Abstract
The objectives of this and the following paper are to identify commonalities and disparities of the extended environment of mononuclear metal sites centering on Cu, Fe, Mn, and Zn. The extended environment of a metal site within a protein embodies at least three layers: the metal core, the ligand group, and the second shell, which is defined here to consist of all residues distant less than 3.5 Å from some ligand of the metal core. The ligands and second-shell residues can be characterized in terms of polarity, hydrophobicity, secondary structures, solvent accessibility, hydrogen-bonding interactions, and membership in statistically significant residue clusters of different kinds. Findings include the following: (i) Both histidine ligands of type I copper ions exclusively attach the Nδ1 nitrogen of the histidine imidazole ring to the metal, whereas histidine ligands for all mononuclear iron ions and nearly all type II copper ions are ligated via the Nɛ2 nitrogen. By contrast, multinuclear copper centers are coordinated predominantly by histidine Nɛ2, whereas diiron histidine contacts are predominantly Nδ1. Explanations in terms of steric differences between Nδ1 and Nɛ2 are considered. (ii) Except for blue copper (type I), the second-shell composition favors polar residues. (iii) For blue copper, the second shell generally contains multiple methionine residues, which are elements of a statistically significant histidine–cysteine–methionine cluster. Almost half of the second shell of blue copper consists of solvent-accessible residues, putatively facilitating electron transfer. (iv) Mononuclear copper atoms are never found with acidic carboxylate ligands, whereas single Mn2+ ion ligands are predominantly acidic and the second shell tends to be mostly buried. (v) The extended environment of mononuclear Fe sites often is associated with histidine–tyrosine or histidine–acidic clusters.
Metals in protein structures serve structural, catalytic, and regulatory purposes ranging from electron transfer, substrate oxidation-reduction, transport processes, and active chemistry in regulation of enzymatic activity. For recent comprehensive reviews, see refs. 1 and 2. In this and the following paper, we define and analyze the extended metal environment centering on the ligands coupled to the “second shell.” The concept of the protein second shell of a metal center has been widely appreciated. For example, in certain proteins the second shell of metal sites is stated to be mainly hydrophobic (3, 4). Others distinguish direct vs. indirect ligands, the indirect ligands connected through hydrogen bonds to one of the direct ligands (5). Our analysis of the residues of the second shell emphasizes similarities and contrasts in residue polarity vs. hydrophobicity, the relative distribution of these residues into α-helices, β-strands, and coils, their degree of solvent accessibility, their hydrogen-bonding networks, and relationships to various statistically significant residue clusters. It is of interest to compare situations where it is clear that second-shell residues are “preorganizing” direct ligands via hydrogen-bonding and other specific interactions, thereby controlling metal-binding affinity, as well as situations in which the second shell appears to have little directing function on the direct ligands. Residues in the second shell may influence a variety of structural and chemical facets of the protein, including hydrogen-bonding interactions to the ligands, pKa values of ligand groups, and the control of the metal center oxidation state and redox potential. In addition to influencing the polarity of the metal environment, neighboring side chains also exert steric and chemical controls over the ability of the metal ion to bind or discriminate substrate and to accommodate conformational changes.
The metal environment of the protein structure emphasizes four facets. (i) Metal core. This term refers to the metal type(s) in mononuclear or multinuclear sites (6).
(ii) Ligand group. Residues of the protein structure that directly bond to the metal(s). Foremost ligands of Cu, Fe, Mn, and Zn ions include an imidazole nitrogen(s) of histidine (H; one-letter code used), carboxylates (D/E), sulfur (C/M), solvent, and carbonyl oxygens. There are differences depending on the metal and function. Thus, Cu ions are never ligated by an acidic residue. Fe ions are predominantly ligated by multiple H residues in conjunction with one or two acidic residues and occasionally Y. The primary ligands of Mn2+ are often only acidic residues. The principal ligands coordinating Zn2+ ions consist of combinations of H, D, E, C, and sometimes Y, N, S, and T.
The two nitrogens in the histidine imidazole ring have the ability to donate ligand electrons to metals via either Nɛ2 or Nδ1 (7). There are bridging bidentate residues that simultaneously coordinate two metal ions or a unibidentate with two bonds (e.g., Oδ1 and Oδ2 of aspartate or side chain plus backbone) to a single metal. Copper coordination rarely includes unibidentates, whereas iron coordination often involves one or more unibidentate residues or molecules. Coordination patterns and metal–ligand bond lengths reflect the flexibility of the coordination geometry, steric constraints, stability in the coordination process, alternative electrostatics, and redox potential.
(iii) Second shell. We define the second shell to consist of all residues and non-amino-acid molecules whose “distance” to any non-hydrogen atom among the ligand group is within 3.5 Å. The distance between a residue pair in the three-dimensional (3D) protein structure is calculated as the minimum distance between side-chain atoms or between side-chain and main-chain atoms (cf. ref. 8).
(iv) Significant residue clusters. Another useful descriptor of metal environments is in terms of a distinctive 3D residue cluster that may highlight the metal ligands and additional neighboring residues (9). Types of 3D residue clusters include the following: concentrations of residues with high net positive charge (positive-charge clusters) designated {KR}, and analogously negative-charge clusters {ED}, or mixed-charge clusters {KRED}; 3D environments rich in histidine (histidine clusters) {H}, or cysteine–histidine {CH}, or histidine–acidic {HED}, or cysteine–histidine–methionine {CHM}. For other types of residue clusters, see refs. 9 and 10.
Methods for Identifying Significant Residue Clusters
Statistically significant 3D residue clusters are identified by appropriately representing a protein structure by a matrix of N (N the length of the primary sequence) linear amino acid sequences (each of length N) and evaluating these sequences for unusual residue sequence clusters (9). Specifically, from each residue ak in a structure, a sequence Sk is generated with respect to a distance measure between residues (9, 10), as follows. Method 1 (M1): in the sequence Sk = {sk,1, sk,2, sk,3, … }, sk,1 is ak, and the jth residues, sk,j, of Sk(j > 1) is the next closest residue to any of the residues sk,1, sk,2, … , sk,j−1. Thus, M1 accumulates residues in Sk, reflecting the shape and density of the structure surrounding the starting residue sk,1. Method 2 (M2): Again sk,1 is ak and the jth residue sk,j(j > 1) is the closest in the 3D structure to the previous residue sk,j−1 that is distinct from sk,1, sk,2, … , sk,j−1. M2 in generating Sk proceeds along a pathway that often reveals channels relevant for transporting suitable molecules or substrates. Method 3 (M3) incorporates residues in the order of increasing distances from ak. Using scoring theory (9), we ascertain whether or not a statistically significant high-scoring segment at 1% level occurs at the start of the sequence Sk generated from sk,1. The test is applied to every sequence Sk of the protein structures, using all three methods. Two significant residue clusters are considered the same if the smaller cluster overlaps the larger cluster in at least 50% of its residues. For precision and elaborations, see ref. 9. Of special interest are distinctive residue clusters overlapping the metal environment. However, other residue clusters separated from the metal may contribute to protein–protein interactions, to substrate channeling, to enhancing stability and/or catalysis at the active site, and to quaternary structure formation (9, 10).
Data sources of the present study are described in the legend to Table 1.
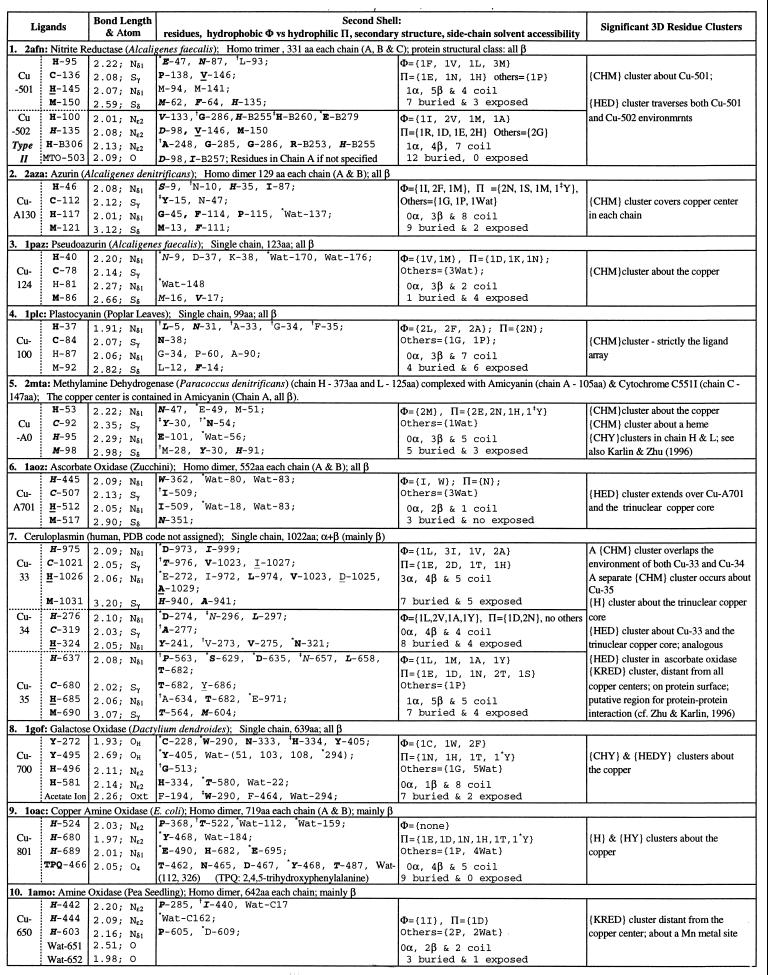
Table 1.
Single copper ions in protein 3D structures
Copper centers 1–7 are classified type I (blue copper); copper centers 8–10 are type II. A ligand residue or a residue in the second shell is shown by the one-letter amino acid code followed by chain identifier and primary sequence position (Protein Data Bank residue number). A chain identifier of a residue is not designated unless necessary. Bond lengths are in Å. Residues are underlined when the residue is in an α-helix, in italic when the residue is in a β-strand, or in ordinary font when the residue is in a coil location. Bold letters indicate that the residue side-chain atoms are buried (side-chain solvent accessibility is less than 10%). A residue in second shells is designated by the symbol ∗ when one of its side-chain atoms forms a hydrogen bond with a side-chain atom of a ligand; the symbol
when one of its main-chain atoms forms a hydrogen bond with a side-chain atom of a ligand; the symbol
when one of its side-chain atoms forms a hydrogen bond with a main-chain atom of ligand. Exclusive main chain and main chain hydrogen bonds are not indicated. Data set: a representative set of protein structures was based on the list of Hobohm and Sander (22), version of December, 1996, with pairwise primary sequence identity less than 25%. Total number of proteins in the list is 443 and those with metal, heme, or iron–sulfur linkage is 129 (29%). The structure data set was augmented with several recent protein structures known to contain metal centers.
Patterns and Contrasts of the Extended Environment for Mononuclear Copper, Iron, Manganese, and Zinc Sites
Table 1 describes the ligand, second-shell composition, and characteristics for representative Cu sites of type I (blue copper motif, electron transfer function) and type II; for general references, see refs. 6 and 11. Table 2 reviews examples of mononuclear Fe (nonheme) extended metal environments; for general references, see refs. 4, 6, and 12. The corresponding listings and descriptions for single Zn metal sites are presented in the following paper (13).
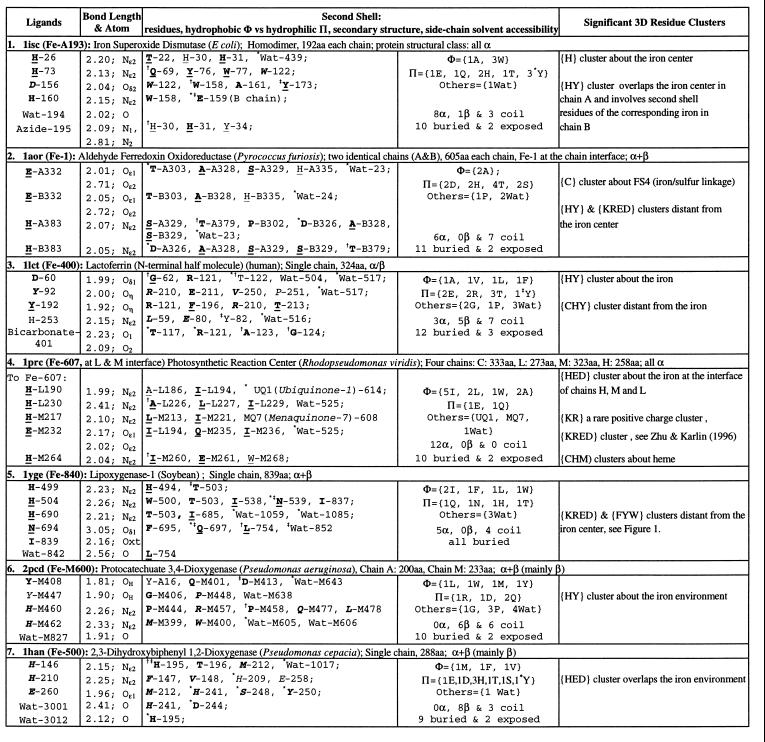
Table 2.
Single iron ions in protein 3D structures
See legend to Table 1.
Tautomeric Histidine–Metal Preferences.
Both histidine ligands of type I Cu ions adopt strictly Nδ1 nitrogen contacts, whereas histidine ligands for all mononuclear Fe ions and nearly all type II Cu ions adopt the Nɛ2 tautomeric conformation (see Tables 1 and 2). By contrast, histidine diiron tautomeric contacts are predominantly Nδ1. In fact, from the five available diiron structures, methane monooxygenase (Protein Data Bank identification number 1mmo), ribonucleotide reductase (1rib), rubrerythrin, Δ9-desaturase (dsa), and hemerythrin (2hmz), the first two and dsa are coordinated by two H and four acidic residues, rubrerythrin is coordinated by one H and four E, and hemerythrin is coordinated with five H and two acidic bridging bidentates. Hemerythrin ligates via Nɛ2 for all its five histidine ligands, whereas the other diiron associations ligate strictly via Nδ1. Histidine tautomeric metal bonding patterns for mononuclear zinc ions are variable (see ref. 13). The bonding of the trinuclear copper complexes of ceruloplasmin (hCP), ascorbate oxidase (1aoz), and the binuclear Cu ions of hemocyanin (1oxy) are ligated in all but one contact (in 1aoz) by histidines via the Nɛ2 geometry (14, 15).
The foregoing contrasts in histidine–metal tautomeric conformations raise challenging questions. There may be electronic and/or chemical differences of Nɛ2 vs. Nδ1 metal contacts—e.g., in the ability to deprotonate the N-H group. Yet, steric differences between Nδ1 and Nɛ2 may be decisive. The Nɛ2–metal interaction has the histidine main-chain displaced radially away from the metal site, whereas for the Nδ1 contact the tautomeric geometry is displaced mostly in a tangential direction. In particular, two histidine Nδ1 ligation contacts would sandwich the metal ion between them and thereby provide a more stable metal–ligand coordination and proximity to the backbone and second-shell environment. This may be a favorable situation for an electron transfer center such as in type I Cu. On the other hand, Nɛ2 ligand contacts afford more space to allow for metal-substrate catalytic interactions such as for O2 transport, O2 detoxification, or substrate oxidation occurring at mononuclear Fe and type II Cu sites. Zinc metalloproteins, including an abundance of hydrolases and transcription factors, do not engage in redox activity, and they manifest quite variable histidine tautomeric patterns (13). Notably, there are to date no examples of metalloproteins having three or more histidine ligands that involve more than one Nδ1 contact for any metal site. In particular, each iron site in diiron structures (except hemerythrin) tends to have a single histidine ligation invariably via Nδ1. The existence of a single Nδ1 contact may enhance stability of the metal–ligand complex and still leave space for other chemical interactions.
Ligand Group and Second Shell.
The familiar type I Cu2+ ion ligands consist of H, C, H, M, occurring in that order in the primary sequence and the latter three ligands subscribing to the motif CXmHXnM, m and n mostly 2–4. At least three different structural motifs occur for the ligand set. The peptides that contain the latter C, H, and M ligands form a rather similar local 3D conformation in nitrite reductase (2afn, Cu-501), ascorbate oxidase (1aoz, Cu-701), and ceruloplasmin (hCP, Cu-33 and Cu-35) (2, 16), where the cysteine is at the C-cap of a β-strand, followed by a loop, and a short α-helix containing the second histidine and the methionine is in the turn following the helix (Table 1; consult secondary structure designation of ligands). A glycine putatively structurally important to form a tight turn is observed before the methionine in the primary sequences. The corresponding ligands in azurin, pseudoazurin, and plastocyanin are all contained in a single loop. However, in amicyanin the ligands are associated with two β-strands sandwiching the Cu ion.
For type I Cu2+ ions, the polar residues of the second shell include generally at least one N (asparagine), often part of an N-S or N-T pair, and one or more acidic residues. The hydrophobic part of the second shell generally involves multiple M residues and/or one or more large aromatic residues. The type I Cu center and its direct ligands are almost always totally buried, whereas the second shell contains about 1/3 solvent-exposed residues. [A residue is considered buried if its side-chain solvent accessibility is ≤10% (17).] The exposed residues may contribute to the electron transfer function by providing pathways to an exogeneous physiological electron transfer acceptor/donor. The blue motif second-shell residues are mostly in coil elements. The solvent-accessible coil residues suggest an intrinsic relationship between the type I Cu metal environment and the protein surface.
Proteins with type II Cu sites include galactose oxidase and two copper amine oxidase structures: 1oac from Escherichia coli and 1amo from pea seedling. An essential organic molecule in these copper amine oxidases is the modified tyrosine cofactor TPQ (2,4,5-trihydroxyphenylalanine quinone), and on this basis these proteins are quinoproteins. The copper ion in chain A is tightly coordinated by H-524, H-526, H-689, and the TPQ residue. Strikingly, the second shell, practically all buried, contains no hydrophobic residues but features eight polar (including two acidic) residues and four water molecules. The two acidic residues, E-490 and E-695, are hydrogen-bonded to the ligand H-689, and Y-468 is hydrogen-bonded to ligand H-526. E-695 and H-689 interact, connecting the side-chain oxygen atom Oɛ1 of E-695 with Nɛ2 of H-689 at the distance about 2.7 Å. 1amo is a dimer homologue of 1oac, each unit containing a single Cu2+ ion and a separate Mn2+ ion not present in 1oac. Each chain of copper amine-oxidase (1oac) contains two separated Ca2+ ions. The ligands of Ca-A802 are D-533, D-535, D-678, L-534, A-679 (L and A coordinating the metal through the carbonyl oxygen), and one water molecule. The second shell consists of Φ = {2Y}, Π = {2K, R, E, N, *W}, one glycine, and two water molecules. K-133 connects with the ligand D-533 by a salt bridge involving the side-chain nitrogen Nζ of K and the side-chain oxygen Oδ2 of D-533 at 2.68 Å distance. The residue W-137 of the second shell hydrogen-bonds with D-535, involving the side-chain atoms (imino nitrogen) Nɛ1 of W and Oδ2 of D-535 (2.69 Å apart). The direct environment of Ca-A803 consists of the coordinating residues E-573, Y-677 (carbonyl ligation), D-670, E-672, and two water molecules; second shell Φ = {F}, Π = {R, E, 2N, T, H}, {1G}, 3 water.
Galactose oxidase (1gof) functions in catalysis of the stereospecific oxidation of a broad range of primary alcohol substrates. The active site is the mononuclear copper cofactor. Apart from two H, the other ligands are two Y residues and an exogenous acetate ion presumed to be the site of substrate binding (11). Eight of nine residues of the second shell are in coils and seven are buried. The nearby C-228 forms a covalent link to a ring carbon of Cu-ligand Y-272. This Y-C moiety constitutes an active-site cofactor that stores a protein oxidizing equivalent critical for catalysis. The second-shell W-290 (hydrogen-bonded to Y-272) may also serve an important role via its π-stacking with Y-272 and C-228 (11). Galactose oxidase also contains a separated sodium (Na-702) ion (ligands: K-29, D-32, N-34, T-37, A-141, E-142 mostly exposed, with the bonding atom for these residues generally the carbonyl oxygen). The second shell consists of seven exposed and two buried residues summarized by Φ = {I, F, L}, Π = {R, D, E}, others {2G, 1P} with its polar residues all charged and most of its residues located in coil elements.
Iron placements occur at the interface of heterodimer structures, aldehyde ferredoxin oxidoreductase (1aor) and the photosynthetic reaction center (1prc), with ligands and second-shell residues contributed from both chains. Lactoferrin (1lct), a member of the transferrin family, which transports iron, and the two dioxygenases 1han and 2pcd are regarded to have multiple domains where the iron placement is near the interface of these domains (cf. refs.4, 12, and 18). Iron-ligand centers tend to be buried, but access to solvent may be vital for substrate binding and catalytic processes. A metal center proximal to an interface may provide flexibility in this respect. The structure of 1aor possesses two unibidentates, E-332A and E-332B, bonded to the iron atom at the interface. Iron superoxide dismutase (1isc) has an exogenous azide bonding via two nitrogen atoms. E-232 of chain M of 1prc is a unibidentate to the Fe ion. In 1lct the bicarbonate-401 molecule bonds to Fe-400 via two oxygen molecules.
Polar residues predominate in the second shell of mononuclear Fe sites. In particular, the second shell contains one or more acidic residues (exception, 1yge). Multiple histidine residues occupy the second shell of 1aor, 1han, and 1isc. A tyrosine hydrogen bonding to an acidic ligand occurs in structures 1han, 2pcd, and 1isc. The second shell of iron superoxide dismutase (1isc) involves about equally “hydrophobic” and polar residues. The hydrophobic component highlights five aromatic residues, three W and two Y of mixed hydrophobic/hydrophilic character but no F residue. Y-173 hydrogen bonds via its hydroxyl group with the carbonyl oxygen of the ligand D-156, and W-158 hydrogen bonds through its main-chain nitrogen atom with the carboxylate of D-156. Aldehyde ferredoxin oxidoreductase (1aor) has two chains, A and B, with a single iron atom at the interface. The second shell of the Fe atom is predominantly polar but strikingly buried.
There are available six examples with a mononuclear manganese metal center (19); for details, see http://gnomic.stanford.edu/~zhu. The ligands of both Mn proteins (2chr, 370 aa, and 2mnr, 357 aa) are purely acidic, exhibiting virtually equal spacings, DX25EX24D and DX25EX25E, respectively. The second shell enclosing the acidic residues have the same hydrophobic residues Φ = {V, W, M} and many common polar residues (Π = {3K, D, 2E, N, H, S} in 2mnr and Π = {3K, E, N, Q} in 2chr). The three lysines suggest that these may help balance the excess net negative charge about the metal–ligand core.
It is interesting to compare the metal environment of the E. coli Fe-sod (1isc), that of the T. thermophilus Mn-sod (1mng), and that of human Mn-sod (1amb). All are homodimers of similar sizes with identical ligand groups consisting of 3H, D, and one bound water. Their secondary structure dispositions are almost identical. The complete primary sequences align in the range of (40–50%) identity. The human Mn-sod (1abm) and E. coli Fe-sod (1isc) significantly differ in the polar composition of the second shell (see Web site). On the other hand, the extended Mn environments of 1isc and 1mng are very similar.
Numbers of Hydrophobic (Φ) vs. Polar (Π) Residues of the Second Shell and Numbers of the Different Types of Closest Atoms.
For each mononuclear metal type, the aggregate residue counts in Φ and Π [glycine (G), proline (P), and water counted separately] are given, and the counts of the closest atoms from second shell residues to ligands [carbon (C), nitrogen (N), oxygen (O), sulfur (S)] are cumulated in Table 3.
Table 3.
Numbers of residues and closest atoms in the metal environment
| Metal | Aggregate no. of
|
Aggregate residue counts
|
No. of closest atoms
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structures | Ligands | 2nd-shell residues | Φ | ∏ | G | P | HOH | C | N | O | HOH | S | |
| Type I Cu | 9 | 35 | 86 | 38 | 34 | 2 | 4 | 8 | 25 | 7 | 52 | 9 | 0 |
| Type II Cu | 4 | 18 | 47 | 10 | 19 | 3 | 3 | 12 | 14 | 3 | 24 | 14 | 1 |
| Single Fe | 7 | 37 | 101 | 30 | 46 | 3 | 5 | 15 | 43 | 17 | 48 | 18 | 0 |
| Single Mn | 7 | 38 | 79 | 18 | 43 | 3 | 1 | 14 | 27 | 28 | 31 | 15 | 0 |
These counts (except for type I Cu) show that the second shell in aggregate favors polar residues, the closest atoms are predominantly oxygen, and in total there are many more O, N, and water contacts than C contacts. The designations in Tables 1 and 2 (see the legend of Table 1) indicate possible hydrogen-bond connections between side-chain atoms and between main-chain and side-chain atoms. The foregoing corroborates the general prevalence of polarity over hydrophobicity of the second shell.
Statistically Significant 3D Residue Clusters in Metalloprotein Structures.
Almost all the blue copper motifs are embedded in {CHM} clusters usually carrying at least two methionine and at least three histidine residues. The homotrimer nitrite reductase (2afn) features a {HED} cluster about the type I copper Cu-A501, consisting of (the notation H-A145, for example, signifies histidine at position 145 in chain A) E-A47, H-A95, Cu-A501, H-A145, C-A136, M-A150, I-A114, H-A135, Cu-A502, H-A100, MTO, H-B306, R-B253, P-B254, M-B260, DA-251, D-A98, E-B279, and H-B255 encompassing all the ligands (double underlined) and residues of the second shell (underlined); MTO is a water molecule bound to Cu-A502. Strikingly, the cluster {HED} simultaneously envelops in chain A the blue copper Cu-501 direct ligands together with the direct ligands of Cu-502 of type II, plus some residues of the second shell and other residues (E and D are not ligands). Thus, these two copper ions interrelate and putatively function cooperatively, allowing electron transfer from Cu-501 to Cu-502, where the nitrite substrate binding and reduction occurs (16).
Azurin (2aza), thought to transfer electrons from cytochrome c-551 to cytochrome oxidase, contains a highly significant {CHM} cluster embodying residues of both chains (from chain B: six M, three H, two C, two Y, three G, P, F, S, and D and from chain A: four M, three H, C, S, and F) and covering the Cu sites of both chains. The large number of methionine residues and aromatic residues in the cluster with many glycine fillers is striking. Azurin is of all β-structure class, yet most of the second shell are residues of coil elements. Pseudoazurin (2paz) and plastocyanin (1plc) contain a significant {CHM} cluster enveloping the blue copper ion. Among type II copper sites galactose oxidase contains a highly significant {CHY} cluster about the copper ion.
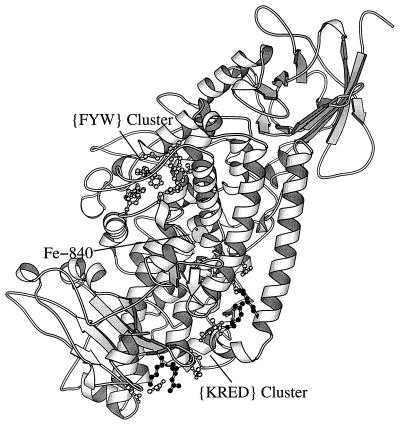
The structure (1aor) contains an iron–sulfur (FeS4) linkage ligated by four cysteines and further possesses a {HY} cluster distant from the Fe site. In this structure, there is a significant mixed charge cluster {KRED} with residues contributed from chains A and B. This mixed-charge cluster is part of the cleft formed by chains A and B, however, with no overlap to the iron environment. Lactoferrin (1lct) possesses a {HY} cluster about the Fe-400. The protein also possesses a cysteine {C} cluster distant from the iron center juxtaposing three disulfide bridges: C-157—C-173, Y-189, P-159, L-172, C-198—C-115, E-187, and C-181—C-170. This may have a role in nucleating the protein conformation and in structural stability (9). The photosynthetic reaction center (1prc) features a {HED} cluster about the iron site. Distant from the Fe center of lipoxygenase (1yge), there is a significant exposed mixed charge cluster {KRED} (Fig. 1). Is it possible that this mixed-charge cluster is a region of protein–protein interaction or a help in mediating the movement of the lipophilic substrate toward the active metal center? An aromatic cluster {FWY} (Fig. 1) may facilitate formation of a hydrophobic cavity or channel for moving the lipid substrate to the active site (6, 20). The dioxygenase structures 2pcd and 1han feature an {HY} and {HED} cluster, respectively, near the iron site.
Figure 1.
Lipooxygenase-1 contains an aromatic {FYW} cluster and a mixed charge {KRED} cluster. Both clusters are distant from the iron center.
Despite the similar metal-ligand environments in {2chr} and {2mnr}, these manganese proteins contrast sharply away from the Mn site. For example, 2chr carries two distinct exposed mixed-charge clusters. Both {KRED} clusters intriguingly consist of exactly the same charged residues two R, one K, two E, and three D in separate regions of the protein surface. By contrast, 2mnr possesses no significant residue cluster of any kind.
Dispelling a Dogma.
There is a commonly held belief that the second shell of metal binding sites is predominantly hydrophobic (e.g, see refs. 3 and 4). However, our analysis shows that the second-shell composition depends on the metal type and the metal multiplicity. A hydrophobic shell putatively inhibits solvent ions and/or water from reaching the active site while accommodating nonpolar groups, whereas a channel coated with polar residues would facilitate movement of ions and/or polar substrates to or from a metal active site. A hydrophobic second shell apparently offers stronger contrasts in the immediate vicinity of the metal by causing changes in electric polarizability and by allowing tuning of the metal center redox potential and electron transfer rate. The detailed structure is important. In some cases polar molecules could concentrate at the surface, but aliphatic portions of these same residues could contribute to a hydrophobic shielding environment. For type I Cu2+ ion environments, we find that nitrite reductase and plastocyanin show a hydrophobic prevalence in the second shell but three others do not. Among the type II copper centers, only the second shell of nitrite reductase (2afn) (Cu-502) at the interface is predominantly hydrophobic. Strikingly, the second shell of the E. coli copper amine oxidase (1oac), although completely buried, is devoid of hydrophobic residues. Of single-iron centers only the photosynthetic reaction center (1prc) has a large majority of hydrophobic residues in the second shell. The mononuclear iron placements of 1aor and of 1prc are both at an interface of two protein units, but these second shells drastically differ in the degree of polarity. More than 80% of zinc-ligand surroundings, including carbonic anhydrase II (2caa), enolase (4enl), and thermolysin (8tln), are predominantly hydrophilic. They are often associated with histidine–acidic clusters and charge residues (13). The hydrophobic component of the second shell of Cu,Zn superoxide dismutase (2sod) is virtually all aromatic, highlighting Y and W residues (data not shown). The second shells of the diiron carboxylate protein structures methane monooxygenase (1mmo), R2 ribonucleotide reductase (1rib), rubrerythrin, Δ9-desaturase, and hemerythrin (2hmz) (cf. ref. 21) are very different. Specifically, the second shell of 1mmo is predominantly polar, that of 1dsa is modestly polar, that of 1rib and 1rub are equally polar vs. hydrophobic, and that of 2hmz carries a preponderance of hydrophobic residues (data not shown).
Conclusion
Extensive comparisons of metalloprotein structures from analysis of x-ray data, especially among metal environments, yield new observations and insights. In particular, we ascertained statistically significant residue clusters that envelop the ligand group and identify other residue clusters separated from the metal. Commonalities and contrasts of tautomeric histidine–metal preferences and composition of the second shell are dependent on metal and functional types. Further analysis and experimental approaches should aid the understanding of how these newly observed 3D elements contribute to protein function.
Acknowledgments
We convey our appreciation for valuable comments on the manuscript by Drs. B. E. Blaisdell, J. Griffin, E. Solomon, and W. Weiss from Stanford University. S.K. was supported in part by National Institutes of Health Grants 5R01GM10452-33 and 5R01HG00335-09 and National Science Foundation Grant DMS9403553-002. K.D.K. was supported in part by National Institutes of Health Grant GM28962.
ABBREVIATION
- 3D
three-dimensional
References
- 1.(1991) Adv. Protein Chem. 42.
- 2.Holm R H, Solomon E I, editors. Chemical Reviews: Bioinorganic Enzymology. 1996; 1996. , Nov. issue. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita M M, Wesson L, Eisenman G, Eisenberg D. Proc Natl Acad Sci USA. 1990;87:5648–5652. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard J B, Rees D C. Adv Protein Chem. 1991;42:199–273. doi: 10.1016/s0065-3233(08)60537-9. [DOI] [PubMed] [Google Scholar]
- 5.Christianson D W, Fierke C A. Acc Chem Res. 1996;29:331–339. [Google Scholar]
- 6.Holm R H, Kennepohl P, Solomon E I. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti P. Protein Eng. 1990;4:57–63. doi: 10.1093/protein/4.1.57. [DOI] [PubMed] [Google Scholar]
- 8.Karlin S, Zuker M, Brocchieri L. J Mol Biol. 1994;239:227–248. doi: 10.1006/jmbi.1994.1365. [DOI] [PubMed] [Google Scholar]
- 9.Karlin S, Zhu Z-Y. Proc Natl Acad Sci USA. 1996;93:8344–8349. doi: 10.1073/pnas.93.16.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z-Y, Karlin S. Proc Natl Acad Sci USA. 1996;93:8350–8355. doi: 10.1073/pnas.93.16.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinman J. Chem Rev. 1996;96:2541–2561. doi: 10.1021/cr950047g. [DOI] [PubMed] [Google Scholar]
- 12.Que L, Jr, Ho R Y N. Chem Rev. 1996;96:2607–2624. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 13.Karlin S, Zhu Z-Y. Proc Natl Acad Sci USA. 1997;94:14231–14236. doi: 10.1073/pnas.94.26.14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messerschmidt A, Ladenstein R, Huber R, Bolognesi M, Avigliano L, Petruzzelli R, Rossi A, Finazzi-Agro A. J Mol Biol. 1992;224:179–205. doi: 10.1016/0022-2836(92)90583-6. [DOI] [PubMed] [Google Scholar]
- 15.Messerschmidt A. Adv Inorg Chem. 1993;40:121–185. [Google Scholar]
- 16.Murphy M E P, Turley S, Kukimoto M, Nishiyama M, Horinouchi S, Sasaki H, Tanokura M, Adman E T. Biochemistry. 1995;34:12107–12117. doi: 10.1021/bi00038a003. [DOI] [PubMed] [Google Scholar]
- 17.Richmond T J, Richards F M. J Mol Biol. 1978;119:537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- 18.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 19.Dismukes G C. Chem Rev. 1996;96:2909–2926. doi: 10.1021/cr950053c. [DOI] [PubMed] [Google Scholar]
- 20.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin J T, Walter R, Axelrod B. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 21.Kurtz D M., Jr J Biol Inorg Chem. 1997;2:159–167. [Google Scholar]
- 22.Hobohm U, Sander C. Protein Sci. 1994;3:522–524. doi: 10.1002/pro.5560030317. [DOI] [PMC free article] [PubMed] [Google Scholar]