Abstract
An important consequence of protein misfolding related to neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), is the formation of proteinaceous inclusions or aggregates within the central nervous system. We have previously shown that several familial ALS-linked copper-zinc superoxide dismutase (SOD1) mutants (A4V, G85R, and G93A) interact and co-localize with the dynein-dynactin complex in cultured cells and affected tissues of ALS mice. In this study, we report that the interaction between mutant SOD1 and the dynein motor plays a critical role in the formation of large inclusions containing mutant SOD1. Disruption of the motor by overexpression of the p50 subunit of dynactin in neuronal and non-neuronal cell cultures abolished the association between aggregation-prone SOD1 mutants and the dynein-dynactin complex. The p50 overexpression also prevented mutant SOD1 inclusion formation and improved the survival of cells expressing A4V SOD1. Furthermore, we observed that two ALS-linked SOD1 mutants, H46R and H48Q, which showed a lower propensity to interact with the dynein motor, also produced less aggregation and fewer large inclusions. Overall, these data suggest that formation of large inclusions depends upon association of the abnormal SOD1s with the dynein motor. Whether the misfolded SOD1s directly perturb axonal transport or impair other functional properties of the dynein motor, this interaction could propagate a toxic effect that ultimately causes motor neuron death in ALS.
Amyotrophic lateral sclerosis (ALS)2 is a progressive and fatal neurodegenerative disease primarily affecting motor neurons in the spinal cord and brainstem. Approximately 10% of ALS cases are inherited, and of these ∼20% are caused by mutations in the Cu,Zn-superoxide dismutase 1 (SOD1) gene (1, 2). To date, more than 100 mutations scattered throughout the SOD1 protein have been identified (3). Many SOD1 mutants retain nearly normal enzymatic activity, and SOD1 knock-out mice do not develop ALS (4, 5). Therefore, it is believed that mutant SOD1s acquire a toxic “gain-of-function” property.
We recently identified dynein as a component of soluble SOD1-containing high molecular weight (HMW) complexes (6), which have been implicated in neuronal toxicity and may be precursors to SOD1 inclusions (7-9). Moreover, co-immunoprecipitation using tissue lysates from transgenic rodents expressing wild-type (WT), G93A, or G85R SOD1, showed that mutant SOD1 interacted with the dynein-dynactin complex to a much greater extent than did WT SOD1. The association was detected in the pre-symptomatic G93A mice (60 days), and the amount of mutant SOD1 interacting with the dynein complex increased over the disease progression (6).
Dynein-mediated microtubule-dependent retrograde transport has been shown to be important for sequestration of misfolded proteins into large inclusions called aggresomes (10, 11). We hypothesized that the interaction between the dynein motor and mutant SOD1 could play a role in the aggregation and formation of inclusions containing mutant SOD1. Several different types of inclusions occur in the spinal cords of both sporadic and familial ALS patients, including axonal spheroids, Bunina bodies, skein-like inclusions, and Lewy body-like hyaline inclusions (LBHI) (12-17). Similar visible aggregates, predominantly LBHI, have also been observed in mutant SOD1 transgenic mice (18-23). These inclusions can be observed in both neurons and astrocytes and often exhibit positive immunostaining for SOD1 (18-23). Whether SOD1-containing inclusions directly contribute to motor neuron injury in ALS has not been established. Several mechanisms by which aggregated SOD1 could be toxic include: (i) sequestration or depletion of other essential proteins via co-aggregation with misfolded SOD1, (ii) overload of protein degradative capacity by excessive misfolded proteins, or (iii) functional disruption of organelles such as mitochondria by aggregates (7, 8, 19, 24-29).
One objective of this study was to test whether dynein-mediated retrograde transport could contribute to mutant SOD1 inclusion formation. We observed that disruption of the dynein-dynactin machinery by overexpression of the dynactin subunit p50 suppressed the interaction between mutant SOD1 and the dynein complex, reduced inclusion formation of SOD1 mutants, and had a beneficial effect on cell survival. While most SOD1 mutants cause rapid progression of ALS symptoms, a minority of mutants, including the H46R variant, show unusually slow disease progression with a typical survival time longer than 10 years after diagnosis (30-36). A second objective, therefore, was to ask whether the extent of mutant SOD1 interaction with dynein correlates with cellular survival or ALS disease progression. In contrast to aggregation-prone mutants such as A4V and G93A, two other SOD1 mutants (H46R and H48Q) did not significantly interact with dynein nor did they readily form HMW complexes or inclusions in our experimental system. We suggest that the mutant SOD1-dynein interaction may contribute to mutant SOD1-induced toxicity and could influence the rate of disease progression in ALS.
MATERIALS AND METHODS
Plasmids, Cell Culture, and Transfection—H46R and H48Q SOD1 constructs tagged with GFP or FLAG were constructed in the same manner as the previously reported GFP-tagged and FLAG-tagged WT, A4V, G85R, and G93A constructs (9). The untagged SOD1 (9), DsRed-tagged p62 (DsRed-p62) and GST-tagged dynein intermediate chain (GST-DIC) constructs have also been previously reported (6, 37). NSC34 and HEK293 cells were cultured at 37 °C under 5% CO2-95% air in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. 80% confluent cells were transfected with the indicated plasmids using Lipofectamine (Invitrogen) following the manufacturer's instructions. Cells were analyzed 24 or 48 h after transfection as indicated.
Immunofluorescence Microscopy—For immunofluorescence staining, cells were grown and transfected on cover slips. Cells were washed with PBS 24 or 48 h after transfection, fixed in 4% paraformaldehyde in 0.1 m PBS for 30 min, and blocked in 10% heat-inactivated FBS in 0.1 m PBS with 0.1% Triton X-100 (PBST) for 30 min. Slides were then incubated with primary antibodies diluted in 2% FBS-PBST overnight at room temperature. The γ-tubulin (T6557, Sigma), Rab5b (sc-598, Santa Cruz Biotechnology), and protein-disulfide isomerase (PDI, SP-890, Stressgen) were used at 1:500, 1:100, and 1:200 respectively. Following primary antibody incubation, sections were washed with PBST and incubated with 4′,6-diamidino-2-phenylindole dihydrochoride (DAPI, Sigma) at 1:7,500 and Alexa Fluor 594 anti-mouse or Alexa Flour 594 anti-rabbit (Molecular Probes) at 1:300 dilution in 10% FBS-PBST at room temperature for 1 h. Sections were then washed with PBST, and then mounted using Vectashield mounting medium. Fluorescence microscopy was carried out using a Leica DM IRBE laser scanning confocal microscope with a ×100 objective. For analysis of microtubule organization center (MTOC) co-localization of inclusions, ∼100 cells with inclusions were counted for each mutant per experiment and data presented are summarized from three independent experiments.
Quantification of Inclusion Formation—HEK293 or NSC34 cells transfected with different SOD1-GFP constructs were visualized by fluorescence microscopy using a Zeiss Axiovert 100 epifluorescent microscope. Images of cells transfected with different SOD1 constructs were taken under the identical microscope and camera settings. To quantify the number of transfected cells with inclusions, the total number of transfected cells and the number of cells with inclusions were counted in 10 random view fields for each SOD1 construct in three independent experiments. Each view field contained ∼50-150 transfected cells. For SOD1 and p62 co-aggregation quantification, the number of transfected cells with DsRed-positive, GFP-positive, and double-positive inclusions were counted in 10 random view fields in three independent experiments.
Dynein GST Pulldown Assays—GST pulldowns were performed as previously described (6). Briefly, HEK293 cells transfected with SOD1-FLAG and GST-DIC constructs were harvested and lysed in radioimmune precipitation assay (RIPA) buffer. Cell lysates were cleared by centrifugation at 1,000 × g for 10 min, and the protein concentration was determined by Bradford assay. Detergent-soluble lysates (1,000 μg) were incubated with 30 μl of a 50% slurry of glutathione-Sepharose 4B (Amersham Biosciences) in a total volume of 1 ml for 3 h at 4 °C with end-over-end rotation. The glutathione beads were then collected in a microcentrifuge at 500 × g for 5 min at 4 °C, and the supernatant was saved for later analysis. The pelleted glutathione beads were then washed three times with ice-cold RIPA buffer. The bound proteins were then released by boiling in 2× SDS sample loading buffer for 5 min and subjected to 12% SDS-PAGE followed by Western blotting.
Subcellular Fractionation—Cells were harvested 48 h after transfection and lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA) supplemented with 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture (P8340, Sigma). After centrifugation at 1,000 × g for 10 min, the cleared lysates were centrifuged at 25,000 × g for 50 min to obtain the supernatant and pellet fractions. The protein concentration of the supernatant fraction was determined using Bradford assay (Bio-Rad), and 30 μg was loaded onto SDS-PAGE. The pellet fraction was re-suspended in 2× SDS-PAGE loading buffer and directly loaded onto SDS-PAGE followed by Western blotting.
Chemical Cross-linking—The chemical cross-linking was performed as previously published (6). Briefly, NSC34 cells were harvested 48 h after being transfected with 1 μg of SOD1-FLAG. Cell were homogenized in 300 μl of 1× PBS containing 1 mm dithiothreitol and 500× diluted protease inhibitor mixture (P8340, Sigma) on ice using a Dounce homogenizer. After 1,000 × g centrifugation for 10 min at 4 °C, the supernatants (protein concentration adjusted to 2 μg/μl with lysis buffer) was incubated with disuccinimidyl glutarate (DSG, Pierce, 1 mm final concentration) for 1 h at room temperature. The reaction was stopped by adding Tris (pH 7.5, 50 mm final concentration). The reaction mixtures were boiled with 6× SDS-PAGE loading buffer, resolved by 10% SDS-PAGE followed by Western blotting.
Western Blotting—For Western blotting analysis, proteins in SDS-PAGE gels were transferred onto nitrocellulose membranes and blocked in 5% milk in TBST (100 mm Tris-buffered saline, pH 7.5, 0.1% Tween-20) for 1 h at room temperature. After primary antibody incubation, membranes were washed 3× with TBST and incubated with the indicated secondary antibodies in 5% milk in TBST for 1 h at room temperature. Membranes were washed 3× with TBST, and the protein of interest was visualized using SuperSignal West Pico Enhanced Chemiluminescent Substrate (ECL) kit (Pierce). Antibodies used were, anti-GFP (sc-8334, Santa Cruz Biotechnology), anti-FLAG (F-3165, Sigma), anti-GST (sc-459, Santa Cruz Biotechnology), anti-SOD1 (sc-11407, Santa Cruz Biotechnology). Intensities of Western blotting bands were quantified using the Kodak 1D software (version 3.6.1).
Propidium Iodide Staining—To visualize dead and dying cells, cells were stained with 12.5 μg/ml propidium iodide (PI) in the cell culture media before the number of PI-positive transfected cells was counted. Ten random view fields for each transfection in three independent experiments were performed.
RESULTS
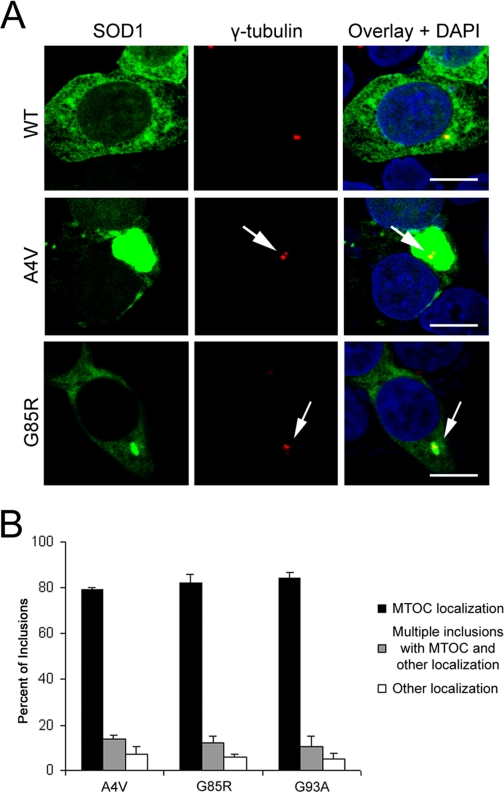
Mutant SOD1 Inclusions Resembled Aggresomes—We reported that the ALS-linked SOD1 mutants A4V, G85R, and G93A interact with the dynein-dynactin complex in cellular and animal models of ALS, while little or no association was observed between WT SOD1 and dynein (6). Dynein-mediated microtubule-dependent transport sequesters misfolded proteins into large inclusions called aggresomes, which can be recognized by their large spherical morphology and close proximity to the microtubule organization center (MTOC) (10, 11, 38, 39). We hypothesized that the interaction between mutant SOD1 and the dynein motor may contribute to the formation of SOD1-containing aggresomes. We therefore initially tested whether the distribution of SOD1 inclusions resembled aggresomes as judged by co-localization of GFP-tagged WT or mutant SOD1 and the MTOC marker γ-tubulin in NSC34 (a mouse motor neuron-like cell line) and HEK293 cells. GFP-tagging of SOD1 was previously shown not to interfere with SOD1 enzymatic activity (9). Fig. 1A shows representative confocal images of GFP-tagged SOD1 and immunostaining of γ-tubulin. WT SOD1 was mainly distributed uniformly within the cytosol while mutant SOD1 formed large inclusions that co-localized with γ-tubulin. Fig. 1B shows the quantitative results of three independent experiments in each of which ∼100 inclusion-containing cells were counted. Approximately 80% of the cells with inclusions contained one large visible protein inclusion that was co-localized with MTOC. In addition, ∼10-15% of the inclusion-forming cells displayed multiple protein inclusions, of which one was co-localized with the MTOC.
FIGURE 1.
Mutant SOD1 inclusions resemble aggresomes. A, 48 h after transfection with SOD1-GFP, HEK293 cells were immunostained for γ-tubulin, a MTOC marker, and analyzed by confocal microscopy. Cells expressing mutant SOD1 developed large inclusions close or around the MTOC. Scale bars are 10 μm. B, quantitative analysis of co-localization of mutant SOD1 inclusions and MTOC. Approximately 100 cells with inclusions were counted for each mutant per experiment, and data from three independent experiments arepresented.
Possible co-localization of mutant SOD1 inclusions with other organelles such as the endoplasmic reticulum (ER) and endosomes was examined similarly by immunofluorescence and confocal microscopy. No consistent co-localization of inclusions with the ER marker PDI or endosomal marker Rab5B was observed (supplemental Fig. S1). Collectively, these data indicated that the majority of mutant SOD1 formed large inclusions near MTOC in these cultured cells, suggesting a similarity to aggresomes.
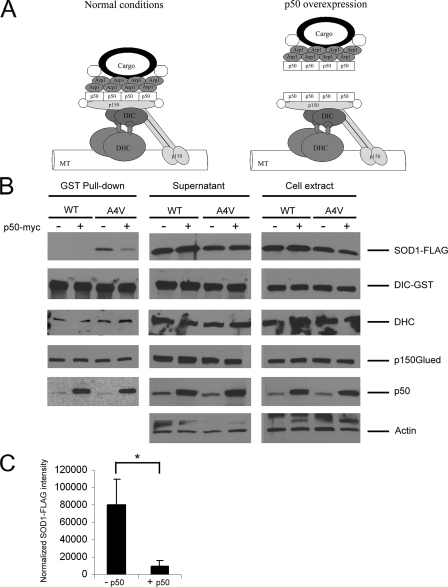
Disruption of Dynein-Dynactin by Overexpression of p50 (Dynamitin) Suppressed the Interaction between Mutant SOD1 and Dynein—The dynactin complex is essential for most dynein functions in vivo; it increases dynein processivity as well as serves as an adaptor between dynein and various cargos (40). Under physiological conditions, dynactin comprises multiple subunits that form a distinct structure: a cargo binding Arp1 filament base and a projecting sidearm linked to the base by a shoulder domain (Fig. 2A, left panel). The projecting sidearm of dynactin is formed by two dimeric p150Glued subunits and is involved in the interaction with both dynein (via DIC) and microtubules. A tetramer of the p50 (also called dynamitin) subunit forms the shoulder of dynactin that links the sidearm to the filament base. Overexpression of p50 competitively dissociates the p150Glued sidearm from the Arp1 filament base, thereby dissociating the cargo from dynein (Fig. 2A, right panel) and inhibiting dynein-mediated retrograde transport (10, 38-42).
FIGURE 2.
Disruption of the dynein-dynactin complex by p50 overexpression blocks the mutant SOD1-dynein interaction. A, schematic drawing of the dynein-dynactin complex under normal and p50 overexpression conditions. Dynein consists of several subunits including two DHC and two DIC. Dynactin also comprises multiple subunits including eight Arp1, four p50, and two p150Glued. Overexpressed exogenous p50 has been reported to dissociate the cargo binding region from the remaining dynactin complex (42). B, GST pulldowns were performed from HEK293 cells transfected with SOD1-FLAG (WT or A4V), GST-DIC and p50-myc or vector control. Western blotting showed that co-precipitation of A4V mutant SOD1 with DIC-GST was abolished in the presence of p50-Myc overexpression. WT-FLAG did not interact with DIC-GST either in the presence or absence of p50-myc overexpression. DHC and p150Glued were co-precipitated with DIC under all conditions. Overexpression of p50-myc increased the amount of p50 co-precipitated with DIC. C, quantitative analysis from three independent experiments showed an approximately eight times reduction in the amount of A4V SOD1 co-precipitated with DIC in the presence of p50-myc. The band intensity of co-precipitated SOD1 was normalized against that of DIC precipitated. *, p < 0.02.
Because the ALS-linked SOD1 mutants were shown to associate with the dynein complex (6), we tested whether inhibition of the dynein-dynactin complex by p50 overexpression would influence the interaction between mutant SOD1 and dynein. HEK293 cells were co-transfected with WT or mutant FLAG-tagged SOD1, GST-DIC or vector control, and p50-myc or vector control. Forty-eight hours after transfection, cell lysates were prepared and subjected to GST pulldowns as previously described (6). As shown in Fig. 2B, the A4V mutant, but not WT SOD1, co-precipitated with DIC in the absence of p50 overexpression, consistent with our previous results. When p50 was overexpressed, however, the amount of A4V mutant co-precipitated by DIC significantly decreased (Fig. 2C). In contrast, dynein heavy chain (DHC) co-precipitated with DIC in all samples, suggesting that the dynein components remained largely intact even during p50 overexpression. Similarly, the p150Glued dynactin subunit, which also binds to DIC, co-precipitated in all samples. An increased association of the p50 dynactin subunit was observed when p50 was overexpressed, consistent with previous reports that exogenous p50 binds to and dissociates the Arp1 cargo binding base from dynactin (40, 42). These results suggested that mutant SOD1 likely interacted with dynein via the dynactin subunits that can be dissociated from dynein by p50 overexpression, which is more characteristic of other dynein motor cargos (40).
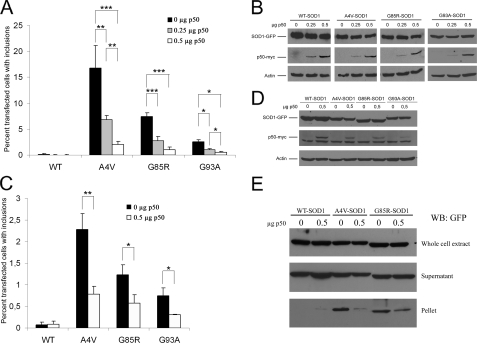
Disrupting the Interaction between Mutant SOD1 and Dynein Reduced Formation of Large Mutant SOD1 Inclusions—We next tested whether disruption of mutant SOD1-dynein interaction by p50 overexpression influenced the formation of mutant SOD1 inclusions. The percentage of cells containing mutant SOD1 inclusions in the absence and presence of p50 overexpression was determined, and the results are shown in Fig. 3. Overexpression of the p50 dynactin subunit prevented the formation of large A4V, G85R, and G93A inclusions in a dose-dependent manner in both HEK293 (Fig. 3A) and NSC34 (Fig. 3C) cells. The expression levels of the SOD1-GFPs were unchanged in the absence and presence of p50 overexpression (Fig. 3, B and D). Consistent with fewer inclusions, cell fractionation experiments also revealed decreased levels of A4V and G85R in the insoluble pellet fraction when p50 was overexpressed (Fig. 3E). The results in Figs. 2 and 3 suggest that an interaction with and transport by the dynein-dynactin motor apparatus contributes to the formation of SOD1-containing inclusions.
FIGURE 3.
Overexpression of p50 prevents inclusion formation by mutant SOD1. A, HEK293 cells were transfected with 0.5 μg of various SOD1-GFP constructs and different amounts of p50-myc plasmid. The total amount of plasmid transfected in each well was kept constant at 1 μg by adding empty vector control for p50-myc. The percentage of transfected cells with inclusions was measured 48 h after transfection. B, Western blotting of protein extracts prepared from cells in A after counting, showing similar levels of SOD1 and increased levels of p50 with increased amounts of transfected p50 plasmid. C, inclusion analysis in NSC34 cells. Experiments were carried out in the same manner as in A, except NSC34 cells were used. D, Western blotting of protein extracts prepared from NSC34 cells in the same manner as in C. E, amount of SOD1 in the whole extract, supernatant, and detergent-resistant pellet fractions in the presence and absence of p50 overexpression. The amounts of mutant A4V and G85R in the pellet fraction was reduced when p50 was overexpressed. WT SOD1 was not or very weakly detected in the pellet fraction under both conditions. Data presented in A and C were obtained from three independent experiments. *, p < 0.04; **, p < 0.004; and ***, p < 0.0004.
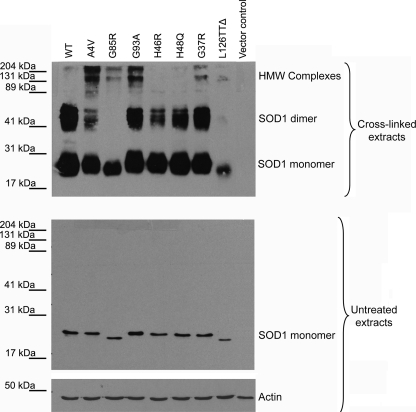
H46R and H48Q SOD1 Mutants Interacted with the Dynein-Dynactin Complex with Low Affinity—Given that interaction of inclusion-forming SOD1 mutants such as A4V with the dynein motor may be important for aggregation, we next hypothesized that SOD1 mutants which are less prone to form HMW complexes or inclusions will interact less efficiently with dynein-dynactin. We used the previously reported chemical cross-linking assay (9) to identify ALS-linked SOD1 mutants with decreased tendency to form HMW complexes. We transfected NSC34 cells with various FLAG-tagged SOD1 mutants, harvested cells 48 h after transfection, and incubated the fresh lysates with an amino-reactive bi-functional cross-linker, disuccinimidyl glutarate (DSG). The reaction proceeded at room temperature for 1 h before being quenched by excess Tris. The reaction mixtures were analyzed by SDS-PAGE and Western blotting using an anti-FLAG antibody. Only a single band corresponding to monomeric SOD1 was observed in the absence of cross-linker for both WT and mutant SOD1s (Fig. 4). In the presence of DSG, species corresponding to both monomeric and dimeric SOD1 were detected, indicating that SOD1 subunits could be covalently cross-linked to stabilize the dimeric state. Moreover, DSG-treated mutants A4V, G85R, G93A, G37R, and L126TTΔ formed HMW complexes (Fig. 4), as did several other mutants (I113T, E100G, and L144F, data not shown). In contrast, mutants H46R and H48Q behaved like WT SOD1 and did not form an appreciable fraction of HMW complexes. In control experiments where DSG was added to fresh cell lysates of NSC34 cells transfected with empty p3xFLAG-CMV14 vector, no FLAG-positive bands could be detected. These results indicated that the two mutants H46R and H48Q differed from other SOD1 mutants under these conditions in that they were significantly less prone to form HMW complexes.
FIGURE 4.
H46R and H48Q form none or few HMW complexes compared with other ALS-linked SOD1 mutants. Fresh NSC34 cell lysates containing WT or various SOD1-FLAG mutants were incubated with the cross-linker DSG for 60 min before the reaction was quenched. The reaction mixtures were resolved in 10% SDS-PAGE and blotted with a FLAG antibody. After cross-linking, WT SOD1 migrated as both monomer and dimer; however, all mutants with the exception of H46R and H48Q showed formation of HMW complexes in addition to monomeric and dimeric forms. As the control in SDS-PAGE, all SOD1-FLAG ran as monomers in the absence of DSG (second panel), and actin blotting showed equal loading of all samples (third panel).
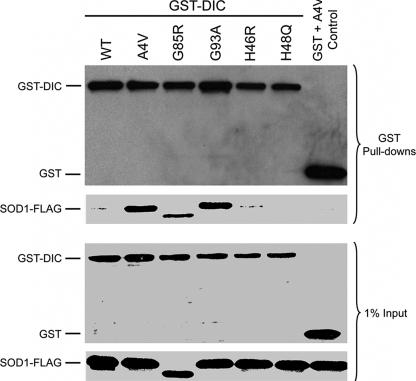
We next tested whether the H46R and H48Q SOD1 mutants showed lower affinity for the dynein-dynactin complex than did mutants that form HMW complexes. GST pulldowns were performed using lysates from cells co-expressing FLAG-tagged WT or mutant SOD1 and GST-tagged dynein intermediate chain (DIC). The GST pulldown samples were subjected to SDS-PAGE and Western blotting analysis. As shown in Fig. 5, mutants A4V, G85R and G93A all co-precipitated with DIC-GST, which is consistent with our previous studies (6). In contrast, H46R and H48Q behaved similar to WT SOD1 and showed no or little co-precipitation with the dynein complex (Fig. 5). As a control, neither WT SOD1 nor any of the mutants were pulled down when GST in the absence of DIC was co-expressed with each SOD1 construct (A4V co-expressed with GST alone was shown at the right lane in Fig. 5, data not shown for other mutants), indicating that DIC is required for the interaction with mutant SOD1 under these conditions.
FIGURE 5.
H46R and H48Q show little affinity for dynein. GST pulldowns were performed from HEK293 cells transfected with GST-DIC and various FLAG-tagged SOD1 constructs. Western blotting for GST and FLAG revealed that WT, H46R, or H48Q SOD1 was not or to very little extent co-precipitated by DIC-GST. However, the inclusion-forming mutants A4V, G85R, and G93A were all readily pulled down together with DIC-GST. Empty GST vector and A4V SOD1 were co-transfected as negative controls showing no co-precipitation of empty GST and A4V SOD1.
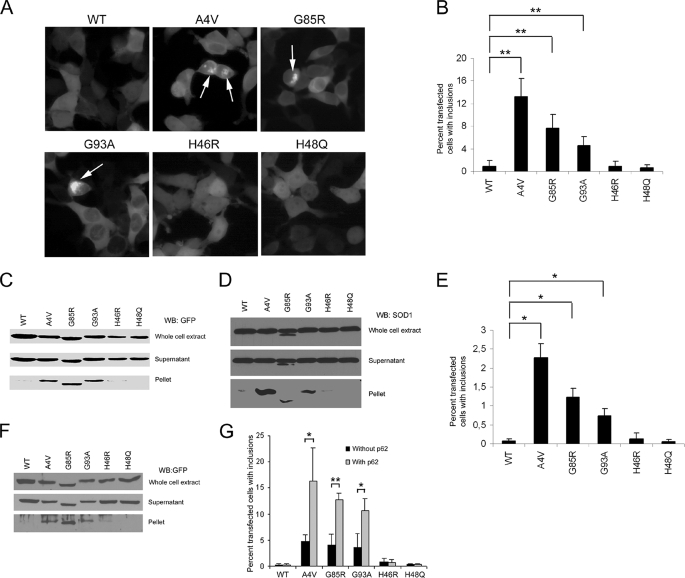
H46R and H48Q Did Not Form Inclusions in Transfected Cells—Because H46R and H48Q SOD1 mutants largely did not interact with the dynein complex, we investigated whether and to what extent H46R and H48Q SOD1 mutants might form insoluble inclusions in live cells. NSC34 or HEK293 cells were transfected with various SOD1-GFP fusion constructs, and the percentage of transfected cells (GFP-positive) with inclusions (indicated by arrows in Fig. 6A) was determined after 48 h. As seen in Fig. 6B, A4V mutant SOD1 showed strong inclusion formation in 13.3 ± 3.2% of the transfected HEK293 cells (p < 0.0003). Similarly, G85R and G93A mutants also showed significantly increased formation of inclusions compared with WT SOD1. G85R and G93A formed inclusions in 7.6 ± 2.4% and 4.6 ± 1.5% of the transfected cells (p < 0.0003), respectively. However, under the same conditions neither H46R nor H48Q showed an increased inclusion formation compared with WT SOD1. Consistent with this, cell lysate fractionation experiments using GFP-tagged (Fig. 6C) or untagged mutant SOD1 (Fig. 6D) revealed no or little H46R or H48Q in the detergent-resistant pellet fractions. In contrast, A4V, G85R and G93A were clearly detectable in the pellet fractions independent of whether GFP-tagged or untagged SOD1 was used in the experiment.
FIGURE 6.
H46R and H48Q mutants behave as WT-SOD1 and do not form inclusions in transfected cells. A, H46R and H48Q do not form inclusions like the A4V, G85R, and G93A mutant SOD1s. HEK293 cells were transfected with SOD1-GFP plasmids and photographed 48 h later using a Nikon Eclipse epifluorescence microscope with identical settings for WT and mutant SOD1s. B, quantification of mutant SOD1 inclusion formation in HEK293 cells. A4V, G85R, and G93A showed a statistically significant increase in inclusion formation versus WT SOD1, whereas H46R and H48Q behaved like WT. **, p < 0.0003. C, fractionation of HEK293 cell lysate 48 h after cells were transfected with SOD1-GFP. A4V, G85R, and G93A were all detected in the pellet fraction, whereas H46R and H48Q remained soluble, similar to WT SOD1. D, fractionation of HEK293 cell lysate 48 h after cells were transfected with untagged SOD1. Increased levels of untagged A4V, G85R, and G93A were observed in pellet fractions compared with WT, H46R, and H48Q. E, quantification of mutant SOD1 inclusion formation in NSC34 cells. A4V, G85R, and G93A, but neither H46R nor H48Q, showed a statistically significant increase in inclusion formation versus WT SOD1. *, p < 0.003. F, fractionation of NSC34 cell lysate as described in C showing increased amounts of A4V, G85R, and G93A in the insoluble pellet fraction. G, quantification of mutant SOD1 inclusion formation in the presence and absence of p62 overexpression. HEK293 cells were transfected with various SOD1-GFP constructs and DsRed-p62 or the vector control. The percentage of transfected cells with inclusions were measured 24 h after transfection. Inclusions formed by H46R or H48Q remained unchanged even with p62 overexpression, whereas inclusions by A4V, G85R, and G93A increased in the presence of p62 overexpression. *, p < 0.05 and **, p < 0.01. Quantitative data in B, E, and G were obtained from three independent experiments.
Parallel experiments were performed using NSC34 cells to assess pulldown with GST-DIC and inclusion formation. Although the absolute percentage of NSC34 cells with inclusions was less compared with HEK293 cells, the relative inclusion formation capabilities of the various mutants were similar in NSC34 cells. As shown in Fig. 6E, A4V, G85R, and G93A all formed more inclusions than WT SOD1 (p < 0.003), whereas H46R and H48Q did not. Furthermore, fractionation experiments using NSC34 cells again revealed an increased presence of A4V, G85R, and G93A, but not WT, H46R, or H48Q in the insoluble pellet fraction (Fig. 6F).
We have recently shown that the p62 protein (also named Sequestosome 1) accumulates and co-localizes with SOD1 in spinal cord inclusions in G93A transgenic mice and that overexpression of p62 increases the formation of large aggregates by A4V and G93A SOD1 mutants in cultured cells (37). We thus tested whether overexpression of p62 could induce formation of large inclusions of H46R or H48Q mutants. We transfected HEK293 or NSC34 cells with DsRed-p62 and H46R-GFP or H48Q-GFP as previously described. As shown in Fig. 6G, p62 overexpression further increased the formation of large inclusions by A4V (p < 0.05), G93A (p < 0.05), and G85R (p < 0.01) mutants. In contrast, H46R or H48Q behaved like WT and rarely formed inclusions even with elevated p62 levels. These data indicate that H46R and H48Q mutants, which do not interact strongly with the dynein-dynactin complex, do not form inclusions under the conditions where other dynein-interacting SOD1 mutants form obvious inclusions. Overall, the correlation between the mutant SOD1-dynein interaction and the ability to form inclusions suggests that the interaction is likely to play an important role in the formation of mutant SOD1 inclusions.
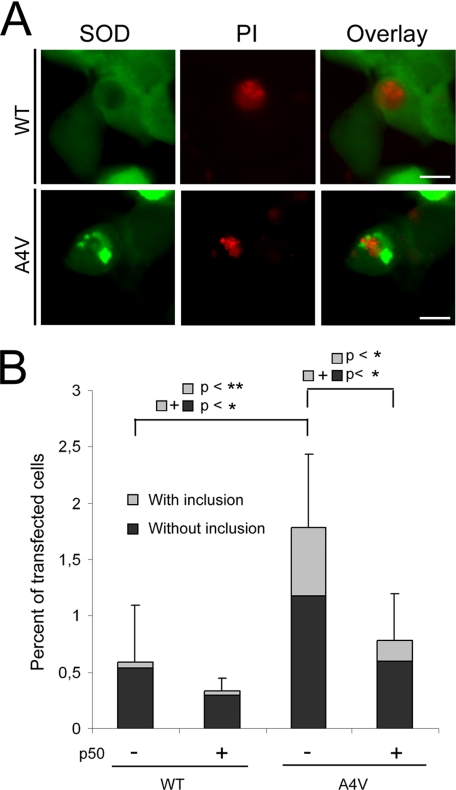
Disruption of the Interaction of Mutant SOD1 with Dynein-affected Cell Viability—Whether or how aggregates containing mutant SOD1 might be toxic to motor neurons has not been established. If aggregation or inclusion formation contributes mechanistically to the pathophysiology, mutants with lower aggregation potential like H46R might be expected to be less toxic than mutants like A4V. The long survival of familial ALS patients carrying H46R mutant SOD1 has been reported (30-36). In Fig. 7, we disrupted the interaction of A4V SOD1 with the dynein motor by co-expressing p50 in HEK293 cells and correlated this to changes in viability and inclusion formation. HEK293 cells were used because a higher percentage of HEK293 cells contained mutant SOD1 inclusions compared with NSC34 cells. Even so, only the highest inclusion-forming mutant A4V showed a statistically significant increase in the total number of PI-positive cells (black plus gray) compared with WT SOD1 (p < 0.05) 48 h after transfection (Fig. 7). Moreover, analysis of the inclusion burden of the PI-positive cells revealed that, compared with WT SOD1-expressing cells, a majority of the cells making up the increased number of A4V-expressing PI-positive cells also contained inclusions (compare gray bars 1 and 3, p < 0.03). In the presence of p50 overexpression, a decrease in the total number of PI-positive cells (compare total bars 3 and 4, p < 0.05) and a decrease in the number of PI-positive inclusion-containing cells (compare gray bars 3 and 4, p < 0.03) were both detected. The results suggest that suppression of mutant SOD1-dynein interaction, decreased inclusion formation, and improved cell survival could be correlated at least in the cell culture system.
FIGURE 7.
Disruption of the interaction of mutant SOD1 with dynein affects cell viability. Cell viability in the absence and presence of p50 overexpression was determined by the percentage of PI-positive transfected cells. A, representative images show PI-positive HEK293 cells expressing WT or A4V SOD1 in the absence of p50 overexpression. B, quantification of cell viability obtained from three independent experiments. The black bars are the percentage of PI-positive cells without inclusions, and the gray bars are the percentage of PI-positive inclusion-containing cells. Standard deviations shown are for the total number of PI-positive cells. *, p < 0.05 and **, p < 0.03.
DISCUSSION
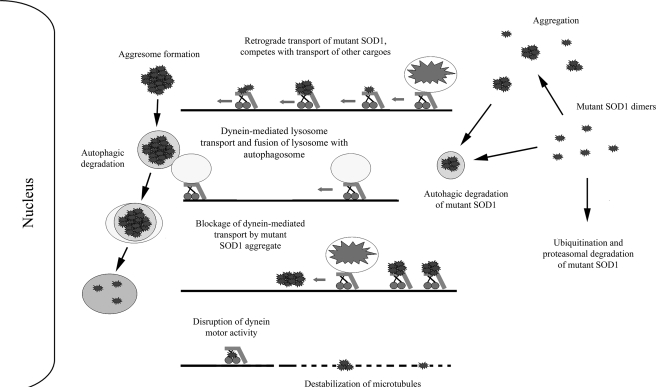
In this study, we show that disruption of the dynein-dynactin complex by overexpressing the p50 dynactin subunit significantly decreased the interaction between dynein and the ALS-linked SOD1 mutants A4V, G85R, and G93A (Fig. 2). Moreover, p50 overexpression also decreased the inclusion formation of these mutants in cultured cells in a dose-dependent manner (Fig. 3). In addition, using cross-linking, cell fractionation, microscopic and pulldown strategies, we identified two specific SOD1 mutants, H46R and H48Q, which showed a significantly lower affinity for interacting with dynein even without p50 overexpression (Fig. 5). We further demonstrated that these two mutants, in contrast to the aggregation-prone mutants, showed minimal tendency to form SOD1 containing HMW complexes and large inclusions (Figs. 4 and 6). These data suggest that the aggregation-prone SOD1 mutants interact with the dynein complex with high affinity and are sequestered by the dynein-mediated transport machinery to form large inclusions. However, the specific form(s) of mutant SOD1 that interact with dynein, whether misfolded monomeric forms or oligomeric intermediates or small aggregates of misfolded mutant SOD1, remain unclear. It is likely that the mutations in SOD1 cause substantial misfolding that can subsequently induce oligomerization of mutant SOD1 after which the dynein complex could facilitate the sequestering of oligomers/mini-aggregates into larger inclusions (Fig. 8).
FIGURE 8.
Potential role(s) of the dynein-dynactin motor complex in mutant SOD1-mediated ALS. Dynein-dynactin can sequester and transport mutant SOD1 to form large aggresome-like inclusions that require autophagic degradation. Dynein is also involved in the autophagosome-lysosome fusion that is critical to the degradation of misfolded protein inclusions in autophagosomes. Interplay of the two dynein-dependent events, the formation and degradation of inclusions, determines the eventual inclusion burden in neurons. Increased levels of mutant SOD1 associated with dynein as the disease progresses could contribute to decreased axonal transport in ALS by multiple mechanisms. These potential mechanisms illustrated here include competitive occupancy of the dynein-mediated transport capacity, disruption of dynein motor activity, destabilization of microtubules, physical blockage of transport by mutant SOD1 aggregates in the axon. Decreased retrograde transport of cargos such as mitochondria and neurotrophic factors as well as increased aggregation burden may determine the toxicity of mutant SOD1 and ALS disease manifestation.
The reduced inclusion formation of H46R and H48Q observed in this study is in agreement with the pathological observations that the ALS patients harboring these mutations fail to show SOD1-positive inclusions (30-36). Another study also showed significantly less H46R and H48Q in the detergent-insoluble fraction compared with other mutants such as A4V and G93A in cultured cells (43). The same study also showed that no large SOD1 positive inclusions could be identified in H46R-H48Q double or H46R-H48Q-H63G-H120H SOD1 quadruple mutant mice using immunohistological techniques. However, smaller insoluble forms of SOD1 could be detected in these mice using a filtration assay (8, 43). Large SOD1-positive inclusions have been observed in end-stage H46R SOD1 transgenic rats (44). Compared with the expression levels of mutant SOD1 in our cell culture system and in ALS patients, the transgenic rats with the paralytic phenotype had ∼25 copies of the human H46R mutant SOD1 transgene and expressed H46R at a level that was estimated to be six times of the endogenous rat SOD1. It is possible that the extremely high expression levels of H46R in the rats contributed to the formation of inclusions in this animal model.
Dynein is the motor protein responsible for retrograde transport that is essential to multiple cellular functions, such as returning damaged membranous organelles from the axon terminal to the cell body and supplying neurotrophic growth factor support for neuronal survival (45, 46). Because this study shows that mutant SOD1 is sequestered to form large inclusions by interacting with dynein, it is logical to propose that the mutant SOD1-dynein interaction could also interfere with the dynein-mediated axonal transport in ALS. Impaired axonal transport, but not a complete inhibition of transport, has been reported in both ALS patients and animal models (47-52). However, the mechanisms underlying this impairment are unclear. Several possibilities of how mutant SOD1 could interfere with retrograde transport could be envisioned (Fig. 8), including competition for dynein transport, disruption of dynein motor activity, physical blockage of dynein transport by mutant SOD1 aggregates, and destabilization of microtubules. A recent study showed that microtubule destabilization is an early event in ALS, and a microtubule stabilization drug had beneficial effects in ALS mice (53). Reduced number of available microtubule tracks in combination with increased levels of misfolded mutant SOD1 bound to dynein could over time lead to severe reduction of transport of other cargos such as neurotrophic factors necessary to promote neuronal survival, thus causing neurotoxicity.
The dependence of aggregate formation on the dynein-mediated transport and the impairment of the dynein-mediated transport by mutant SOD1, which may appear to be contradictory to each other, can be two distinct consequences of the mutant SOD1-dynein interaction. Although the aberrant interaction between mutant SOD1 and dynein may contribute to the reduced transport by multiple mechanisms as discussed above, the remaining transport can conceivably be utilized to sequester mutant SOD1 to large inclusions. In turn, the more transport capacity is utilized to form inclusions, the less capacity will be available to transport other cargos. The expected consequence over time is both the appearance of large inclusions and the impaired axonal transport.
In addition to transporting organelles and neurotrophic factors, dynein-mediated transport has also been implicated in several steps of autophagic degradation of misfolded proteins. In the autophagic degradation pathway, large protein aggregates are engulfed by membranous structures to form autophagosomes, which are subsequently fused with lysosomes containing hydrolytic enzymes (54). Dynein is believed to collect and transport misfolded proteins to inclusion structures called aggresomes that are suggested to be degraded by autophagic degradation (10, 39). In addition, dynein is also important for the fusion of the autophagosome and the lysosome (Refs. 38, 41, see Fig. 8). Disruption of dynein function may interfere with the sequestering of misfolded proteins to inclusions, as well as impair the autophagosome-lysosome fusion and the degradation of misfolded proteins. Thus, disruption of dynein function may affect the eventual aggregation burden in cells differentially in different systems depending on which factor is more important. A recent study showed that reduced dynein function caused by a dynein heavy chain mutation (Legs at odd angle, Loa) resulted in decreased autophagic clearance of mutant Huntingtin protein by preventing autophagosome-lysosome fusion. The study also showed that the reduced dynein function caused increased inclusion burden and toxicity in a Huntington disease model (38). It is possible that a high propensity of mutant huntingtin to self-aggregate could make the role of dynein-mediated sequestration less important. Thus, inhibition of dynein function could more affect the role of dynein in the autophagosome-lysosome fusion and the degradation of inclusions. In contrast, we demonstrate in this study that disruption of the dynein-dynactin motor complex could suppress the formation of mutant SOD1 inclusions, i.e. the dynein-mediated transport is critical to the formation of mutant SOD1 inclusions in the first place.
The role(s) of dynein in ALS disease has proven to be more complex (see reviews in Refs. 55, 56). As discussed earlier, reduced dynein-mediated transport can contribute to deficiencies in the turnover of damaged organelles and/or insufficient neurotrophic support to neurons. Surprisingly, crossing G93A ALS mice with the dynein mutant Loa mice had a positive effect on ALS disease onset and progression (57) in contrast to the negative effect on Huntington disease in the other study (38). Similarly another dynein heavy chain mutation, Cra1 (Cramping 1), also had a positive effect on the ALS disease progression when crossed with G93A mice (58). In line with these data, disruption of mutant SOD1-dynein interaction, suppression of mutant SOD1 inclusion formation and improved cell survival appeared to be correlated in the cell culture system in this study. Furthermore, it is noted that patients carrying the H46R mutation, which does not interact with dynein and is not prone to form inclusions in this study, survive 6-8 times longer than patients carrying the A4V mutation that shows strong interaction with dynein and high aggregation capacity (30-36). Analysis of various dynein-dynactin mutant mice have revealed that different dynein-dynactin functions, such as long distance retrograde transport and dynein-dependent degradative pathways can be affected independently of each other (59, 60). Although blockage of the role of dynein in inclusion formation might be beneficial, dynein is also essential to reduce toxicity by clearing away inclusions once inclusions form. It remains to be determined experimentally how dynein functions in the autophagic degradation of mutant SOD1 inclusions. Pharmacological inhibition of autophagy by 3-methyladenine or inhibition of lysosomal proteolysis by ammonium chloride was shown to increase mutant SOD1 toxicity and cell death in Neuro2A cells expressing mutant SOD1 (61). However, this study did not measure the inclusion formation or address the role of dynein directly. It is expected that in these cells with functional dynein, large inclusions should form and the blockage of autophagy would interfere with clearance of the inclusions thus causing increased toxicity.
In summary, we have shown that only SOD1 mutants with high propensity to form HMW complexes or inclusions interact with dynein and that blockage of the mutant SOD1-dynein interaction reduces inclusion formation and has a positive effect on cell survival. These data suggest that the intrinsic misfolding caused by each mutation may help to determine whether a specific mutant interacts with the dynein complex and that this interaction could play a role in sequestering mutant SOD1 into larger inclusion structures. The dependence of mutant SOD1 inclusions on the dynein-mediated retrograde transport demonstrated in this study, and the impairment of the axonal transport and potential interference with other essential dynein functions are likely to be different outcomes of the aberrant mutant SOD1-dynein interactions. These events could contribute to an aggravating cycle that may determine the mutant SOD1 toxicity in ALS.
Supplementary Material
Acknowledgments
We thank Dr. Joan S. Valentine at UCLA for providing the WT and mutant SOD1 cDNAs and Dr. Richard Valle at Columbia University for providing the p50-myc plasmid.
This work was supported, in whole or in part by National Institutes of Health Grants R01-NS49126 (to H. Z.) and R01-NS44170 (to L. J. H.). This work was also supported by the Muscular Dystrophy Association (to L. J. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: ALS, amyotrophic lateral sclerosis; SOD, copper-zinc superoxide dismutase; FBS, fetal bovine serum; PBS, phosphate-buffered saline; GST, glutathione S-transferase; WT, wild type; ER, endoplasmic reticulum; HMW, high molecular weight; MTOC, microtubule organization center; PI, propidium iodide; DIC, dynein intermediate chain; DSG, disuccinimidyl glutarate.
References
- 1.Deng, H. X., Hentati, A., Tainer, J. A., Iqbal, Z., Cayabyab, A., Hung, W. Y., Getzoff, E. D., Hu, P., Herzfeldt, B., Roos, R. P., Warner, C., Deng, G., Soriano, E., Smyth, C., Parge, H. E., Ahmed, A., Roses, A. D., Hallewell, R. A., Pericak-Vance, M. A., and Siddique, T. (1993) Science 261 1047-1051 [DOI] [PubMed] [Google Scholar]
- 2.Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., Donaldson, D., Goto, J., O'Regan, J. P., Deng, H. X., Rahmani, Z., Krizus, A., McKenna-Yasek, D., Cayabyab, A., Gaston, S. M., Berger, R., Tanzi, R. E., Halperin, J. J., Herzfeldt, B., Vandenbergh, R., Hung, W.-Y., Bird, T., Deng, G., Mulder, D. W., Smyth, C., Laing, N. G., Soriano, E., Pericak-Vance, M. A., Haines, J., Rouleau, G. A., Gusella, J. S., Horvitz, H. R., and Brown, R. H. (1993) Nature 362 59-62 [DOI] [PubMed] [Google Scholar]
- 3.Gaudette, M., Hirano, M., and Siddique, T. (2000) Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1 83-89 [DOI] [PubMed] [Google Scholar]
- 4.Borchelt, D. R., Lee, M. K., Slunt, H. S., Guarnieri, M., Xu, Z. S., Wong, P. C., Brown, R. H., Jr., Price, D. L., Sisodia, S. S., and Cleveland, D. W. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8292-8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reaume, A. G., Elliott, J. L., Hoffman, E. K., Kowall, N. W., Ferrante, R. J., Siwek, D. F., Wilcox, H. M., Flood, D. G., Beal, M. F., Brown, R. H., Jr., Scott, R. W., and Snider, W. D. (1996) Nat. Genet. 13 43-47 [DOI] [PubMed] [Google Scholar]
- 6.Zhang, F., Strom, A. L., Fukada, K., Lee, S., Hayward, L. J., and Zhu, H. (2007) J. Biol. Chem. 282 16691-16699 [DOI] [PubMed] [Google Scholar]
- 7.Johnston, J. A., Dalton, M. J., Gurney, M. E., and Kopito, R. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12571-12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, J., Xu, G., Gonzales, V., Coonfield, M., Fromholt, D., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2002) Neurobiol. Dis. 10 128-138 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, F., and Zhu, H. (2006) Biochim. Biophys. Acta 1760 404-414 [DOI] [PubMed] [Google Scholar]
- 10.Johnston, J. A., Illing, M. E., and Kopito, R. R. (2002) Cell Motil. Cytoskeleton 53 26-38 [DOI] [PubMed] [Google Scholar]
- 11.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, C. Z., and Hays, A. P. (2004) J. Neurol. Sci. 217 47-54 [DOI] [PubMed] [Google Scholar]
- 13.Hirano, A. (1996) Neurology 47 S63-66 [DOI] [PubMed] [Google Scholar]
- 14.Ikemoto, A., Hirano, A., and Akiguchi, I. (2000) Amyotroph Lateral Scler Other Motor Neuron Disord. 1 97-104 [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, S., Kusaka, H., Ito, H., Shibata, N., Asayama, T., and Imai, T. (1996) Clin. Neuropathol. 15 41-46 [PubMed] [Google Scholar]
- 16.Piao, Y. S., Wakabayashi, K., Kakita, A., Yamada, M., Hayashi, S., Morita, T., Ikuta, F., Oyanagi, K., and Takahashi, H. (2003) Brain Pathol. 13 10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata, N., Hirano, A., Kobayashi, M., Sasaki, S., Kato, T., Matsumoto, S., Shiozawa, Z., Komori, T., Ikemoto, A., Umahara, T., and Asayama, K. (1994) Neurosci. Lett. 179 149-152 [DOI] [PubMed] [Google Scholar]
- 18.Bruijn, L. I., Becher, M. W., Lee, M. K., Anderson, K. L., Jenkins, N. A., Copeland, N. G., Sisodia, S. S., Rothstein, J. D., Borchelt, D. R., Price, D. L., and Cleveland, D. W. (1997) Neuron 18 327-338 [DOI] [PubMed] [Google Scholar]
- 19.Bruijn, L. I., Houseweart, M. K., Kato, S., Anderson, K. L., Anderson, S. D., Ohama, E., Reaume, A. G., Scott, R. W., and Cleveland, D. W. (1998) Science 281 1851-1854 [DOI] [PubMed] [Google Scholar]
- 20.Kato, S., Sumi-Akamaru, H., Fujimura, H., Sakoda, S., Kato, M., Hirano, A., Takikawa, M., and Ohama, E. (2001) Acta Neuropathol. (Berl) 102 233-238 [DOI] [PubMed] [Google Scholar]
- 21.Shibata, N., Hirano, A., Kobayashi, M., Dal Canto, M. C., Gurney, M. E., Komori, T., Umahara, T., and Asayama, K. (1998) Acta Neuropathol. (Berl) 95 136-142 [DOI] [PubMed] [Google Scholar]
- 22.Watanabe, M., Dykes-Hoberg, M., Cizewski Culotta, V., Price, D. L., Wong, P. C., and Rothstein, J. D. (2001) Neurobiol. Dis. 8 933-941 [DOI] [PubMed] [Google Scholar]
- 23.Watanabe, Y., Yasui, K., Nakano, T., Doi, K., Fukada, Y., Kitayama, M., Ishimoto, M., Kurihara, S., Kawashima, M., Fukuda, H., Adachi, Y., Inoue, T., and Nakashima, K. (2005) Brain Res. Mol. Brain Res. 135 12-20 [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., Lillo, C., Jonsson, P. A., Vande Velde, C., Ward, C. M., Miller, T. M., Subramaniam, J. R., Rothstein, J. D., Marklund, S., Andersen, P. M., Brannstrom, T., Gredal, O., Wong, P. C., Williams, D. S., and Cleveland, D. W. (2004) Neuron 43 5-17 [DOI] [PubMed] [Google Scholar]
- 25.Menzies, F. M., Cookson, M. R., Taylor, R. W., Turnbull, D. M., Chrzanowska-Lightowlers, Z. M. A., Dong, L., Figlewicz, D. A., and Shaw, P. J. (2002) Brain 125 1522-1533 [DOI] [PubMed] [Google Scholar]
- 26.Pasinelli, P., Belford, M. E., Lennon, N., Bacskai, B. J., Hyman, B. T., Trotti, D., and Brown, R. H., Jr. (2004) Neuron 43 19-30 [DOI] [PubMed] [Google Scholar]
- 27.Urushitani, M., Kurisu, J., Tsukita, K., and Takahashi, R. (2002) J. Neurochem. 83 1030-1042 [DOI] [PubMed] [Google Scholar]
- 28.Wong, P. C., Pardo, C. A., Borchelt, D. R., Lee, M. K., Copeland, N. G., Jenkins, N. A., Sisodia, S. S., Cleveland, D. W., and Price, D. L. (1995) Neuron 14 1105-1116 [DOI] [PubMed] [Google Scholar]
- 29.Zhu, S., Stavrovskaya, I. G., Drozda, M., Kim, B. Y., Ona, V., Li, M., Sarang, S., Liu, A. S., Hartley, D. M., Wu du, C., Gullans, S., Ferrante, R. J., Przedborski, S., Kristal, B. S., and Friedlander, R. M. (2002) Nature 417 74-78 [DOI] [PubMed] [Google Scholar]
- 30.Aoki, M., Ogasawara, M., Matsubara, Y., Narisawa, K., Nakamura, S., Itoyama, Y., and Abe, K. (1993) Nat. Genet. 5 323-324 [DOI] [PubMed] [Google Scholar]
- 31.Arisato, T., Okubo, R., Arata, H., Abe, K., Fukada, K., Sakoda, S., Shimizu, A., Qin, X. H., Izumo, S., Osame, M., and Nakagawa, M. (2003) Acta Neuropathol. (Berl) 106 561-568 [DOI] [PubMed] [Google Scholar]
- 32.Ohi, T., Saita, K., Takechi, S., Nabesima, K., Tashiro, H., Shiomi, K., Sugimoto, S., Akematsu, T., Nakayama, T., Iwaki, T., and Matsukura, S. (2002) J. Neurol. Sci. 197 73-78 [DOI] [PubMed] [Google Scholar]
- 33.Shaw, C. E., Enayat, Z. E., Powell, J. F., Anderson, V. E., Radunovic, A., al-Sarraj, S., and Leigh, P. N. (1997) Neurology 49 1612-1616 [DOI] [PubMed] [Google Scholar]
- 34.Cudkowicz, M. E., McKenna-Yasek, D., Sapp, P. E., Chin, W., Geller, B., Hayden, D. L., Schoenfeld, D. A., Hosler, B. A., Horvitz, H. R., and Brown, R. H. (1997) Ann. Neurol. 41 210-221 [DOI] [PubMed] [Google Scholar]
- 35.Ohi, T., Nabeshima, K., Kato, S., Yazawa, S., and Takechi, S. (2004) J. Neurol. Sci. 225 19-25 [DOI] [PubMed] [Google Scholar]
- 36.Aoki, M., Abe, K., and Itoyama, Y. (1998) Cell Mol. Neurobiol. 18 639-647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gal, J., Strom, A. L., Kilty, R., Zhang, F., and Zhu, H. (2007) J. Biol. Chem. 282 11068-11077 [DOI] [PubMed] [Google Scholar]
- 38.Ravikumar, B., Acevedo-Arozena, A., Imarisio, S., Berger, Z., Vacher, C., O'Kane, C. J., Brown, S. D., and Rubinsztein, D. C. (2005) Nat. Genet. 37 771-776 [DOI] [PubMed] [Google Scholar]
- 39.Taylor, J. P., Tanaka, F., Robitschek, J., Sandoval, C. M., Taye, A., Markovic-Plese, S., and Fischbeck, K. H. (2003) Hum. Mol. Genet. 12 749-757 [DOI] [PubMed] [Google Scholar]
- 40.Schroer, T. A. (2004) Annu. Rev. Cell Dev. Biol. 20 759-779 [DOI] [PubMed] [Google Scholar]
- 41.Burkhardt, J. K., Echeverri, C. J., Nilsson, T., and Vallee, R. B. (1997) J. Cell Biol. 139 469-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melkonian, K. A., Maier, K. C., Godfrey, J. E., Rodgers, M., and Schroer, T. A. (2007) J. Biol. Chem. 282 19355-19364 [DOI] [PubMed] [Google Scholar]
- 43.Wang, J., Slunt, H., Gonzales, V., Fromholt, D., Coonfield, M., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2003) Hum. Mol. Genet. 12 2753-2764 [DOI] [PubMed] [Google Scholar]
- 44.Nagai, M., Aoki, M., Miyoshi, I., Kato, M., Pasinelli, P., Kasai, N., Brown, R. H., Jr., and Itoyama, Y. (2001) J. Neurosci. 21 9246-9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vale, R. D. (2003) Cell 112 467-480 [DOI] [PubMed] [Google Scholar]
- 46.Guzik, B. W., and Goldstein, L. S. (2004) Curr. Opin. Cell Biol. 16 443-450 [DOI] [PubMed] [Google Scholar]
- 47.Breuer, A. C., and Atkinson, M. B. (1988) Cell Motil Cytoskeleton 10 321-330 [DOI] [PubMed] [Google Scholar]
- 48.Breuer, A. C., Lynn, M. P., Atkinson, M. B., Chou, S. M., Wilbourn, A. J., Marks, K. E., Culver, J. E., and Fleegler, E. J. (1987) Neurology 37 738-748 [DOI] [PubMed] [Google Scholar]
- 49.Collard, J. F., Cote, F., and Julien, J. P. (1995) Nature 375 61-64 [DOI] [PubMed] [Google Scholar]
- 50.Ligon, L. A., LaMonte, B. H., Wallace, K. E., Weber, N., Kalb, R. G., and Holzbaur, E. L. (2005) Neuroreport 16 533-536 [DOI] [PubMed] [Google Scholar]
- 51.Sasaki, S., and Iwata, M. (1996) Neurology 47 535-540 [DOI] [PubMed] [Google Scholar]
- 52.Williamson, T. L., and Cleveland, D. W. (1999) Nat. Neurosci. 2 50-56 [DOI] [PubMed] [Google Scholar]
- 53.Fanara, P., Banerjee, J., Hueck, R. V., Harper, M. R., Awada, M., Turner, H., Husted, K. H., Brandt, R., and Hellerstein, M. K. (2007) J. Biol. Chem. 282 23465-23472 [DOI] [PubMed] [Google Scholar]
- 54.Mizushima, N. (2007) Genes Dev. 21 2861-2873 [DOI] [PubMed] [Google Scholar]
- 55.Strom, A. L., Gal, J., Shi, P., Kasarskis, E. J., Hayward, L. J., and Zhu, H. (2008) J. Neurochem. 106 495-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banks, G. T., and Fisher, E. M. (2008) Genome Biol. 9 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieran, D., Hafezparast, M., Bohnert, S., Dick, J. R., Martin, J., Schiavo, G., Fisher, E. M., and Greensmith, L. (2005) J. Cell Biol. 169 561-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teuchert, M., Fischer, D., Schwalenstoecker, B., Habisch, H. J., Bockers, T. M., and Ludolph, A. C. (2006) Exp. Neurol. 198 271-274 [DOI] [PubMed] [Google Scholar]
- 59.Chevalier-Larsen, E. S., Wallace, K. E., Pennise, C. R., and Holzbaur, E. L. (2008) Hum. Mol. Genet. 17 1946-1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai, C., Lin, X., Chandran, J., Shim, H., Yang, W. J., and Cai, H. (2007) J. Neurosci. 27 13982-13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabuta, T., Suzuki, Y., and Wada, K. (2006) J. Biol. Chem. 281 30524-30533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.