-deficient rad52 rad1 and rad52 rad14 double mutants (Figure 7B). Thus, these observations suggest that the enhanced resistance to high MMS concentrations conferred by dot1 does not result solely from an increased activity of the BER and NER pathways.
Dot1 negatively regulates TLS repair by the Polζ and Rev1 polymerases:
In addition to the repair mechanisms mentioned above, cells possess DNA damage tolerance pathways, such as TLS and template switching, which are also essential for maintaining viability in response to genotoxic agents. TLS polymerases can replicate past lesions due to a more relaxed catalytic center, allowing the cell to “tolerate” the damage and thus avoid replicative arrest, although at the cost of introducing errors. To investigate the possibility that the increased resistance of the dot1 mutant to MMS could be the consequence of an elevated DNA damage tolerance, we combined dot1 with mutants defective in different TLS polymerases, such as rad30 (Polη), rev3 (catalytic subunit of Polζ), and rev1 (Kunz et al. 2000), or altered in post-replication repair by template switching, such as rad5 (Blastyak et al. 2007).
The original W303 strains carry a rad5-G535R mutation (Fan et al. 1996), which confers MMS sensitivity (supplemental Figure S3), but deletion of DOT1 increased the MMS resistance of both the rad5-defective and RAD5-converted versions of W303 (supplemental Figure S3), suggesting that the Rad5-dependent pathway of post-replication repair is not regulated by Dot1. However, when we combined dot1 with TLS mutants, we observed that at low doses of MMS (0.005 and 0.01%), at which dot1 is only slightly more resistant than the wild type and the rev3 and rev1 single mutants are slightly more sensitive, the rev3 dot1 and rev1 dot1 double mutants were significantly more sensitive to MMS (Figure 8A). In contrast, rad30 and rad30 dot1 did not display altered sensitivity at these low MMS doses (Figure 8A), but at a higher concentration (0.03%), the rad30 dot1 double mutant was slightly more resistant than rad30 (Figure 8B). Thus, the increased MMS resistance conferred by the absence of Dot1 requires Polζ and Rev1 activity.
To determine whether the partial suppression of the rad52 MMS sensitivity by dot1 was dependent on Polζ and Rev1 function, we compared the rad52 rev3 and rad52 rev1 double mutants with the rad52 rev3 dot1 and rad52 rev1 dot1 triple mutants, respectively. The rad52 rev3 and rad52 rev1 double mutants were extremely sensitive to MMS; therefore, we used very low MMS concentrations (0.0005%) to be able to detect growing colonies, but deletion of DOT1 did not alleviate the sensitivity (Figure 8C), confirming that the attenuation of MMS sensitivity by the absence of Dot1 depends on Rev3- and Rev1-mediated TLS.
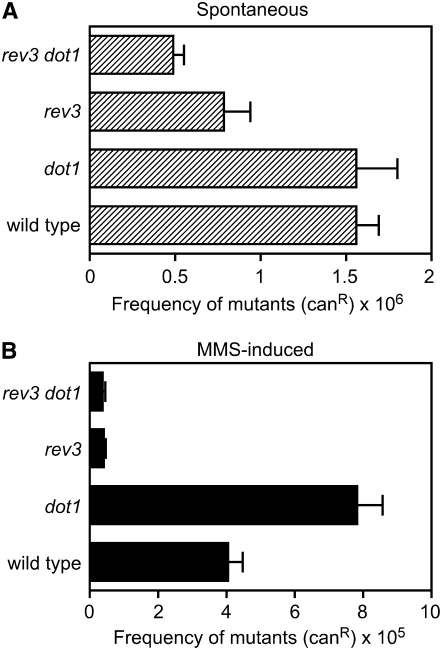
Taken together, these observations suggest that Dot1 is a negative regulator of the Rev1- and Polζ-dependent TLS pathway and that the MMS resistance of the dot1 mutant could be explained, at least in part, by the increased activity of this DNA damage tolerance mechanism. TLS by Polζ, in collaboration with Rev1, results in error-prone repair because incorrect bases are often inserted opposite the damaged sites (Kunz et al. 2000; Prakash et al. 2005); therefore, we reasoned that if Dot1 inhibits the action of these polymerases, the frequency of damage-induced mutagenesis should be increased in the absence of Dot1. We examined the frequency of forward mutation at the CAN1 locus in response to MMS treatment and observed that it increased about twofold in the dot1 mutant compared to the wild type (Figure 9). Moreover, this elevated mutagenesis frequency of dot1 depends entirely on Polζ, because the rev3 dot1 double mutant and the rev3 single mutant displayed the same reduced levels (Figure 9). Thus, Dot1 is required to limit the action of error-prone TLS in the presence of genotoxic agents such as MMS.
Figure 9.—
MMS-induced mutagenesis at the CAN1 locus is increased in the dot1 mutant. The mutagenesis frequency, expressed as the proportion of CanR colonies that appeared in cultures from cells growing in the absence (A) or in the presence of 0.005% MMS (B), is represented. The average and standard deviation from four independent experiments is shown. Strains are BY4741 (wild type), BY4741-dot1Δ (dot1), BY4741-rev3Δ (rev3), and YP1082 (rev3 dot1).
DISCUSSION
To gain further insight into how histone modifications impact the cellular response to DNA damage, we have investigated the role of the S. cerevisiae Dot1 protein, which is the only methyltransferase responsible for methylation of H3K79. In particular, we have studied the response of the dot1 mutant to the widely used alkylating agent MMS. By analyzing the MMS sensitivity of dot1 combined with mutants defective in the DNA damage checkpoint, such as rad9 and rad24, we found that rad9 and dot1 are in the same epistasis group. Consistent with this interpretation, the rad24 dot1 double mutant is more sensitive to MMS than the rad24 single mutant. A similar result was observed in a previous report where IR sensitivity was examined (Wysocki et al. 2005). One of the essential functions of the DNA damage checkpoint is the establishment and maintenance of cell cycle arrest upon genome injuries, although the checkpoint also controls DNA repair (Harrison and Haber 2006); thus, in principle, Dot1 could be influencing either one or both of these checkpoint functions. It has been reported that Dot1 is required to slow down cell cycle progression after treatment with UV, MMS, or IR at least during the G1 and S phases, because it is involved in the Rad9-dependent activation of Rad53 (Giannattasio et al. 2005; Wysocki et al. 2005; Grenon et al. 2007). Nevertheless, either MMS sensitivity was not analyzed in those studies (Giannattasio et al. 2005; Wysocki et al. 2005) or, at the single MMS concentration tested, the dot1 mutant was similar to the wild type (Grenon et al. 2007). However, our results indicate that, despite its checkpoint defect, the dot1 mutant does not show sensitivity to this genotoxic agent and, surprisingly, the absence of Dot1 actually increases resistance to MMS and attenuates the sensitivity of certain DNA repair mutants. In contrast, the dot1 mutant displays mild sensitivity to other genotoxic agents, such as UV (Bostelman et al. 2007) or IR (Game et al. 2005, 2006), although other reports found very little or no difference in IR sensitivity of dot1 compared to the wild type (Wysocki et al. 2005; Toh et al. 2006). The different yeast strain background (BY4742 and W303) used in those studies may explain the different IR effect observed. The sensitivity of dot1 to certain types of DNA damage (UV, IR) could be easily explained by the inability of the dot1 mutant to delay cell cycle progression, although a more direct role for Dot1 in DNA repair has also been proposed. However, the increased MMS resistance of dot1 is more difficult to reconcile with the checkpoint defect and implies that Dot1 performs different roles in the response to DNA damage, depending on the type of lesions.
MMS has traditionally been used as a radiomimetic agent because mutants defective in DSB repair by HR are extremely MMS sensitive; NHEJ mutants are also MMS sensitive but to a much lesser extent (Milne et al. 1996). However, there is no evidence indicating that MMS directly forms DSBs, and the requirement for DSB repair pathways, especially HR, to maintain viability after MMS treatment can be due to recombinogenic lesions generated from the processing of the alkylated damaged sites, such as single-strand breaks that could be converted into DSBs during replication, or from stalled replication forks (Wyatt and Pittman 2006). We have found that the absence of Dot1 attenuates the MMS sensitivity of mutants lacking both HR and NHEJ, which are unable to repair DSBs.
In principle, one of the simplest interpretations of this result could be that the different chromatin structure generated by the lack of H3K79 methylation somehow obstructs the access of MMS to the DNA, resulting in the production of fewer lesions. Although this possibility cannot be completely ruled out until direct MMS-induced methylation of DNA is measured, several lines of evidence argue against the existence of less MMS-induced damage in the dot1 mutant. First, chromosome fragmentation detected by PFGE is similar in dot1 compared to the wild type after a short period (15 min) of MMS treatment. It has been reported that MMS methylation produces heat-labile sites in DNA that result in chromosome fragmentation during incubation at 50° in the PFGE procedure (Lundin et al. 2005); thus, the extent of breakage serves as an indirect indication of the in vivo MMS-promoted methylation. Second, MMS treatment triggers the formation of Rad52 repair foci in vivo (Lisby et al. 2003) and promotes histone H2A phosphorylation (Downs et al. 2000; Prado et al. 2004). Regardless of whether these events reflect only the formation of DSBs or are also promoted by other types of lesions, the dot1 mutant both shows increased frequency of MMS-induced Rad52 foci and maintains high levels of phosphorylated H2A for more prolonged periods. These observations, together with the modestly increased DNA breakage detected after 30 min of MMS exposure, are more compatible with a defect in repair by HR, rather than with the existence of fewer MMS-induced lesions. It has been reported that, during DSB repair, multiple lesions are recruited to a single or a few repair centers (Lisby et al. 2003). Strikingly, we found that dot1 shows not only an increased incidence of repair centers, that is, a higher proportion of cells containing Rad52 foci, but also a higher number of foci per cell. The latter observation could be simply a consequence of the accumulation of unrepaired lesions, but also may reflect a special chromatin structure requirement for the congregation of several lesions into a single repair center, suggesting a possible role for Dot1-dependent H3K79 methylation in this gathering mechanism.
Another possibility for explaining the attenuated MMS sensitivity of DSB repair mutants conferred by dot1 could be that Dot1 limits the action of other(s) pathway(s) involved in the processing and elimination of MMS-induced damage. In the absence of Dot1, the increased activity of this pathway(s) would result in increased MMS resistance. MMS initially provokes methylation of DNA that leads to the formation of abasic (AP) sites as intermediates in the BER process, which removes damaged bases (Boiteux and Guillet 2004, 2006). NER can also act as a backup activity in the repair of AP sites (Xiao and Chow 1998; Torres-Ramos et al. 2000). Cleavage of AP sites results in the formation of single-strand breaks that can be converted into DSBs during DNA replication (Caldecott 2001; Guillet and Boiteux 2002) (Figure 10). If the number of breaks resulting from incomplete BER or NER were lower in the dot1 mutant as a consequence of an enhanced activity of these pathways, that would result in fewer lesions to be repaired by HR and, therefore, increased MMS resistance. However, mutation of DOT1 still partially suppresses MMS sensitivity of BER and NER single mutants as well as double mutants with rad52, indicating that the increased MMS resistance observed in dot1 cells does not require active BER and NER pathways. The enhanced DNA damage resistance of dot1 and the attenuation of rad52 sensitivity appear to be specific to genotoxic agents, such as MMS, that can generate DSBs indirectly when a replication fork encounters the original or the processed lesion. In fact, dot1 does not suppress the sensitivity of rad52 to “clean” DSBs generated by the HO endonuclease (Figure 4), and dot1 is sensitive to, not more resistant to, IR (Game et al. 2005, 2006).
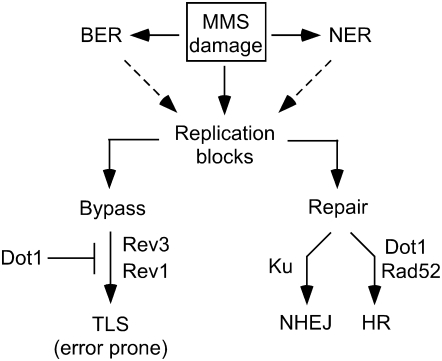
Figure 10.—
Model for the role of Dot1 in the response to alkylating damage. MMS-induced damage can cause the stall of replication forks either directly or as a consequence of incomplete BER or NER. Stalled replication forks result in recombinogenic lesions, which can be repaired mainly by Rad52-dependent HR, with NHEJ acting as a minor repair pathway for DSBs. Replication blocks can also be bypassed by the action of TLS pathways. Dot1 functions as a negative regulator of Polζ- and Rev1-dependent TLS and is also required for efficient HR. In the absence of Dot1, the enhanced TLS activity results in fewer stalled replication forks, leading to increased MMS resistance.
To promote viability after DNA damage, in addition to DNA repair mechanisms, eukaryotic cells also possess mechanisms that avoid the stall of replication forks and permit the replication of damaged DNA by TLS (Lopes et al. 2006). We found that the increased MMS resistance of dot1 depends on the Polζ and Rev1 TLS polymerases; indeed, the dot1 rev3 and dot1 rev1 double mutants are even more sensitive to MMS than the single mutants. We propose that Dot1 limits the activity of TLS and that, in the absence of Dot1, the enhanced TLS activity is responsible for the increased MMS resistance (Figure 10). The fact that the rev3 dot1 (or rev1 dot1) double mutant is more sensitive to MMS than rev3 (or rev1) implies that Dot1 is also somehow involved in another repair pathway that becomes more important for viability in the absence of TLS (Figure 10). This pathway is likely HR because the rev3 rad52 or rev1 rad52 double mutants are extremely sensitive to MMS and the rev3 rad52 dot1 or rev1 rad52 dot1 triple mutants show the same sensitivity as rev3 rad52 or rev1 rad52, respectively. Indeed, we have observed that Dot1 is required for efficient DSB repair by sister-chromatid recombination (F. Conde, V. Cordón-Preciado, F. Cortés-Ledesma, E. Refolio, L. Aragón, A. Aguilera and P. San-Segundo, unpublished results). The increased number of Rad52 foci observed in dot1 (Figure 6) and its sensitivity to IR (Game et al. 2005, 2006) are also consistent with a role for Dot1 in HR. Polζ, in cooperation with Rev1, is the main TLS polymerase involved in damage-induced mutagenesis (Lawrence 2002). Consistent with the hypothesis that Dot1 inhibits the action of these polymerases, we found an increased frequency of Rev3-dependent MMS-induced mutagenesis in the dot1 mutant.
How could Dot1 regulate TLS? Dot1-mediated H3K79 methylation is involved in chromatin silencing by establishing the boundaries between euchromatin and heterochromatin and it has been also linked to transcription elongation (Shilatifard 2006). The MMS resistance of dot1 appears to be independent of the silencing function defect because, in contrast to dot1, we found that deletion of SIR2 or SIR3 enhances the MMS sensitivity of rad52 or rad52 yku80 and, in addition, dot1 still confers increased MMS resistance in the absence of Sir3. Nevertheless, it is possible that Dot1 represses the transcription of individual TLS genes, perhaps REV3 or REV1, in a silencing-independent manner, accounting for the augmented TLS activity in the dot1 mutant. However, the fact that we do not observe an increased frequency of spontaneous mutagenesis in the dot1 mutant (Figure 9) suggests that the effect of Dot1 on TLS may be related to some aspect of the DNA damage response. Interestingly, it has been described that cells expressing an HA-tagged version of RAD53, which results in reduced levels of this checkpoint kinase, are sensitive to hydroxyurea but, like dot1, display increased resistance to MMS. Strikingly, the MMS-induced mutagenesis at the CAN1 gene is also about twofold higher in rad53-HA cells compared to the wild type (Cordon-Preciado et al. 2006). Because Dot1 is required for full activation of Rad53 after MMS treatment (Giannattasio et al. 2005), it is tempting to speculate that the effect of the lack of Dot1 in MMS resistance is due to the reduced levels of Rad53 kinase activity. PCNA ubiquitination regulates TLS by promoting the switching of replicative polymerases for TLS polymerases at stalled replication forks (Ulrich 2005; Jansen et al. 2007). In response to UV damage, physical interaction with the PCNA-like 9-1-1 checkpoint complex and Mec1-dependent phosphorylation is required for chromatin association of Polζ-Rev1 (Sabbioneda et al. 2005, 2007); however, the effect of MMS has not been analyzed. Future experiments will be required to examine how Dot1 and Rad53 may regulate TLS, particularly in response to alkylating damage.
It is believed that many of the mutations generated by genotoxic agents do not arise from the initial damage per se, but from the subsequent mutagenic processing by TLS polymerases (Pages and Fuchs 2002). Therefore, it is important for the cell to keep these tolerance mechanisms under strict control to avoid nucleotide misincorporation in undamaged templates. Furthermore, understanding the mechanisms regulating TLS has implications in cancer therapy, because suppression of TLS polymerase activity may aid in minimizing secondary mutations produced during treatments with antitumoral drugs. Given the evolutionary conservation of Dot1 in higher eukaryotes, studies in yeast may shed light on the mechanisms that contribute to maintaining genomic stability in humans.
In summary, our identification of this novel role for Dot1 in the DNA damage response adds another level of complexity to the already complicated picture of the regulation of this cellular response by histone modifications. Our results imply that H3K79 methylation may have different functions, depending not only on the phase of the cell cycle when the cell is injured, but also on the specific type of lesions produced.
Acknowledgments
We are indebted to Shirleen Roeder, in whose lab the initial observation of dot1 MMS resistance was made. We thank R. Rothstein, S. Jackson, J. Haber, S. Moreno, F. Prado, A. Aguilera, T. Weinert, and S. Roeder for providing plasmids and/or strains. We are grateful to A. Aguilera and F. Prado for invaluable discussions and suggestions throughout this work and to S. Erdman and F. Prado for critical reading of the manuscript. We thank A. Calzada for technical assistance with PFGE experiments and S. Meihoff and D. Ontoso for help in strain construction. F.C. was supported in part by a predoctoral fellowship from the Ministry of Education and Science of Spain. This work was supported by grant BFU2005-00955 from the Ministry of Education and Science of Spain and grant CSI05A07 from Junta de Castilla y León (Spain), to P.S.-S.
References
- Bardwell, A. J., L. Bardwell, A. E. Tomkinson and E. C. Friedberg, 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265 2082–2085. [DOI] [PubMed] [Google Scholar]
- Blastyak, A., L. Pinter, I. Unk, L. Prakash, S. Prakash et al., 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux, S., and M. Guillet, 2004. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3 1–12. [DOI] [PubMed] [Google Scholar]
- Boiteux, S., and M. Guillet, 2006. Use of yeast for detection of endogenous abasic lesions, their source, and their repair. Methods Enzymol. 408 79–91. [DOI] [PubMed] [Google Scholar]
- Bostelman, L. J., A. M. Keller, A. M. Albrecht, A. Arat and J. S. Thompson, 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair 6 383–395. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. a Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. b Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott, K. W., 2001. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. BioEssays 23 447–455. [DOI] [PubMed] [Google Scholar]
- Chien, C. T., S. Buck, R. Sternglanz and D. Shore, 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75 531–541. [DOI] [PubMed] [Google Scholar]
- Chlebowicz, E., and W. J. Jachymczyk, 1979. Repair of MMS-induced DNA double-strand breaks in haploid cells of Saccharomyces cerevisiae, which requires the presence of a duplicate genome. Mol. Gen. Genet. 167 279–286. [DOI] [PubMed] [Google Scholar]
- Cordon-Preciado, V., S. Ufano and A. Bueno, 2006. Limiting amounts of budding yeast Rad53 S-phase checkpoint activity results in increased resistance to DNA alkylation damage. Nucleic Acids Res. 34 5852–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ruiz, M. A., C. M. Green and N. F. Lowndes, 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17 2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J. A., N. F. Lowndes and S. P. Jackson, 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408 1001–1004. [DOI] [PubMed] [Google Scholar]
- Downs, J. A., M. C. Nussenzweig and A. Nussenzweig, 2007. Chromatin dynamics and the preservation of genetic information. Nature 447 951–958. [DOI] [PubMed] [Google Scholar]
- Fan, H. Y., K. K. Cheng and H. L. Klein, 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics 142 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst et al., 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12 1052–1058. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O., A. Lee, M. Nussenzweig and A. Nussenzweig, 2004. H2AX: the histone guardian of the genome. DNA Repair 3 959–967. [DOI] [PubMed] [Google Scholar]
- Game, J. C., M. S. Williamson and C. Baccari, 2005. X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics 169 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game, J. C., M. S. Williamson, T. Spicakova and J. M. Brown, 2006. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics 173 1951–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert, M., 1996. A new view of V(D)J recombination. Genes Cells 1 269–275. [DOI] [PubMed] [Google Scholar]
- Giannattasio, M., F. Lazzaro, P. Plevani and M. Muzi-Falconi, 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280 9879–9886. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Grenon, M., T. Costelloe, S. Jimeno, A. O'Shaughnessy, J. Fitzgerald et al., 2007. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24 105–119. [DOI] [PubMed] [Google Scholar]
- Guillet, M., and S. Boiteux, 2002. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 21 2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32 561–599. [DOI] [PubMed] [Google Scholar]
- Harrison, J. C., and J. E. Haber, 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40 209–235. [DOI] [PubMed] [Google Scholar]
- Jansen, J. G., M. I. Fousteri and N. de Wind, 2007. Send in the clamps: control of DNA translesion synthesis in eukaryotes. Mol. Cell 28 522–529. [DOI] [PubMed] [Google Scholar]
- Klein, H. L., 2007. Reversal of fortune: Rad5 to the rescue. Mol. Cell 28 181–183. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., 2004. DNA replication fidelity. J. Biol. Chem. 279 16895–16898. [DOI] [PubMed] [Google Scholar]
- Kunz, B. A., A. F. Straffon and E. J. Vonarx, 2000. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat. Res. 451 169–185. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., 2000. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 451 91–105. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. W., 2002. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair 1 425–435. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner et al., 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94 399–409. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., F. Paques, J. Sylvan and J. E. Haber, 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9 767–770. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., P. Pasero, A. Bensimon and E. Schwob, 2001. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 29 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby, M., R. Rothstein and U. H. Mortensen, 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby, M., U. H. Mortensen and R. Rothstein, 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5 572–577. [DOI] [PubMed] [Google Scholar]
- Lisby, M., J. H. Barlow, R. C. Burgess and R. Rothstein, 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699–713. [DOI] [PubMed] [Google Scholar]
- Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani et al., 1997. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16 5216–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lopes, M., M. Foiani and J. M. Sogo, 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21 15–27. [DOI] [PubMed] [Google Scholar]
- Lundin, C., M. North, K. Erixon, K. Walters, D. Jenssen et al., 2005. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33 3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall, D., and T. Weinert, 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270 1488–1491. [DOI] [PubMed] [Google Scholar]
- Lydall, D., and S. Whitehall, 2005. Chromatin and the DNA damage response. DNA Repair 4 1195–1207. [DOI] [PubMed] [Google Scholar]
- Martin, S. G., T. Laroche, N. Suka, M. Grunstein and S. M. Gasser, 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97 621–633. [DOI] [PubMed] [Google Scholar]
- Milne, G. T., S. Jin, K. B. Shannon and D. T. Weaver, 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst et al., 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg, K. A., R. J. Michelson, C. W. Putnam and T. A. Weinert, 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36 617–656. [DOI] [PubMed] [Google Scholar]
- Pages, V., and R. P. Fuchs, 2002. How DNA lesions are turned into mutations within cells? Oncogene 21 8957–8966. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., and J. Cote, 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18 602–616. [DOI] [PubMed] [Google Scholar]
- Prado, F., F. Cortes-Ledesma and A. Aguilera, 2004. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 5 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451 13–24. [DOI] [PubMed] [Google Scholar]
- Prakash, S., R. E. Johnson and L. Prakash, 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74 317–353. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11 2600–2621. [DOI] [PubMed] [Google Scholar]
- Rudner, A. D., B. E. Hall, T. Ellenberger and D. Moazed, 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda, S., B. K. Minesinger, M. Giannattasio, P. Plevani, M. Muzi-Falconi et al., 2005. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous pol zeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280 38657–38665. [DOI] [PubMed] [Google Scholar]
- Sabbioneda, S., I. Bortolomai, M. Giannattasio, P. Plevani and M. Muzi-Falconi, 2007. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair 6 121–127. [DOI] [PubMed] [Google Scholar]
- Sancar, A., 1996. DNA excision repair. Annu. Rev. Biochem. 65 43–81. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P. A., and G. S. Roeder, 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97 313–324. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P. A., and G. S. Roeder, 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11 3601–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R. H., and S. Prakash, 1988. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol. 8 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard, A., 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75 243–269. [DOI] [PubMed] [Google Scholar]
- Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson et al., 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks and R. Sternglanz, 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, G. W., A. M. O'Shaughnessy, S. Jimeno, I. M. Dobbie, M. Grenon et al., 2006. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair 5 693–703. [DOI] [PubMed] [Google Scholar]
- Torres-Ramos, C. A., R. E. Johnson, L. Prakash and S. Prakash, 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20 3522–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, H. D., 2005. The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chembiochem 6 1735–1743. [DOI] [PubMed] [Google Scholar]
- van Attikum, H., and S. M. Gasser, 2005. The histone code at DNA breaks: A guide to repair? Nat. Rev. Mol. Cell Biol. 6 757–765. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109 745–756. [DOI] [PubMed] [Google Scholar]
- Wyatt, M. D., and D. L. Pittman, 2006. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 19 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki, R., A. Javaheri, S. Allard, F. Sha, J. Cote et al., 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25 8430–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., and B. L. Chow, 1998. Synergism between yeast nucleotide and base excision repair pathways in the protection against DNA methylation damage. Curr. Genet. 33 92–99. [DOI] [PubMed] [Google Scholar]
- Zhou, B. B., and S. J. Elledge, 2000. The DNA damage response: putting checkpoints in perspective. Nature 408 433–439. [DOI] [PubMed] [Google Scholar]