Abstract
This review compares the folding behavior of proteins and RNAs. Topics covered include the role of topology in the determination of folding rates, major folding events including collapse, properties of denatured states, pathway heterogeneity, and the influence of the mode of initiation on the folding pathway.
Keywords: protein structure/folding, RNA folding
Proteins and RNAs must fold into specific structures in order to carry out their functions within the cell. For both biomolecules, the stability and specificity of the native fold results from the burial of hydrophobic and aromatic side groups, and the formation of hydrogen bonds. Nevertheless, their folding behavior differs in many important ways (Thirumalai and Hyeon 2005)(Table 1). Proteins have a higher folding cooperativity with secondary structure (SecStr) and collapse occurring in the same kinetic event. In contrast, RNA SecStr typically forms at low ionic conditions with collapse and tertiary structure (TertStr) formation occurring only upon the addition of counterions that shield the repulsion of the negatively charged RNA phosphodiester backbone. Specific Mg2+ binding often helps assemble the preformed RNA SecStrs into tertiary architectures (Cate et al. 1997; Batey et al. 1999; Klein et al. 2004). As a result, the protein folding landscape has fewer stable intermediates than the RNA folding landscape (Thirumalai and Woodson 1996; Treiber et al. 1998; Thirumalai et al. 2001; Treiber and Williamson 2001; Das et al. 2003; Baird et al. 2007).
Table 1.
Properties of proteins and RNA
Folding rates and chain topology
When interpreting data and identifying the critical folding events, one must distinguish between the intrinsic (unavoidable) kinetic barriers from the optional barriers, which arise on some pathways but not on others. Proteins and RNAs can fold directly to the native state or be trapped by kinetic barriers related to undoing errors formed early in the pathway (Bryngelson and Wolynes 1989; Abkevich et al. 1994a; Sosnick et al. 1994; Guo and Thirumalai 1995; Thirumalai and Woodson 1996; Pan and Sosnick 1997; Pan et al. 1997, 1999b; Treiber and Williamson 1999; Russell et al. 2002). The nature of the errors include premature collapse and nonnative secondary structure formation. These optional barriers obscure the intrinsic barriers, those unavoidable steps that determine folding rates in the absence of error correction.
Protein folding
The error-free folding of proteins had been proposed to be a nucleation process where the chain attains a coarse version of the native topology in the transition state (Abkevich et al. 1994b; Guo and Thirumalai 1995; Sosnick et al. 1995, 1996; Englander et al. 1998). This proposal is supported by the correlation between ln kf and the relative contact order (RCO) (Fig. 1A; Plaxco et al. 1998) and other metrics of the protein's topological complexity (Fig. 2A; Goldenberg 1999; Ivankov et al. 2003; Bai et al. 2004). To better understand the origin of the correlation, knowledge of the structure of the transition state ensemble (TSE) is required.
Figure 1.
Correlation between relative contact order and folding rates of two-state proteins. (A) The RCO values of the three proteins, ubiquitin, acyl phosphatase, and BdpA, studied using ψ-analysis span the RCO range. (B) Native and TS models of the three proteins.
Figure 2.
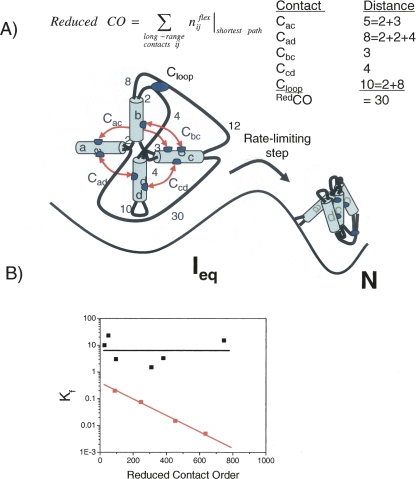
RNA folding rates and redCO. (A) RNA folding is multistate with highly structured intermediates that accumulate prior to the rate-limiting step. A sample calculation of RedCO is depicted for a model RNA with five tertiary contacts. RedCO is the sum of the length of the shortest paths between each contact. Numbers denote residue length of each unstructured segment. (B) The measured or extrapolated folding rates at 37°C and ∼10 mM Mg2+ for 10 RNAs. The black and red lines identify the fast and slow folding classes, respectively.
We developed ψ analysis in part to provide such structural information. This complement to mutational φ analysis (Matthews 1987; Fersht et al. 1992; Goldenberg 1992) proceeds by introducing relatively benign bi-Histidine (biHis) metal ion-binding sites at specific positions on the protein surface. Upon the addition of metal ions, these sites stabilize secondary and tertiary structures, because an increase in the metal ion concentration stabilizes the interaction between the two histidine partners. The metal-induced stabilization of the TSE relative to the native state, as represented by the ψ-value, directly reports on the proximity of the two partners in the TSE. The ψ-value depends on the degree to which the biHis site is formed in the TSE. Values of zero or one indicate that the biHis site is absent or fully native-like, respectively. Fractional values indicate that the biHis site recovers only part of the binding-induced stabilization of the native state. This situation can occur when the site is formed in only a subpopulation with a native-like binding affinity, or when the site has binding affinity weaker than in the native state.
By virtue of the binding site being defined by two known partners, ψ analysis is particularly well suited for identifying the side chain to side chain contacts that define the TS's topology. The mutational counterpart, φ analysis, reports on the energetic influence of side-chain alteration and can underestimate the structural content of the TS (Bulaj and Goldenberg 2001; Krantz and Sosnick 2001; Krantz et al. 2004; Sosnick et al. 2004) due to chain relaxation and accommodation, or nonnative interactions (Feng et al. 2004b; Neudecker et al. 2006). In addition, the ψ-value is calculated in the limit of no added metal, thereby reporting on the partners' proximity prior to the perturbation, whereas φ-values often are obtained using data after a destabilizing core mutation.
We applied ψ analysis to two α/β proteins, ubiquitin (Ub) (Krantz et al. 2004) and acyl phosphatase (Acp) (Pandit et al. 2006), and a small three-helix bundle, B domain of protein A (BdpA) (M. Baxa, K.F. Freed, T.R. Sosnick, in prep.). These proteins were chosen because their RCO values span the observed range for which the correlation with k f applies. To help characterize the TS of BdpA, we also determined the total helical hydrogen bond content of BdpA using kinetic H/D amide isotope effects (Krantz et al. 2000, 2002b) and utilized mutational φ-analysis data by Fersht and coworkers (Sato et al. 2004). The TS ensembles of these three proteins are found to share a common and high fraction of their native topology, RCOTS ≈ 0.7 · RCON (Fig. 1B). This commonality suggests that the universal k f versus RCO correlation could be rationalized if all the TSs of the proteins obeying the correlation also have a similarly high fraction of their native RCO value.
Other works points to a similar conclusion. Wallin and Chan used a Cα Gō-like model and find the TSEs of 13 proteins have 0.7 · RCON (Wallin and Chan 2006). Similarly, in Vendruscolo et al.'s all-atom simulations of 10 proteins, their TSEs share a common, albeit lower fraction, 0.5 · RCON (Paci et al. 2005), potentially due to the incorporation of φ-values that may underestimate chain–chain contacts. Likewise, Bai et al. (2004) find that a universal 78% value for the total contact distance of the TS produced the best correlation between the critical nucleation size of the TS and ln kf for 41 proteins.
The RCOTS ∼ 0.7·RCON relationship is predictive, and provides a useful guide for interpreting previous studies and for modeling the TSs of other proteins. For example, the high RCOTS/RCON fraction restricts the degree to which a TS can be small and polarized, as has been inferred from φ-value data for some proteins that obey the RCO correlation (Grantcharova et al. 1998; Gruebele and Wolynes 1998; Riddle et al. 1999; Klimov and Thirumalai 2001; Lindberg et al. 2002; Yi et al. 2003; Garcia-Mira et al. 2004; Guo et al. 2004). In addition, this high fraction limits the degree of possible TS heterogeneity, as defined by the participation of different subsets of helices or strands. Minimal experimental evidence exists for TS heterogeneity in small proteins at this broad level. Generally, a single folding nucleus is likely to be a general phenomenon in the folding of small proteins unless there is a strong symmetry to the protein (Kim et al. 2000; Grantcharova et al. 2001; Krantz and Sosnick 2001; Klimov and Thirumalai 2005; Shen et al. 2005; Lindberg and Oliveberg 2007; Olofsson et al. 2007).
To characterize a protein's topology, we have utilized the RCO metric, in part because of its broad usage. Other metrics (Goldenberg 1999; Ivankov et al. 2003; Bai et al. 2004) produce a similar conclusion. An advantage of the RCO metric is that topologically similar TS structures have similar RCO values even when there is local “microscopic heterogeneity” such as frayed helices or hairpins (Krantz et al. 2004; Pandit et al. 2006). In these situations, the frayed portions have contacts that have approximately the same average sequence separation as their neighbors. Hence, the RCO value, which is normalized to the number of contacts, remains unchanged upon fraying.
Regardless, all such metrics are just proxies for the key properties of the TS. At the TS, the chain is pinned at enough points that further structure formation is thermodynamically favorable. The precise RCO value at which this occurs varies according to the type and arrangement of secondary structure elements in any individual protein. Nevertheless, our results indicate that a native-like topology will be a general property of the TS for many proteins.
RNA folding
In an effort to provide a similar framework for the determinants of RNA folding rates, we considered the relationship between kf and the topological complexity of the RNA (Sosnick and Pan 2004). The relationship between folding rates and topology is quite different for protein and RNAs. For proteins that fold in a two-state manner, the path from the denatured state to the TS involves a conformational search involving most of the chain. The RCO metric is an appropriate metric, as it reflects the global nature of the search. RNA folding, however, is multistate, with well-populated, structured intermediates. In these intermediates, a majority of the residues are involved in stable structures and do not change conformation during the rate-limiting step (RLS).
To account for the presence of these highly structured RNA folding intermediates, we introduced a new metric, Reduced Contact Order (RedCO). RedCO is defined as the number of residues outside of Watson-Crick base-paired regions along the shortest path between each pair of long-range contacts (Fig. 2A). A plot of RedCO versus kf for 10 RNAs suggested a separation into two distinct classes: A fast folding class with little dependence on RedCO, and a slow class with some dependence on RedCO (Fig. 2B).
To test this separation, we measured the relative folding rates for circular permuted libraries of an RNA selected from the slow and fast classes (Sosnick and Pan 2004). In these libraries, the wild-type 5′ and 3′ ends were covalently linked, followed by the generation of new 5′ and 3′ ends at nearly all other positions in the chain. The representatives of the slow and the fast class were the full-length Bacillus subtilis P RNA and its C-domain, respectively. These RNAs were chosen because the folding of the full-length P RNA is limited by the disruption of misfolded structure, whereas the limiting step for the C-domain folding is a small-scale event, the consolidation of a metal ion-binding site, and not a kinetic trap (Fang et al. 2002). Folding rates of the circular permuted library of P RNA exhibited a 10-fold variation, consistent with the sensitivity of kf to RedCO for the slow class. Folding rates of the entire circular permuted library of the C-domains were within 1.2-fold, in spite of their large variation in RedCO. This result correlates well with the null correlation between folding rate and RedCO conjectured for the fast class.
The separation of RNAs into different classes is not surprising, as the RLS can be significantly different for various RNAs (Treiber and Williamson 2001; Sosnick and Pan 2003). The nature of the RLS ranges from conformational changes leading to the formation of a metal ion-binding site (Sosnick and Pan 2003) to the disruption of nonnative or prematurely formed native structures (Treiber and Williamson 2001). When the RLS involves only a small portion of the chain, such as the formation of a metal ion-binding site, the folding rates are expected to be independent of the RedCO, as observed for the fast folding class. But, when the RLS involves structural disruption, the folding rates can vary with chain connectivity, for example, by circular permutation (Pan et al. 1999b; Woodson 2002; Heilman-Miller and Woodson 2003). The correlation between slower folding and increased complexity now reflects an increase in the probability for misfolding, rather than a more time-consuming conformational search involving the whole RNA.
These issues apply to protein folding as well. Increased complexity of the fold results in slower folding irrespective of whether RLS is a productive conformational search or an error correction event. This similar dependence may explain why a correlation between k f and RCO exists across both classes (Ivankov et al. 2003; Kamagata and Kuwajima 2006) even though the natures of the rate-limiting barriers are different.
Folding events and pathways
Denatured states and collapse in protein folding
The importance of residual structure in the denatured state has been a topic of considerable debate (Shortle and Ackerman 2001; Fitzkee and Rose 2004). Nevertheless, the chemically denatured state appears to be well described by a statistical coil (Kohn et al. 2004). Models have been developed (Bernado et al. 2005; Jha et al. 2005a) that reproduce both the global and local structure of urea-denatured apoMb (Mohana-Borges et al. 2004). In these models, each amino acid adopts φ,ψ dihedral angles according to their statistical frequencies and correlations with the angles and type of neighboring residues from a coil library (a subset of the PDB for residues outside of regular SecStr). In the coil library, dihedral angles consistent with polyproline II, β-sheet, and helical geometries are observed at a 4:3.5:3 ratio (Jha et al. 2005b). The predominance of polyproline II conformers is consistent with model peptide studies (Shi et al. 2002; Chellgren and Creamer 2004).
The removal of denaturant does subtly alter the distribution of backbone dihedral angles in the denatured state. The distribution most likely results in a shift from the polyproline II region to the helical region of the Ramachandran map (Jacob et al. 2004; Jha et al. 2005a,b). In addition, some local helical or turn structure may form (Religa et al. 2007). Whether this process represents the denatured state under native conditions or a distinct intermediate is determined by the presence of a distinct barrier between the species and the denatured state under denaturing conditions. Many ultrafast folding signals can be explained by the barrier-free situation where the signal change simply is a baseline adjustment of the denatured chain to the new solvent condition, rather than the formation of a distinct folding intermediate (Gutin et al. 1995; Sosnick et al. 1996, 1997; Qi et al. 1998; Krantz et al. 2002a; Jacob et al. 2004).
The amount of collapse upon denaturant dilution is another subject of considerable debate. As intrachain interactions increase at lower denaturant, it is commonly expected that the chain will rapidly collapse. However, no distinct early collapse phase was observed in small-angle X-ray scattering measurements on protein L (Plaxco et al. 1999), ubiquitin, acyl-phosphotase (Jacob et al. 2004), and reduced RNase A (Jacob et al. 2007; Wang et al. 2008). Furthermore, the Rg of reduced Rnase A in the absence of denaturant is constant between 20°C and 90°C. This paradigmatic unfolded polypeptide is behaving as a statistical coil in the noninteracting, high-temperature limit.
Evidently, stable collapse is challenging and requires extensive organization. For the aforementioned proteins, water appears to be as good a solvent as that with high denaturant concentrations when considering the overall dimensions of the denatured state. Some intermediate length-scale behavior, however, may be slightly different (Wang et al. 2008). Even under aqueous conditions, generic intrachain contacts do not outweigh protein–solvent interactions to produce a collapsed denatured state. The origins of these observations are due to thermodynamic considerations including backbone desolvation, chain stiffness and entropy, and hydrogen bonding requirements. For example, hydrogen bond formation and surface burial are concomitant processes in the TS according to kinetic amide isotope effects (Krantz et al. 2000, 2002b).
In contrast to the SAXS results, FRET-based methods often report a compaction of the denatured state at low denaturant concentrations (Magg and Schmid 2004; Kuzmenkina et al. 2005; Sinha and Udgaonkar 2005; Moglich et al. 2006). The origin for the discrepancy between the two methods is unknown. In the FRET studies, the contraction may be related to the addition of the large aromatic or hydrophobic chromophores. These moieties may interact with the side chains on the denatured protein to produce a net contraction. In support of the SAXS results is the tautology that a distinct kinetic phase representing stable chain collapse cannot occur for two-state folding reactions, in particular for those satisfying the chevron criteria (Jackson and Fersht 1991; Krantz et al. 2002a). If there was a distinct phase, the reactions would not be two-state, and all of the energy and surface burial between U and N would not be accounted for in the observed reaction. Further work is needed to clarify this discrepancy between the SAXS and the FRET measurements.
For two-state reactions, intermediates do not accumulate, and hence any species stable relative to the unfolded state must be located after the major barrier. That is, the formation of a stable intermediate is the RLS in two-state folding (Sosnick et al. 1996). In multistate folding, there is an analogous “initial barrier” leading to the formation of the first stable collapsed intermediate. This species accumulates due to a larger, second barrier often related to error correction.
Denatured states and collapse in RNA folding
The amount of residual SecStr in denatured RNAs strongly depends on solvent conditions. Hairpins and other structures can be disrupted by urea (Fang et al. 2000). The addition of monovalent ions can increase the amount of SecStr present. In some RNAs, nonnative structures can form at intermediate ionic conditions and bias the RNA to fold through kinetically trapped intermediates (Pan et al. 1999b; Russell et al. 2002).
The RNA collapse process can be described as follows. From the chemically denatured state devoid of base pairing, the formation of SecStr including long-range duplexes results in a high degree of chain contraction (Fang et al. 2000). Further collapse can occur upon nonspecific charge neutralization of the RNA backbone by monovalent or divalent cations (Das et al. 2003; Tan and Chen 2006). The RNA continues to contract as divalent cations stabilize specific TertStrs, which eventually results in a solvent-excluded core (Chauhan et al. 2005). The final stage of collapse occurs with the cooperative folding transition to the native structure. The change in Rg for this final step is relatively small for the C-domain of P RNA (Fang et al. 2002) and Group I introns (Das et al. 2003; Chauhan et al. 2005). Hence, this transition likely is dominated by local consolidation or rearrangement of pre-existing structures.
Steps in protein folding
What are the fundamental folding steps that lead to the native structure? Native state hydrogen exchange (NSHX) studies have shown that subglobal unfolding events occur in many proteins (Bai and Englander 1996; Chamberlain et al. 1996; Feng et al. 2004a). These openings represent the unfolding of individual or groups of SecStr elements and highlight that protein folding is not completely cooperative even for proteins with apparent two-state kinetics. These subglobal openings generally correspond to events on the unfolding pathway leading back up to the TS (Bai 2003; Feng et al. 2003; Kato et al. 2007). When viewed from the folding direction, these events represent the sequential stabilization of SecStr elements or “foldons,” wherein pre-existing structure provides a foundation on which unfolded regions can dock (Bai et al. 1995; Maity et al. 2005; Krishna and Englander 2007). These events are located after the initial collapse-related TS.
A growing amount of evidence argues that the foldons form in a well-defined sequence. This result is due to chain connectivity and sequential stabilization. For many protein architectures, the addition of foldons can only be accomplished, or is strongly preferred, in a specific order due to a structural hierarchy. At points along a pathway when two foldons can be added independently, or with comparable energy, a pathway can temporarily bifurcate (Krantz et al. 2004; Krishna et al. 2006, 2007; Sosnick et al. 2006).
The principles that point to an ordered pathway down from the TS to the native state are likely to be operational on the pathway up to the TS as well (Krantz et al. 2004; Sosnick et al. 2006). The early folding process includes a search through a multitude of mostly unproductive conformations. These conformations are unstable and repeatedly fall apart. Nevertheless, some of the more accessible conformations provide a suitable foundation for the addition of new SecStrs. The energetic biases for certain pathways and structural hierarchy may not be as pronounced as for the post-TS pathways. Nevertheless, we expect the uphill steps to be a largely linear accretion of structure as SecStr elements layer on top of pre-existing structure.
We proposed a detailed folding pathway for ubiquitin (Fig. 3) using this reasoning along with experimental results including ψ analysis (Krantz et al. 2004; Sosnick et al. 2006). Each major step in the pathway represents the addition or consolidation of pieces of SecStr. The steps occur with a commensurate level of hydrogen bond formation and surface burial, as suggested by the aforementioned kinetic amide isotope effects (Krantz et al. 2000, 2002b). The native topology is established incrementally through the formation of a series of intermediates with native-like structure, rather than by the initial formation of long-range side chain contacts, followed by SecStr formation. Whether this type of analysis, which points to a largely sequential pathway, is applicable to other proteins remains an open issue, requiring more experimental and theoretical studies.
Figure 3.
Proposed Ub folding pathway. Ub's folding pathway is described by a largely sequential pathway with a conserved nucleus. The β-hairpin, the most stable of isolated SecStr, forms first. Other initial structures are less plausible, such as the α-helix (low intrinsic helicity) or the parallel association between the two terminal strands (big loop closure penalty). The sequence proximity and increased hydrophobic burial for helix association suggests that it generally folds prior to the carboxy-terminal strand (upper route preferred). The addition of the fourth β-strand completes the TS, although some “microscopic heterogeneity” may exist involving frayed strands and helices. The two remaining elements, the 310 helix and fifth strand, add on the way down. Native-state HX suggests that the 310 helix folds first.
Steps in RNA folding
For Mg2+-induced folding, the pathway largely is a hierarchal process with most of the SecStr forming before the TertStr (Fig. 4). The final Ieq ↔ N transition often is very cooperative and can be fit with a Hill-type analysis (Zarrinkar and Williamson 1996; Doherty and Doudna 1997; Pan and Sosnick 1997). The Hill constant is the differential ion binding term, and typically is between 2 and 7. It reflects the uptake of additional specific and nonspecific ions upon the formation of the more compact native structure that has higher charge density.
Figure 4.
RNA folding pathways. (A) Folding pathway of the S-domain from Bacillus subtilis illustrated with the native crystal structure, Ieq, and two hypothetical late kinetic intermediates. (B) Reaction scheme for the Ieq-to-N macroscopic transition with two kinetic intermediates and three microscopic transitions. (C) Observed relaxation rates plotted as a function of final [Mg2+] for folding (●) and unfolding measurements (○) generate a Mg2+ chevron. (D) Corresponding free energy surfaces change with [Mg2+]. At [Mg2+] >> KMg (dashed line), folding rates saturate at k f due to the rapid formation of a folding intermediate, I1k, followed a Mg2+-independent barrier. At low [Mg2+ ] << KMg (dotted line), unfolding rates saturate at k u due to the formation of an unfolding intermediate, I2k. At [Mg2+] ∼ KMg, the observed rate reflects the relative population of the kinetic intermediate times the rate of going over the Mg2+-independent barrier. Although the RLS does not involve an uptake of additional metal ions for the bulk solution, it is sensitive to cation type. These results indicate that the RLS represents a small-scale structural consolidation around one or more prebound metal ions.
The Mg dependence of the relaxation rates of the Ieq ↔ N transition produces a Mg chevron (Fig. 4B; Fang et al. 1999), similar to the denaturant chevron familiar in protein folding. For the S-domain and C-domains of P RNA, the transition has three microscopic steps with an I1k folding and an I2k unfolding intermediate. For the C-domain of P RNA, properties of the rate-limiting step indicate that it represents a small-scale structural consolidation around one or more pre-bound metal ions (Fig. 4; Fang et al. 2002). The height of the kinetic barrier is associated with the difficulty of driving the negatively charged RNA backbone into close enough proximity that it can bind a partially desolvated ion.
Support for this model can be seen in other studies. Specific ion binding sites can have partially dehydrated cations with up to five RNA ligands (Klein et al. 2004). The RLS in the folding of the Group II intron is the binding of Mg2+ and the capture of the κ–ζ ion-binding pocket (Waldsich and Pyle 2008). In single-molecule force-unfolding measurements on the Tetrahymena thermaphila ribozyme, a series of SecStr “rips” are followed by the disruption of a brittle tertiary interaction (Liphardt et al. 2001; Onoa et al. 2003). This brittle interaction presumably represents the loss of a specific Mg2+-binding site.
The principles that point to defined pathways in protein folding are applicable to RNA, being based on chain connectivity and sequential stabilization. In the D135 Group II ribozyme, the formation of a small region is an early event that controls the collapse and overall folding rate (Waldsich and Pyle 2008). This behavior is indicative of a sequential pathway. However, the independent stability of RNA motifs generally reduces the importance of sequential stabilization and increases the frequency of alternative pathways. For example, the single-molecule pulling studies have observed different unfolding routes, with unfolding events being biased to occur at the points of applied tension (Liphardt et al. 2001; Onoa et al. 2003).
Ionic conditions also can alter the order of structure formation. Monovalent ions promote the formation of isolated SecStr, whereas divalent ions promote TertStr for RNAs with specific binding sites. This differential dependence on mono- and divalent ions can invert the order of structure formation for RNAs with long-range helices. These helices can form prior to any TertStr under high ionic conditions, whereas they may form after some TertStr formation under lower ionic conditions (Shelton et al. 2001).
Modes of initiation
Three different modes have been used to initiate the folding reaction: a change in solvent condition, chain synthesis, and the application or release of force typically applied to the ends of the molecule. Generally, RNA folding is more sensitive to the mode of initiation, in part due to the intrinsic stability of its SecStr and the influence of ionic conditions.
Protein and RNA folding often is initiated by the dilution of denaturant or the addition of Mg2+, respectively. The resulting pathway is likely to be similar to the pathway for synthesis-initiated folding for small proteins but not for RNAs. Small proteins generally do not form stable structures until nearly the entire chain has been synthesized due to their high degree of folding cooperativity (De Prat Gay et al. 1995), particularly for proteins whose chain termini make long-range contacts (Krishna and Englander 2005).
However, RNA hairpins can fold as soon as they have been synthesized. For synthesis at high Mg2+ concentration, tertiary modules can form once the relevant portions have been synthesized (Pan and Sosnick 2006). This behavior contrasts with the SecStr → TertStr paradigm for folding induced by the addition of Mg2+. Long-range RNA helices pose a particular folding problem, as the duplexes cannot form until both halves are synthesized. This lag leaves the 5′ end available for long-lived, nonnative interactions. For some RNAs, transient and specific nonnative structure forms to sequester the 5′ regions until their 3′ partners are formed (Wong et al. 2007). In addition, folding behavior is modulated transcriptional pausing (Pan et al. 1999a; Wickiser et al. 2005; Wong et al. 2005, 2007; Pan and Sosnick 2006). A recent report suggests translational pausing may also influence protein folding (Kimchi-Sarfaty et al. 2007).
Forced unfolding samples another region of the folding landscape. During unfolding, tension is placed on molecules, and structure preferentially is lost at the points of attachment (Liphardt et al. 2001; Carrion-Vazquez et al. 2003; Onoa et al. 2003). Hence, the unfolding pathway may not be the same as the pathway induced by a change in solvent condition. In ubiquitin, for example, the amino and carboxy termini are associated in the denaturant-induced TS (Krantz et al. 2004) but not in the forced-induced TS when the protein is pulled by the termini (Carrion-Vazquez et al. 2003).
A similar disparity can occur between Mg2+-induced and forced-induced RNA unfolding. Mg2+-induced folding is hierarchical, with the SecStr forming before the TertStr. Conversely, in the unfolding direction induced by lowering [Mg2+], the native structure unfolds to a conformation where essentially all of the secondary structures are present. In the forced-induced unfolding pathway, however, unfolding occurs with the alternating disruption of elements of secondary and tertiary structure (Liphardt et al. 2001; Onoa et al. 2003). Hence, the intermediates on the forced unfolding pathway generally are quite different from those observed on a pathway sampled by the reduction in [Mg2+]. This difference also implies that the stabilities derived from the two types of measurements are different, as their thermodynamic reference states are different.
Concluding thoughts
Being one of a few researchers with a foot in both the protein and RNA worlds, I have often contemplated the parallels between their folding behavior—which tools and principles could be transferred from one field to the other? My first study with Walter Englander dealt with fast and slow protein folding and misfolding. Within 3 years, Tao Pan and I confirmed that these principles also applied to RNA folding. We demonstrated that folding rates increased with urea concentration, a strategy borrowed from protein folding studies. Relatively early on, Walter and I began thinking that obtaining a coarse version of the native topology was a critical element of the two-state protein folding TS. But, the relationship between topology and RNA-folding rates was unclear. We needed better tools to characterize RNA pathways and kinetic barriers. We adapted the denaturant-dependent protein-folding chevron to RNA folding (Fang et al. 1999) and concluded that the rate-limiting step on an error-free pathway was a small-scale event (Fang et al. 2002), rather than the global search relevant to protein folding. The intellectual flow reversed directions when the RNA strategy of measuring folding as a function of [metal] became the preferred way of implementing ψ analysis.
NSHX has proven to be an extremely powerful tool for dissecting protein structure, energetics, and pathways. Chemical footprinting methods can provide residue level information even for very large RNAs. However, these methods have limited dynamic range as the rate of first backbone cut is measured, while the more interesting, higher energy fluctuations escape detection. Whether RNA folding can take full advantage of the power of NSHX remains to be determined (Vermeulen et al. 2005; Lee and Pardi 2007).
Undoubtedly, each field will benefit from technical advances in the other field. One overlapping area is RNA–protein interactions, particularly when one or both partners undergo a conformational change upon binding. Another overlapping, and extremely challenging area, is electrostatics. Divalent ions often need to be treated explicitly, particularly in regions of high electrostatic potential where ion–ion correlations are significant. In addition, dehydration and binding of ions to nucleophiles must be accurately treated both in RNA folding and ion channel conductance. I look forward to progress in these and other synergistic areas.
Acknowledgments
I thank my long-time collaborators in the RNA and protein worlds, T. Pan, S.W. Englander, K. Freed, N. Kallenbach, and P. Thiyagarajan, and other colleagues along with present and past members of my laboratory for engaging discussions, insights, and good data. This work is supported by research grants from the NIH (GM55694 and GM57880, T. Pan).
Footnotes
Reprint requests to: Tobin R. Sosnick, University of Chicago, 929 East 57th Street, GCIS W107E, Chicago, IL 60637, USA; e-mail: trsosnic@uchicago.edu; fax: (773) 702-0439.
Abbreviations: Acp, acyl phosphatase; BdpA, B-domain of protein A; biHis, bi-histidine; CD, circular dichroism; GdmCl, guanidinium chloride; K Mg, Mg2+ concentration at the midpoint of an RNA folding transition; n, the Hill constant; NSHX, native state hydrogen exchange; Ieq, equilibrium intermediate; N, native; RLS, rate-limiting step; RedCO, reduced contact order; RCO, relative contact order; SAXS, small-angle X-ray scattering; SecStr, secondary structure; TertStr, tertiary structure; TSE, transition state ensemble; Ub, ubiquitin; U, unfolded.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036319.108.
References
- Abkevich, V.I., Gutin, A.M., Shakhnovich, E.I. Free energy landscape for protein folding kinetics: Intermediates, traps, and multiple pathways in theory and lattice model simulations. J. Chem. Phys. 1994a;101:6052–6062. [Google Scholar]
- Abkevich, V.I., Gutin, A.M., Shakhnovich, E.I. Specific nucleus as the transition state for protein folding: Evidence from the lattice model. Biochemistry. 1994b;33:10026–10036. doi: 10.1021/bi00199a029. [DOI] [PubMed] [Google Scholar]
- Bai, Y. Hidden intermediates and Levinthal paradox in the folding of small proteins. Biochem. Biophys. Res. Commun. 2003;305:785–788. doi: 10.1016/s0006-291x(03)00800-3. [DOI] [PubMed] [Google Scholar]
- Bai, Y., Englander, S.W. Future directions in folding: The multi-state nature of protein structure. Proteins. 1996;24:145–151. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Bai, Y., Sosnick, T.R., Mayne, L., Englander, S.W. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y., Zhou, H., Zhou, Y. Critical nucleation size in the folding of small apparently two-state proteins. Protein Sci. 2004;13:1173–1181. doi: 10.1110/ps.03587604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, N.J., Srividya, N., Krasilnikov, A.S., Mondragon, A., Sosnick, T.R., Pan, T. Structural basis for altering the stability of homologous RNAs from a mesophilic and a thermophilic bacterium. RNA. 2006;12:598–606. doi: 10.1261/rna.2186506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, N.J., Fang, X.W., Srividya, N., Pan, T., Sosnick, T.R. Folding of a universal ribozyme: The ribonuclease P RNA. Q. Rev. Biophys. 2007;40:113–161. doi: 10.1017/S0033583507004623. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Doudna, J.A. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Bernado, P., Blanchard, L., Timmins, P., Marion, D., Ruigrok, R.W., Blackledge, M. A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc. Natl. Acad. Sci. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryngelson, J.D., Wolynes, P.G. Intermediates and barrier crossing in a random energy model with application to protein folding. J. Phys. Chem. 1989;93:6902–6915. [Google Scholar]
- Bulaj, G., Goldenberg, D.P. φ-Values for BPTI folding intermediates and implications for transition state analysis. Nat. Struct. Biol. 2001;8:326–330. doi: 10.1038/86200. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., Li, H., Lu, H., Marszalek, P.E., Oberhauser, A.F., Fernandez, J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Hanna, R.L., Doudna, J.A. A magnesium ion core at the heart of a ribozyme domain. Nat. Struct. Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- Chamberlain, A.K., Handel, T.M., Marqusee, S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat. Struct. Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- Chauhan, S., Caliskan, G., Briber, R.M., Perez-Salas, U., Rangan, P., Thirumalai, D., Woodson, S.A. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Chellgren, B.W., Creamer, T.P. Short sequences of non-proline residues can adopt the polyproline II helical conformation. Biochemistry. 2004;43:5864–5869. doi: 10.1021/bi049922v. [DOI] [PubMed] [Google Scholar]
- Das, R., Kwok, L.W., Millett, I.S., Bai, Y., Mills, T.T., Jacob, J., Maskel, G.S., Seifert, S., Mochrie, S.G., Thiyagarajan, P., et al. The fastest global events in RNA folding: Electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- De Prat Gay, G., Ruiz-Sanz, J., Neira, J.L., Itzhaki, L.S., Fersht, A.R. Folding of a nascent polypeptide chain in vitro: Cooperative formation of structure in a protein module. Proc. Natl. Acad. Sci. 1995;92:3683–3686. doi: 10.1073/pnas.92.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, E.A., Doudna, J.A. The P4-P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry. 1997;36:3159–3169. doi: 10.1021/bi962428+. [DOI] [PubMed] [Google Scholar]
- Englander, S.W., Sosnick, T.R., Mayne, L.C., Shtilerman, M., Qi, P.X., Bai, Y. Fast and slow folding in cytochrome c . Accts. Chem. Res. 1998;31:737–744. [Google Scholar]
- Fang, X., Pan, T., Sosnick, T.R. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- Fang, X., Littrell, K., Yang, X., Henderson, S.J., Siefert, S., Thiyagarajan, P., Pan, T., Sosnick, T.R. Mg2+-dependent compaction and folding of yeast tRNA(Phe) and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering. Biochemistry. 2000;39:11107–11113. doi: 10.1021/bi000724n. [DOI] [PubMed] [Google Scholar]
- Fang, X.W., Golden, B.L., Littrell, K., Shelton, V., Thiyagarajan, P., Pan, T., Sosnick, T.R. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc. Natl. Acad. Sci. 2001;98:4355–4360. doi: 10.1073/pnas.071050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, X.W., Thiyagarajan, P., Sosnick, T.R., Pan, T. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc. Natl. Acad. Sci. 2002;99:8518–8523. doi: 10.1073/pnas.142288399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H., Takei, J., Lipsitz, R., Tjandra, N., Bai, Y. Specific nonnative hydrophobic interactions in a hidden folding intermediate: Implications for protein folding. Biochemistry. 2003;42:12461–12465. doi: 10.1021/bi035561s. [DOI] [PubMed] [Google Scholar]
- Feng, H., Vu, N.D., Bai, Y. Detection and structure determination of an equilibrium unfolding intermediate of Rd-apocytochrome b562: Native fold with non-native hydrophobic interactions. J. Mol. Biol. 2004a;343:1477–1485. doi: 10.1016/j.jmb.2004.08.099. [DOI] [PubMed] [Google Scholar]
- Feng, H., Vu, N.D., Zhou, Z., Bai, Y. Structural examination of φ-value analysis in protein folding. Biochemistry. 2004b;43:14325–14331. doi: 10.1021/bi048126m. [DOI] [PubMed] [Google Scholar]
- Fersht, A.R., Matouschek, A., Serrano, L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- Fitzkee, N.C., Rose, G.D. Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mira, M.M., Boehringer, D., Schmid, F.X. The folding transition state of the cold shock protein is strongly polarized. J. Mol. Biol. 2004;339:555–569. doi: 10.1016/j.jmb.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Goldenberg, D.P. Mutational analysis of protein folding and stability. In: Creighton T.E., editor. Protein folding. W. H. Freeman; New York: 1992. pp. 353–403. [Google Scholar]
- Goldenberg, D.P. Finding the right fold. Nat. Struct. Biol. 1999;6:987–990. doi: 10.1038/14866. [DOI] [PubMed] [Google Scholar]
- Grantcharova, V.P., Riddle, D.S., Santiago, J.V., Baker, D. Important role of hydrogen bonds in the structurally polarized transition state for folding of the src SH3 domain. Nat. Struct. Biol. 1998;5:714–720. doi: 10.1038/1412. [DOI] [PubMed] [Google Scholar]
- Grantcharova, V., Alm, E.J., Baker, D., Horwich, A.L. Mechanisms of protein folding. Curr. Opin. Struct. Biol. 2001;11:70–82. doi: 10.1016/s0959-440x(00)00176-7. [DOI] [PubMed] [Google Scholar]
- Gruebele, M., Wolynes, P.G. Satisfying turns in folding transitions. Nat. Struct. Biol. 1998;5:662–665. doi: 10.1038/1354. [DOI] [PubMed] [Google Scholar]
- Guo, F., Cech, T.R. Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nat. Struct. Biol. 2002;9:855–861. doi: 10.1038/nsb850. [DOI] [PubMed] [Google Scholar]
- Guo, Z.Y., Thirumalai, D. Kinetics of protein-folding: Nucleation mechanism, timescales, and pathways. Biopolymers. 1995;36:83–102. [Google Scholar]
- Guo, W., Lampoudi, S., Shea, J.E. Temperature dependence of the free energy landscape of the src-SH3 protein domain. Proteins. 2004;55:395–406. doi: 10.1002/prot.20053. [DOI] [PubMed] [Google Scholar]
- Gutin, A.M., Abkevich, V.I., Shakhnovich, E.I. Is burst hydrophobic collapse necessary for protein folding? Biochemistry. 1995;34:3066–3076. doi: 10.1021/bi00009a038. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller, S.L., Woodson, S.A. Perturbed folding kinetics of circularly permuted RNAs with altered topology. J. Mol. Biol. 2003;328:385–394. doi: 10.1016/s0022-2836(03)00304-8. [DOI] [PubMed] [Google Scholar]
- Ivankov, D.N., Garbuzynskiy, S.O., Alm, E., Plaxco, K.W., Baker, D., Finkelstein, A.V. Contact order revisited: Influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.E., Fersht, A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Jacob, J., Krantz, B., Dothager, R.S., Thiyagarajan, P., Sosnick, T.R. Early collapse is not an obligate step in protein folding. J. Mol. Biol. 2004;338:369–382. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- Jacob, J., Dothager, R.S., Thiyagarajan, P., Sosnick, T.R. Fully reduced ribonuclease A does not expand at high denaturant concentration or temperature. J. Mol. Biol. 2007;367:609–615. doi: 10.1016/j.jmb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Jha, A.K., Colubri, A., Freed, K.F., Sosnick, T.R. Statistical coil model of the unfolded state: Resolving the reconciliation problem. Proc. Natl. Acad. Sci. 2005a;102:13099–13104. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, A.K., Colubri, A., Zaman, M.H., Koide, S., Sosnick, T.R., Freed, K.F. Helix, sheet, and polyproline II frequencies and strong nearest neighbor effects in a restricted coil library. Biochemistry. 2005b;44:9691–9702. doi: 10.1021/bi0474822. [DOI] [PubMed] [Google Scholar]
- Kamagata, K., Kuwajima, K. Surprisingly high correlation between early and late stages in non-two-state protein folding. J. Mol. Biol. 2006;357:1647–1654. doi: 10.1016/j.jmb.2006.01.072. [DOI] [PubMed] [Google Scholar]
- Kato, H., Vu, N.D., Feng, H., Zhou, Z., Bai, Y. The folding pathway of T4 lysozyme: An on-pathway hidden folding intermediate. J. Mol. Biol. 2007;365:881–891. doi: 10.1016/j.jmb.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.E., Fisher, C., Baker, D. A breakdown of symmetry in the folding transition state of protein L. J. Mol. Biol. 2000;298:971–984. doi: 10.1006/jmbi.2000.3701. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty, C., Oh, J.M., Kim, I.W., Sauna, Z.E., Calcagno, A.M., Ambudkar, S.V., Gottesman, M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., Steitz, T.A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov, D.K., Thirumalai, D. Multiple protein folding nuclei and the transition state ensemble in two-state proteins. Proteins. 2001;43:465–475. doi: 10.1002/prot.1058. [DOI] [PubMed] [Google Scholar]
- Klimov, D.K., Thirumalai, D. Symmetric connectivity of secondary structure elements enhances the diversity of folding pathways. J. Mol. Biol. 2005;353:1171–1186. doi: 10.1016/j.jmb.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Kohn, J.E., Millett, I.S., Jacob, J., Zagrovic, B., Dillon, T.M., Cingel, N., Dothager, R.S., Seifert, S., Thiyagarajan, P., Sosnick, T.R., et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz, B.A., Sosnick, T.R. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nat. Struct. Biol. 2001;8:1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- Krantz, B.A., Moran, L.B., Kentsis, A., Sosnick, T.R. D/H amide kinetic isotope effects reveal when hydrogen bonds form during protein folding. Nat. Struct. Biol. 2000;7:62–71. doi: 10.1038/71265. [DOI] [PubMed] [Google Scholar]
- Krantz, B.A., Mayne, L., Rumbley, J., Englander, S.W., Sosnick, T.R. Fast and slow intermediate accumulation and the initial barrier mechanism in protein folding. J. Mol. Biol. 2002a;324:359–371. doi: 10.1016/s0022-2836(02)01029-x. [DOI] [PubMed] [Google Scholar]
- Krantz, B.A., Srivastava, A.K., Nauli, S., Baker, D., Sauer, R.T., Sosnick, T.R. Understanding protein hydrogen bond formation with kinetic H/D amide isotope effects. Nat. Struct. Biol. 2002b;9:458–463. doi: 10.1038/nsb794. [DOI] [PubMed] [Google Scholar]
- Krantz, B.A., Dothager, R.S., Sosnick, T.R. Discerning the structure and energy of multiple transition states in protein folding using ψ-analysis. J. Mol. Biol. 2004;337:463–475. doi: 10.1016/j.jmb.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Krishna, M.M., Englander, S.W. The N-terminal to C-terminal motif in protein folding and function. Proc. Natl. Acad. Sci. 2005;102:1053–1058. doi: 10.1073/pnas.0409114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna, M.M., Englander, S.W. A unified mechanism for protein folding: Predetermined pathways with optional errors. Protein Sci. 2007;16:449–464. doi: 10.1110/ps.062655907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna, M.M., Maity, H., Rumbley, J.N., Lin, Y., Englander, S.W. Order of steps in the cytochrome C folding pathway: Evidence for a sequential stabilization mechanism. J. Mol. Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Krishna, M.M., Maity, H., Rumbley, J.N., Englander, S.W. Branching in the sequential folding pathway of cytochrome c . Protein Sci. 2007;16:1946–1956. doi: 10.1110/ps.072922307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmenkina, E.V., Heyes, C.D., Nienhaus, G.U. Single-molecule Forster resonance energy transfer study of protein dynamics under denaturing conditions. Proc. Natl. Acad. Sci. 2005;102:15471–15476. doi: 10.1073/pnas.0507728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Pardi, A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, M.O., Oliveberg, M. Malleability of protein folding pathways: A simple reason for complex behaviour. Curr. Opin. Struct. Biol. 2007;17:21–29. doi: 10.1016/j.sbi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Lindberg, M., Tangrot, J., Oliveberg, M. Complete change of the protein folding transition state upon circular permutation. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- Liphardt, J., Onoa, B., Smith, S.B., Tinoco, I.J., Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- Magg, C., Schmid, F.X. Rapid collapse precedes the fast two-state folding of the cold shock protein. J. Mol. Biol. 2004;335:1309–1323. doi: 10.1016/j.jmb.2003.11.050. [DOI] [PubMed] [Google Scholar]
- Maity, H., Maity, M., Krishna, M.M., Mayne, L., Englander, S.W. Protein folding: The stepwise assembly of foldon units. Proc. Natl. Acad. Sci. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, C.R. Effects of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- Moglich, A., Joder, K., Kiefhaber, T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana-Borges, R., Goto, N.K., Kroon, G.J., Dyson, H.J., Wright, P.E. Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings. J. Mol. Biol. 2004;340:1131–1142. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Neudecker, P., Zarrine-Afsar, A., Choy, W.Y., Muhandiram, D.R., Davidson, A.R., Kay, L.E. Identification of a collapsed intermediate with non-native long-range interactions on the folding pathway of a pair of Fyn SH3 domain mutants by NMR relaxation dispersion spectroscopy. J. Mol. Biol. 2006;363:958–976. doi: 10.1016/j.jmb.2006.08.047. [DOI] [PubMed] [Google Scholar]
- Olofsson, M., Hansson, S., Hedberg, L., Logan, D.T., Oliveberg, M. Folding of S6 structures with divergent amino acid composition: Pathway flexibility within partly overlapping foldons. J. Mol. Biol. 2007;365:237–248. doi: 10.1016/j.jmb.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Onoa, B., Dumont, S., Liphardt, J., Smith, S.B., Tinoco I., Jr, Bustamante, C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci, E., Lindorff-Larsen, K., Dobson, C.M., Karplus, M., Vendruscolo, M. Transition state contact orders correlate with protein folding rates. J. Mol. Biol. 2005;352:495–500. doi: 10.1016/j.jmb.2005.06.081. [DOI] [PubMed] [Google Scholar]
- Pan, T., Sosnick, T.R. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- Pan, T., Sosnick, T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- Pan, J., Thirumalai, D., Woodson, S.A. Folding of RNA involves parallel pathways. J. Mol. Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- Pan, T., Artsimovitch, I., Fang, X., Landick, R., Sosnick, T.R. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. 1999a;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T., Fang, X., Sosnick, T.R. Pathway modulation, circular permutation and rapid RNA folding under kinetic control. J. Mol. Biol. 1999b;286:721–731. doi: 10.1006/jmbi.1998.2516. [DOI] [PubMed] [Google Scholar]
- Pandit, A.D., Jha, A., Freed, K.F., Sosnick, T.R. Small proteins fold through transition states with native-like topologies. J. Mol. Biol. 2006;361:755–770. doi: 10.1016/j.jmb.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Plaxco, K.W., Simons, K.T., Baker, D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- Plaxco, K.W., Millett, I.S., Segel, D.J., Doniach, S., Baker, D. Chain collapse can occur concomitantly with the rate-limiting step in protein folding. Nat. Struct. Biol. 1999;6:554–556. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
- Qi, P.X., Sosnick, T.R., Englander, S.W. The burst phase in ribonuclease A folding and solvent dependence of the unfolded state. Nat. Struct. Biol. 1998;5:882–884. doi: 10.1038/2321. [DOI] [PubMed] [Google Scholar]
- Religa, T.L., Johnson, C.M., Vu, D.M., Brewer, S.H., Dyer, R.B., Fersht, A.R. The helix-turn-helix motif as an ultrafast independently folding domain: The pathway of folding of Engrailed homeo domain. Proc. Natl. Acad. Sci. 2007;104:9272–9277. doi: 10.1073/pnas.0703434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D.S., Grantcharova, V.P., Santiago, J.V., Alm, E., Ruczinski, I.I., Baker, D. Experiment and theory highlight role of native state topology in SH3 folding. Nat. Struct. Biol. 1999;6:1016–1024. doi: 10.1038/14901. [DOI] [PubMed] [Google Scholar]
- Russell, R., Zhuang, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S., Herschlag, D. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Religa, T.L., Daggett, V., Fersht, A.R. Testing protein-folding simulations by experiment: B domain of protein A. Proc. Natl. Acad. Sci. 2004;101:6952–6956. doi: 10.1073/pnas.0401396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton, V.M., Sosnick, T.R., Pan, T. Altering the intermediate in the equilibrium folding of unmodified yeast tRNA(Phe) with monovalent and divalent cations. Biochemistry. 2001;40:3629–3638. doi: 10.1021/bi002646+. [DOI] [PubMed] [Google Scholar]
- Shen, T., Hofmann, C.P., Oliveberg, M., Wolynes, P.G. Scanning malleable transition state ensembles: Comparing theory and experiment for folding protein U1A. Biochemistry. 2005;44:6433–6439. doi: 10.1021/bi0500170. [DOI] [PubMed] [Google Scholar]
- Shi, Z., Woody, R.W., Kallenbach, N.R. Is polyproline II a major backbone conformation in unfolded proteins? Adv. Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- Shortle, D., Ackerman, M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- Sinha, K.K., Udgaonkar, J.B. Dependence of the size of the initially collapsed form during the refolding of barstar on denaturant concentration: Evidence for a continuous transition. J. Mol. Biol. 2005;353:704–718. doi: 10.1016/j.jmb.2005.08.056. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R., Pan, T. RNA folding: Models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R., Pan, T. Reduced contact order and RNA folding rates. J. Mol. Biol. 2004;342:1359–1365. doi: 10.1016/j.jmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R., Mayne, L., Hiller, R., Englander, S.W. The barriers in protein folding. Nat. Struct. Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R., Mayne, L., Hiller, R., Englander, S.W. The barriers in protein folding. In: DeGrado W.F., editor. Peptide and protein folding workshop. International Business Communications; Philadelphia, PA: 1995. pp. 52–80. [Google Scholar]
- Sosnick, T.R., Mayne, L., Englander, S.W. Molecular collapse: The rate-limiting step in two-state cytochrome c folding. Proteins. 1996;24:413–426. doi: 10.1002/(SICI)1097-0134(199604)24:4<413::AID-PROT1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R., Shtilerman, M.D., Mayne, L., Englander, S.W. Ultrafast signals in protein folding and the polypeptide contracted state. Proc. Natl. Acad. Sci. 1997;94:8545–8550. doi: 10.1073/pnas.94.16.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnick, T.R., Dothager, R.S., Krantz, B.A. Differences in the folding transition state of ubiquitin indicated by φ and ψ analyses. Proc. Natl. Acad. Sci. 2004;101:17377–17382. doi: 10.1073/pnas.0407683101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnick, T.R., Krantz, B.A., Dothager, R.S., Baxa, M. Characterizing the protein folding transition state using ψ analysis. Chem. Rev. 2006;106:1862–1876. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- Tan, Z.J., Chen, S.J. Ion-mediated nucleic acid helix–helix interactions. Biophys. J. 2006;91:518–536. doi: 10.1529/biophysj.106.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai, D., Woodson, S.A. Kinetics of folding of proteins and RNA. Acc. Chem. Res. 1996;29:433–439. [Google Scholar]
- Thirumalai, D., Hyeon, C. RNA and protein folding: Common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., Lee, N., Woodson, S.A., Klimov, D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K., Williamson, J.R. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K., Williamson, J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K., Rook, M.S., Zarrinkar, P.P., Williamson, J.R. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- Vermeulen, A., McCallum, S.A., Pardi, A. Comparison of the global structure and dynamics of native and unmodified tRNAval. Biochemistry. 2005;44:6024–6033. doi: 10.1021/bi0473399. [DOI] [PubMed] [Google Scholar]
- Waldsich, C., Pyle, A.M. A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin, S., Chan, H.S. Conformational entropic barriers in topology-dependent protein folding: Perspectives from a simple native-centric polymer model. J. Phys. Condens. Matter. 2006;18:S307–S328. [Google Scholar]
- Wang, Y., Trewhella, J., Goldenberg, D.P. Small-angle X-ray scattering of reduced ribonuclease A: Effects of solution conditions and comparisons with a computational model of unfolded proteins. J. Mol. Biol. 2008;377:1576–1592. doi: 10.1016/j.jmb.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser, J.K., Winkler, W.C., Breaker, R.R., Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wong, T., Sosnick, T.R., Pan, T. Mechanistic insights on the folding of a large ribozyme during transcription. Biochemistry. 2005;44:7535–7542. doi: 10.1021/bi047560l. [DOI] [PubMed] [Google Scholar]
- Wong, T., Sosnick, T.R., Pan, T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, S.A. Folding mechanisms of group I ribozymes: Role of stability and contact order. Biochem. Soc. Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- Yi, Q., Rajagopal, P., Klevit, R.E., Baker, D. Structural and kinetic characterization of the simplified SH3 domain FP1. Protein Sci. 2003;12:776–783. doi: 10.1110/ps.0238603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar, P.P., Williamson, J.R. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding [see comments] Nat. Struct. Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]