Abstract
Pancreatic-duodenal homeobox factor-1 (Pdx1) is highly enriched in islet β cells and integral to proper cell development and adult function. Of the four conserved 5′-flanking sequence blocks that contribute to transcription in vivo, Area II (mouse base pairs -2153/-1923) represents the only mammalian specific control domain. Here we demonstrate that regulation of β-cell-enriched Pdx1 expression by the MafA and MafB transcription factors is exclusively through Area II. Thus, these factors were found to specifically activate through Area II in cell line transfection-based assays, and MafA, which is uniquely expressed in adult islet β cells was only bound to this region in quantitative chromatin immunoprecipitation studies. MafA and MafB are produced in β cells during development and were both bound to Area II at embryonic day 18.5. Expression of a transgene driven by Pdx1 Areas I and II was also severely compromised during insulin+ cell formation in MafB-/- mice, consistent with the importance of this large Maf in β-cell production and Pdx1 expression. These findings illustrate the significance of large Maf proteins to Pdx1 expression in β cells, and in particular MafB during pancreatic development.
Much effort is currently being directed to define the biochemical pathways essential to islet β-cell formation, with the hope that such insight will aid in the development of treatment strategies aimed at reducing β-cell dysfunction in diabetic patients. A transcription factor that is critical to both β-cell development and function is pancreatic-duodenal homeobox-1 (Pdx1,5 IPF-1 in humans), the first pancreas-enriched gene product expressed in budding epithelium at embryonic day 8.5 (E8.5) (1, 2). Pdx1 is produced in early pancreatic endocrine, exocrine, and ductal progenitors, and complete loss in humans and mice results in an apancreatic phenotype (3–5). Later in development, islet β cells can be distinguished from pancreatic exocrine and ductal cells by their high Pdx1 levels (1, 2). In mature pancreas, Pdx1 is principally localized to β cells (1), with specific removal leading to a severe diabetic phenotype due to β-cell dysfunction in mice (6, 7). Pdx1 is also one of few genes associated with an autosomal dominant form of diabetes in humans (5) and is viewed as a master regulator of β-cell formation and function (8, 9).
Endogenous Pdx1 expression is predominately controlled by four conserved 5′-flanking sequence domains, referred to as Area I (mouse AI, base pairs (bp) -2761/-2457), AII (bp -2153/-1923), AIII (bp -1879/-1600), and AIV (bp -6529/-6047) (10–12). AI-III mediates pancreas-specific expression, as early targeted removal of these regions from the endogenous gene profoundly reduces pancreas formation in vivo, while leaving Pdx1 expression in the stomach and duodenum intact (13). Several transgenic reporter lines encompassing AI, AII, and AIII have been developed to more precisely define their roles in pancreatic expression. Interactions between AIII and AI/II have been shown to be necessary in the early and broad transcription of Pdx1 in acinar, endocrine, and ductal progenitors (14). In contrast, AI and AII cooperate to selectively induce high Pdx1 expression in the insulin+ cells produced after E13.5 that populate the adult islet (10, 15, 16). Notably, AII is the only Pdx1 control region unique to mammals and capable of independently directing transgenic expression to (a fraction of) islet β cells in vivo (17).
Islet β-cell-enriched expression of Pdx1 appears to be principally regulated by AII, and several key islet transcription factors bind to and activate through conserved cis-acting elements in this mammalian-specific control region, including FoxA2, Pax6, MafA, and MafB (11, 15–19). These closely related large Maf proteins are both expressed in β cells during development, with MafA found uniquely in adult islet β cells and only MafB compromising Pdx1 expression andβ-cell formation in knock-out mice (20–22). Here we first identified a new conserved MafA and/or MafB binding site in AII. Chromatin immunoprecipitation (ChIP) analysis then illustrated that MafA binding within AII was highly enriched over AI, AIII, and AIV in islet β cell lines, while transfection-based reporter assays showed that the MafA/MafB binding elements contributed to activation. Both MafA and MafB were bound in vivo to Area II in E18.5 pancreata, and AI/AII-driven transgene activity was dramatically reduced during β-cell development in MafB-/- mice. Collectively, these data support an essential role for MafA and MafB specifically in AII-mediated activation of Pdx1 transcription, and highlight the significance of MafB regulation in developing β cells.
EXPERIMENTAL PROCEDURES
Electrophoretic Mobility Shift Assay (EMSA)—βTC3, MIN6, HeLa, and NIH3T3 cell nuclear extracts (NE) were prepared using lysis buffer (100 mm Tris-HCl, pH 8.0, 0.5% Igepal CA-630 (Sigma-Aldrich), 2 mm sodium orthovanadate, 2 mm sodium fluoride, and protease inhibitor mixture (Roche Applied Science)), followed by pellet resuspension in nuclear extraction buffer (20 mm Hepes, pH 7.9, 400 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 2 mm sodium orthovanadate, 2 mm sodium fluoride, and protease inhibitor mixture). The Area II double-stranded oligonucleotide probes were polynucleotide kinase-labeled with [γ32P]dATP. Reactions (20 μl, total volume) were conducted with 10 μg of nuclear extract, 0.4 pmol of 32P probe, and 1 μg of poly(dGdC) at room temperature for 10 min in binding buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 4 mm dithiothreitol, 2 mm EDTA, and 20% glycerol). The conditions for the competition analyses were the same, except that a 10–100-fold excess of the unlabeled specific competitor DNA was included. To identify protein-DNA complexes, MafA (Bethyl Labs, Montgomery, TX), MafB (Bethyl Labs), or Pax6 (Covance, Dedham, MA) antibody was added to the indicated binding reactions. Samples were resolved on a 6% nondenaturing polyacrylamide gel in TGE buffer (50 mm Tris, 380 mm glycine, 2 mm EDTA, pH 8.5) before drying and autoradiography. The commercially generated (IDT, Coralville, IA) probe and competitor sequences are shown in supplemental Table S1.
Transfection Construct—Pdx1 reporter constructs were generated using mouse and human sequences, cloned directly upstream of the herpes simplex virus thymidine kinase (TK) minimal promoter in a chloramphenicol acetyltransferase (CAT) expression vector, pTK(An):CAT (23). The previously generated Pdx1:pTK constructs are as follows: PstBst:pTK (mouse -2917/-1918 bp, (16)); huAII:pTK (human -2141/-1890bp, (17)); mAI:pTK (mouse -2761/-2457 (12)); mAII: pTK (mouse -2153/-1923 (12)); mAIV:pTK (mouse -6529/-6010 (12)); and mAI/II:pTK ((AI) 2759/-2439 and (AII) -2200/-1923(15)). The Pdx1:pTK mutants were generated using the QuikChange mutagenesis kit (Stratagene): PstBst: pTK at B2 (-2126/-2115) (17), B4/5 (-2101/-2086, (18), and B2–5 (-2126/2086); huAII:pTK at B2-TGC and -CAC (Fig. 2D); mAI/II:pTK TGC mutations at 5′ (-2180/-2178), B2 (-2120/-2118), B4 (-2098/-2096), and B13 (-2009/-2007). MafA and MafB expression constructs were described previously (24). EnMafB:pcDNA3.1 expresses a fusion protein of the Drosophila Engrailed (En) N-terminal repressor domain (coding sequence for amino acids 1–298) and mouse MafB C-terminal DNA binding domain (coding sequence for amino acids 82–324). These sequences were cloned into pcDNA3.1 (Invitrogen) along with a DNA fragment encoding an IRES and nuclear EGFP (25). All construct sequences were confirmed by restriction enzyme digestion and partial sequence analysis.
FIGURE 2.
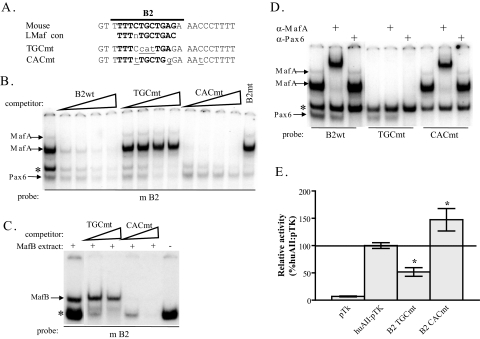
MafA stimulates B2-mediated activity, but not Pax6. A, shown is the relationship of the TGC and CAC mutations to the large Maf consensus. The mutated nucleotides are underlined. B, competition assays (10–100-fold excess) were performed to analyze for MafA and Pax6 binding to the TGC and CAC mutants of B2. The TGCmt specifically competes for Pax6 binding inβTC3 nuclear extracts, while only MafA binding is lost with the CACmt. C, nuclear extracts from MafB-transfected Hela cells were used in competition assays (10–100-fold excess) to analyze for MafB binding to the TGC and CAC mutants of B2. As observed with MafA, the TGC mutation does not compete for MafB binding while the CAC mutant effectively competes for MafB. A control lane with vector-transfected Hela nuclear extract is included to demonstrate the MafB-DNA complex band. D, binding assays were conducted with wild-type and mutant B2 probes to further illustrate the difference in TGCmt and CACmt binding to Pax6 and MafA, respectively. The location of MafA and Pax6 complexes were illustrated by addition of α-MafA and α-Pax6 antibodies to the βTC3 nuclear extract binding reactions. E, βTC3 cells were transfected with wild-type and MafA (TGCmt) or Pax6 (CACmt) binding site mutant versions of B2 in huAII:pTK. The activity of the pTK vector is also shown. Significantly, huAII:pTK activity was only compromised in the MafA binding mutant (n = 8–11; *, p < 0.005 versus huAII:pTK).
Cell Transfections—Monolayer cultures of mouse pancreatic insulinoma βTC3 cells and rat kidney KNRK cells were grown in Dulbecco modified Eagle's medium containing 17% horse serum or 5% calf serum, respectively. Rous sarcoma virus enhancer-driven firefly luciferase (pRSV:LUC) was cotransfected as an internal control with Pdx1:pTK using Lipofectamine (Invitrogen, Carlsbad, CA). Extracts were prepared 40–48 h after transfection in lysis buffer (100 mm Tris-HCl, pH 8.0, 0.5% Igepal CA-630, and protease inhibitor mixture (Roche Applied Science) and assayed for CAT (26) and LUC (27) activity. Transfections were conducted with at least two different experimental plasmid preparations. Statistical analysis was performed by one-way analysis of variance followed by Fisher's PSLD, and p < 0.05 was considered significant.
ChIP Assays—βTC3 cells and disrupted mouse E18.5 pancreata were formaldehyde cross-linked, and the sonicated protein-DNA complexes were isolated under conditions described previously (19). 100 μg of cross-linked βTC3 DNA or 5 μg of cross-linked E18.5 pancreata DNA were incubated with 10 μg or 5 μg, respectively, of rabbit anti-MafA, anti-MafB, or normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C, and the antibody/protein/DNA complexes were isolated with protein A-agarose blocked with salmon sperm DNA (Upstate, Charlottesville, VA). Real time PCR analysis was performed on the purified, immunoprecipitated βTC3 DNA using SyBr green PCR master mix (Applied Biosciences) and the ABI Prism 7000. Input DNA was utilized to generate a standard curve, and only reactions with 100 ± 20% efficiency (E = 10-1/slope) were included in the analysis. The enrichment of Pdx1 control sequences per sample was calculated relative to the inactive PEPCK promoter by employing the Pfaffl method (28) and the enrichment of each Pdx1 control sequence was compared by one-way analysis of variance followed by Fisher's PSLD. The mouse primer sets were generated with the ABI software and provided in supplemental Table S2. The purified immunoprecipitated E18.5 pancreata DNA was analyzed by PCR with Pdx1 Area II and PEPCK transcriptional control region primers as described previously (19). Amplified products were resolved on a 1.4% agarose gel in Tris acetate/EDTA buffer containing ethidium bromide.
Immunohistochemistry—The mouse Pdx1AI/AII hsp-LacZ transgenic line (15) was crossed into the MafB+/- line (29) to generate wild-type and MafB-/- littermates that contain the Pdx-1AI/AII hsp-LacZ transgene. The day of vaginal plug discovery was designated E0.5 and tissue was harvested on E18.5, as MafB-/- mice die at birth because of central apnea (29). Abdominal organs and pancreata were fixed, dehydrated, embedded in paraffin, and 6-μm serial sections were mounted on glass slides for immunohistochemical analysis. The primary antibodies were rabbit α-β galactosidase (1:500; Molecular Probes), guinea pig α-insulin (1:2000, Linco Research, ST Charles, MO), and goat α-Pdx1 (1:10,000, Chris Wright, Vanderbilt University Medical School). The β-gal signal was enhanced with biotin-conjugated α-rabbit (1:500, Jackson ImmunoResearch, West Grove, PA) and Cy5-conjugated streptavidin (1:1000 Jackson ImmunoResearch). Fluorescence-conjugated secondary antibodies used were Cy3-α-guinea pig and Cy2-α-goat (1:500, Jackson ImmunoResearch). Fluorescent images were captured with a Zeiss LSM 510 confocal microscope using an optical depth of 1 μm. The percentage of insulin+ cells co-staining for β-galactosidase was determined by counting cells in at least 20 random fields throughout the pancreas, and mean differences were tested for statistical significance with the Student's two tailed t test.
RESULTS
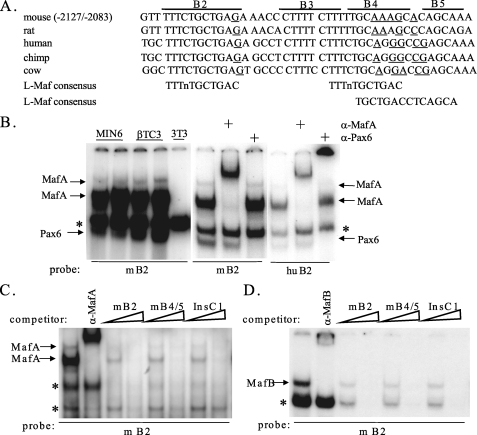
The AII B2 Element Binds MafA, MafB, and Pax6 in Vitro—Mutation analysis of conserved AII sequences revealed several activator sites in β cell line-based reporter assays (17). Upon examination of these sequences, Block 2 (B2, bp -2124/-2113 in mice) was found to have high identity with the recently described large Maf binding consensus (Ref. 30 and Fig. 1A). Three bands were specifically detected in gel shift experiments performed with β-cell nuclear extracts using either mouse or human B2 probes, with the top two supershifted with MafA antibody (compare MIN6 and βTC3 to NIH3T3 in Fig. 1B). Antibody screening studies demonstrated that the other band contains the Pax6 transcription factor, another activator of islet cell development and β-cell function (31, 32). However, the interaction between Pax6 and B2 appears to be weak, because the Pax6-DNA complex was disrupted by a supershift competent Pax6 antibody (Fig. 1B), and only a very small amount of Pax6 was bound when compared with a Pax6 consensus site probe (data not shown). In contrast, B2 had high affinity for MafA as judged using bona fide large Maf competitors, Insulin C1 (24, 33) and AII B4/5 (17) (Fig. 1C). (AII B4/5 resides just 14 bp from B2 (Fig. 1A)). Using the same large Maf competitors, MafB was also observed to bind the B2 element with the same specificity as MafA (Fig. 1D). These data identify the B2 element of Area II as large Maf and Pax6 binding sites in EMSA, albeit Pax6 relatively poorly.
FIGURE 1.
MafA and Pax6 bind to the AII B2 element in vitro. A, sequence identity within the B2, B4, and B5 region of Pdx1 between various mammalian species. The underlined nucleotides represent differences with the two reported large Maf consensus sequences (30, 47). B, sequences spanning mouse and human B2 were used in gel shift binding assays with β-(Min6 and βTC3) and non-β-cell (NIH3T3) nuclear extract. Three β-cell-specific B2 complexes were detected, with the upper two supershifted by α-MafA and the lower disrupted with α-Pax6 antibodies. The asterisk denotes a nonspecific complex, as concluded from B2 mutant analysis (see Fig. 2). C, specificity of MafA-DNA complex formation in βTC3 extracts was determined by competition with a 10–100-fold molar excess of unlabeled probe and large Maf binding competitors, the Pdx1 AII B4/5 element and insulin C1 element. Notably, mB2 competes as effectively as mB4/5 and InsC1 for MafA binding. D, MafB-mB2 complex in MafB-transfected HeLa cell extracts had similar binding properties to MafA in competitions with 10–100-fold molar excess of the probe and large Maf binding competitors. The anti-MafB antibody supershift illustrates the location of the MafB complex formed with mB2.
Only MafA or MafB Stimulate B2 Activity—Gel shift assays were performed with a series of B2 mutants to more precisely localize large Maf and Pax6 protein binding. Two unique 3-bp mutants were identified that distinguished their binding (Fig. 2). Hence, changing the TGC core in the large Maf consensus completely eliminates MafA and MafB binding to B2 without impacting Pax6 association in competition and direct binding assays (Fig. 2, B–D, see TGCmt). In contrast, mutation of individual base pairs outside of the large Maf consensus disrupted Pax6 binding without impairing MafA or MafB binding (Fig. 2, B–D, see CACmt).
The effect of the large Maf and Pax6 distinguishing mutations in B2 on human AII-driven activity was analyzed in β cell line transfection assays, a context wherein only MafA and Pax6 are expressed. Selectively eliminating MafA binding in βTC3 cells reduces AII activity, while the Pax6 binding mutant slightly increases activity (Fig. 2E). It is possible that AII B2-CAC mutant activity was potentiated due to either removal of a repressor (i.e. Pax6) or enhanced MafA activator binding. We favor the latter, as Pax6 actually binds very weakly to this site in relation to MafA/B in gel shift assays, and there is no precedence for this factor acting in a repressive manner. In addition, MafA binding was increased in the Pax6 binding mutant, as observed in concentration-dependent competition gel shift assays (Fig. 2B).
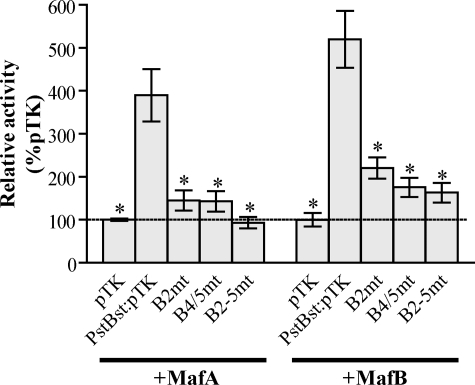
Although normal islet β cells and islet β cell lines only produce MafA, insulin+/Pdx1+ progenitors express MafA and MafB during development (34, 35). To directly test the ability of MafA and MafB to stimulate AII-directed activity, each was coexpressed with an AI/AII-spanning (PstBst) reporter in the non-β KNRK cell line. PstBst:pTK reporter activity alone is minimal in these cells (data not shown), but significantly stimulated by MafA or MafB expression (Fig. 3). Activation by these factors was compromised upon preventing large Maf binding at either B2 or B4/5. Collectively, these experiments strongly suggest that MafA and MafB, but not Pax6, stimulate AII B2 activity.
FIGURE 3.
MafA and MafB activate through the Area II large Maf binding sites. MafA or MafB were cotransfected into KNRK cells with pTK, PstBst:pTK, or AII mutant versions of PstBst:pTK. Preventing large Maf binding within B2 or B4/5 significantly reduced the ability of MafA or MafB to activate PstBst:pTK (n = 4–6; *, p < 0.0001 versus PstBst:pTK).
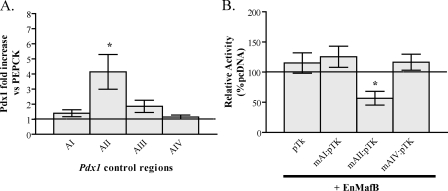
The Large Maf Binding Sites of the Pdx1 Gene Are Clustered within AII—Many Pdx1 transcription factors influence more than one control region. For example, FoxA2 acts on AIV ((12), AII (11), and AI (11), while Pdx1 and Nkx2.2 both function through AIV (12) and AI (11, 15). ChIP analysis was performed in βTC3 cells to determine the relative distribution of MafA binding sites between conserved Pdx1 transcriptional control regions. Real-time PCR analysis was used to quantitatively compare the enrichment of these regions to that of a non-transcribed gene, PEPCK. AII sequences were highly enriched relative to AI, AIII, and AIV (Fig. 4A). Although the AI and AIII signals were slightly above the PEPCK control, this was not significant and likely represents randomly sheared chromosomal DNA (primarily 100–500 bp) encompassing AI/AII or AII/III. As expected from their close proximity, AIII, located only 45 bp from AII, had a higher signal than AI, which is separated by 300 bp.
FIGURE 4.
AII contains the only large Maf control elements within the Pdx1 gene. A, formaldehyde cross-linked chromatin from βTC3 cells was incubated with antibodies specific to MafA and the precipitate quantitatively analyzed for AI, AII, AIII, AIV, and PEPCK sequences by Real-time PCR. AII was highly enriched over the other Pdx1 control regions. The signals were normalize to the inactive PEPCK gene (n = 4). B, βTC3 cells were cotransfected with the EnMafB repressor chimera and pTK, mAI:pTK, mAII:pTK, or mAIV:pTK. EnMafB inhibited mAII-mediated activity, but not Area I or IV (n = 4–5; *, p ≤ 0.01 versus each pTK reporter + pcDNA).
To further assess functional regulation by MafA and MafB through the Pdx1 control regions, an Engrailed (En) repressor domain and MafB DNA binding domain chimera (EnMafB) was cotransfected with AI-, AII-, and AIV-driven reporters in βTC3 cells. (The inactivity of AIII-driven reporters in β cell lines (11) precluded analysis.) EnMafB was expected to compete with endogenous MafA and repress transcription. As predicted, EnMafB significantly reduced AII-driven activity (Fig. 4B), while not affecting AI- or AIV-mediated expression. This and the ChIP data are complementary and indicate that large Maf factors bind to and regulate specifically through Area II.
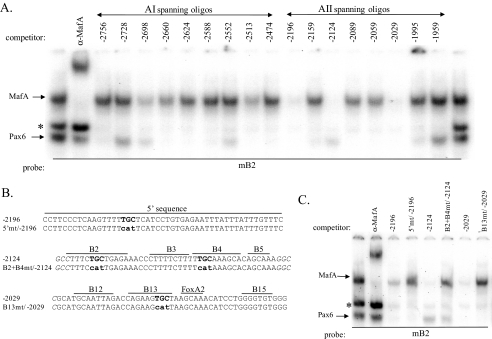
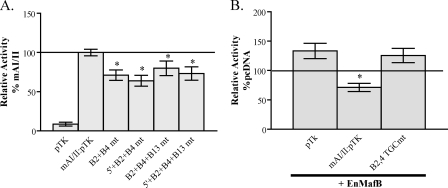
B2 and B4/5 Are the Principal Large Maf Binding Sites within AII—Because large Maf binding was localized to the AII control domain of the Pdx1 gene, we screened for additional large Maf binding elements within this region using a comprehensive series of overlapping oligonucleotides in gel shift competition assays. AI competitors served as negative controls in these experiments, and, as expected, did not significantly influence MafA binding to B2 in βTC3 nuclear extracts (Fig. 5A). However, 3 different AII competitors did impact MafA binding, with the -2124 competitor containing both B2 and B4/5 (Fig. 5B). In contrast, the -2196 competitor spanned sequences located 5′ to the highly conserved AII domain, while the conserved B12–15 elements were in -2029. Neither the -2196 nor -2029 competitors had high sequence identify with the large Maf consensus, although mutating their common TGC motif eliminated MafA binding (Fig. 5C). Preventing MafA binding in the -2196 or -2029 sequences did not further compromise B2 and B4/5 mutant activity in βTC3 cells (see the 5′ and B13 mutants (mt) in B2 + B4mt mAI/II:pTK mt of Fig. 6A), nor did EnMafB suppress B2 + B4mt mAI/II:pTK activity (Fig. 6B). These results suggest that MafA and/or MafB stimulation of AII in developing and adult β cells is primarily through B2 and B4/5 element binding.
FIGURE 5.
AII contains three distinct large Maf binding sites. A, competitor oligonucleotides (40–50 bp) spanning mouse AI and AII were used in mB2 probe gel shift binding assays in βTC3 nuclear extract. The competitors are labeled by their 5′-end bp. B, wild type and mutant versions of the three AII MafA competitors are shown. Denoted is also the location of their conserved sequence blocks (17), with one block labeled by the transcription factor associated with control, FoxA2 (16). C, the loss of MafA binding in the -2196, -2124, and -2029 mutants is illustrated in mB2 gel shift assays performed with a 100-fold molar excess of unlabeled wild-type or mutant competitor in βTC3 nuclear extract. The MafA antibody addition demonstrates the location of this protein/B2 complex.
FIGURE 6.
B2 and B4/5 represent the only functional large Maf binding sites. A, βTC3 cells were transfected with pTK, mAI/II:pTK, and AII TGC mutant versions of mAI/II:pTK. The 5′ and/or B13 mutants do not impact B2+ B4 mt mAI/II:pTK activity (n = 13–21; *, p < 0.005 versus mAI/II). B, EnMafB (or pcDNA) was cotransfected into βTC3 cells with pTK, mAI/II:pTK, or B2 + B4mt mAI/II:pTK. Notably, repression by EnMafB is eliminated in the B2 and B4 mt (n = 4–8; p < 0.01 versus each pTK reporter + pcDNA).
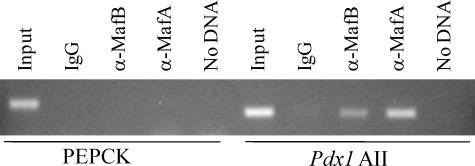
MafA and MafB Bind to AII during β-Cell Development—High Pdx1 levels demarcate β cell progenitors from other exocrine and endocrine cell populations during the second and principal wave of islet cell expansion (1, 2). As a consequence, ChIP studies were performed to determine whether MafA and/or MafB bound to AII in wild-type E18.5 pancreata (Fig. 7). Anti-MafA and -MafB antibodies precipitated this Pdx1 region but not PEPCK, a gene not expressed in the pancreas. This analysis supports a direct role of these large Maf proteins in Pdx1 transcription during β-cell maturation.
FIGURE 7.
MafA and MafB bind to Pdx1 AII in developing pancreata at E18.5. ChIP analysis was performed on E18.5 pancreata treated with anti-MafA and -MafB antibodies. The precipitated chromatin was analyzed by PCR for Pdx1 AII and PEPCK control region sequences. As controls, PCR reactions were run with input chromatin (1:200 dilution), DNA obtained after treatment with rabbit IgG, and reactions with no chromatin DNA.
MafB Regulates AI/II-mediated Transgene Expression during β-Cell Development—A notable phenotype of MafB-/- mice is the reduction in second wave insulin+ cell production from E13.5. Interestingly, although Pdx1 is detected in E15.5 hormone-negative endocrine progenitors, it is no longer present in these cells by E18.5 (20), suggesting that MafB is required for maintenance of Pdx1 transcription late in β-cell formation. In contrast, neither Pdx1 expression nor islet cell formation is impacted in MafA-/- mice during development (45).
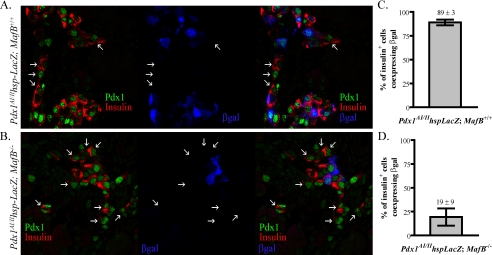
To examine in vivo whether MafB regulates AII-mediated transcription, we crossed the Pdx1AI/II-LacZ reporter transgenic mouse line (15) into a MafB-/- background. LacZ expression was compared in wild-type and MafB-null littermates at E18.5. Pdx1AI/II-LacZ expression is primarily restricted to the β-cell population and labels 89 ± 3% of the insulin+ cells in wild-type and MafB+/- mice (Fig. 8), a transgene penetrance level comparable to that in adult islet β cells (15). In addition, transgene activity was only detected in insulin+ cells in MafB-/- littermates (Fig. 8B), recapitulating the pattern of endogenous expression in the mutant (20). However, while Pdx1 was present in essentially all insulin+/MafA+ cells in MafB-/- mice (89%, Ref. 20), Pdx1AI/II-LacZ expression was observed in a smaller fraction of insulin+ cells (19 ± 9%, Fig. 8 and supplemental Fig. S1). Presumably, activator(s) and/or coactivator(s) levels are not sufficient in most mutant insulin+ cells to allow comprehensive Pdx1AI/II-LacZ activity. Notably, these data support a direct role for MafB in AII-directed activation of Pdx1 transcription in the developing β-cell population.
FIGURE 8.
MafB regulates AI/II-mediated transgenic expression during β-cell development. The Pdx1AI/II-LacZ transgenic line was crossed with MafB+/- mice to generate MafB wild-type (Pdx1AI/II-LacZ;MafB+/+and Pdx1AI/II-LacZ;MafB+/-) or null (Pdx1AI/II-LacZ; MafB-/-) littermates. Representative images ofβ-gal (blue), insulin (red), and Pdx1 (green) staining in wild type (A) and MafB-null (B) transgenic pancreata at E18.5 are shown. Arrows indicate insulin+Pdx1+ cells that do not produce detectable β-gal. The percentage of insulin+ cells (±S.E.) costaining with β-gal in wild type (C) and MafB-null (D) was quantitated across multiple random sections of three independent pancreata.
DISCUSSION
Pdx1 is essential for pancreas development and adult β-cell function. It is expressed broadly and dynamically within the developing foregut, but then is most abundantly produced in β cells late in development and in adulthood. The factors controlling the unique transcription pattern of Pdx1 are largely unknown, but will likely be independently linked to organ specification, endocrine/exocrine cell differentiation, and β-cell function. The Pdx1 AII control domain represents the core regulator of β-cell-enriched expression, as concluded from Pdx1 transgenic (17) and endogenous AI-II-III knock-out analysis (13) in mice. Our results demonstrate that large Maf proteins activate Pdx1 transcription by binding to AII and illustrate how expression is compromised in developing β cells in the specific absence of MafB. This work also supports MafA regulation of Pdx1, with a prominent role presumably in the adult islet β-cell. The control mechanisms described here are likely to be generally utilized by large Maf transcription factors, each of which have been implicated in multiple developmental processes (e.g. (NRL), eye rod formation (36); c-Maf, chondrocyte differentiation (37–39): MafB, hindbrain segmentation (40, 41), and podocyte differentiation in the kidney (42)).
AII was determined to be the exclusive large Maf binding region within the Pdx1 5′-flanking region in quantitative ChIP and Pdx1-driven transfection experiments. A new AII large Maf control element, B2, was also discovered that lies directly upstream of the previously characterized B4/5 site (18). Two other potential binding sites were revealed in Area II by gel shift analysis (termed 5′ and B13, Fig. 5), but activation appears to principally be through B2 and B4/5 elements, as revealed in experiments performed with the EnMafB suppressor and 5′ and B13 mutants in β cell lines (Fig. 6B). In vitro binding assays also recognized the ability of B2 to bind the Pax6 activator, albeit weakly. Specific B2 mutations that differentially eliminated large Maf versus Pax6 binding demonstrated that only large Maf proteins stimulate Pdx1 transcription in β cell lines. Although the Pax6 interaction is weak and no detectable effect on activity was observed, it is conceivable that Pax6 could function in vivo under certain circumstances to potentiate or even competitively limit large Maf-mediated regulation.
MafA and MafB exhibit very similar properties in B2 and B4/5 binding and transcription-based reporter assays (Figs. 1, 2, 3). However, these factors have distinct expression patterns during β-cell development. Hence, MafA is only detected in the β cells produced during the massive wave of α- and β-cell differentiation that starts at around E13.5 (21), termed the “secondary” transition. In contrast, MafB is produced in both glucagon+ and insulin+ cells during the “primary” (i.e. <E13) and “secondary” transition (34, 35). The developmental expression pattern of MafA is unique in relation to all other islet-enriched transcription factors, at first suggesting a critical part in inducing high Pdx1 levels in β-cell progenitors. However, β-cell formation was just impacted in MafB-/- mice, resulting in a specific reduction in insulin+ (and glucagon+) cell numbers without changing the overall level of endocrine cells (20). Strikingly, MafA was only detected in insulin+ cells while high Pdx1 levels were in both insulin+ and hormone- progenitors until E15.5 in MafB-/- mice, although Pdx1 was clearly reduced in hormone- cells by E18.5 (20). Pdx1AI/II-LacZ activity was also specifically detected in insulin+ cells in MafB-/- mice at E18.5 (Fig. 8). Significantly, these results demonstrated that MafB is necessary for sustaining high Pdx1 levels during β-cell maturation. The observed occupancy of MafB within Pdx1 AII in the developing pancreas and the compromised activity of Pdx1AI/II-LacZ in MafB-/- mice supports a direct activation mechanism in developing β cells (Figs. 7 and 8).
The signals necessary for initiating and sustaining high Pdx1 in β-cell progenitors are found within AI/AII, as a transgene driven by these regions alone is exclusively expressed during the secondary transition (15). Notably, the presence of Pdx1AI/II-LacZ activity exclusively in MafB-/- insulin+ cells recapitulated the pattern of the endogenous gene. However, the relatively poor penetrance of the transgene relative to endogenous Pdx1 in the mutant cell population likely reflects heterogeneity in expression of other key trans-acting factors. It is unclear whether this results from differential distribution of a novel transcriptional activator(s) or, perhaps even a co-regulator(s). For example, the kinases that regulate the steady-state level and activity of MafA may not be uniformly distributed within MafB-/- insulin+ cells (43–45), even though the activator is found throughout. Similar regulatory processes may also explain why the GLUT2 transporter is only found in a comparably small fraction of mutant insulin+ cells (2). The comprehensive expression of endogenous Pdx1 in these mutant cells presumably reflects compensation by trans-regulators acting on non-AI/AII control sequences, possibly AIII or AIV.
Collectively, our results demonstrate that AII alone contains the large Maf control elements necessary for (at least) MafB-mediated activation of Pdx1 transcription in developing β cells. As MafA is the only other family member expressed in mature β cells (21), this transcription factor is expected to be critical to Pdx1 expression in adults. Interestingly, AII is found only within mammalian Pdx1 genes, implying that regulation by factors like the large Mafs may be related to specific differences in the timing and/or level of Pdx1 expression in non-mammalian species. For example, differential expression of Pdx1 may impact formation of the two kinds of islets uniquely described in the avian pancreas, which contain different proportions of glucagon and insulin cells (46). Future efforts focused on identifying AII regulatory factors will provide additional insight into the novel mechanisms involved in regulating Pdx1 transcription and pancreas development in mammals, and significantly, will likely also help to identify heritable defects that cause insulin deficiency and diabetes.
Supplementary Material
Acknowledgments
We thank Dr. C. V. E. Wright for critically reading this manuscript and for providing the goat α-Pdx1 antibody. Min Guo provided technical support very valuable to many of these studies.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 DK42502 & DK50203 (to R. S.) and 5T32 DK07563-20 METP postdoctoral training (to A. V.). This work was also supported in part by the American Diabetes Association (7-04-RA-116, to R. S.) and the Juvenile Diabetes Research Foundation (Advanced Postdoctoral 10-2006-5, to I. A.). Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (Public Health Service Grant P60 DK20593). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: Pdx, pancreatic-duodenal homeobox factor; ChIP, chromatin immunoprecipitation assay; EMSA, electrophoretic mobility shift assay; β-gal, β-galactosidase.
References
- 1.Guz, Y., Montminy, M. R., Stein, R., Leonard, J., Gamer, L. W., Wright, C. V., and Teitelman, G. (1995) Development 121 11-18 [DOI] [PubMed] [Google Scholar]
- 2.Ohlsson, H., Karlsson, K., and Edlund, T. (1993) EMBO J. 12 4251-4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonsson, J., Carlsson, L., Edlund, T., and Edlund, H. (1994) Nature 371 606-609 [DOI] [PubMed] [Google Scholar]
- 4.Offield, M. F., Jetton, T. L., Labosky, P. A., Ray, M., Stein, R. W., Magnuson, M. A., Hogan, B. L., and Wright, C. V. (1996) Development 122 983-995 [DOI] [PubMed] [Google Scholar]
- 5.Stoffers, D. A., Zinkin, N. T., Stanojevic, V., Clarke, W. L., and Habener, J. F. (1997) Nat. Genet. 15 106-110 [DOI] [PubMed] [Google Scholar]
- 6.Ahlgren, U., Jonsson, J., Jonsson, L., Simu, K., and Edlund, H. (1998) Genes Dev. 12 1763-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gannon, M., Ables, E. T., Crawford, L., Lowe, D., Offield, M. F., Magnuson, M. A., and Wright, C. V. (2008) Dev. Biol. 314 406-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J. D., Bernal-Mizrachi, E., Alejandro, E. U., Han, Z., Kalynyak, T. B., Li, H., Beith, J. L., Gross, J., Warnock, G. L., Townsend, R. R., Permutt, M. A., and Polonsky, K. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19575-19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller, D. M., McWeeney, S., Arsenlis, A., Drouin, J., Wright, C. V., Wang, H., Wollheim, C. B., White, P., Kaestner, K. H., and Goodman, R. H. (2007) J. Biol. Chem. 282 32084-32092 [DOI] [PubMed] [Google Scholar]
- 10.Gannon, M., Gamer, L. W., and Wright, C. V. (2001) Dev. Biol. 238 185-201 [DOI] [PubMed] [Google Scholar]
- 11.Gerrish, K., Gannon, M., Shih, D., Henderson, E., Stoffel, M., Wright, C. V., and Stein, R. (2000) J. Biol. Chem. 275 3485-3492 [DOI] [PubMed] [Google Scholar]
- 12.Gerrish, K., Van Velkinburgh, J. C., and Stein, R. (2004) Mol. Endocrinol. 18 533-548 [DOI] [PubMed] [Google Scholar]
- 13.Fujitani, Y., Fujitani, S., Boyer, D. F., Gannon, M., Kawaguchi, Y., Ray, M., Shiota, M., Stein, R. W., Magnuson, M. A., and Wright, C. V. (2006) Genes Dev. 20 253-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiebe, P. O., Kormish, J. D., Roper, V. T., Fujitani, Y., Alston, N. I., Zaret, K. S., Wright, C. V., Stein, R. W., and Gannon, M. (2007) Mol. Cell. Biol. 27 4093-4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Velkinburgh, J. C., Samaras, S. E., Gerrish, K., Artner, I., and Stein, R. (2005) J. Biol. Chem. 280 38438-38444 [DOI] [PubMed] [Google Scholar]
- 16.Wu, K. L., Gannon, M., Peshavaria, M., Offield, M. F., Henderson, E., Ray, M., Marks, A., Gamer, L. W., Wright, C. V., and Stein, R. (1997) Mol. Cell. Biol. 17 6002-6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samaras, S. E., Cissell, M. A., Gerrish, K., Wright, C. V., Gannon, M., and Stein, R. (2002) Mol. Cell. Biol. 22 4702-4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samaras, S. E., Zhao, L., Means, A., Henderson, E., Matsuoka, T. A., and Stein, R. (2003) J. Biol. Chem. 278 12263-12270 [DOI] [PubMed] [Google Scholar]
- 19.Gerrish, K., Cissell, M. A., and Stein, R. (2001) J. Biol. Chem. 276 47775-47784 [DOI] [PubMed] [Google Scholar]
- 20.Artner, I., Blanchi, B., Raum, J. C., Guo, M., Kaneko, T., Cordes, S., Sieweke, M., and Stein, R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3853-3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka, T. A., Artner, I., Henderson, E., Means, A., Sander, M., and Stein, R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2930-2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, C., Moriguchi, T., Kajihara, M., Esaki, R., Harada, A., Shimohata, H., Oishi, H., Hamada, M., Morito, N., Hasegawa, K., Kudo, T., Engel, J. D., Yamamoto, M., and Takahashi, S. (2005) Mol. Cell. Biol. 25 4969-4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacoby, D. B., Zilz, N. D., and Towle, H. C. (1989) J. Biol. Chem. 264 17623-17626 [PubMed] [Google Scholar]
- 24.Matsuoka, T. A., Zhao, L., Artner, I., Jarrett, H. W., Friedman, D., Means, A., and Stein, R. (2003) Mol. Cell. Biol. 23 6049-6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megason, S. G., and McMahon, A. P. (2002) Development 129 2087-2098 [DOI] [PubMed] [Google Scholar]
- 26.Nordeen, S. K., Green, P. P., 3rd, and Fowlkes, D. M. (1987) Dna 6 173-178 [DOI] [PubMed] [Google Scholar]
- 27.de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., and Subramani, S. (1987) Mol. Cell. Biol. 7 725-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchi, B., Kelly, L. M., Viemari, J. C., Lafon, I., Burnet, H., Bevengut, M., Tillmanns, S., Daniel, L., Graf, T., Hilaire, G., and Sieweke, M. H. (2003) Nat. Neurosci. 6 1091-1100 [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, T., Ohkumo, T., Ishibashi, S., and Yasuda, K. (2005) Nucleic Acids Res. 33 3465-3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St-Onge, L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A., and Gruss, P. (1997) Nature 387 406-409 [DOI] [PubMed] [Google Scholar]
- 32.Ashery-Padan, R., Zhou, X., Marquardt, T., Herrera, P., Toube, L., Berry, A., and Gruss, P. (2004) Dev. Biol. 269 479-488 [DOI] [PubMed] [Google Scholar]
- 33.Olbrot, M., Rud, J., Moss, L. G., and Sharma, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6737-6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Artner, I., Le Lay, J., Hang, Y., Elghazi, L., Schisler, J. C., Henderson, E., Sosa-Pineda, B., and Stein, R. (2006) Diabetes 55 297-304 [DOI] [PubMed] [Google Scholar]
- 35.Nishimura, W., Kondo, T., Salameh, T., El Khattabi, I., Dodge, R., Bonner-Weir, S., and Sharma, A. (2006) Dev. Biol. 293 526-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., Sieving, P. A., and Swaroop, A. (2001) Nat. Genet. 29 447-452 [DOI] [PubMed] [Google Scholar]
- 37.Kim, J. I., Li, T., Ho, I. C., Grusby, M. J., and Glimcher, L. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3781-3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacLean, H. E., Kim, J. I., Glimcher, M. J., Wang, J., Kronenberg, H. M., and Glimcher, L. H. (2003) Dev. Biol. 262 51-63 [DOI] [PubMed] [Google Scholar]
- 39.Ring, B. Z., Cordes, S. P., Overbeek, P. A., and Barsh, G. S. (2000) Development 127 307-317 [DOI] [PubMed] [Google Scholar]
- 40.Cordes, S. P., and Barsh, G. S. (1994) Cell 79 1025-1034 [DOI] [PubMed] [Google Scholar]
- 41.Sadl, V. S., Sing, A., Mar, L., Jin, F., and Cordes, S. P. (2003) Dev. Dyn. 227 134-142 [DOI] [PubMed] [Google Scholar]
- 42.Sadl, V., Jin, F., Yu, J., Cui, S., Holmyard, D., Quaggin, S., Barsh, G., and Cordes, S. (2002) Dev. Biol. 249 16-29 [DOI] [PubMed] [Google Scholar]
- 43.Han, S. I., Aramata, S., Yasuda, K., and Kataoka, K. (2007) Mol. Cell. Biol. 27 6593-6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocques, N., Abou Zeid, N., Sii-Felice, K., Lecoin, L., Felder-Schmittbuhl, M. P., Eychene, A., and Pouponnot, C. (2007) Mol. Cell 28 584-597 [DOI] [PubMed] [Google Scholar]
- 45.Zhao, L., Cissell, M. A., Henderson, E., Colbran, R., and Stein, R. (2000) J. Biol. Chem. 275 10532-10537 [DOI] [PubMed] [Google Scholar]
- 46.Rawdon, B. B. (1998) Microsc. Res. Tech. 43 292-305 [DOI] [PubMed] [Google Scholar]
- 47.Kerppola, T. K., and Curran, T. (1994) Oncogene 9 675-684 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.