Abstract
Signal Transducer and Activator of Transcription 3 (STAT3) is a transcription factor that plays a crucial role in interleukin-6 (IL-6) signaling, mediating the acute-phase induction of the human Angiotensinogen (hAGT) gene in hepatocytes. We showed earlier that IL-6 induces acetylation of the STAT3 NH2-terminus by the recruitment of the p300 coactivator. We had also observed a physical interaction of STAT3 and Histone Deacetylase1 (HDAC1) in an IL-6-dependent manner that leads to transcriptional repression. In this study, we sought to elucidate the mechanism by which HDAC1 controls STAT3 transcriptional activity. Here, we mapped the interacting domains of both STAT3 and HDAC1 and found that the COOH-terminal domain of HDAC1 is necessary for IL-6-induced STAT3 transcriptional repression, whereas the NH2-terminal acetylation domain of STAT3 is required for HDAC1 binding. Interestingly, over expression of HDAC1 in HepG2 cells leads to significantly reduced amounts of nuclear STAT3 after IL-6 induction, whereas silencing of HDAC1 resulted in accumulation of total and acetylated STAT3 in the nucleus. We have found that HDAC1 knockdown also interferes with the responsiveness of the STAT3-dependent MCP1 target gene expression to IL-6, as confirmed by real-time RT–PCR analysis. Together, our study reveals the novel functional consequences of IL-6-induced STAT3-HDAC1 interaction on nucleocytoplasmic distribution of STAT3.
INTRODUCTION
The signal transducers and activators of transcription (STATs) are a family of latent cytoplasmic transcription factors mediating target gene activation in response to cytokines and growth factor stimulation (1,2). Seven STAT family members (and their alternative splice products) have been identified, with each member being activated by a distinct spectrum of cytokines (3). Like other STAT transcription factors, STAT3 is predominantly cytoplasmic in resting cells, a feature which facilitates the ability of STAT3 to directly transduce signals from cell surface associated cytokine receptor to target genes in the nucleus. In response to cytokine stimulation, STATs become tyrosine phosphorylated at their COOH-terminus by receptor and receptor-associated tyrosine kinases. Activated STATs form homo- or heterodimers through intermolecular src homology domain 2 (SH2)-phosphotyrosine interactions, disengage from the liganded receptor and subsequently translocate into the nucleus where they bind enhancer sequences [5′-TT(N4-6)AA-3′] of target genes (4,5). Once the activated STAT dimer recognizes a target promoter, the transcription rate from this promoter is dramatically increased, reflecting the ability of STAT transcriptional activation domains to recruit nuclear coactivators that mediate chromatin decondensation and communicate with proteins binding the core promoter.
Although phosphorylation is a crucial posttranslational modification that regulates the activities of different proteins, there are many others including methylation (6), ubiquitination (7), sumoylation (8), isgylation (9) and acetylation (10). Indeed, it has been found that different STAT 1 and 3 isoforms are inducibly acetylated, a modification yielding a variety of consequences for target gene transcription. For example, we had recently shown that IL-6-induced acetylation of STAT3 NH2-terminus is required for recruitment of the p300 coactivator and is necessary for target gene expression through a novel mechanism involving acetylation/deacetylation (11,12). We further showed that the STAT3 NH2-terminal acetylation is necessary for target gene transcription by stabilizing the STAT3–p300 complex. Others have found that STAT3 dimerization is regulated by reversible acetylation of lysine at 685 in the SH2 domain of STAT3 (13). STAT1 is also acetylated by the CBP coactivator, a modification that regulates NF-κB activity leading to induction of apoptosis (14). Together, these observations indicate that site-specific acetylation of STAT3 is an important regulatory modification that influences protein–protein interaction.
Histone and non-histone protein acetylation is a reversible reaction controlled by the steady state level of histone acetyltransferases (HATs) and histone deacetylases (HDACs). In humans, HDACs are divided into three categories: class I RPD3-like proteins (HDAC1, HDAC2, HDAC3 and HDAC8); the class II HDA1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10); and the class III SIR2-like proteins (15,16). Class I HDACs are ubiquitously expressed while the expression of many class II HDACs are tissue-specific. Recently class IV HDACs, a group consisting of HDA-C11–related enzymes has also been described (17,18). HDAC1, 2 and 8 are predominantly nuclear proteins while HDAC 3, 4, 5, 7 and 9 shuttle between the nucleus and cytoplasm (15).
The class I enzyme HDAC1 is a nuclear protein and can heterodimerize with the closely related deacetylase HDAC2 (19). Two important functional regions of HDAC1 proteins have been identified in mouse (20): the NH2-terminus contains a motif required for HDAC1 homo-oligomerization and for hetero-oligomerization of HDAC1 with HDAC2 and HDAC3. The COOH-terminal lysine rich sequence (amino acid residues 438–482), on the other hand, is crucial for the nuclear localization of HDAC1. Both HDAC1 and 2 are found in three major multiprotein complexes, named Sin3, NuRD, and CoREST (21,22). HDAC1 can repress gene transcription either directly or as part of these multiprotein complexes when recruited to the promoter by a variety of transcriptional regulators (15,23). One emerging concept is that acetylation–deacetylation reactions play a role in cellular processes independent of transcription. For example, acetylation regulates protein stability, protein–protein interactions and nuclear localization (24). Acetylation of HNF4 and E1A adenovirus-transforming protein, is known to enhance nuclear retention by increasing nuclear import and blocking nuclear export of these transcription factors (25,26). Conversely, deacetylation of SRY, a Y chromosome-encoded DNA-binding protein, by HDAC3 induces cytoplasmic delocalization of the protein from the nuclear compartment (27). HDAC mediated deacetylation of proteins, in many cases is known to be prerequisite for subsequent ubiquitination and degradation (16,24).
In this study we show for the first time that acetylation of STAT3 and its subsequent binding with HDAC1 is involved in control of its nucleocytoplasmic distribution. We present evidence that the NH2-terminal domain of STAT3 interacts with HDAC1, and that STAT3 acetylation is required for this interaction. Overexpression of HDAC1 in HepG2 cells leads to significantly reduced amount of IL-6-induced nuclear STAT3, whereas HDAC1 silencing through RNA interference results in STAT3 accumulation in the nucleus. Consistent with this finding, we have found that HDAC1 knockdown also interferes with the responsiveness of the STAT3-dependent reporter gene and target gene expression to IL-6 stimulation. Together, these findings suggest a novel mechanism of how protein–protein interaction coupled with chemical modification regulate STAT3 transcription.
MATERIALS AND METHODS
Cell culture and reagents
Human hepatoblastoma HepG2 cells were obtained from ATCC (Manassas, VA) and cultured as previously described (28). Human colon carcinoma HCT 116 cells (ATCC) were grown at 37°C in 5% CO2 in McCoy 5A medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 μg/ml). Recombinant IL-6 was from Calbiochem (San Diego, CA) (12). Human acute monocytic leukemia THP-1 cells (ATCC) were cultured at 37°C in 5% CO2 in RPMI medium 1640 with 25 mM HEPES and 2 mM l-glutamine (GIBCO) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 µg/ml), 4.5 g/l glucose, and 1 mM sodium pyruvate.
Plasmids and transfection
The WT STAT3, NH2- and COOH-terminal truncated human STAT3 expression plasmids [amino acids (aa) 1–770, 130–770, 1–688, 1–585] were constructed by PCR using wild type STAT3 as a template and gene-specific primers (11). Plasmid V5-tagged STAT3 (aa 1–130) were constructed using the primer pairs as follows: 5′-GGAAATGGCCCAATGGAATCAGCTACAG-3′ and 5′-GTTGGCCTGGCCCCCTTGCTG-3′ and cloned into pEF6/V5-His. STAT3 Lys-to-Arg mutations at residues 49 and 87 were performed by PCR synthesis by overlap extension using STAT3 WT cDNA as a template and with the mutagenic primer pairs (11).
FLAG-tagged STAT3 WT (1–124), STAT3 K49/87Q (1–124) and STAT3 K49/87R (1–124) were made in pECFP-Nuc by PCR reaction using full length template STAT3 WT, STAT3K49/87Q, STAT3 K49/87R in pEF6-V5, respectively. The primer pairs used were: sense primer 5′ CATCGATGGATCCATGGACTACAAAGACGATG ACGATAAGGCCCAATGGAATCAGCTACAG 3′ and antisense primer 5′ CGTACCTCTAGACTACTGGGCCGCAGTGGCTGCAGTCTG 3′.
Fusion protein Gal4-STAT3(1–770) and UAS-LUC reporter were previously described (12).
FLAG- tagged wild-type HDAC class I expression vectors and reporter plasmids (hAPRE1)5-LUC were previously described (11,28). FLAG-tagged HDAC1 deletion mutants (aa 1–482, 1–140 and 141–482) were constructed in pcDNA3 FLAG expression plasmid using the primer pairs as follows: For HDAC1 (aa 1–482), 5′-TGCTCAGGATCCGCGCAGACGCAGGGCACCCGG-3′ and 5′-GCTACTAAGCTTTCAGGCCAACTTGACCTCCTC-3′ were used, for HDAC1 (aa 1–140), 5′-TGCTCAGGATCCGCGCAGACGCAGGGCACCCGG-3′ and 5′-GCTACTAAGCTTTCACAGGCCCCCAGCCCAATT-3′ were used and for HDAC1 (aa 141–482), 5′-TGCTCAGGATCCCACCATGCAAAGAAGTCCGAG-3′ and 5′- GCTACTAAGCTTTCAGGCCAACTTGACCTCCTC-3′ were used.
Transient transfections in exponentially growing HepG2 were performed using Lipofectamine PLUS reagent (Life Technologies, Inc.). One microgram of indicated promoter reporter was cotransfected with the transfection efficiency control plasmid pSV2PAP, and indicated expression plasmids into 6-well plates (2.5 × 105 cells). Twelve hours later, cells were stimulated with IL-6 (8 ng/ml, 24 h) before harvest and luciferase assay and alkaline phosphatase activity were measured. All transfections were carried out in triplicate plates in three independent experiments.
siRNA transfection
Control and HDAC1 siRNA (Dharmacon Smart Pools) were transfected into HepG2 cells by TransIT-siQUEST transfection reagent (Mirus, Madison, WI) at 50 nmol/l final concentration, following the manufacturer's instruction. After 48–72 h, cells were stimulated with IL-6 for different time periods as indicated.
Antibodies and immunoprecipitation
Sources of primary antibody (Ab) included Santa Cruz Biotechnology for anti-STAT3 (K15 and C20) and anti-phospho-Tyr STAT3 (B7); New England Bio Labs for anti-HDAC1 Ab; anti-V5 and anti-FLAG Ab were obtained from Invitrogen and Sigma respectively. Anti-acetyl K87STAT3 Ab was generated and affinity purified as described earlier (11).
For immunoprecipitation and Western blot analysis, either whole cell extracts (WCEs) or Nuclear Extracts (NEs) were used as indicated. WCEs were prepared by lysing HepG2 cells in modified RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.25% sodium deoxycholate, 1% Nonidet P-40, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4, and 1 µg/ml each of aprotinin, leupeptin, and pepstatin). Sucrose cushion-purified NEs were prepared by lysing HepG2 cells with nonionic detergent (0.5% IGEPAL-60) and centrifugation over a sucrose cushion as described (28). Extracts were precleared with protein A-Sepharose 4B (Sigma) for 10 min at 4°C and the cleared lysate incubated with primary Ab for 2–12 h at 4°C. Immune complexes were captured by adding 30 µl of protein A-Sepharose beads (50% slurry) and rotated for 1 h at 4°C. Beads were washed three times for 5 min with cold PBS, immune complexes eluted by incubation in SDS–PAGE loading buffer and fractionated by 10% SDS–PAGE. Proteins were transferred to polyvinylidine difluoride membranes (Millipore, Bedford, Mass.) and blocked with 5% milk for 1 h followed by primary antibody treatment for overnight at 4°C. Membranes were washed in TBST (0.1%) tween and incubated with secondary antibody for 1 h. Signals were detected by the enhanced chemiluminescence assay (ECL, Amersham) or visualized by the Odyssey Infrared Imaging system (LICOR Biosciences, Lincoln, NE).
Real-time RT–PCR analysis
Total cellular RNA was extracted by Tri Reagent (Sigma). One microgram of RNA was used for reverse transcription using iScript cDNA Synthesis Kit (Bio Rad). Three microliters of cDNA products was amplified in 20 μl reaction system containing 10 μl iQ SYBR Green Super Mix (Bio Rad) and 400 nM primer mix. All the primers were designed by Primer Express version 2.0 software. For Human MCP-1 mRNA expression, forward primer 5′-CATTGTGGCCAAGGAGATCTG-3′ and reverse primer 5′-CTTCGGAGTTTGGGTTTGCTT-3′ were used. All reactions were processed in MyiQ Single-Color Real-Time PCR Detection System (Bio Rad) and results were analyzed by IQ5 program (Bio Rad). To normalize template input, GAPDH (endogenous control) transcript level was measured for each sample. Data are expressed as fold change relative to unstimulated, after normalizing to GAPDH.
Immunofluorescence staining
Cells were acetone-methanol fixed, rinsed in PBS containing 50 mM NH4Cl for 10 min, and permeabilized in PBS containing 0.1% Triton X-100. Cells were then blocked with pre-immune heterologous serum (diluted 1 : 10 in PBS) for 30 min, washed, and incubated with primary and secondary (FITC and Rhodamine (Rd)-conjugated) Ab. After staining, cells were mounted in antifading mounting solution (DAKO Inc. Carpentaria, CA). Confocal microscopy was performed on a Zeiss LSM510 META System using the 488 nm line of the Argon-laser for excitation of FITC and Helium-Neon 543 nm line for excitation of Rd, combined with appropriate dichroic mirrors and emission band filters to discriminate between green and red fluorescence. Images were captured at a magnification of ×60 (60× oil immersion objective numerical aperture 1.4). Co-localization was visualized by superimposition of green and red images using MetaMorph software Version 4.6r9 (Universal Imaging Corp).
In vitro deacetylation assay
Wild type (aa 1–770) and NH2-terminal deleted (aa 130–770) V5-tagged STAT3 plasmids were expressed in HepG2 cells and in vitro deacetylation assay (UPSTATE Biochemicals) was performed following the protocol as recommended by the supplier.
RESULTS
HDAC1 interferes with STAT3-dependent transactivation
Earlier we showed by nondenaturing coimmunoprecipitation assay that STAT3 binds HDAC1 in an IL-6-dependent manner and interferes with hAGT expression (11).
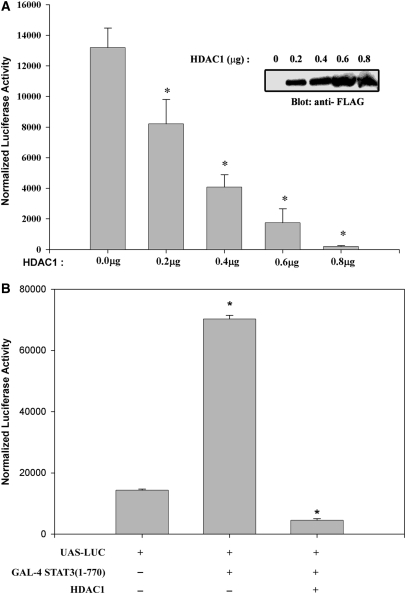
To gain further insights into how IL-6-induced STAT3–HDAC1 interaction could control target gene repression, we examined the effect of increasing doses of ectopically expressed HDAC1 on IL-6 inducible transcription using a high affinity STAT3-binding site termed (hAPRE1)5-LUC reporter plasmid, in HepG2 cells (28). Relative to empty vector control, cells transfected with HDAC1 showed significant inhibition of IL-6-mediated hAGT promoter activity in a dose-dependent manner (Figure 1A). A significant reduction of promoter activity was seen with transfection of only 0.2 µg of HDAC1 expression plasmid and a >90% reduction with 0.8 µg. This confirms our previous observation of the inhibitory effect of HDACs in IL-6-induced transactivation (11).
Figure 1.
(A) Dose-dependent inhibition of IL-6-mediated target gene expression by histone deacetylase 1 (HDAC1). HepG2 cells were transiently transfected with (hAPRE1)5-LUC reporter plasmid, pSV2-PAP internal control, and indicated amounts of HDAC1 expression vectors (200, 400, 600 and 800 ng, respectively). Twenty four hours after transfection, cells were stimulated with IL-6 (8 ng/ml). Luciferase activity was measured after 24 h of stimulation and normalized to alkaline phosphatase activity. Amount of transfected DNA was kept equivalent using an empty expression plasmid. *P < 0.05 relative to empty vector, t-test. Ectopic expression of FLAG-tagged HDAC1 in transient transfection is shown by Western blot (inset). (B) Transrepression of STAT3 with HDAC1. HepG2 cells were transfected with UAS-LUC and either Gal4-STAT3 (full length, 1–770) expression plasmid or Gal4- STAT3 and HDAC1 together and stimulated with IL-6. HDAC1 repressed transactivation of UAS-LUC by GAL4- STAT3. Luciferase activity was measured after 24 h of IL-6 stimulation and normalized to alkaline phosphatase activity. *P < 0.05 relative to empty vector, t-test.
To further confirm that HDAC1 inhibits STAT3 mediated transactivation, we first assayed whether Gal4-STAT3 (1–770) could transactivate Gal4-binding sites (UAS-LUC) in HepG2 cells (Figure 1B). We observed that a 4.6-fold increase in UAS-LUC promoter activity mediated by Gal4-STAT3 (1–770), is completely inhibited by ectopic HDAC1 expression (although HDAC1 alone has no effect on UAS-LUC, data not shown), indicating the role of HDAC1 as a repressor in STAT3 transactivation (11).
The COOH-terminal domain of HDAC1 is required for target gene repression
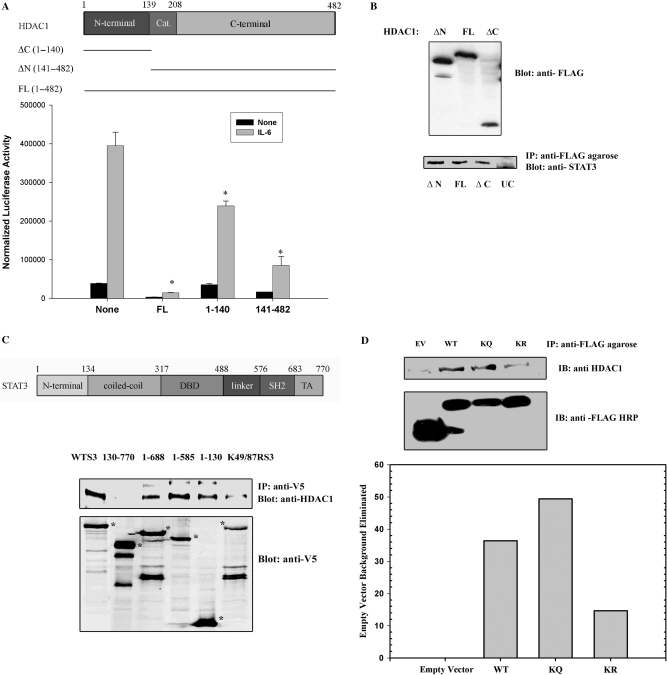
To identify the domains required for transcriptional inhibition, FLAG epitope-tagged HDAC1 deletion mutants were expressed and effects were tested on STAT3-dependent transcription. For this purpose, HepG2 cells were cotransfected with (hAPRE1)5-LUC reporter plasmid and expression plasmids encoding either with FLAG-tagged HDAC1 full length (FL, aa 1–482) or the HDAC1 NH2-terminal regulatory domain (ΔC, aa 1–140) or the HDAC1 carboxy-terminal domain (ΔN, aa 141–482). In absence of HDAC1, IL-6 strongly induced (hAPRE1)5-LUC reporter activity by 8-fold (Figure 2A). Cotransfection of HDAC1 full length strongly inhibited both basal and IL-6-induced reporter activity. By contrast, the HDAC1 NH2-terminal regulatory domain had little effect. Expression of the HDAC1 COOH-terminal decaetylase and phosphorylation domain showed a strong inhibitory effect on STAT3 dependent transcription. This result suggested to us that the histone deacetylation activity of HDAC1 is closely linked to its transcriptional repression activity.
Figure 2.
(A) HDAC1 COOH-terminal domain is required for IL-6 induced repression of target gene. Schematic diagram of HDAC1 domains (top panel). Amino acid residues (1–139) represents NH2-terminal domain, aa residues (140–208) represents catalytic domain and aa residues (209–482) represents COOH-terminal domain. HepG2 cells were transfected with (hAPRE1)5-LUC reporter plasmid and either with HDAC1 full-length expression vector (FL, aa 1–482), or COOH-terminal deleted HDAC1 (ΔC aa 1–140) or NH2-terminal deleted HDAC1 (ΔN aa 141–482) expression vectors were transfected (bottom panel). Twenty four hours after transfection cells were simulated with IL-6. Luciferase activity was measured after 24 h. *P < 0.05 relative to empty vector, t-test. (B) HDAC1 deleted mutants (ΔC and ΔN) binds to STAT3. HCT 116 cells were transfected with FLAG-tagged HDAC1 FL, or HDAC1 ΔN or HDAC1 ΔC deletion mutants. IL-6 stimulated WCEs were immmunoprecipitated with anti-FLAG conjugated agarose Ab followed by Western immunoblot with anti-STAT3 Ab (lower panel), UC represents untransfected control lane. Upper panel shows the expression of FLAG-tagged HDAC1 mutants. (C) Mapping of HDAC1-binding site on STAT3. Top panel shows the schematic diagram of STAT3 domains. HCT 116 cells were transfected with either full-length STAT3-V5 expression vector, (aa 1–770) or with different deletion mutants of STAT3 (aa 130–770, 1–130, 1–585, 1–688) or with acetylation-deficient STAT3 mutant (STAT3K49R/K87R). IL-6 stimulated WCEs were immunoprecipitated with anti-V5 Ab and Western immunoblots were performed with anti-HDAC1 Ab (upper panel). Lower panel shows expression of different V5-tagged STAT3 mutants. *P < 0.05 relative to empty vector, t-test. (D) Association of FLAG-tagged WT/KQ/KR STAT3 (1–124) with endogenous HDAC1. HCT 116 cells were transfected with FLAG-tagged STAT3 WT (1–124), STAT3 K49/87Q (1–124) and STAT3 K49/87R (1–124). WCE were immunoprecipitaed with FLAG-agarose conjugate and immune-complexes were detected with anti-HDAC1 antibody. Lower bar diagram shows the quantitation of bands (from upper panel) that indicate immunoprecipitated HDAC1. IB represents immune blot. EV represents empty vector.
We next performed coimmunoprecipitation studies to localize the domains of HDAC1 that bind to endogenous STAT3. FLAG-epitope tagged HDAC1 FL (aa 1–482), HDAC1 ΔC (aa 1–140) and HDAC1 ΔN (aa 141–482) were transfected in HCT 116 cells and IL-6 stimulated WCEs were immunoprecipitated with anti-FLAG conjugated agarose beads. Western immunoblot analysis with anti-STAT3 antibody shows that both NH2-terminal domain and COOH-terminal domain of HDAC1 bind to endogenous STAT3, although these domains had differential repression effect on STAT3 transcription (Figure 2B).
HDAC1 binds to the NH2-terminus of STAT3
To map the STAT3 domains involved in HDAC1 complex formation, we transfected HepG2 cells with different V5-tagged deletion mutants of STAT3 (aa 1–770, aa 1–688, aa 1–585, aa 1–130 and aa 130–770). Whole cell extracts were isolated and subjected to non denaturing immunoprecipitation with anti-V5 Ab. After SDS-PAGE fractionation, Western immunoblots were done with anti-HDAC1 Ab to detect associated endogenous HDAC1. Full-length STAT3 (aa 1–770) and the COOH-terminal deletion mutants (aa 1–130), (aa 1–585), (aa 1–688), all bind endogenous HDAC1 whereas the STAT3 NH2 terminal deletion mutant (aa 130–770) failed to bind (Figure 2C). This finding suggested that the NH2-terminal domain of STAT3 was necessary and sufficient for HDAC1 interaction.
NH2-terminal acetylation of STAT3 is necessary for HDAC1 binding
Next, we tested the requirement of STAT3 NH2-terminal acetylation for its in vivo interaction with HDAC1 by transfecting HepG2 cells with a full-length STAT3 acetyl-deficient mutant (STAT3 K49R/K87R). Figure 2C, lane 6 shows that efficient binding of HDAC1 with STAT3 requires acetylation of STAT3 since STAT3 K49R/K87R apparently weakly binds to endogenous HDAC1. However, because the expression of STAT3 K49R/K87R were reduced relative to those of wild type (WT) STAT3 we conducted further experiments with FLAG epitoped tagged NH2 terminal domain of STAT3, where the level of expression was comparable. For this experiment, FLAG-WT STAT3 (1–124), pseudoacetylated STAT3 (K49/87Q) (1–124) and acetyl deficient STAT3 (K49/87R) (1–124) were transfected and complexes were immunoprecipitated with anti-FLAG agarose beads (Figure 2D). We noted that endogenous HDAC1 bound with the WT- and pseudoacetylated STAT3, whereas the acetyl mutant STAT3 bound very weakly.
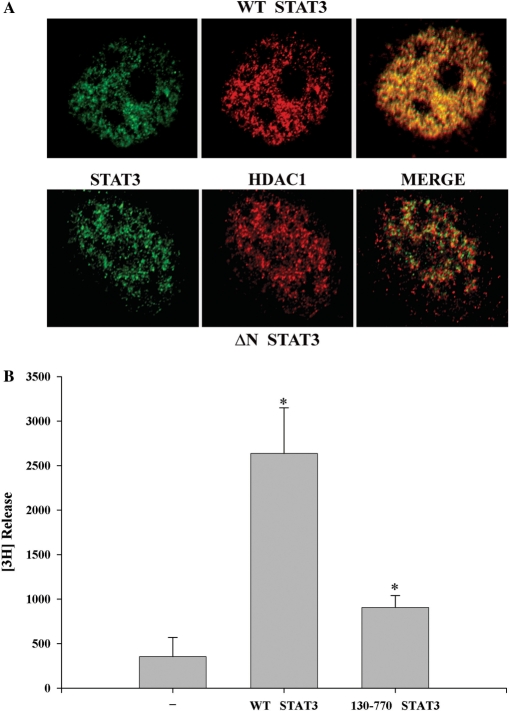
Next, we performed confocal immunohistochemistry to study colocalization of WT STAT3 and NH2-terminal deleted STAT3 (130–770) with endogenous HDAC1. Expression vectors encoding WT-STAT3 and STAT3 (130–770) were transfected in HepG2 cells and stimulated with IL-6. Both HDAC1 (red) and STAT3 (green) were distributed throughout the nucleus and are excluded from the nucleoli (Figure 3A). The merged image showed that WT STAT3 colocalized with endogenous HDAC1, whereas the NH2-terminal deleted STAT3 mutant poorly localized, further confirming requirement of the NH2-terminal STAT3 in HDAC1 binding. Together, these results indicate that NH2-terminal domain of STAT3 as well as its acetylation is necessary for stable complex formation with HDAC1.
Figure 3.
(A) Localization of STAT3 with HDAC1. HepG2 cells were transfected with either V5-tagged WT STAT3 (1–770) (upper panel) or NH2-terminal deleted STAT3 (130–770) (lower panel) and stimulated with IL-6 for 30 min. Cells were fixed and incubated with anti-V5 Ab and antibody to HDAC1. Binding of primary antibody was detected by FITC- or Texas red-labeled secondary antibody. Confocal microscopy was performed on a Zeiss LSM510 META System using the 488 nm and 543 nm excitation for FITC and Texas red, respectively. Images were captured at a magnification of ×60. Co-localization was visualized (in the right panel, merge) by superimposition of green and red images using MetaMorph software. (B) Histone deacetylase activity is associated with STAT3 immune complex. HepG2 cells were transfected with either control empty vector, or full-length V5-tagged STAT3 or STAT3 NH2-terminal deleted mutant (aa 130–770) that weakly binds HDAC1. IL-6 stimulated WCEs were immunoprecipitated with anti-V5 Ab and these immune complexes were used for in vitro deacetylation assay to decaetylate [3H]-labeled histone H4 peptide (substrate), as described in the deacetylation assay kit protocol (UPSTATE Biochemicals). *P < 0.05 relative to empty vector control extract, t-test.
We next sought to determine whether the STAT3–HDAC1 complex is associated with HDAC activity. For this purpose, we developed an immunoprecipitation-in vitro deacetylation assay. Wild type (aa 1–770) V5-tagged STAT3 and NH2-terminal deleted STAT3 (aa 130–770) were cotransfected with HDAC1 expression vector and IL-6 stimulated HepG2 WCEs were immunoprecipitated with V5 antibody. The washed immune complexes were then incubated in vitro with [3H]-labeled histone H4 (aa 2–24) peptide and deacetylation activity was measured by [3H] release. We observed strong histone deacetylase activity in the immunoprecipitated V5-tagged full length STAT3 complexes from IL-6 stimulated HepG2 cells. Most strikingly, the 130 aa NH2-terminal deletion of STAT3 (130–770), a mutation that does not bind HDAC1 (Figures 2C, 2D and 3A), showed very little HDAC activity. This data further indicated that the STAT3 NH2-terminus is essential for the functional recruitment of histone deacetylases (Figure 3B).
HDAC1 controls nuclear STAT3 abundance
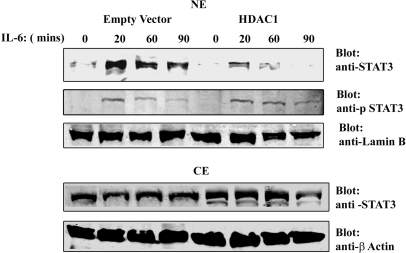
To verify the role of HDAC1 in STAT3 transcriptional activity, IL-6 responses were evaluated in cells where HDAC1 expression level was either enhanced or reduced. For this, HepG2 cells were transfected with HDAC1 expression vector or empty vector as control and stimulated with IL-6 for 0, 20, 60 and 90 min. Nuclear and cytoplasmic distribution of endogenous STAT3 protein was then determined by Western immunoblot analysis with anti-STAT3 Ab (Figure 4). In empty vector transfected cells, IL-6 induced a rapid accumulation of STAT3 within 20 min of stimulation, concomitant with the nuclear appearance of phospho-Tyr705 STAT3. In contrast, HDAC1 overexpression reduced nuclear STAT3 (total) abundance after IL-6 stimulation compared to empty vector control although the phospho-Tyr705 STAT3 level remained the same. Because HDAC1 reduced the nuclear accumulation of STAT3 and enhanced its cytoplasmic abundance, these data suggested to us that HDAC1 is involved in STAT3 nucleocytoplasmic partitioning.
Figure 4.
Rapid nuclear export of endogenous STAT3 after HDAC1 overexpression. HepG2 cells were transfected with either empty vector or HDAC1 expression vector. Twenty-four hours after transfection, cells were stimulated with IL-6 for indicated times. NEs (top panel) and CEs (bottom panel) were separated by SDS–PAGE and Western immunoblot analysis were performed with anti-STAT3 Ab and anti-phospho-Tyr705 STAT3 Ab (NE only). Lamin B and β Actin Abs were used as nuclear and cytoplasmic marker, respectively.
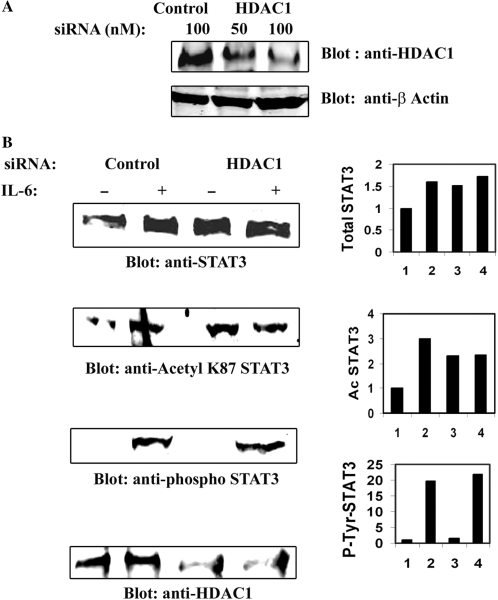
HDAC1 knockdown accumulates acetylated STAT3 in the nucleus
To further prove the involvement of HDAC1 in STAT3 nucleocytoplasmic distribution, the effect of reducing endogenous HDAC1 level was investigated. For this purpose, RNA interference was used to reduce HDAC1 expression in HepG2 cells, where HDAC1 levels were reduced by more than 70%, compared to control siRNA (Figure 5A). IL-6-dependent nuclear translocation of STAT3 was then determined by Western immunoblot analysis after HDAC1 knockdown. HepG2 cells were transfected with either control siRNA or HDAC1 specific siRNA and IL-6 stimulated prior to NE preparation. Western blots of NEs were probed with either anti-STAT3 Ab, or anti- phospho-STAT3 Ab or with anti-acetyl K87 STAT3 Abs (11). Figure 5B indicates that, in the presence of control siRNA, STAT3 is efficiently translocated from cytoplasm into the nucleus. In presence of HDAC1 siRNA, both the total STAT3 and acetylated STAT3 accumulated in the nucleus even in the absence of stimulation. Interestingly, we did not observe any change in abundance of phospho-Tyr705 STAT3 in the nucleus in the presence of HDAC1 siRNA. This above result of nuclear retention along with that from the HDAC1 overexpression study (Figure 4) strongly implicate that HDAC1 plays an important role in nucleocytoplasmic partitioning of STAT3.
Figure 5.
(A) HDAC1 knockdown in HepG2 cells by specific siRNA. HepG2 cells were transfected with indicated amounts of control and HDAC1 siRNA (Smart Pool from Dharmacon Inc.). WCEs were isolated 72 h after transfection and Western immunoblots were performed with anti-HDAC1 Ab (upper panel). Lower panel shows expression of β Actin as internal control. (B) HDAC1 knockdown accumulates STAT3 in the nucleus. HepG2 cells were transfected with either control siRNA or HDAC1 siRNA for 72 h. IL-6 stimulated (20 min) NEs were prepared and separated by SDS–PAGE followed by Western immunoblot analysis with anti-STAT3 Ab, anti-AcLys87 STAT3 Ab, or anti-phospho-Tyr705 Ab. Lower panel shows expression of HDAC1 after HDAC1 siRNA knockdown. Band intensity were plotted and shown in right-hand side panel.
Subcellular localization of STAT3 after HDAC1 knockdown
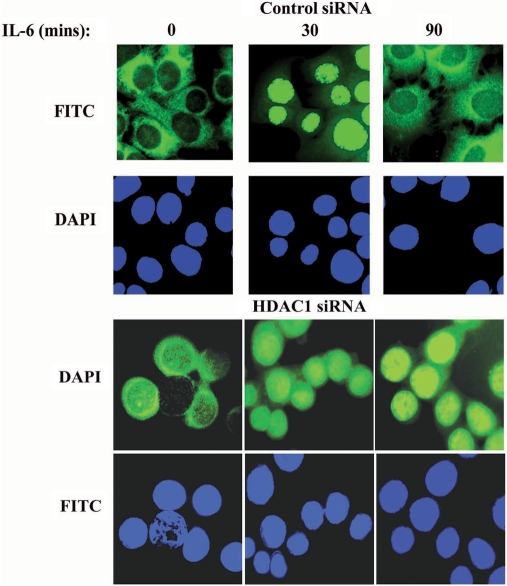
We next assessed the dynamics of cytoplasmic-nuclear translocation of STAT3 by confocal microscopic imaging on HepG2 cells transfected either with control siRNA or with HDAC1 siRNA, in presence or absence of IL-6. In control siRNA-treated unstimulated cells, the majority of STAT3 was detected in the cytoplasm (Figure 6). Treatment of cells with IL-6 for 30 min altered the equilibrium between the nuclear and cytoplasmic STAT3 and induced a complete nuclear translocation of STAT3 and which again redistributed back into the cytoplasm after 90 min of stimulation. Similarly, with HDAC1 siRNA treatment, although we observed some basal level of STAT3 in the nucleus and in the perinuclear region of the resting cells, STAT3 rapidly translocated in the nucleus from cytoplasmic compartment after 30 min of IL-6 stimulation, and it remained accumulated in the nucleus even after 90 min of IL-6 treatment, a time when STAT3 normally exported back into the cytoplasm (upper panel). This data further supports our above conclusion that HDAC1 is involved in STAT3 nucleocytoplasmic partitioning.
Figure 6.
Accumulation of nuclear STAT3 after HDAC1 knockdown. HepG2 cells were transfected with either control siRNA or HDAC1 siRNA. Seventy-two hours later cells were stimulated with IL-6 for indicated time. Cells were fixed and stained with either fluorescein isothiocyanate (FITC)-anti-STAT3 Ab. Staining with FITC-anti-AcK87 STAT3 Ab (data not shown) had similar result. Nuclei were stained with DAPI. Confocal immunofluorescence of representative cells is shown.
HDAC1 knockdown interferes with the responsiveness of STAT3-dependent target gene activation
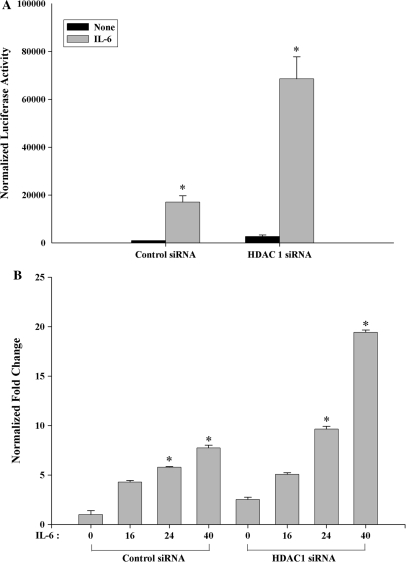
To determine HDAC1's role in IL-6 inducible transcription, cells transfected with HDAC1 siRNA were subsequently re-transfected with STAT3-dependent (hAPRE1)5-LUC reporter. As seen in Figure 7A, IL-6 treatment increased STAT3 dependent (hAPRE1)5-LUC luciferase activity by 10–12-fold in control siRNA transfected cells. In the HDAC1 knockdown cells, however, IL-6 inducible reporter activity was further increased to 30-fold, indicating that endogenous HDAC1 levels normally repress IL-6-inducible transcription. We interpret these data to mean that the effect of HDAC1 knockdown on hAGT reporter is due to reduced nuclear export, leading to nuclear accumulation of acetyl STAT3 and enhanced target gene transcription.
Figure 7.
(A) HDAC1 knockdown activates IL-6 stimulated hAGT promoter activity in HepG2 cells. HepG2 cells were transfected with either control siRNA or HDAC1 siRNA along with (hAPRE1)5-LUC reporter. Forty-eight hours after transfection cells were stimulated with IL-6 for 24 h and then harvested for luciferase reporter assay. *P < 0.05 relative to control siRNA, t-test. (B) Silencing of HDAC1 activates IL-6 regulated MCP1 expression in human monocytic THP1 cells. THP1 cells were transfected with either control siRNA or HDAC1 siRNA. Thirty-six hours after transfection, cells were stimulated with IL-6 for indicated time and total RNA was extracted at the same time for real-time RT–PCR analysis. *P < 0.05 relative to control siRNA, t-test.
Next, to determine whether the effect of HDAC1 on STAT3 transcription is cell type-specific or global phenomena, we measure more quantitatively the effect of HDAC1 knockdown on IL-6-inducible STAT3 target gene by real-time RT–PCR analysis. For this purpose we used human acute monocytic leukemia THP-1 cells (29) to measure the abundance of Monocyte Chemotactic Protein-1 (MCP1) mRNA after IL-6 induction for different time periods. MCP1 is known to be an IL-6-induced STAT3 downstream target gene and is expressed in human promonocytic cell lines (30). In contrast to control siRNA, the induction of MCP1 mRNA after HDAC1 knockdown was significantly increased after 24 h and 40 h of IL-6 treatment (Figure 7B).
From these results, we concluded that HDAC1 is required for the termination of IL-6-dependent transcription by STAT3 and subsequent redistribution from the nucleus.
DISCUSSION
STAT3 is the central mediator of IL-6-induced gene expression which is phosphorylated at Tyr705 by the receptor associated Janus kinases, an essential prerequisite for dimerization and nuclear translocation (1,3). In addition, an acetylation/deacetylation cascade of STAT3 has been recently identified (11,13), which is essential for its enhanced transcriptional competence. The duration and degree of gene activation in response to cytokine signals is primarily dictated by the subcellular distribution and nucleocytoplasmic shuttling of STAT3. STAT3 is predominately cyotplasmic in unstimulated resting cells. Upon cytokine stimulation, it quickly translocates to the nucleus, where it mediates the induction of immediate early genes. To control transient signal transmission by activated STAT3, re-export of STAT3 back to the cytoplasm is also regulated. Thus nuclear export of STAT3 serves to prepare the cells for its next round of signaling. Although the mechanism controlling the nuclear import of STAT3 upon cytokine stimulation is known (2,3), the mechanism by which activated STAT3 export from the nucleus is not well defined. Previous studies have shown that nuclear STAT3 is regulated by two distinct export of pathways controlled by three distinct nuclear export signal (NES) elements with various potencies, located at residues 306–318 (NES1), 404–414 (NES2), and 524–535 (NES3) (31). Among these, NES1 and NES2 are conserved with STAT1, whereas NES3 being unique to the STAT3 isoform. NES1 appears to be important for rapid post stimulation nuclear export, whereas NES2 and NES3 play important roles in controlling the basal nuclear export pathways.
Acetylation of histones and non-histone proteins are reversed by a large number of HDACs, which are ubiquitous in eukaryotic cells. Human HDAC1 and HDAC2 exist together in at least three distinct multi-protein co-repressors complexes called the Sin3, NURD/Mi2, and CoREST complexes (21,22). Many known transcriptional repressors such as YY1 and RB associate and recruit the HDAC1/HDAC2 and mSin3A corepressor complexes to target gene promoters, thereby repressing transcription (22). Although, majority of HDAC1 target genes showed reduced expression accompanied by recruitment of HDAC1 and local reduction in histone acetylation at regulatory regions, a small subset of HDAC1 target genes (Gja1, Irf1 and Gbp1) were also found that require HDAC1 activity and recruitment for their transcriptional activation (32). This regulation occurs either by direct recruitment of HDAC1 to the promoter or by indirect deacetylation of acetylated transcription factors thereby modulating their activity. Recently, posttranslationaly modified HDAC1 was found to associate with hormone-activated glucocorticiod receptor (GR) (33). Here, hormone activation of GR leads to progressive acetylation of HDAC1, an event that inhibits the deacetylase activity of the enzyme required for promoter activation.
In contrast to our observation where HDAC1 acts as a repressor for IL-6-mediated STAT3 transactivation, it has also been demonstrated that the deacetylase activity of HDAC1 (and also 2 and 3) is required for STAT1 dependent gene activation (34,35). Treatment of HDAC inhibitors (HDACi), or silencing of HDAC1, 2 and 3 negatively regulate STAT1-dependent transcriptional activation. These findings suggest that hyperacetylation of histones may negatively regulate STAT1-dependent gene activation. Consistent with these findings, it has been recently demonstrated that STAT1 undergoes acetylation upon HDACi treatment in human melanoma cell lines, which alters its interaction with HDACs, CBP and NF-κB. Acetylated STAT1 can only interact with NF-κB p65 and inhibits its DNA binding, nuclear localization and hence expression of anti-apoptotic genes (14).
In this study, we provide the first evidence for the involvement of HDAC1 on nucleocytoplasmic distribution of STAT3. We have also delineated a molecular mechanism by which STAT3 acetylation-mediated HDAC1 interaction controls partitioning of STAT3. Our earlier studies showed that IL-6-dependent acetylation of two Lys residues (Lys49 and Lys87) in the STAT3 NH2-terminus plays a critical role in transactivation of the human AGT gene (11). We also showed that HDAC1 stably interacts with STAT3 in IL-6-dependent manner and downregulates hAGT transactivation. In this study, we found that the NH2-terminal domain (aa residues 1–130) of STAT3 is required for HDAC1 binding and acetylation of this domain at Lys49 and Lys87 are necessary for HDAC1 interaction. It is interesting to note that the NH2-terminus of STAT3 is an independently folded modular domain that is highly conserved across the various STAT family members (36). The NH2-terminus is thought to play an important role in DNA-binding and protein–protein interaction. For example, NH2-terminus of both STAT1 and STAT3 have been shown to interact with the p300 acetyltransferase (13,37). In our previous study we have found that NH2-terminal acetylation of STAT3 is also required for its stable interaction with p300 (11). This evidence indicates that the HDAC1 deacetylase and the p300 acetylase interact with STAT3 in the same NH2-terminal region (aa 1–130). Together, we suggest that HDAC1 negatively regulates STAT3 transcription not only by its catalytic activity indirectly, but also inhibiting interaction with p300 and playing a key role in STAT3 transcription.
We showed earlier that the acetylation deficient STAT3 mutant also has a qualitative defect in cytoplasmic redistribution (11). This raised the possibility that acetylation of STAT3 may controls its nucleocytoplasmic distribution. In this present study, our observation of acetylation-deficient mutant STAT3 had reduced binding with HDAC1 can explain our previous observation why the STAT3 acetyl mutant had delayed nuclear export.
Three different assays were used to investigate the role of HDAC1 in the nucleocytoplasmic partitioning of STAT3. First, over expression of HDAC1 in HepG2 cells, followed by IL6-stimulation, resulted in a rapid nuclear export and hence less nuclear STAT3 abundance. Whereas in empty vector transfection, STAT3 efficiently translocates into the nucleus within 20 min of IL-6 stimulation and starts to redistribute back into the nucleus in 60–90 min, the kinetics that are markedly accelerated with overexpressed HDAC1. Second, siRNA mediated knock down of endogenous HDAC1 prolonged STAT3 nuclear translocation. Here we found accumulation of total and AcSTAT3 in unstimulated NE (because of constitutive shuttling) and this level remained the same even after IL-6 stimulation. Interestingly, we did not observe any accumulation of phospho-Tyr705 STAT3 in the nucleus of unstimulated cells indicating that dephosphorylated and acetylated STAT3 accumulated in the nucleus of resting cells in presence of HDAC1 siRNA. Although dephosphorylation of Tyr705 of STAT3 was proposed to be involved in nuclear export to a certain extent (38,39), our observation that HDAC1 overexpression/knockdown does not change the phosphorylated form of STAT3 in the nucleus, indicates that HDAC1 is not involved in the dephosphorylation of STAT3. Third, immunofluoresence studies with HDAC1 knockdown cells showed a slightly increased basal amount of STAT3 in the nucleus of unstimulated cells which increased upon IL-6 stimulation (30 min). Importantly, there was a delayed re-export of nuclear STAT3 after 90 min of IL-6 stimulation. These data further support our conclusion that HDAC1 is required for STAT3 distribution.
STATs are nucleocytoplasmic shuttling proteins that uses of both carrier-independent and carrier-mediated translocation mechanism (40). The exportin CRM1 supports nuclear export of STATs to a minor extent (39,40). Functional studies with GFP-tagged STAT3 and previously identified NESs by Bhattacharya et al. indicated that these elements (NES1, 2 and 3) direct a bonafide CRM1-dependent nuclear export. The crystal structure of STAT3 bound to DNA indicated that the NES2 (404–414) and NES3 (524–535) is either buried or partially exposed. It is likely that these elements will only regulate export after the STAT complex is dissociated from DNA or binding of others factors that affect the conformation of the protein. Thus it appears likely that structural changes associated with binding of HDAC1 to AcSTAT3 may play an important role in binding of CRM1 with nuclear export signal (NES) (31). Alternatively, it is possible that after transactivation, AcSTAT3 binds with HDAC1 for deacetylation that enhances its interaction with nuclear exporter CRM1 or with other HDACs (HDAC3 or other type II HDACs that can shuttle between cytoplasm to nucleus (41,42). However, it is interesting to note that acetylation sites of STAT3 are not located in either of these NESes. Therefore, how the AcSTAT3–HDAC1 association couples with the rapid post-stimulation nuclear export pathway will require further investigation.
In conclusion, we provide evidence that HDAC1 regulates STAT3 nucleocytoplasmic distribution at least in part by binding with acetylated STAT3 in IL-6-dependent manner. Because STAT3 is of pathological importance, and HDAC inhibitors are emerging as promising therapeutic agent against numerous diseases, their ability to modify cytokine-dependent transcriptional regulation is an important clinical consideration that might guide the development of second generation deacetylase inhibitors.
ACKNOWLEDGEMENTS
This project was supported by American Heart Association grant (0665129Y) (to S.R.) and National Heart Lung and Blood Institute R01 HL070925 (to A.R.B.). Core Laboratory support was from National Institute of Environmental Health Sciences grant P30 ES06676 (to J.Halpert).
Conflict of interest statement. None declared.
REFERENCES
- 1.Darnell JE., Jr STATs and Gene Regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JEJ. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcritptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.Horvath CM, Wen Z, Darnell J.E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 5.Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 6.D'Alessio AC, Weaver IC, Szyf M. acetylation induced transcription is required for active DNA demethylation, in methylation silenced genes. Mol. Cell Biol. 2007;27:7462–7474. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol. Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang DE, Kim KI. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol. 2005;398:491–499. doi: 10.1016/S0076-6879(05)98040-3. [DOI] [PubMed] [Google Scholar]
- 10.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 11.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 12.Ray S, Sherman CT, Lu M, Brasier AR. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol. Endocrinol. 2002;16:824–836. doi: 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- 13.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 14.Krämer OH, Baus D, Knauer SK, Stein S, Jäger E, Stauber RH, Grez M, Pfitzner E, Heinzel T. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ruijter AJM, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 17.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 18.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol. Cell Biol. 2007;13:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taplick J, Kurtev V, Kroboth K, Posch M, Lechner T, Seiser C. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol. 2001;308:27–38. doi: 10.1006/jmbi.2001.4569. [DOI] [PubMed] [Google Scholar]
- 21.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 23.Seto E, Sengupta N. Regulation of histone deacetylase activities. J. Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 24.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Madison DL, Yaciuk P, Kwok RP, Lundblad JR. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J. Biol. Chem. 2002;277:38755–38763. doi: 10.1074/jbc.M207512200. [DOI] [PubMed] [Google Scholar]
- 26.Soutoglou E, Viollet B, Vaxillaire M, Yaniv M, Pontoglio M, Talianidis I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001;20:1984–1992. doi: 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thevenet L, Mejean C, Moniot B, Bonneaud N, Galeotti N, Aldrian-Herrada G, Poulat F, Berta P, Benkirane M, Boizet-Bonhoure B. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO J. 2004;23:3336–3345. doi: 10.1038/sj.emboj.7600352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman CT, Brasier AR. Role of signal transducers and activators of transcription1 and 3 in inducible regulation of the human angiotensinogen gene by interleukin-6. Mol. Endocrinol. 2001;15:441–457. doi: 10.1210/mend.15.3.0609. [DOI] [PubMed] [Google Scholar]
- 29.Biswas P, Delfanti F, Bernasconi S, Mengozzi M, Cota M, Polentarutti N, Mantovani A, Lazzarin A, Sozzani S, Poli G. Interleukin-6 induces monocyte chemotactic protein-1 in peripheral blood mononuclear cells and in the U937 cell line. Blood. 1998;91:258–265. [PubMed] [Google Scholar]
- 30.Chen SC, Chang YL, Wang DL, Cheng JJ. Herbal remedy magnolol suppresses IL-6-induced STAT3 activation and gene expression in endothelial cells. Br. J. Pharmacol. 2006;148:226–232. doi: 10.1038/sj.bjp.0706647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J. Clin. Invest. 2003;111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol. Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J. Biol. Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 35.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl Acad. Sci. USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinkemeier U, Moarefi I, Darnell JE, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell J.E., Jr Two contact regions between Stat1 and CBP/p300 in interferon. Proc. Natl Acad. Sci. USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J. Biol. Chem. 2004;279:18998–19007. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 39.Lodige I, Marg A, Wiesner B, Malecova B, Oelgeschlager T, Vinkemeier U. Nuclear export determines the cytokine sensitivity of STAT transcription factors. J. Biol. Chem. 2005;280:43087–43099. doi: 10.1074/jbc.M509180200. [DOI] [PubMed] [Google Scholar]
- 40.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 2004;165:823–833. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escaffit F, Vaute O, Chevillard-Briet M, Segui B, Takami Y, Nakayama T, Trouche D. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol. Cell Biol. 2007;27:554–567. doi: 10.1128/MCB.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]