Abstract
A large-scale transcriptome analysis has been conducted using μPEACH1.0 microarray on nectarine (Prunus persica L. Batsch) fruit treated with 1-methylcyclopropene (1-MCP). 1-MCP maintained flesh firmness but did not block ethylene biosynthesis. Compared with samples at harvest, only nine genes appeared to be differentially expressed when fruit were sampled immediately after treatment, while a total of 90 targets were up- or down-regulated in untreated fruit. The effect of 1-MCP was confirmed by a direct comparison of transcript profiles in treated and untreated fruit after 24 h of incubation with 106 targets differentially expressed. About 30% of these targets correspond to genes involved in primary metabolism and response processes related to ethylene, auxin, and other hormones. In treated fruit, altered transcript accumulation was detected for some genes with a role in ripening-related events such as softening, colour development, and sugar metabolism. A rapid decrease in flesh firmness and an increase in ethylene production were observed in treated fruit maintained for 48 h in air at 20 °C after the end of the incubation period. Microarray comparison of this sample with untreated fruit 24 h after harvest revealed that about 45% of the genes affected by 1-MCP at the end of the incubation period changed their expression during the following 48 h in air. Among these genes, an ethylene receptor (ETR2) and three ethylene-responsive factors (ERF) were present, together with other transcription factors and ethylene-dependent genes involved in quality parameter changes.
Keywords: ACC synthase, ethylene biosynthesis, ethylene receptors, gene expression, microarray, 1-methylcyclopropene, post-harvest
Introduction
In climacteric fruit, ethylene is required for ripening and the control of this developmental process mainly relies on the possibility of affecting ethylene production and/or action. This can be achieved through genetic manipulation, by modulating environmental parameters (temperature and atmosphere composition) during storage, or using specific ethylene inhibitors. In general, the result of these approaches is an altered gene transcription pattern and, as a consequence, modified ripening behaviour. 1-methylcyclopropene (1-MCP) is thought to interact with ethylene receptors thus preventing ethylene-dependent processes (Sisler and Serek, 1997). Research carried out using 1-MCP report that the chemical, by affecting ethylene production, respiration, softening, colour change, and aroma production, delays ripening and increases the post-harvest life of numerous climacteric fruits. However, treatment efficacy is variable and depends on several factors including the concentration of 1-MCP used, storage condition and duration, and maturity of the fruit before application (for a review on 1-MCP effects see Watkins, 2006). In addition, 1-MCP induces variable responses in different climacteric fruit species and even in varieties of the same species (Blankenship and Dole, 2003), thus suggesting the presence of different mechanisms controlling ripening in climacteric fruit. This is particularly evident when comparing two important fruit species such as apple and peach. Following 1-MCP treatment, ethylene production is strongly reduced, ripening is inhibited or delayed for many days, and storage prolonged in apples (Watkins et al., 2000; Fan and Mattheis, 2002; Jiang and Joyce, 2002; Bai et al., 2005). On the other hand, the effects of 1-MCP treatments on peaches and nectarines are limited to the incubation period and a few hours afterwards, when the maintenance of flesh firmness is associated with an altered gene expression pattern as observed for endo-polygalacturonase (endo-PG) (Dong et al., 2001; Dal Cin et al., 2005). Recovery of ripening occurs within a few days after the end of treatment in both peaches and nectarines which soften rapidly, especially at room temperature (Mathooko et al., 2001; Fan et al., 2002; Ziliotto et al., 2003). These features make apple and peach (both belonging to the Rosaceae family) interesting models for elucidating the molecular mechanisms responsible for the different ripening patterns and responses to ethylene and its inhibitors. In a comparative work, Dal Cin et al. (2006) pointed out that, whereas ethylene biosynthesis is markedly inhibited by 1-MCP in apples, its production in peaches was not reduced by the chemical. This suggests that the variable responses to the inhibitor of ethylene perception might be due to differences in terms of ratio, expression pattern, turnover of the ethylene receptors, and/or mechanisms leading to altered chemical binding of 1-MCP.
The genomics approach and the development of high throughput technologies for large-scale analyses of transcriptomes represent powerful tools for unravelling the molecular mechanisms of complex processes, such as fruit ripening, and elucidating, on a large-scale basis, the role and effects of endogenous and/or exogenous factors. In particular, microarray analyses have been performed for transcriptome profiling during the transition from the pre-climacteric to the climacteric stage and the role of ethylene in ripening pear (Fonseca et al., 2004), tomato (Alba et al., 2005), and peach (Trainotti et al., 2006) fruit. In this last species, a dramatic up-regulation has been detected for genes encoding transcription factors and others involved in ethylene biosynthesis, perception, and signal transduction. Using the same microarray, Trainotti et al. (2007) identified genes involved in biosynthesis, transport, and signalling of auxin that show, in peach mesocarp, an increased expression at ripening, and demonstrated the existence of an important cross-talk between auxin and ethylene, with genes in the auxin domain regulated by ethylene, and genes in the ethylene domain regulated by auxin.
The results of a transcriptomic approach, undertaken with the aim of elucidating molecular mechanisms and identifying candidate genes involved in the responses of nectarine fruit to the inhibitor of ethylene action 1-MCP are reported here.
Materials and methods
Plant material and experimental design
Nectarine (Prunus persica L. Batsch, cv. Super Crimson Gold) fruit were harvested at commercial ripening stage (about 60 N flesh firmness) and immediately transferred to the Post-harvest Laboratory of the Faculty of Agriculture, University of Padova, Italy, where they were maintained at room temperature (20 °C) throughout the experiments. They were then divided in two groups of 40 fruit each: the first group was enclosed in gas-tight glass jars and treated with 1 μl l−1 of 1-MCP for 24 h. The second group (control) was enclosed for 24 h in sealed jars of the same volume without 1-MCP. To avoid excessive CO2 accumulation, KOH was added to the jars. At the end of the 24 h incubation period fruit were removed from the jars and transferred to air at 20 °C for a further 48 h (72 h from harvest, 72MCP and 72AIR). Samplings were performed at the beginning of the experiment (T0), at 24 (24MCP and 24AIR, end of the incubation period), 48 and 72 (72MCP and 72AIR, 48 h from the end of the incubation period) hours after harvest.
At each sampling time, ethylene production of 10 individual fruit was determined using a gas chromatograph (DANI 3200) as described by Tonutti et al. (1997) and flesh firmness measured with a penetrometer (TR, Forlì, Italy) equipped with a 8 mm probe. For each sampling time (T0, 24AIR, 24MCP, 72AIR and 72MCP) two representative fruit (biological replicates) have been used for transcript analyses.
RNA extraction, microarray preparation, and hybridization
Total RNA was extracted using the protocol described by Ruperti et al. (2001). RNA yield and purity was checked by means of UV absorption spectra, whereas RNA integrity was ascertained by means of electrophoresis in agarose gels.
Total RNA (20 μg) from T0, air-, and 1-MCP-treated fruits was converted into target cDNA by reverse transcription using the SuperScript™ Indirect cDNA Labeling System (Invitrogen, USA) following the manufacturer's instructions.
The features, preparation, and hybridization protocols of the peach microarray μPEACH 1.0 are described in Trainotti et al. (2006).
Data analysis
The microarrays were scanned as described by Trainotti et al. (2006). The TM4 (www.tm4.org) package developed at TIGR (www.tigr.org; Saeed et al., 2003) was used to analyse microarray data. The images were processed using the Spotfinder 2.2.3. software by means of the Otsu algorithm. The expression data extracted by Spotfinder were normalized by MIDAS 2.18 using the LOWESS (Locally Weighted Regression Scatter Plot Smoothing; Cleveland, 1979) algorithm with the ‘block mode’, keeping the Cy3 channel as reference. After normalization, data from each slide were split in two as each probe is spotted twice on μPEACH1.0. Thereafter, each spot value was considered to be independent. Normalized split data were loaded in MeV 3.1 and for each comparison (24MCPvsT0, 24AIRvsT0, 24MCPvsAIR; 72MCPvs24AIR, at least three experiments) a 66% cut-off was imposed to select genes differentially expressed by one-class unpaired SAM (Significance Analysis of Microarrays; Tusher et al., 2001) analysis. Clones with significant changes in expression (threshold ratios, expressed as log2, higher than 1 and lower than –1 for up- and down-regulation, respectively) were identified at delta values giving a 0% of false discovery rate (FDR).
Quantification of mRNA via qRT-PCR
To validate microarray data, transcript accumulation of AUX/IAA, β-carotene hydroxylase, catalase, ethylene receptor ETR2, ERF-like, and trehalose-6-phosphatase genes was evaluated via qRT-PCR. The cDNA single strand used for RT-PCR was obtained as follows: 50 μg of total RNA were treated with 10 units of RQ1 RNase-Free DNase (Promega) and 1 unit of RNAguard (RNase INHIBITOR, GE Healthcare) for 30 min, then purified by phenol–chloroform. One microgram of total DNA-free RNA was reverse-transcribed with 200 U of M-MLV Reverse Transcriptase (Promega), 1 unit of RNAguard (RNase INHIBITOR) and 2.5 μM oligo-dT12–18 as primer at 37 °C for 90 min in a final volume of 20 μl, as described in Sambrook et al. (1989). The single strand cDNA obtained (1 μl) was subjected to real-time PCR in a final volume of 10 μl containing 2× Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, USA) and specific primers (3 pmol) (see Supplementary Table S1 at JXB online). Three technical replicates for each sample were run on an ABI 7500 Real Time PCR System Sequence Detection machine (PE Applied Biosystems) programmed to heat for 10 min at 95 °C, followed by cycling conditions (melting step for 30 s at 95 °C, annealing for 30 s at 64 °C, and extension for 35 s at 72 °C) repeated for 40 cycles. An additional dissociation cycle (15 s at 95 °C, 1 min at 60 °C, and 15 s at 95 °C) was carried out. The amplified cDNA fragments were cloned into pBluescript II KS+ vector (Fermentas International, Burlington, Canada) and sequenced. The expression values were calculated following the mathematical model proposed by Livak and Schmittgen (2001) using 18S as housekeeping.
Results
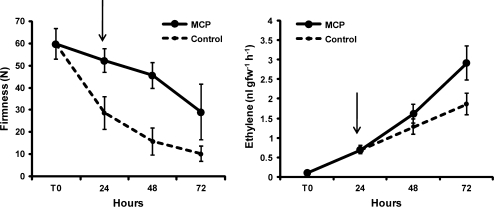
Compared with the control, which softened rapidly, 1-MCP-treated fruit maintained higher flesh firmness at the end of the 24 h incubation period (Fig. 1). A difference between samples was still present after transferring treated fruit to air for 24 h and 48 h (48 h and 72 h from harvest, respectively) when treated fruit softened. A similar increasing pattern of ethylene biosynthesis was observed in both samples throughout the experiment. At the end of the incubation period and 24 h afterwards, treated fruit synthesized ethylene at the same level as the control, while at 72 h a higher production was detected in treated fruit (Fig. 1).
Fig. 1.
Flesh firmness (left panel) and ethylene biosynthesis (right panel) in 1-MCP-treated (solid line) and control (dotted line) nectarine fruit. Arrows indicate the end of the incubation period. Data are means of 10 fruit. Vertical bars represent ±SD.
Transcript profiling at the end of 1-MCP treatment
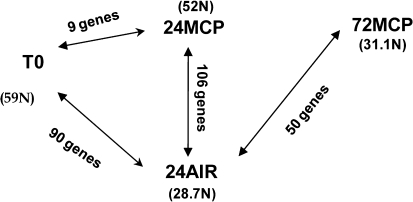
A first set of microarray hybridizations was carried out using a simple loop/flipped dye design (Fig. 2), to compare T0, 24MCP, and 24AIR samples. In the 24AIRvsT0 comparison, a total of 215 targets were significant at SAM analysis, whereas 212 targets were identified in both 24MCPvsT0 and 24MCPvs24AIR comparisons. According to the threshold log ratio (>1 for induced; <–1 for repressed), 90 (43 induced and 47 repressed) and nine unigenes (three induced and six repressed) were expressed differentially in 24AIRvsT0 and 24MCPvsT0, respectively. Compared with the 24AIR sample, a total of 106 targets showed different (54 greater and 52 lower) transcript accumulation in treated fruit (24MCP) (Fig. 2). This clearly indicates that the presence of 1-MCP in the atmosphere surrounding nectarine fruit is effective in altering the expression of specific genes, resulting in a block of the ripening process as demonstrated by the maintenance of high flesh firmness values (Fig. 1).
Fig. 2.
Experimental design of microarray analyses. In the simple loop, six slides (12 technical replicates) were used for the comparison 24MCPvsT0 and three (six technical replicates) for 24AIRvsT0, 24MCPvs24AIR, and 72MCPvs24AIR. Dye flip was performed for each comparison (three for 24MCPvsT0 and one for 24AIRvsT0, 24MCPvs24AIR, and 72MCPvs24AIR). For each comparison the number of genes showing differential expression, identified by SAM analysis, is reported. Flesh firmness value (N) is also indicated.
A list of targets that were significant by SAM analysis and those showing differential expression and/or transcript accumulation is reported in Table S2 at JXB online. Among targets that were more abundant in 24MCP than in 24AIR, two are involved in auxin metabolism (ctg_57 and ctg_1741), and two correspond to genes related to cell wall metabolism (expansin PpEXP2, ctg_941; and mannan endo-1,4 β-mannosidase, ctg_1954). Higher transcript levels of ctg_1024 and ctg_5424, corresponding to a catalase and a lipoxygenase, respectively, and ctg_1751, representing a glucose acyltransferase were also observed at the end of 1-MCP treatment. Of the 54 genes that more were expressed in 1-MCP-treated fruit, 36 (66.6%) appeared down-regulated in the 24AIRvsT0 comparison microarray (see Supplementary Table S2 at JXB online), suggesting that these genes are involved in the ripening process of nectarines.
Among the 52 targets showing significantly lower (log ratio <–1) transcript accumulation in the 24MCPvs24AIR comparison (see Supplementary Table S2 at JXB online), specific cell wall-related genes are present as endo-PG (ctg_420), pectin acetylesterase (PAE) precursor (ctg_1816), one putative pectin methylesterases (PME) (ctg_4533), expansin PpEXP3 (ctg_676), and EGase (PpEG1, ctg_2197). Genes related to quality parameters such as colour, flavour development, and sugar metabolism have also been identified: β-carotene hydroxylase (ctg_711), omega-6 fatty acid desaturase (ctg_835), putative invertase inhibitor (ctg_4499), pyruvate decarboxylase (ctg_112), sucrose synthase (ctg_61), glucose acyltransferase (ctg_1752), and trehalose-6-phosphate phosphatase, TPPA (ctg_4621). With respect to hormone metabolism, not only two auxin-related genes (AUX/IAA protein, ctg_768, auxin response factor, ARF, ctg_1991) and a 9-cis-epoxycarotenoid dioxygenase2 (ctg_2980) involved in ABA biosynthesis, but a number of genes with a role in ethylene production, perception, and signal transduction accumulated lower transcripts in 1-MCP-treated fruit: 1-aminocyclopropane-1-carboxylate (ACC) oxidase (ACO1, ctg_64), ethylene receptor ETR2 (ctg_4109), ethylene-response factors (ERF1, ctg_3350; ERF2, ctg_2116; ERF-like, ctg_2757). Two other transcription factors, a MADS-box homologue (ctg_1511), and HD-ZIP protein (ctg_2443) were identified, as well as three targets corresponding to pathogenesis-related proteins (ctg_1026, ctg_1069, and ctg_4524) and one ripening-related protein (ctg_938). Of the 52 genes showing lower transcript accumulation at the end of the 1-MCP incubation period, 27 (51.9%) appeared up-regulated in the 24AIRvsT0 comparison (see Supplementary Tables S2 at JXB online). This implies a role for these genes in nectarine ripening.
As reported above, only nine targets (three induced and six repressed) resulted as being differentially expressed in 24MCP compared with the T0 sample. Of the three targets up-regulated, one putatively corresponds to a sucrose synthase gene, and one of the six repressed genes refers to an auxin-induced protein (data not shown).
Transcript profiling in the post-treatment phase
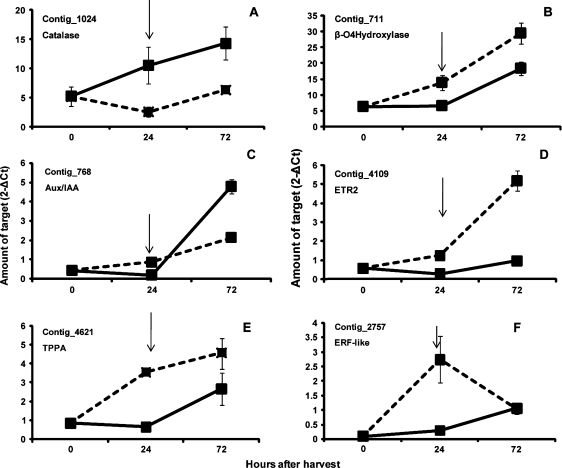
After transferring fruit to air at the end of the 24 h incubation period, rapid softening occurred at 72 h (Fig. 1). This was accompanied by a marked increase in ethylene biosynthesis. To elucidate this behaviour at a molecular level, the microarray hybridization design was implemented to perform an additional comparison: 72MCP (31.1 N flesh firmness) versus 24AIR (28.7 N flesh firmness). Of the 54 targets showing greater transcript accumulation in the 24MCPvs24AIR comparison, 12 appeared to be expressed at a similar level in 72MCP and 24AIR samples, indicating a decreasing transcription activity during the post-treatment phase, whereas 36 targets (66.6%) still displayed a higher transcript accumulation 48 h after the end of 1-MCP treatment (see Supplementary Table S2 at JXB online). Among these targets, catalase (ctg_1024) was selected to validate, via qRT-PCR, the microarray data: as reported in Fig. 3A, expression analysis confirmed that 1-MCP induced an increased expression of this gene at the end of the incubation period and this behaviour was also observed 48 h later. Considering the 52 targets showing a reduced transcript accumulation at the end of the 24 h treatment period (24MCPvs24AIR), only 13 still maintained a reduced transcript level in the 72MCPvs24AIR comparison, and for 35 (67.3%) log ratio values higher than <–1 were detected (see Supplementary Table S2 at JXB online). This would indicate that a number of targets negatively affected by 1-MCP at the end of the 24 h incubation period recovered their expression 48 h later. The lower amount of transcript accumulation in 1-MCP-treated samples at the end of the incubation period and the recovery pattern at 72 h was confirmed by qRT-PCR analysis of ctg_711 (β-carotene hydroxylase), ctg_768 (AUX/IAA protein), ctg_4109 (ETR2), and ctg_4621 (TPPA) (Fig. 3B, C, D, E). A marked reduced transcript level at 24 h and slight recovery at 72 h was also observed via qRT-PCR in treated fruit for ctg_2757 (ERF-like) (Fig. 3F), one of the 13 targets still maintaining log ratio value <–1 in the 72MCPvs24AIR comparison.
Fig. 3.
Expression profiles throughout the experiment of selected genes showing higher (A, catalase) or lower (B, β-carotene hydroxylase; C, AUX/IAA; D, ETR2; E, TPPA; F, ERF-like) transcript accumulation in the 24MCPvs24AIR comparison microarray. Solid lines indicate 1-MCP-treated fruit and dotted lines control fruit. Bars represent ±SE of three independent replicates. Arrows indicate the end of 1-MCP treatment.
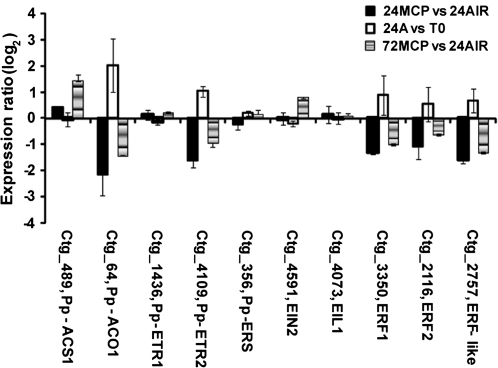
With the aim of better describing molecular aspects that characterize the response of nectarine fruit to 1-MCP, targets present in the μPEACH1.0 array (and being shown as significant with SAM analysis) were grouped according to their putative function and microarray data resulting from different comparisons (24MCPvs24AIR, 24AIRvsT0, and 72MCPvs24AIR) were plotted together. Considering genes involved in ethylene physiology (Fig. 4), as stated above, lower (<–1) transcript accumulation was detected for ACO1 (ctg_64), ETR2 (ctg_4109), ERF1 (ctg_3350), ERF2 (ctg_2116), and ERF-like (ctg_2757) genes in 24MCP samples compared with 24AIR, whereas no significant difference was observed for ACC synthase (ACS1, ctg_489), ETR1 (ctg_1436), ERS (ctg_356), EIN2 (ctg_4591), and EIL1 (ctg_4073). All the targets showing lower transcript accumulation at 24 h in treated fruit displayed a recovering expression at 72 h (higher log ratio value in 72MCPvs24AIR than in 24MCPvs24AIR). This behaviour was demonstrated by qRT-PCR analyses for ctg_4109 (ETR2) and ctg_2757 (ERF-like) (Fig. 3D, F). Interestingly, an increase in transcript accumulation at 72 h was also detected for ACS1 and, although not statistically significant, for EIN2 (Fig. 4). Worthy of note is the fact that at the early ripening in air (24AIRvsT0) a significant increase of transcript was only observed for ACO1 and ETR2.
Fig. 4.
Hybridization intensity ratio, reported as log2, of probes spotted on μPEACH1.0 and significant at SAM analysis corresponding to genes involved in ethylene biosynthesis, perception, and signal transduction. The ratio has been calculated by comparing, in three microarray experiments, the hybridization signal of cDNAs corresponding to transcripts of fruit sampled at harvest (T0), after 24 h in air (24A) or in 1-MCP (24MCP), and 48 h in air from the end of 1-MCP treatment (72MCP). Log value is the average of a minimum of four replicates. Bars indicate standard deviation. Abbreviations: ACO1, 1-aminocyclopropane-1-carboxylate oxidase; ACS1, 1-aminocyclopropane-1-carboxylate synthase; EIL, EIN3-like protein; ERF, ethylene response factors; ERS, ethylene receptor ERS-like; ETR, ethylene receptor ETR type.
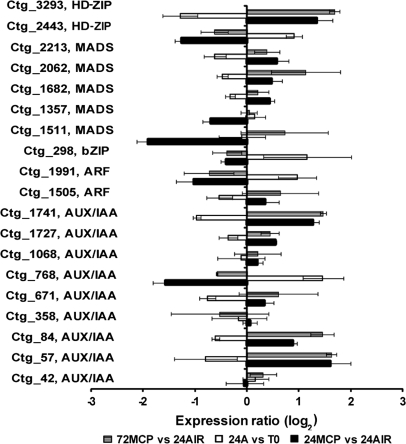
Besides the ethylene-related Transcription Factors (TFs), several other targets corresponding to TFs are present in the μPEACH1.0 array and some of these appeared to be affected by 1-MCP. The effect of the ethylene inhibitor appeared to be variable: at the end of the incubation period, it significantly reduced the accumulation of transcripts of a MADS box homolog (ctg_1511), ARF (ctg_1991), AUX/IAA (ctg_768), and an HD-ZIP (ctg_2443), and increased that of another HD-ZIP (ctg_3293) and two AUX/IAA TFs (ctg_57 and ctg_1741) (Fig. 5). The effect of 1-MCP was maintained at 72 h for ctg_57, ctg_1741, and ctg_3293, whereas a recovery of gene expression was detected for ctg_1511, ctg_2443, and ctg_768, which displayed a dramatic increase in transcript accumulation (Fig. 5), as demonstrated for ctg_768 by qRT-PCR analysis (Fig. 3C). In addition, higher transcript levels in treated fruit at 72 h were detected for ctg_84 (AUX/IAA) (Fig. 5).
Fig. 5.
Hybridization intensity ratio, reported as log2, of probes spotted on μPEACH1.0 and significant at SAM analysis corresponding to members belonging to transcriptional regulator gene families. The ratio has been calculated by comparing, in three microarray experiments, the hybridization signal of cDNAs corresponding to transcripts of fruit sampled at harvest (T0), after 24 h in air (24A) or in 1-MCP (24MCP), and 48 h in air from the end of 1-MCP treatment (72MCP). Log value is the average of a minimum of four replicates. Bars indicate standard deviation.
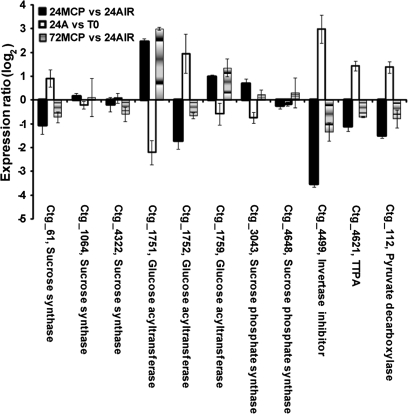
Three other groups of targets involved in quality-related metabolic processes were identified. The first deals with sugar metabolism (Fig. 6): four targets (ctg_112, pyruvate decarboxylase; ctg_1752, glucose acyltransferase; ctg_4499, putative invertase inhibitor; ctg_4621, TTPA) were up-regulated during early ripening and significantly less represented in treated fruit at the end of the 1-MCP incubation period, showed a recovery of their expression at 72 h (as demonstrated for ctg_4621 by qRT-PCR analysis, Fig. 3E). Another putative glucose acyltransferase gene (ctg_1751) appeared to be down-regulated during the early ripening stage and showed a higher transcript level in treated fruit at both 24 h and 72 h.
Fig. 6.
Hybridization intensity ratio, reported as log2, of probes spotted on μPEACH1.0 and significant at SAM analysis corresponding to genes involved in sugar metabolism. The ratio has been calculated by comparing, in three microarray experiments, the hybridization signal of cDNAs corresponding to transcripts of fruit sampled at harvest (T0), after 24 h in air (24A) or in 1-MCP (24MCP), and 48 h in air from the end of 1-MCP treatment (72MCP). Log value is the average of a minimum of four replicates. Bars indicate standard deviation. Abbreviations: TPPA, trehalose-6-phosphate phosphatase.
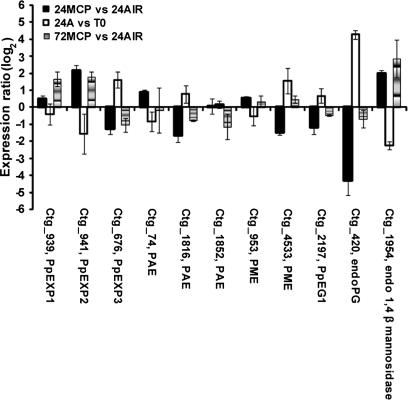
The second group comprises targets involved in cell-wall metabolism (Fig. 7): with the exception of ctg_676 (PpEXP3), all the targets displaying lower transcript accumulation at 24 h in treated fruit and an up-regulation during the early stages of ripening (ctg_420, endo-PG; ctg_2197, PpEG1; ctg_4533, PME; ctg_1816, PAE) were characterized by a significant recovery of expression at 72 h. Interestingly, PpEXP2 (ctg_941), down-regulated at early ripening stage and showing higher transcript levels in treated fruit at 24 h, did not undergo any expression change at 72 h. A similar behaviour has been observed for ctg_1954 representing a mannan endo-1,4-β-mannosidase (Fig. 7).
Fig. 7.
Hybridization intensity ratio, reported as log2, of probes spotted on μPEACH1.0 and significant at SAM analysis corresponding to genes involved in cell wall metabolism. The ratio has been calculated by comparing, in three microarray experiments, the hybridization signal of cDNAs corresponding to transcripts of fruit sampled at harvest (T0), after 24 h in air (24A) or in 1-MCP (24MCP), and 48 h in air from the end of 1-MCP treatment (72MCP). Log value is the average of a minimum of four replicates. Bars indicate standard deviation. Abbreviations: EG, endo-β-1,4-glucanase; endoPG, endopolygalacturonase; EXP, expansin; PAE, pectin acetylesterase; PME, pectin methylesterase.
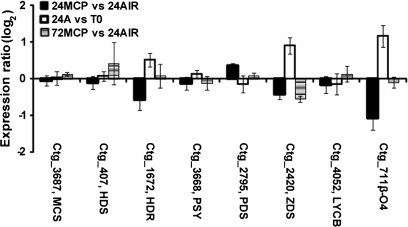
The last selected group includes transcripts corresponding to genes involved in the isoprenoid pathway and the development of typical yellow pigmentation in this nectarine variety (Fig. 8). The only gene appearing significantly affected by 1-MCP at the end of the 24 h incubation period was β-carotene hydroxylase (ctg_711), up-regulated during the early ripening stages. The effect of the ethylene inhibitor on this gene was confirmed by qRT-PCR (Fig. 3B): this analysis pointed out, as also indicated by microarray data, a recovery of the expression of this gene at 72 h.
Fig. 8.
Hybridization intensity ratio, reported as log2, of probes spotted on μPEACH1.0 and significant at SAM analysis corresponding to genes involved in isoprenoid pathway. The ratio has been calculated by comparing, in three microarray experiments, the hybridization signal of cDNAs corresponding to transcripts of fruit sampled at harvest (T0), after 24 h in air (24A) or in 1-MCP (24MCP), and 48 h in air from the end of 1-MCP treatment (72MCP). Log value is the average of a minimum of four replicates. Bars indicate standard deviation. Abbreviations: β-O4: β-carotene hydroxylase; HDR, 14-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; HDS, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; LCYB, lycopene β cyclase; MCS, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; PDS, phytoene dehydrogenase; PSY, phytoene synthase; ZDS, ζ-carotene desaturase.
Discussion and conclusions
The discovery of 1-MCP as a powerful antagonist of ethylene action is providing great opportunities for scientists to gain insight into the fundamental mechanisms involved in many plant processes including fruit ripening. It is confirmed here that, differently from other climacteric species, the responses of peaches and nectarines to 1-MCP treatment are confined to the incubation period and a few hours thereafter. The large-scale transcriptome analysis performed using μPEACH1.0 indicates that the quick ripening observed after transferring treated peaches to air is not due to a limited physiological effect of 1-MCP during incubation. In fact, at the end of the 24 h treatment period, the maintenance of high firmness (52.0 N) is accompanied by marked changes in transcript profiling if compared to control fruit (24AIR). Only nine genes showed significant changes in 24MCPvsT0 microarray analysis compared with 90 targets displaying differential expression during 24 h ripening in air (24AIRvsT0). Such results indicate that the presence of 1-MCP, by altering ethylene perception, induces a block of ripening as demonstrated by the maintenance of flesh firmness value. This physiological effect is confirmed by the 106 genes that were differentially expressed when comparing 24MCP and 24AIR samples, and the fact that a number of affected targets correspond to genes with a role in hormone (ethylene, but also auxin and ABA) metabolism and in regulating transcription. Considering that 66.6% and 51.9% of transcripts displaying greater and lower transcript accumulation, respectively, at the end of the 1-MCP incubation period show an opposite trend in ripening fruit kept in air (24AIRvsT0 comparison), it might be hypothesized that these targets correspond to ripening-related and ethylene-dependent genes.
Cell wall-related genes
The maintenance of high firmness values at the end of the incubation period appears to be the result of a down-regulation of cell wall-related genes, involved, in particular, in pectin metabolism, such as endo-PG. Microarray data on endo-PG confirm specific expression analysis (Ziliotto et al., 2003; Dal Cin et al., 2005), showing a reduction of endo-PG transcript accumulation at the end of 1-MCP treatment and further emphasize the crucial role played by endo-PG in peach fruit softening, as clearly demonstrated in non-melting (Callahan et al., 2004; Morgutti et al., 2006) and stony hard (Hayama et al., 2006a, b; Begheldo et al., 2008) genotypes.
Considering the strict relationship existing between endo-PG and ethylene (Sitrit and Bennett, 1998), the marked reduction of endo-PG transcript accumulation in treated fruit (log ratio value –4.29, the second lowest value among affected targets) indicates that ethylene action is markedly inhibited by 1-MCP in ripening nectarines. One of the targets showing higher transcript level in 1-MCP-treated fruit is PpEXP2 and this would confirm that this gene, down-regulated by ethylene, is related to peach fruit growth and, differently from PpEXP3 (ctg_676), which displayed a reduced level of transcript accumulation in 1-MCP treated fruit, is not responsible for softening as reported by Hayama et al. (2001, 2003).
Regulation of ethylene biosynthesis
When evaluating the list of targets showing lower transcript accumulation at the end of the 1-MCP incubation period, it is interesting to notice that, of the two elements involved in the last ethylene biosynthetic steps, only ACO1 is present, whereas ACS1 does not appear to be negatively affected by 1-MCP. In addition, microarray data of the 24AIRvsT0 comparison confirm that, in P. persica fruit, ACO1 is induced earlier than ACS1 at the onset of ripening as previously reported (Tonutti et al., 1997; Begheldo et al., 2008). Considering ethylene receptors, ETR2 but not ETR1 and ERS1 show lower transcript levels after 24 h treatment with 1-MCP. These data are in agreement with results published by Dal Cin et al. (2006) indicating that ACS1, ETR1, and ERS1 expressions are not or only slightly affected by 1-MCP in peaches, and suggest that ETR2, induced at the very early steps of climacteric (Fig. 4) and displaying a ripening-related expression pattern (Trainotti et al., 2006), plays a crucial role in the modulation of ethylene responses in peach fruit. This hypothesis is reinforced by the fact that, in ripening peaches, a great inductive effect on ETR2 is yielded by exogenous ethylene treatment (Trainotti et al., 2007).
As specifically pointed out by Mathooko et al. (2001) and Dal Cin et al. (2006), ACS1 expression and activity are not affected by 1-MCP in ripening peaches. Considering that ACO1 gene expression and activity appear reduced but not blocked, as reported by our microarray data and by Mathooko et al. (2001), the result is that ethylene production is not inhibited by 1-MCP in P. persica fruit at the end of 1-MCP treatment. The higher level of ethylene production observed in the post-treatment phase (72 h), paralleled by a significant increase in ACS1 transcript accumulation, confirms previously published data (Dong et al., 2001; Rasori et al., 2002; Ziliotto et al., 2003; Dal Cin et al., 2006). ACO1 gene expression is also only slightly affected in apples but, unlike peaches, ACS1 transcript accumulation is suppressed by 1-MCP (Dal Cin et al., 2006; Tatsuki et al., 2007). Taken together, these data would indicate that ACS1 might represent one crucial factor in the modulation of responses to 1-MCP application. Trainotti et al. (2007) report that many genes involved in biosynthesis, transport, and signalling of auxin have an increased expression in peach mesocarp during ripening and some ripening-related genes, including ACS1, are more strongly influenced by exogenous auxin (NAA) than by ethylene. The same authors suggest that, in addition to the independent role played by auxins, an active cross-talk between auxin and ethylene modulates peach ripening. Besides the well-known inductive effects of auxin on ACS1 and the ethylene climacteric (Abel and Theologis, 1996; Bleecker and Kende, 2000), the hypothesis of a crucial interplay between auxin and ethylene action is supported by our microarray experiments showing significant transcription changes at early ripening and an effect of 1-MCP on the expression of some auxin metabolism-related genes (ctg_57, ctg_768, ctg_1741, and ctg_1991). Whether the limited effect of 1-MCP in delaying peach fruit ripening is the result of this interplay between auxin and ethylene remains to be elucidated.
Genes involved in ethylene perception and signal transduction
The assumption that 1-MCP irreversibly binds to the ethylene receptors explains the effects of the chemical during the incubation period and this occurs either in systems where ethylene production is blocked (e.g. apple) or not, as in nectarines, where, at the end of the treatment, the delay in ripening is accompanied by altered transcript profiles. It has been postulated that, after 1-MCP treatment, the return of ethylene sensitivity is due to the appearance of new receptors (Sisler and Serek, 1997; Blankenship and Dole, 2003). This appears to be the case for nectarines where the transcription of ETR1 and ERS1 appears to be unaffected, in our experimental conditions, by 1-MCP (Fig. 4) and ETR2 expression recovers when treated fruit are transferred to air (Fig. 3D). In fact, the rapid ripening observed at 72 h (48 h from the end of 1-MCP treatment) is associated with the increased transcript accumulation of well-known ethylene-dependent genes, such as endo-PG, and others (ctg_2197, PpEG1; and ctg_4533, PME) showing an up-regulation during peach fruit ripening and/or following ethylene treatment (Trainotti et al., 1997, 2003). This occurs in parallel with the accumulation of EIN2 transcripts (ctg_4591) and recovering expression of specific ERFs at 72 h (ctg_2116, ctg_2757, and ctg_3350; Figs 3F, 4). Transcript accumulation of EIN2, absolutely required for ethylene signalling (Chen and Bleeker, 1995; Alonso et al., 1999), has been associated with ripening peaches (Trainotti et al., 2006; Begheldo et al., 2008). Considering ERFs, proteins that specifically bind to the so-called GCC box in promoters of ethylene-responsive genes, the up-regulation observed in ripening fruit (24AIRvsT0), and the reduced amount of transcripts detected at 24 h in treated fruit confirm results obtained in apple and plum, where ERF1 expression sharply increases with the climacteric peak and 1-MCP treatment inhibits transcript accumulation (El-Sharkawy et al., 2007; Wang et al., 2007). An up-regulation of ERF2 has been associated with peach fruit ripening and to the presence of ethylene (Trainotti et al., 2007). The relationship between ERF2 and ethylene is confirmed by our data showing the presence of reduced amounts of specific transcripts in 1-MCP treated fruit. Even though expression of ERFs appears to be regulated not exclusively by ethylene in ripening fruit (Wang et al., 2007), the increasing expression trend of the three selected ERFs observed at 72 h (Figs 3F, 4) might be interpreted as the result of the recovered perception of ethylene and the reactivation of the signal transduction mechanisms leading to the induction of ethylene-dependent processes.
Colour development and sugar metabolism
Considering the biosynthetic pathway of xanthophylls, pigments responsible for the yellow colour in peaches and nectarines, microarray data indicate that β-carotene hydroxylase is the earliest gene induced at the onset of ripening and appears to be, among the selected targets, the only one significantly affected by 1-MCP (Fig. 8). Ethylene-dependent expression of β-carotene hydroxylase has been observed in orange fruit, another species accumulating xanthophylls at ripening (Rodrigo and Zacarias, 2007), but not in tomato, where lycopene is the main pigment and this gene, together with phytoene synthase (PSY), is expressed in an ethylene-independent manner (Alba et al., 2005). Considering these aspects, it might be hypothesized that different ethylene-related regulatory mechanisms of carotenoid biosynthesis operate in ripening fruit, depending on the genetic background, determining the accumulation pattern and abundance of specific compounds (lycopene, β-carotene, xanthophylls), rather than the presence or not of the ethylene climacteric at ripening. The increasing trend of transcript accumulation detected at 72 h in treated fruit confirms that carotenoid genes are expressed in a ripening-related manner in P. persica fruit (Trainotti et al., 2006) and suggests the presence of different factors, including development and ethylene, modulating their transcription. The complex regulation of carotenoid biosynthesis in Prunus spp. species is supported by evidence on P. mume, where PSY is constitutively expressed, but ethylene accelerates its induction (Kita et al., 2007), and on P. armeniaca, where different effects of 1-MCP have been detected on PSY, phytoene desaturase, and ζ-carotene desaturase (Marty et al., 2005). In addition, stony hard peaches, characterized by a lack of ethylene production at ripening, undergo colour changes during the last developmental stages (Haji et al., 2003) and, if treated with exogenous ethylene, develop a more pronounced yellow colour (P. Tonutti et al., unpublished data), reinforcing the hypothesis that both development and ethylene synergistically contribute to the changes in ground colour of ripening peaches. Considering sugar metabolism, our data indicate that ethylene may induce variable effects even on genes belonging to the same family, as in the case of glucose acyltransferases. Five targets appear to be significantly influenced by 1-MCP: in particular, the expression of a putative invertase inhibitor (ctg_4499), markedly induced at ripening, is strongly affected by the ethylene antagonist. Inhibitor proteins often regulate the activity of enzymes involved in carbohydrate metabolism that are secreted out of the cytoplasmic compartment into the apoplast or to the vacuole (Juge et al., 2004). Examples are represented by invertase and PME inhibitors that are members of a large family of proteins named PMEI-related proteins (Bellincampi et al., 2004; Hothorn et al., 2004). Neutral invertase activity and gene expression (PpNI1) decline during peach fruit ripening in parallel with an increase of sucrose, a decrease of glucose+fructose content (Vizzotto et al., 1996; Nonis et al., 2007) and a dramatic up-regulation of a putative invertase inhibitor (Fig. 6). In grape berries, a putative invertase/PME inhibitor protein is highly represented at the prevéraison stage but not during the later stages of fruit development when, differently from peaches, glucose+fructose but not sucrose accumulate (da Silva et al., 2005). Whether the putative invertase inhibitor and the ethylene-dependent modulation of its expression play a major role in sugar metabolism of ripening peaches and nectarines remains to be elucidated.
In conclusion, the genomics approach described in this paper has shed light on mechanisms involved in the limited responses to 1-MCP of nectarine fruit, characterized by a recovery of ripening after the end of the incubation period, and to identify new genes potentially implicated in the ripening process and modulated by ethylene. The quick recovery of nectarine ripening following 1-MCP treatment seems to be related to the limited inhibition of ethylene synthesis by 1-MCP: in this context, ACS1 and its regulation appear to play a crucial role. However, other mechanisms appear to modulate the expression of ethylene-dependent genes involved in ripening of nectarines. If for some genes (e.g. endo-PG, ETR2) a marked expression recovery has been observed in the post-treatment phase, and this could be imputed to the presence of ethylene, for others (e.g. PpEXP2, PpEXP3, HD-ZIP ctg_3293) no expression changes have been detected after transferring fruit to air for 48 h indicating that the ethylene inhibitor is still effective. Targeted studies and comparative genomics approaches in Prunus species characterized by a wide range of ripening phenotypes with different responses to ethylene and its inhibitors will be of great help to elucidate these complex physiological mechanisms.
Supplementary data
Supplementary data are available at JXB online. Supplementary Table 1 lists the oligonucleotides used in the qRT-PCR experiments. Supplementary Table 2 reports data of microarray experiments for all the 4805 probes present on μPEACH1.0.
Supplementary Material
Acknowledgments
This research has been funded by the Italian Ministry of Research and University (MIUR), Cofin (PRIN) project no. 2006072159 co-ordinated by PT.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiology. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Bai JH, Baldwin EA, Goodner KL, Mattheis JP, Brecht JK. Response of four apple cultivars to 1-methylcyclopropene treatment and controlled atmosphere storage. Horticultural Science. 2005;40:1534–1538. [Google Scholar]
- Begheldo M, Manganaris GA, Bonghi C, Tonutti P. Different post-harvest conditions modulate ripening and ethylene biosynthetic and signal transduction pathway in stony hard peaches. Post-harvest Biology and Technology. 2008;48:84–91. [Google Scholar]
- Bellincampi D, Camardella L, Delcour JA, et al. Potential physiological role of plant glycosidase inhibitors. Biochimica et Biophysica Acta. 2004;1696:265–274. doi: 10.1016/j.bbapap.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Blankenship SM, Dole JM. 1-methylcyclopropene: a review. Post-harvest Biology and Technology. 2003;28:1–25. [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annual Review of Cell and Developmental Biology. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Callahan AM, Scorza R, Bassett C, Nickerson M, Abeles FB. Deletions in an endopolygalacturonase gene correlate with non-melting flesh texture in peach. Functional Plant Biology. 2004;3:159–168. doi: 10.1071/FP03131. [DOI] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB. Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiology. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS. Robust locally weighed regression and smoothing scatterplots. Journal of the American Statistical Association. 1979;74:829–836. [Google Scholar]
- Da Silva FG, Iandolino A, Al-Kayal F, et al. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiology. 2005;139:574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin V, Rizzini FM, Botton A, Tonutti P. The ethylene biosynthetic and signal transduction pathways are differently affected by 1-MCP in apple and peach fruit. Post-harvest Biology and Technology. 2006;42:125–133. [Google Scholar]
- Dal Cin V, Rizzini FM, Botton A, Ziliotto F, Danesin M, Tonutti P. Different response of apple and peach fruit to 1-MCP: a case of different sensitivity to ethylene? Acta Horticulturae. 2005;682:321–327. [Google Scholar]
- Dong L, Zhou HW, Sonego L, Lers A, Lurie S. Ethylene involvement in the cold storage disorder of ‘Flavortop’ nectarine. Post-harvest Biology and Technology. 2001;23:105–115. [Google Scholar]
- El-Sharkawy I, Kim WS, El-Kereamy A, Jayasankar S, Svircev AM, Brown DCW. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.) Journal of Experimental Botany. 2007;58:3631–3643. doi: 10.1093/jxb/erm213. [DOI] [PubMed] [Google Scholar]
- Fan X, Argenta L, Mattheis JP. Interactive effects of 1-MCP and temperature on ‘Elberta’ peach quality. Horticultural Science. 2002;37:134–138. [Google Scholar]
- Fan X, Mattheis JP. Impact of 1-methylcyclopropene and methyl jasmonate on apple volatile production. Journal of Agricultural and Food Chemistry. 2002;47:2847–2853. doi: 10.1021/jf990221s. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Hackler L, Jr, Zvara A, Ferriera S, Baldé A, Dudits D, Pais MS, Puskas LG. Monitoring gene expression along pear fruit development, ripening and senescence using cDNA microarrays. Plant Science. 2004;167:457–469. [Google Scholar]
- Haji T, Yaegaki H, Yamaguchi M. Softening of stony hard peach by ethylene and the induction of endogenous ethylene by 1-aminocyclopropane-1-.carboxylic acid (ACC) Journal of the Japanese Society for Horticultural Science. 2003;72:212–217. [Google Scholar]
- Hayama H, Ito A, Moriguchi T, Kashimura Y. Identification of a new expansin gene closely associated with peach fruit softening. Post-harvest Biology and Technology. 2003;29:1–10. [Google Scholar]
- Hayama H, Shimada T, Fujii H, Ito A, Kashimura Y. Ethylene-regulation of fruit softening and softening-related genes in peach. Journal of Experimental Botany. 2006a;57:4071–4077. doi: 10.1093/jxb/erl178. [DOI] [PubMed] [Google Scholar]
- Hayama H, Shimada T, Ito A, Yoshioka H, Kashimura Y. Changes in the levels of mRNAs for putative cell wall-related genes during peach fruit development. Scientia Horticulturae. 2001;91:239–250. [Google Scholar]
- Hayama H, Tatsuki M, Ito A, Kashimura Y. Ethylene and fruit softening in the stony hard mutation in peach. Post-harvest Biology and Technology. 2006b;41:16–21. [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. The Plant Cell. 2004;16:3437–3447. doi: 10.1105/tpc.104.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Joyce DC. 1-Methylcyclopropene treatment effects on intact and fresh-cut apple. Journal of Horticultural Sciences and Biotechnology. 2002;77:19–21. [Google Scholar]
- Juge N, Svensson B, Henrissat B, Williamson G. Plant proteinaceous inhibitors of carbohydrate-active enzymes. Biochimica et Biophysica Acta. 2004;1696:141. [Google Scholar]
- Kita M, Kato M, Ban Y, Honda C, Yaegaki H, Ikoma Y, Moriguchi T. Carotenoid accumulation in Japanese apricot (Prunus mume Siebold & Zucc.): molecular analysis of carotenogenic gene expression and ethylene regulation. Journal of Agricultural and Food Chemistry. 2007;55:3414–3420. doi: 10.1021/jf063552v. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marty I, Bureau S, Sarkissian G, Gouble B, Audergon JM, Albagnac G. Ethylene regulation of carotenoid accumulation and carotenogenic gene expression in colour-contrasted apricot varieties (Prunus armeniaca) Journal of Experimental Botany. 2005;56:1877–1886. doi: 10.1093/jxb/eri177. [DOI] [PubMed] [Google Scholar]
- Mathooko FM, Tsunasima Y, Owino W, Kubo Y, Inaba A. Regulation of genes encoding ethylene biosynthesis in peach fruit by carbon dioxide and 1-methylcyclopropene. Post-harvest Biology and Technology. 2001;21:265–281. [Google Scholar]
- Morgutti S, Negrini N, Nocito FF, Ghiani A, Bassi D, Cocucci M. Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phytologist. 2006;171:315–328. doi: 10.1111/j.1469-8137.2006.01763.x. [DOI] [PubMed] [Google Scholar]
- Nonis A, Ruperti B, Falchi R, Casatta E, Enferadi ST, Vizzotto G. Differential expression and regulation of a neutral invertase encoding gene from peach (Prunus persica): evidence for a role in fruit development. Physiologia Plantarum. 2007;129:436–446. [Google Scholar]
- Rasori A, Ruperti B, Bonghi C, Tonutti P, Ramina A. Characterization of two putative ethylene receptor genes expressed during peach fruit development and abscission. Journal of Experimental Botany. 2002;53:2333–2339. doi: 10.1093/jxb/erf097. [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Zacarias L. Effect of post-harvest ethylene treatment on carotenoid accumulation and the expression of carotenoid biosynthetic genes in the flavedo of orange (Citrus sinensis L. Osbeck) fruit. Post-harvest Biology and Technology. 2007;43:14–22. [Google Scholar]
- Ruperti B, Bonghi C, Rasori A, Ramina A, Tonutti P. Characterization and expression of two members of the peach 1-aminocyclopropane-1-carboxylate oxidase gene family. Physiologia Plantarum. 2001;111:336–344. doi: 10.1034/j.1399-3054.2001.1110311.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J. TM4: a free, opensource system for microarray. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sisler EC, Serek M. Inhibitors of ethylene responses in plants at the receptor level: recent developments. Physiologia Plantarum. 1997;100:577–582. [Google Scholar]
- Sitrit Y, Bennett A. Regulation of tomato fruit polygalacturonase mRNA accumulation by ethylene: a re-examination. Plant Physiology. 1998;116:1145–1150. doi: 10.1104/pp.116.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Endo A, Ohkawa H. Influence of time from harvest to 1-MCP treatment on apple fruit quality and expression of genes for ethylene biosynthesis enzymes and ethylene receptors. Post-harvest Biology and Technology. 2007;43:28–35. [Google Scholar]
- Tonutti P, Bonghi C, Ruperti B, Tornielli GB, Ramina A. Ethylene evolution and 1-aminocyclopropane-1-carboxylate oxidase gene expression during early development and ripening of peach fruit. Journal of the American Society for Horticultural Science. 1997;122:642–647. [Google Scholar]
- Trainotti L, Bonghi C, Ziliotto F, Zanin D, Rasori A, Casadoro G, Ramina A, Tonutti P. The use of microarray μPEACH1.0 to investigate transcriptome changes during transition from pre-climacteric to climacteric phase in peach fruit. Plant Science. 2006;170:606–613. [Google Scholar]
- Trainotti L, Spolaore S, Ferrarese L, Casadoro G. Characterization of ppEG1, a member of a multigene family which encodes endo-β-1,4-glucanase in peach. Plant Molecular Biology. 1997;34:791–802. doi: 10.1023/a:1005884429760. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Tadiello A, Casadoro G. The involvement of auxin in the ripening of climacteric fruits comes to age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. Journal of Experimental Botany. 2007;58:3299–3308. doi: 10.1093/jxb/erm178. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Zanin D, Casadoro G. A cell wall-oriented genomic approach reveals a new and unexpected complexity of the softening in peaches. Journal of Experimental Botany. 2003;389:1821–1832. doi: 10.1093/jxb/erg198. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences, USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzotto G, Pinton R, Varanini Z, Costa G. Sucrose accumulation in developing peach fruit. Physiologia Plantarum. 1996;96:225–230. [Google Scholar]
- Wang J, Chen G, Hu Z, Chen X. Cloning and characterization of the EIN2-homology gene LeEIN2 from tomato. DNA Sequence. 2007;18:33–38. doi: 10.1080/10425170600986738. [DOI] [PubMed] [Google Scholar]
- Watkins CB. The use of 1-methylcyclopropene (1-MCP) on fruits and vegetables. Biotechnology Advances. 2006;24:389–409. doi: 10.1016/j.biotechadv.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Watkins CB, Nock JF, Whitaker BD. Responses of early, mid and late season apple cultivars to post-harvest application of 1-methylcyclopropene (1-MCP) under air and controlled atmosphere storage conditions. Post-harvest Biology and Technology. 2000;19:17–32. [Google Scholar]
- Ziliotto F, Botton A, Bonghi C, Tonutti P. Effect of 1-MCP on nectarine fruit post-harvest physiology. In: Vendrell M, Klee H, Pech JC, Romojaro F, editors. Biology and biotechnology of the plant hormone ethylene. III. IOS Press; 2003. pp. 457–458. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.