Abstract
Syntaxin 1, synaptobrevins or vesicle-associated membrane proteins, and the synaptosome-associated protein of 25 kDa (SNAP-25) are key molecules involved in the docking and fusion of synaptic vesicles with the presynaptic membrane. We report here the molecular, cell biological, and biochemical characterization of a 32-kDa protein homologous to both SNAP-25 (20% amino acid sequence identity) and the recently identified SNAP-23 (19% amino acid sequence identity). Northern blot analysis shows that the mRNA for this protein is widely expressed. Polyclonal antibodies against this protein detect a 32-kDa protein present in both cytosol and membrane fractions. The membrane-bound form of this protein is revealed to be primarily localized to the Golgi apparatus by indirect immunofluorescence microscopy, a finding that is further established by electron microscopy immunogold labeling showing that this protein is present in tubular-vesicular structures of the Golgi apparatus. Biochemical characterizations establish that this protein behaves like a SNAP receptor and is thus named Golgi SNARE of 32 kDa (GS32). GS32 in the Golgi extract is preferentially retained by the immobilized GST–syntaxin 6 fusion protein. The coimmunoprecipitation of syntaxin 6 but not syntaxin 5 or GS28 from the Golgi extract by antibodies against GS32 further sustains the preferential interaction of GS32 with Golgi syntaxin 6.
INTRODUCTION
Soluble N-ethylmaleimide–sensitive factor (NSF) and soluble NSF attachment proteins (SNAPs) play a central role along the secretory pathway and in other trafficking events (Graham and Emr, 1991; Pryer et al., 1992; Rothman, 1994; Rothman and Wieland, 1996; Whiteheart and Kubalek, 1995; Schekman and Orci, 1996). The action of SNAPs and NSF is mediated by SNAP receptors (SNAREs) on the membrane (Rothman, 1994; Whiteheart and Kubalek, 1995). The SNARE hypothesis is proposed to account for the specificity of vesicle transport and predicts that specific interaction between vesicle-associated SNAREs with the cognate SNAREs (t-SNAREs) on the target membrane (Rothman, 1994; Whiteheart and Kubalek, 1995) is central for vesicle docking onto and fusion with the correct target membrane. Synaptobrevin or vesicle-associated membrane proteins (VAMP 1) are vesicle-associated SNAREs associated with synaptic vesicles, whereas syntaxin 1 and the synaptosome-associated protein of 25 kDa (SNAP-25) are t-SNAREs associated with the presynaptic membrane. The specific interaction of synaptobrevin or VAMP 1 with a complex of syntaxin 1 and SNAP-25 is involved in the docking and fusion of synaptic vesicles with the presynaptic membrane (Söllner et al., 1993; Ferro-Novick and Jahn, 1994; Rothman, 1994; Rothman and Warren, 1994; Scheller, 1995; Südhof, 1995; Weber et al., 1998).
In addition to involvement in synaptic vesicle docking and fusion, syntaxin-like proteins have also been implicated in transport events that occur on the plasma membrane and in intracellular organelles. In yeast, Sso1p and Sso2p are involved in the docking and fusion of Golgi-derived vesicles with the plasma membrane (Aalto et al., 1993), whereas Sed5p is involved in the ER–Golgi transport (Hardwick and Pelham, 1992). Ufe1p participates in retrograde transport from the Golgi back to the ER (Lewis and Pelham, 1996), and Pep12 and Vam3p are involved in the transport from the late Golgi to the vacuolar system (Becherer et al., 1996; Darsow et al., 1998; Odorizzi et al., 1998; Sato et al., 1998). Recently, two other syntaxin-like yeast proteins (Tlg1p and Tlg2p) have been shown to be involved in the endosomal pathway (Holthuis et al., 1998). Several syntaxins have been identified in mammalian cells, possibly up to 14 distinct syntaxins (Bock and Scheller, 1997; Tang et al., 1998). Although syntaxin 1 participates specifically in synaptic vesicle docking and fusion and regulated exocytosis (Bennett et al., 1993; Söllner et al., 1993; Scheller, 1995; Südhof, 1995), syntaxins 2, 3, and 4 may be involved in docking and/or fusion on the plasma membrane (Bennett et al., 1992, 1993; Gaisano et al., 1996; Low et al., 1996; Peng et al., 1997). Syntaxins 5 and 6 are two Golgi-associated syntaxins (Bennett et al., 1993; Banfield et al., 1994; Bock et al., 1996). Syntaxin 5 is the mammalian counterpart of Sed5p and is involved in the ER–Golgi transport and possibly in the intra-Golgi transport (Bennett et al., 1993; Dascher et al., 1994; Fernandez and Warren, 1998). The precise role played by syntaxin 6 still remains to be established, but it is known to be associated with the trans-Golgi (Bock et al., 1997; Hay et al., 1997).
Similarly, synaptobrevin 1– or VAMP 1–like proteins have been shown to participate in diverse transport events, and up to eight different synaptobrevins or VAMPs may exist in mammalian cells (Bock and Scheller, 1997). Cellubrevin is ubiquitously expressed and may participate in trafficking events in the endocytotic pathway (McMahon et al., 1993; Galli et al., 1994). TI-VAMP, another ubiquitously expressed VAMP, is implicated in exocytotic processes at the apical plasma membrane of epithelial cells (Galli et al., 1998). VAMP 4 and VAMP 5 are likely to be enriched in the Golgi and the cell surface, respectively (Advani et al., 1998; Zeng et al., 1998). Endobrevin has recently been identified and is enriched in the early endosome (Wong et al., 1998b). Distantly related synaptobrevin- or VAMP-like proteins, such as Bet1p, Sec22p, and Bos1p and their mammalian counterparts, are involved in the ER–Golgi transport (Newman et al., 1990, 1992; Dascher et al., 1991; Ossig et al., 1991; Shim et al., 1991; Lian and Ferro-Novick, 1993; Sögaard et al., 1994; Xu et al., 1997; Zhang et al., 1997), whereas another synaptobrevin- or VAMP-like protein, Sft1p, is implicated in the intra-Golgi retrograde transport (Banfield et al., 1995). Ykt6p, a synaptobrevin- or VAMP-like protein, is associated with the Golgi system and may be involved in the ER–Golgi and/or the intra-Golgi transport (Sögaard et al., 1994; Lupashin et al., 1997; McNew et al., 1997)
In contrast to syntaxin-like and synaptobrevin- or VAMP-like proteins, SNAP-25–like proteins are implicated to date only in trafficking events on the plasma membrane. SNAP-25 is involved mainly in the synaptic vesicle docking and/or fusion and in regulated exocytosis (Trimble et al., 1988; Elferink et al., 1989; Oyler et al., 1989; Südhof et al., 1989; Bark and Wilson, 1994; Bock et al., 1996), whereas SNAP-23 has been proposed to be a ubiquitously expressed counterpart of SNAP-25 and is associated with the plasma membrane (Ravichandran et al., 1996; Araki et al., 1997; Wong et al., 1997). Sec9p, a SNAP-25–like yeast protein, has been shown to participate in the docking and/or fusion of Golgi-derived vesicles with the plasma membrane (Brennwald et al., 1994). Except for syntaxin 6 and 10 that have some homologies to SNAP-25 at the C-terminal coiled-coil domain (Bock et al., 1996), no other SNAP-25–like proteins have yet been shown to be associated with any intracellular organelles or to participate in other trafficking events. Even in the case of syntaxin 6 and 10, despite their amino acid sequence homology with SNAP-25, it displays structural characteristics very similar to the members of the syntaxin family, with two potential coiled-coil regions and a C-terminal membrane–anchored domain (Bock et al., 1996). These observations raised the possibility that, unlike syntaxin-like and synaptobrevin- or VAMP-like proteins, SNAP-25–like proteins may be restricted to the docking and/or fusion events on the plasma membrane but not that in intracellular organelles (Weimbs et al., 1998).
In this manuscript, we have molecularly characterized a Golgi SNARE of 32 kDa (GS32) that is significantly related to SNAP-25 and SNAP-23. Furthermore, we show that GS32 is specifically associated with the Golgi apparatus by indirect immunofluorescence microscopy as well as by electron microscopy (EM) immunogold labeling. Phylogenetics analysis based on conserved coiled-coil domains classifies GS32 in the same group as SNAP-23, SNAP-25, and Sec9p but segregates GS32 away from the syntaxins. We demonstrate here that GS32 behaves like a SNARE and preferentially interacts with syntaxin 6.
MATERIALS AND METHODS
Materials
All cell lines were obtained from the American Type Culture Collection (Rockville, MD). Synthetic oligonucleotides were purchased from Oligos Etc (Wilsonville, OR). Antibodies against RhoGDI and EEA1 were obtained from Santa Cruz (Santa Cruz, CA) and Transduction Laboratories (Lexington, KY), respectively. Antibody against rat transferrin receptor (anti-rat CD71) was purchased from PharMingen (San Diego, CA). The rat brain λZAP cDNA library and Pyrococcus furiosus DNA polymerase were from Stratagene (La Jolla, CA). The rat mRNA multiple tissues Northern filter was purchased from Clontech (Palo Alto, CA). The oligolabeling kit and glutathione Sepharose 4B beads were from Pharmacia (Uppsala, Sweden). Fluorescein isothiocyanate–conjugated goat anti-mouse immunoglobulin (IgG) and rhodamine-conjugated goat anti-rabbit IgG were purchased from Boehringer Mannheim (Indianapolis, IN). Brefeldin A (BFA) was from Epicentre Technologies (Madison, WI).
cDNA Cloning and Sequencing
A human expressed-sequence tag (EST) clone (accession number, R51970) encoding an open reading frame that is homologous to SNAP-25 was revealed during database searches using the BLAST program. Two oligonucleotides, primer 1 (5′-GGGAATTCTAAAGATCGACAGCAACCTAGATG) and primer 2 (5′-GGGTCTAGATCAGAGTTGTCGAACTTTTCTTTCTG), were used to polymerase chain react a 196-bp DNA fragment from this EST clone, which was 32P-labeled and used to screen a rat brain λZAP cDNA library as described (Lowe et al., 1996). One full-length clone with an insert size of ∼1.5 kb was sequenced.
Northern Blot Analysis
The DNA sequence coding for the full-length protein produced by polymerase chain reaction (PCR) using primers 3 (5′-CGGGATCCATGTCTGGCTATCCTAAAAGC) and 4 (5′-TCCCCCGGGCTAGAGTTGCCGCACCTT) was used as a probe and hybridized to a rat mRNA multiple tissues Northern filter blot of poly(A)+ mRNA.
Expression and Purification of Recombinant Proteins
For GS32 expression as a HisX6-tagged protein, the PCR product using primers 3 and 4 was digested with BamHI and SmaI restriction enzymes, ligated into the pQE30 vector (Qiagen, Hilden, Germany), and transformed into the Escherichia coli M15[pREP4] strain. For HisX6-tagged syntaxin 6, the PCR product derived from primers A (5′-GCTCTCCATGGAGGACCCCTTCTTTGTAGTG-3′) and B (5′-CTCTGGATCCGCGCCGATCACTGGTCATGTGAGA-3′), encoding for the cytoplasmic domain of syntaxin 6, was inserted into the NcoI and BamHI sites of the pQE60 vector (Qiagen) and then transformed into E. coli M15[pREP4]. Recombinant protein was produced and purified as described previously (Subramaniam et al., 1996). For the preparation of GST fusion proteins of syntaxins 3, 4, 5, and 6 (the entire cytoplasmic domain of the syntaxins was fused to the C-terminus of GST), PCR products from primers 5 (5′-GGGAATTCTAATGAAGGACCGACTGG) and 6 (5′-GGCTCTAGATCATTTCTTTCGAGCCTGACCCTG) (for syntaxin 3), primers 7 (5′-GGGAATTCTAATGCGCGACAGGACCCATG) and 8 (5′-GGCTC-TAGATCACCTCGCCTTCTTCTGATTCTCTAG) (for syntaxin 4), primers 9 (5′-GGGAATTCTAATGTCCTGCCGGGATCGG) and 10 (5′-GGCTCTAGATCATTTGACCATGAGCCACCGATTG) (for syntaxin 5), and primers 11 (5′-GGGAATTCTAATGTCCATGGAGGA-CCCCTTCTTTG) and 12 (5′-GGCTCTAGATCAGCGCCGATCACTGGTCATG) (for syntaxin 6) were digested with EcoRI and XbaI and then ligated with the EcoRI/XbaI-digested pGEX-KG vector (Guan and Dixon, 1991). Ligation mixtures were then transformed into DH5α cells and screened for colonies expressing GST fusion proteins as described (Sambrook et al., 1989). Expression and purification of GST recombinant proteins were performed as described elsewhere (Lowe et al., 1996).
Preparation and Purification of Polyclonal Antibodies
Rabbits were each injected with 300 μg of purified recombinant proteins (GST–syntaxin 5, –HisX6-tagged GS32, and –syntaxin 6) emulsified in complete Freund’s adjuvant. Booster injections containing similar amounts of the antigen emulsified in incomplete Freund’s adjuvant were administered every 2 wk. Rabbits were bled 10 d after the second and subsequent booster injections. For affinity purification, serum was diluted twice with phosphate-buffered saline (PBS) and incubated with cyanogen bromide–activated Sepharose coupled with the antigen, and specific antibodies were eluted from the beads.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described previously (Subramaniam et al., 1995; Lowe et al., 1996). For treatment of cells with BFA or nocodazole, normal rat kidney (NRK) cells were incubated with 10 μg/ml of either BFA or nocodazole for 60 min at 37°C before being processed for immunofluorescence microscopy. For the treatment of cells with digitonin, epithelial lung cells from rat (L2 cells) on a coverslip (either untreated or treated with BFA or nocodazole) were permeabilized with 25 μg/ml of digitonin in PBS containing 1 mM MgCl2 and 1 mM CaCl2 for 5 min on ice. The coverslip was then washed five times with ice-cold PBS containing 1 mM MgCl2 and 1 mM CaCl2 before being processed for immunofluorescence microscopy.
Immunogold Labeling
Cryosections and EM immunogold double labeling were performed as described previously (Slot et al., 1991; Griffiths, 1993; Griffiths et al., 1994).
Immunoblot Analysis of Cytosol and Membrane Fractions
NRK cells grown in T-25 flasks were trypsinized and washed twice with DMEM media containing 10% fetal bovine serum. The cell pellet was resuspended in 200 μl of PBS and lysed by sonication. Nuclei and unbroken cells were removed by spinning the lysate at 6000 × g for 10 min. The postnuclear supernatant was then centrifuged at 100,000 × g for 30 min to separate the cytosol (supernatant) from the total membrane (pellet). The pellet was then resuspended in 200 μl of PBS containing 1% Triton X-100 and was incubated on ice for 1 h. Equal fractions of the supernatant and the pellet were separated by SDS-PAGE and analyzed by immunoblot (Subramaniam et al., 1995; Lowe et al., 1996).
Preparation of Golgi-enriched Membranes
Preparation and subfractionation of membranes were performed as described previously (Subramaniam et al., 1992; Wong et al., 1998b). Briefly, livers from Harlan Sprague Dawley (Indianapolis, IN) rats were homogenized in homogenization buffer (25 mM HEPES, pH 7.3, 5 mM MgCl2, 1 mM PMSF) containing 0.25 M sucrose and were centrifuged at 10,000 × g for 10 min. The supernatants were then recentrifuged at 100,000 × g for 1 h, and the total membrane pellet was resuspended in a minimal volume of homogenization buffer containing 0.25 M sucrose. The membrane suspension, adjusted to a final concentration of 1.25 M sucrose, was overlaid with step gradients of 10 ml of 1.1 M sucrose, 10 ml of 1.0 M sucrose, and 5.0 ml of 0.5 M sucrose in homogenization buffer and then was centrifuged at 28,000 rpm for 3 h in a Beckman (Fullerton, CA) SW 28 rotor. The Golgi at the 0.5 M/1.0 M sucrose interphase was collected and used for the subsequent experiments.
Treatment of Membranes with Salts and Detergents
Preparation and subfractionation of membranes were performed as described previously (Subramaniam et al., 1992; Wong et al., 1998b). Golgi membrane (500 μg) was extracted on ice for 1 h in 100 μl of either PBS, 2 M KCl, 2.5 M urea, 0.15 M sodium bicarbonate (pH 12.0), 1% Triton X-100, or 1% Nonidet P-40 (NP-40) and then was centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was collected, and the pellet was resuspended in 100 μl of 1× SDS sample buffer. Equal fractions (20 μl) from both the supernatant as well as the pellet were separated by SDS-PAGE and analyzed by immunoblotting.
Protease Protection Analysis using Golgi Membranes
Protease treatment of Golgi membranes was performed as described previously (Subramaniam et al., 1995). Briefly, Golgi membranes (100 μg in 0.25 M sucrose and 25 mM HEPES, pH 7.3) were incubated in the presence or absence of trypsin (2 mg/ml) at 4°C for 1 h. The reactions were stopped by the addition of 2 mM PMSF, separated by SDS-PAGE, and analyzed by immunoblotting.
Formation of 20-S SNARE Complex
This was performed as described (Wilson et al., 1992; Söllner et al., 1993; Subramaniam et al., 1995).
In Vitro Binding of Golgi Extract with Immobilized GST–α-SNAP and GST–Syntaxins
Golgi-enriched membranes (1 mg) were extracted in 500 μl of incubation buffer (100 mM KCl, 20 mM HEPES, pH 7.3, 2 mM EDTA, 2 mM DTT, 0.2 mM ATP) containing 1% Triton X-100 and then were diluted with 500 μl of incubation buffer without Triton X-100. The extracted proteins were separated from the membrane debris by centrifugation. GST–α-SNAP and GST–retinoblastoma protein (the entire polypeptide of retinoblastoma protein [RB] fused to the C-terminus of glutathione S-transferase protein) fusion proteins immobilized on beads (2 μg) were washed twice with incubation buffer containing 0.5% Triton X-100 and then were incubated with increasing amounts of Golgi extracts in a total volume of 100 μl at 4°C for 3 h with agitation. Beads were then washed twice with incubation buffer containing 0.5% Triton X-100, once with incubation buffer containing 0.1% Triton X-100, and twice with incubation buffer without Triton X-100. The beads were then processed for immunoblot analysis to detect GS32. For interaction with syntaxins, Golgi extracts were incubated with immobilized GST–syntaxins 3, 4, 5, and 6 (8 μg) as described above. GS32 retained by the beads was analyzed by immunoblot.
Immunoprecipitation
GS32- and GS28-specific antibody and rabbit IgG (100 μg) bound to protein A Sepharose beads (Pharmacia) were each incubated overnight with 500 μg of Golgi extracts in incubation buffer (20 mM HEPES, pH 7.2, 100 mM KCl, 1 mM DTT, 10 mM EDTA, 0.2 mM ATP, 1% Triton X-100) at 4°C. Beads were then washed twice in buffer A (identical to incubation buffer except that it contains 0.5% Triton X-100) and twice in buffer B (identical to incubation buffer except that it contains 0.2% Triton X-100) before being resuspended in SDS sample buffer. Immunoprecipitated proteins and 20% of the supernatant were separated on SDS-PAGE and analyzed by immunoblot using antibodies against GS32, syntaxin 5, and syntaxin 6.
RESULTS
GS32, a Protein Related to SNAP-25 and SNAP-23
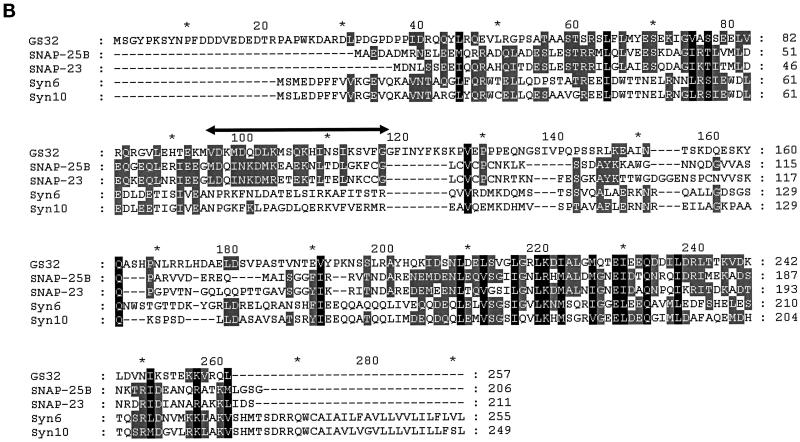
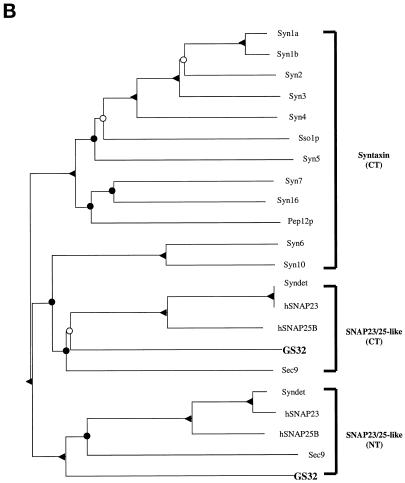
A human EST clone (accession number, R51970) that could encode a protein fragment having amino acid sequence similarity with SNAP-25 was revealed by the BLAST program using the amino acid sequence of SNAP-25 to search the EST database. The PCR product of this EST clone was used to screen a rat brain λZAP cDNA library. Several clones were isolated and partially sequenced, and one full-length clone was completely sequenced. The nucleotide and the deduced amino acid sequences of GS32 are shown in Figure 1A. The BLAST search of the National Center for Biotechnology Information’s NR protein Database and Beauty using the full-length GS32 amino acid sequence as the query retrieved SNAP-25A, SNAP-25B, SNAP-23, and syndet as well as Sec9 at the high score range. Figure 1B shows that the deduced 257-amino acid sequence of GS32 is ∼20, 19, 9, and 10% identical to SNAP-25B, SNAP-23, syntaxin 6, and syntaxin 10, respectively (Trimble et al., 1988; Elferink et al., 1989; Oyler et al., 1989; Südhof et al., 1989; Bark and Wilson, 1994; Bock et al., 1996; Ravichandran et al., 1996; Wong et al., 1997; Tang et al., 1998). GS32 is related to SNAP-25 evenly throughout the entire protein with an overall similarity of 32%. Furthermore, phylogenetic analysis, based on the conserved coiled-coil domains of GS32 and t-SNAREs such as SNAP-25, SNAP-23, and the syntaxins, shows that GS32 is closely related to the SNAP-23/25 family but segregates away from the syntaxins (Figure 2, A and B).
Figure 1.
(A) The nucleotide (upper) and deduced amino acid (lower) sequences of GS32. Sequence numbering is shown on the left. (B) The alignment of the GS32 amino acid sequence with that of SNAP-25B, SNAP-23, syntaxin (Syn)6, and syntaxin 10. Amino acid residues shaded gray and black indicate >40 and 80% similarity, respectively. There exists a region (residues 95–118; double-headed horizontal arrow) in the primary sequence of GS32 that is only homologous to SNAP-25 and SNAP-23 but not to syntaxins.
Figure 2.
(A) Sequence alignments of the N-terminal and C-terminal coiled-coil regions of GS32, SNAP-25, SNAP-23, and the syntaxins. The protein names, species, and accession numbers are as follows: Syn1a (Rattus norvegicus; P32851), Syn1b (Rattus norvegicus; P32853), Syn2 (Homo sapiens; P32856), Syn3 (Homo sapiens; X90581), Syn4 (Homo sapiens; U07158), Sso1p (Saccharomyces cerevisiae; P32867), Syn5 (Homo sapiens; U26648), Pep12p (Saccharomyces cerevisiae; P32854), Syn16 (Homo sapiens; AF038897), Syn7 (Homo sapiens; U77942), Syn6 (Homo sapiens; AJ002078), Syn10 (Homo sapiens; AF035531), Sec9 (Saccharomyces cerevisiae; P40357), hSNAP25B (Homo sapiens; P13795), hSNAP23 (Homo sapiens; U55936), and Syndet (Mus musculus; U73143). NT and CT refer to the N-terminal and C-terminal homology domain of SNAP-25–related proteins, respectively. (B) Phylogenetic tree (nearest-neighbor dendrogram) based on the conserved coiled-coil domains of GS32, SNAP-25, SNAP-23, and the syntaxins. Phylogenetic analysis was performed according to the method described previously (Weimbs et al., 1997). Bifurcation points confirmed in 90–100% of bootstrap replicates (out of 1000) are marked by solid triangles, in 50–90% of replicates are marked by solid circles, and in 30–50% of replicates are marked by open circles.
GS32 Is Widely Expressed
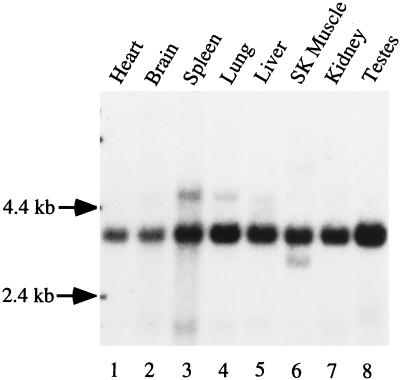
To understand whether GS32 is involved in a cellular process in a general manner or whether its function is restricted to certain tissues, we performed Northern blot analysis to examine the levels of mRNA in various rat tissues (Figure 3). A major mRNA species of ∼3.8 kb was detected and is present at high levels in all the tissues examined, suggesting that it may participate in a general cellular process. Despite the fact that the size of GS32 mRNA is significantly longer than that of the cloned cDNA, the derived amino acid sequence is of full-length because the open reading frame is flanked in-frame with a strong initiation Met codon (Kozak, 1984) at the 5′-end and a termination codon at the 3′-end. Furthermore, there is an in-frame termination codon upstream from the initiation Met codon at nucleotide position 16.
Figure 3.
Northern blot analysis of GS32 mRNA.
GS32 Is Detected in Both the Cytosol and Membranes
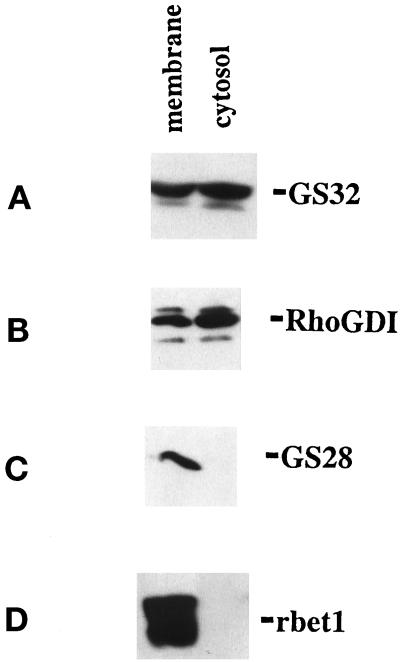
The amino sequence of GS32 suggests that GS32 could be a soluble protein because of the absence of a putative transmembrane region. To determine this point, we fractionated the postnuclear supernatant of homogenized NRK cells and analyzed equal percentages of the cytosol and membrane fractions on SDS-PAGE. Immunoblot analysis of these fractions with affinity-purified anti-GS32 showed that GS32, having an apparent size of 32 kDa, exists both in the cytosol (∼60%) and membrane (∼40%) fractions (Figure 4A). In another set of experiments, RhoGDI (Boivin and Beliveau, 1995), a soluble protein that has been shown previously to exist both in the cytosol as well as in the membrane (Marshansky et al., 1996), is also detected more in the cytosol (70%) as compared with the membrane (30%) fractions (Figure 4B). On the other hand, transmembrane proteins GS28 (Subramaniam et al., 1995) and rbet1 (Zhang et al., 1997) are detected only in the membrane fraction (Figure 4, C and D). Because rbet1 has been implicated as present on vesicles (Newman et al., 1990, 1992; Stones et al., 1997; Zhang et al., 1997; Rowe et al., 1998), its specific detection in the membranes but not in the cytosol fraction clearly excluded the possibility that GS32 detected in the cytosol fraction could be contributed to a certain extent by contamination from unpelleted small vesicles.
Figure 4.
GS32 is a 32 kDa protein associated with both the cytosol and membrane fractions. The cytosol and total membrane fractions derived from NRK cells were resolved by SDS-PAGE and then processed for immunoblot analysis using antibodies against GS32 (A), RhoGDI (B), GS28 (C), and rbet1 (D).
GS32 Associates Tightly with Membranes
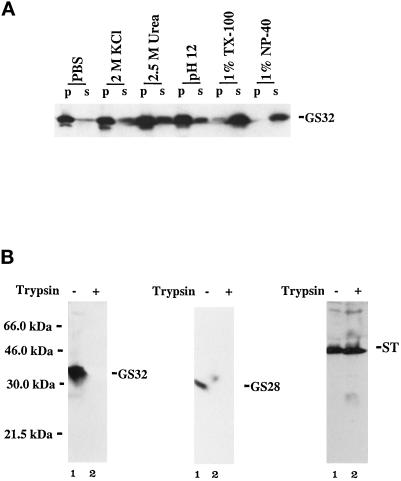
The enrichment of GS32 in the cytosol fraction and the absence of a hydrophobic membrane anchor domain suggest that GS32 may be a soluble protein that associates with membranes. To determine this point, we extracted the membrane fraction with PBS, 2 M KCl, 2.5 M urea, 0.15 M sodium bicarbonate (pH 12.0), 1% Triton X-100, or 1% NP-40. Figure 5A shows that GS32 is not solubilized in PBS or 2 M KCl, is solubilized partially in 2.5 M urea and 0.15 M sodium bicarbonate (pH 12.0), and is solubilized effectively by 1% Triton X-100 and 1% NP-40. The ability of high pH and urea to partially extract GS32 from membranes shows that GS32 is a soluble protein that associates strongly with membranes. This stable high-affinity association with membranes indicates that GS32 could be interacting with a membrane receptor. To examine further whether GS32 is associated with the membranes on the side facing the cytosol, we performed a protease-protecting analysis as described previously (Subramaniam et al., 1995). The rational for this analysis is that trypsin will digest polypeptides that are exposed only on the cytosolic surface of the Golgi membranes (the association of GS32 with Golgi membranes is demonstrated below), whereas those inside (luminal) are protected. Golgi membranes were either untreated or treated with trypsin and then were analyzed by immunoblotting to detect GS32, GS28, and α-2,6-sialyltransferase (ST). As shown in Figure 5B, GS32 could be detected only in untreated Golgi membranes, and its detection was completely abolished after treatment with trypsin. As a control, GS28, a SNARE anchored to the Golgi by its C-terminal membrane anchor (Subramaniam et al., 1995, 1996), was also not detected after treatment with trypsin. On the other hand, ST, with nearly all of its polypeptide (C-terminal) in the luminal side of the Golgi apparatus (Colley et al., 1989; Wong et al., 1992), was protected from trypsin. However, in the presence of both trypsin and 1% Triton X-100, ST could not be detected (our unpublished observations). Taken together, these results establish that GS32 is associated with the cytoplasmic side of the Golgi membranes.
Figure 5.
The membrane pool of GS32 is associated stably on the cytosolic side of Golgi membranes. (A) Golgi membranes were extracted with a range of different reagents as indicated and were separated by high-speed centrifugation into pellet (p) and supernatant (s) fractions. Aliquots (100 μg of these fractions) were analyzed by SDS-PAGE and immunoblot, and GS32 was detected. (B) Golgi membranes (100 μg) were either treated (lane 2) or untreated (lane 1) with trypsin (2 mg/ml) at 4°C for 1 h and were analyzed by SDS-PAGE and immunoblot analysis using antibodies against GS32, GS28, and ST.
GS32 Is Localized to the Golgi Apparatus
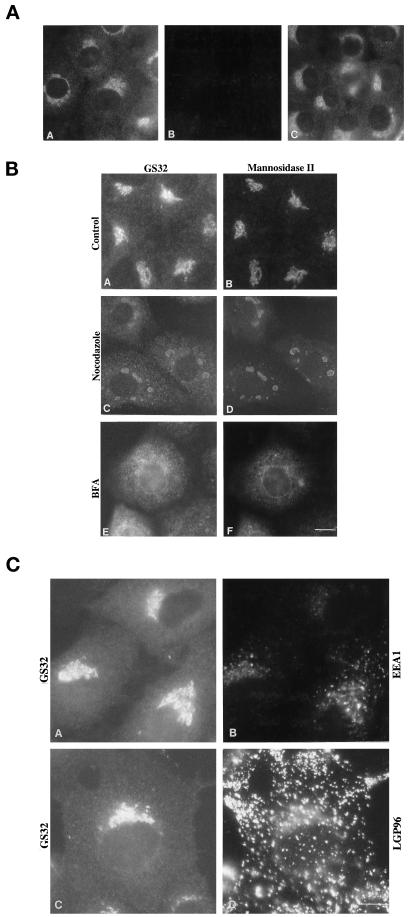
To investigate the specific membrane with which GS32 is associated, indirect immunofluorescence microscopy was used to examine the cellular localization of GS32. As shown in Figure 6A, panel A, affinity-purified polyclonal antibodies against GS32 clearly labeled the perinuclear Golgi-like region (Louvard et al., 1982) and the cytosol in NRK cells. The labeling by GS32 antibodies is specific because both cytosolic and perinuclear Golgi-like labelings were abolished by preincubating the antibodies with the recombinant GS32 (Figure 6A, panel B) but not with the recombinant syntaxin 6 (panel C). Figure 6B, panels A and B, shows that the perinuclear labeling of GS32 colocalized well with that of Golgi mannosidase II. Colocalization of GS32 with Golgi mannosidase II could still be seen on punctate structures after treatment of cells with nocodazole (Figure 6B, panels C and D). In BFA-treated cells, both GS32 and Golgi mannosidase II are redistributed to the ER-like structures (Figure 6B, panels E and F), a typical characteristic of protein localized to the Golgi apparatus. The colocalization of the GS32 and Golgi mannosidase II labeling in the nocodazole- and BFA-treated and in untreated cells suggests that GS32 is associated with the Golgi apparatus.
Figure 6.
GS32 is associated with the Golgi apparatus. (A) NRK cells were processed for indirect immunofluorescence microscopy using antibodies against GS32 (panel A). The specific Golgi-like and cytosolic labelings were abolished by preincubation of the antibodies with the recombinant GS32 (panel B) but not with the recombinant syntaxin 6 (panel C). (B) NRK cells were untreated (panels A and B) or treated with either nocodazole (panels C and D) or BFA (panels E and F) and then were stained for GS32 (panels A, C, and E) or mannosidase II (panels B, D, and F). (C) Double labeling of GS32 (panels A and C) with either EEA1 (panel B) or LGP96 (panel D) in NRK cells is shown. Bars, 10 μm.
Some fine punctate structures were seen throughout the diffuse cytosolic labeling of GS32 other than its Golgi labeling. These punctate structures could represent local concentrated areas of GS32 in the cytoplasm of intact cells or induced in chemically fixed cells. Alternatively, these structures could represent association of GS32 with vesicular structures of the endosomal pathway. Two independent experiments were performed to distinguish between these two possibilities. First, cells were double labeled with polyclonal antibodies against GS32 and with monoclonal antibodies against markers of the endosomal pathway. As shown in Figure 6C, the fine punctate structures marked by GS32 colocalized neither with the early endosomal structures marked by EEA1 (Mu et al., 1995; Stenmark et al., 1996; Patki et al., 1997) nor with the late endosomal and lysosomal structures marked by LGP96 (Okazaki et al., 1992; Cuervo and Dice, 1996), suggesting that labeling of these fine punctate structures does not represent endosomal structures.
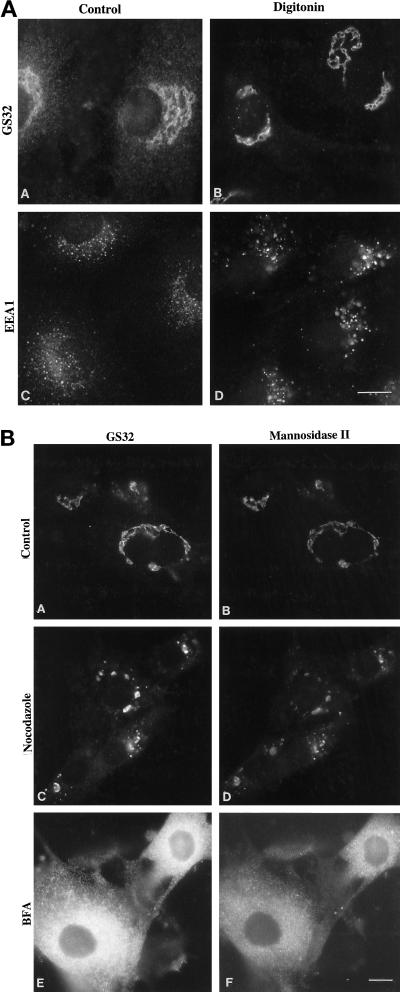
To distinguish further whether these fine punctate structures are membrane-associated or cytosolic-concentrated areas of GS32, we first selectively permeabilized the plasma membrane with low concentrations of digitonin and followed with a brief washing with a mild buffer such as PBS to remove the cytosolic pool of GS32. The cells were than fixed and processed for indirect immunofluorescence microscopy. As shown in panels A and B of Figure 7, A and B, when the cytosolic pool of GS32 was first removed by this procedure, GS32 was seen associated only with the Golgi apparatus marked by Golgi mannosidase II. Although it is highly unlikely, it remains a possibility that peripheral proteins associated with the endosomal system are selectively removed by digitonin permeabilization and by PBS washing. To eliminate this possibility, we examined the distribution of early endosomal EEA1 (Mu et al., 1995; Stenmark et al., 1996; Patki et al., 1997) in control cells and in cells that had undergone digitonin permeabilization and PBS washing (Figure 7A, panels C and D). As shown, distinct punctate and some diffuse cytosolic labeling for EEA1 was observed in control cells (Figure 7A, panel C). In cells that had undergone digitonin permeabilization and PBS washing, the punctate labeling became more prominent, while the cytosolic labeling was selectively removed (Figure 7A, panel D), establishing that the procedure of digitonin permeabilization and PBS washing removes selectively the cytosolic pool of peripheral membrane proteins and has no effect on their association with structures of the endosomal pathway. Taken together, these observations suggest that while the cytosolic pool of GS32 was selectively removed, GS32 is associated only with the Golgi apparatus marked by Golgi mannosidase II.
Figure 7.
GS32 is only localized to the Golgi apparatus in digitonin-permeabilized L2 cells. (A) L2 cells were either untreated (panels A and C) or treated with digitonin (panels B and D) and were labeled for either GS32 (panels A and B) or EEA1 (panels C and D). (B) L2 cells were either untreated (panels A and B) or treated with either nocodazole (panels C and D) or BFA (panels E and F) before permeabilization with digitonin and double labeling for either GS32 (panels A, C, and E) or mannosidase II (panels B, D, and F). Bars, 10 μm.
We next determined the distribution of GS32 in digitonin-permeabilized L2 cells that have been either untreated or pretreated with nocodazole or BFA. As shown in Figure 7B, GS32 (panel A) is colocalized perfectly with Golgi mannosidase II (panel B) in the Golgi apparatus of untreated digitonin-permeabilized L2 cells. Nocodazole treatment that fragments the Golgi apparatus marked by mannosidase II (Figure 7B, panel D) also caused an effect similar to GS32 (panel C), and GS32 is associated with the fragmented-Golgi structures. BFA treatment redistributes both the GS32 and mannosidase II to the ER-like structures (Figure 7B, panels E and F). Under the BFA condition, the labeling of GS32 colocalized with that of mannosidase II. Taken together, these results suggest that GS32, in addition to its presence in the cytosol, is associated with the Golgi apparatus.
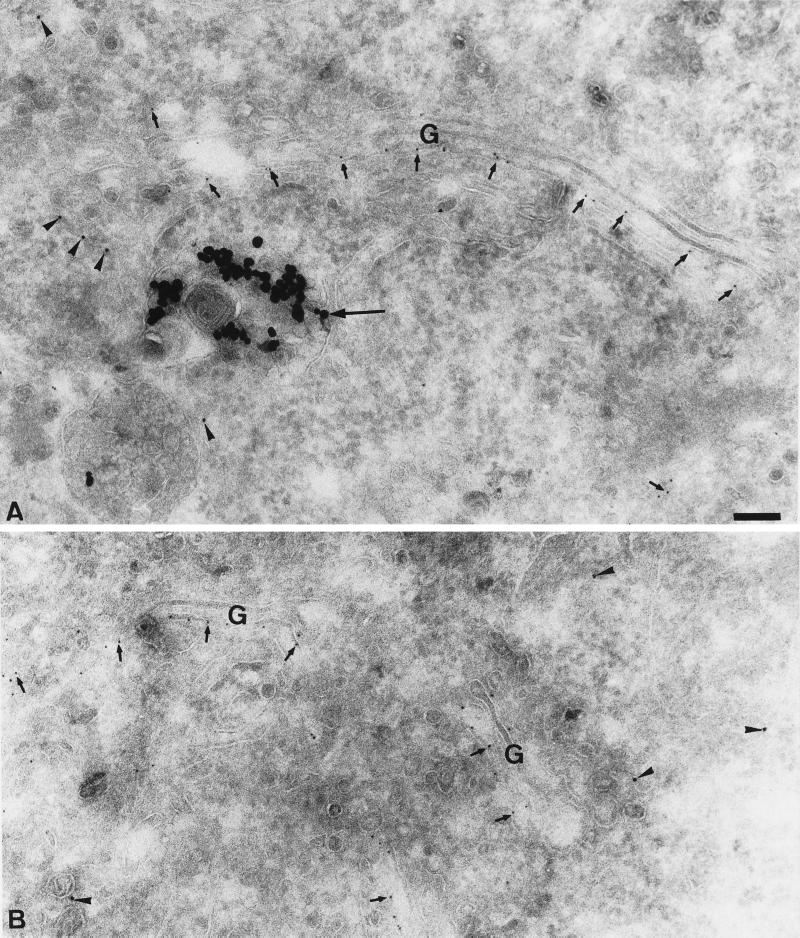
To support further the conclusion that GS32 is associated with the Golgi apparatus and to define the structures of the Golgi apparatus, EM immunogold labeling was performed in cryosections of Hela (SA:48) cells that were stably expressing the human ST tagged on the lumenal side with the P5D4 epitope from the cytoplasmic domain of the vesicular stomatitis virus glycoprotein (Kreis, 1986; Griffiths et al., 1994). As shown in Figure 8, A and B, GS32 (arrowheads) is observed on tubular and vesicular profiles of the Golgi apparatus (small arrows). The Golgi cisternae marked by ST contain no GS32 labeling. A recent study has shown that ST is associated mainly with the cisternae of the Golgi apparatus (Lovelock and Lucocq, 1998). In addition, GS32 is not detected in the late endosomes and/or lysosomes structure marked by the 15 nm gold particles that were internalized from the cell surface for 30 min (Figure 8A, large arrow). These results establish that GS32 is associated preferentially with the tubular-vesicular structures of the Golgi apparatus.
Figure 8.
GS32 is enriched in the Golgi apparatus. (A and B) Cryosections of Hela SA:48 cells were processed for EM immunogold labeling to detect G-protein tail for the epitope-tagged ST (small arrows) and GS32 (arrowheads). The large arrow in A indicates 15 nm gold in the late endosomes and/or lysosomes. Bar, 100 nm.
GS32 as a Novel Golgi SNARE
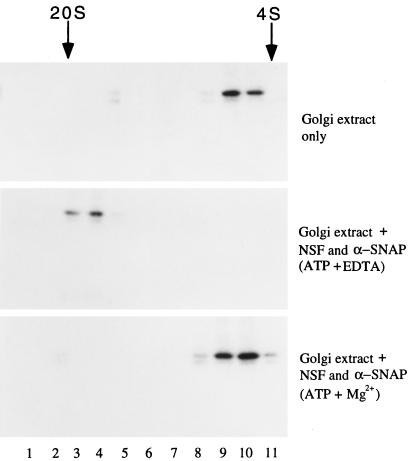
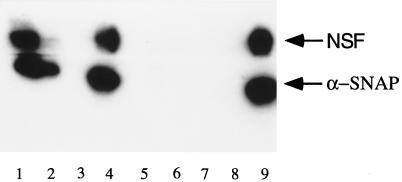
From the analysis of the amino acid sequences as well as the phylogenetic analysis of GS32, SNAP-25, SNAP-23, and the syntaxins, we are tempted to suggest that GS32 could be a SNARE and may share similarities with SNAP-25. We then investigated whether GS32 is a new SNARE associated with the Golgi apparatus. As shown in Figure 9, when Golgi extract was sedimented on a glycerol gradient, GS32 had a sedimentary coefficient of ∼6 S (Figure 9, top). When incubated in the presence of recombinant α-SNAP and NSF in the buffer that promotes the formation of the 20-S SNARE complex, GS32 was shifted to the 20-S fractions (Figure 9, middle). This shift did not occur when Golgi extract was incubated with equal amounts of α-SNAP and NSF in the buffer that promotes disassembly of the SNARE complex (Figure 9, bottom). To determine whether GS32 is indeed associated with α-SNAP and/or NSF in the 20-S complex (Figure 10), we immunoprecipitated 20-S SNARE complex–containing fractions with GS32 antibodies (lanes 1–4) or control rabbit IgG (lanes 5–8). The immunoprecipitates were eluted with either assembly buffer (Figure 10, lanes 1, 3, 5, and 7) or disassembly buffer (lanes 2, 4, 6, and 8). The beads (Figure 10, lanes 1, 2, 5, and 6) and eluates (lanes 3, 4, 7, and 8), together with 100 ng of NSF and α-SNAP (lane 9), were processed for immunoblot analysis to detect NSF and α-SNAP. Both α-SNAP and NSF were coimmunoprecipitated by GS32 antibodies (Figure 10, lane 1) and could be released from the immunoprecipitate in disassembly conditions (lane 4) but not in assembly conditions (lane 3). GS32 thus exists in an authentic 20-S SNARE complex that contains NSF and α-SNAP. Furthermore, when increasing amounts of Golgi extract were incubated with immobilized GST–α-SNAP, increasing amounts of GS32 were retained by the beads until saturation (Figure 11, top). Under identical conditions, GS32 was not retained by immobilized GST–RB (the entire polypeptide of retinoblastoma protein fused to the C-terminus of glutathione S-transferase protein) (Figure 11, bottom). Taken together, these results suggest that GS32 in the Golgi extract can interact specifically with α-SNAP and established that GS32 is a SNARE.
Figure 9.
GS32 could be incorporated into a 20-S SNARE complex with NSF and α-SNAP in conditions that promote SNARE complex formation. Golgi extract in assembly buffer (top) and Golgi extract incubated with NSF and α-SNAP in assembly buffer (middle) or in disassembly buffer (bottom) were resolved by a glycerol gradient in assembly buffer (top and middle) or disassembly buffer (bottom). Fractions were collected from the bottom of the centrifuge tube (starting at lane 1) and were analyzed by immunoblot to detect GS32.
Figure 10.
GS32 exists in a 20-S complex that contains NSF and α-SNAP. The 20-S fractions were immunoprecipitated with antibodies against GS32 (lanes 1–4) or control rabbit IgG (lanes 5–8). The immunoprecipitates were eluted with assembly buffer (lanes 1, 3, 5, and 7) or disassembly buffer (lanes 2, 4, 6, and 8). The beads (lanes 1, 2, 5, and 6) and eluates (lanes 3, 4, 7, and 8), together with 100 ng of NSF and α-SNAP (lane 9), were analyzed by immunoblot to detect NSF and α-SNAP. NSF and α-SNAP were associated with the beads (lane 1) but not the eluate (lane 3) in assembly buffer. In disassembly buffer, the majority of NSF and α-SNAP was released into the eluate (lane 4), with a small portion remaining associated with the beads (lane 2).
Figure 11.
GS32 in Golgi extracts can interact with immobilized α-SNAP. The indicated amounts (below horizontal arrow) of the Golgi extract were incubated with 2 μg of GST–α-SNAP (top) or GST–RB (bottom) immobilized on beads. After extensive washing, the amounts of GS32 retained were analyzed by immunoblot.
Interaction of GS32 with Golgi Syntaxin 6
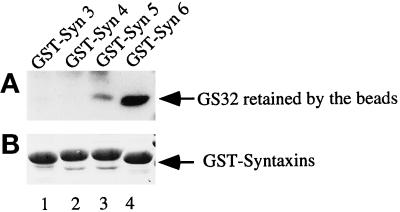
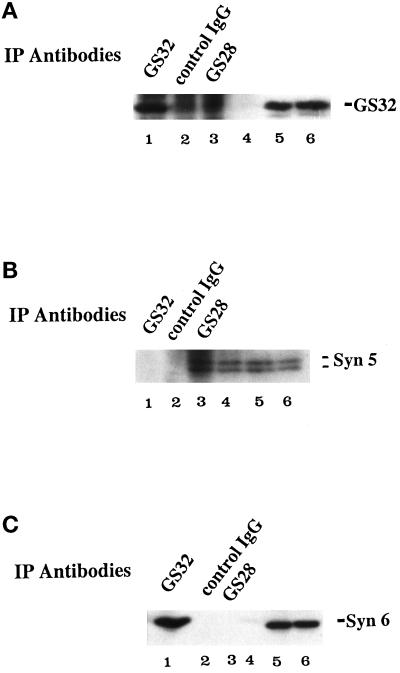
Because SNAP-25 and SNAP-23 interact with several surface syntaxins, the Golgi association of GS32 indicates that it may also interact with a syntaxin(s) of the Golgi apparatus. Syntaxins 5 and 6 are the only two characterized syntaxins associated with the Golgi apparatus (Bennett et al., 1993; Banfield et al., 1994; Bock et al., 1996, 1997), and thus we proceeded to examine whether GS32 in the Golgi extract could interact with immobilized GST–syntaxin 5 or GST–syntaxin 6. GST–syntaxin 3 and GST–syntaxin 4 were used as controls for surface syntaxins. When Golgi extracts were incubated with immobilized GST–syntaxins 3, 4, 5, and 6, GS32 was preferentially retained by GST–syntaxin 6 (Figure 12, lane 4), although some GS32 was also retained, to a much lesser extent, by GST–syntaxin 5 (lane 3). We next examined whether GS32 and syntaxin 6 exist in a protein complex in the Golgi extract. Golgi extracts were immunoprecipitated with antibodies against GS32 and GS28 and with control rabbit IgG. Immunoprecipitates were analyzed by immunoblot using antibodies against GS32, syntaxin 5, and syntaxin 6. GS28 is a SNARE that has been demonstrated previously to exist together with syntaxin 5 in a SNARE complex that is required for ER–Golgi transport (Hay et al., 1997; Subramaniam et al., 1997). As shown in Figure 13, anti-GS32 antibodies immunoprecipitate both GS32 (A) and syntaxin 6 (C) but not syntaxin 5 (B). On the other hand, anti-GS28 antibodies immunoprecipitate only syntaxin 5 (Figure 13, B) and not GS32 (A) or syntaxin 6 (C). Neither GS32, syntaxin 5, nor syntaxin 6 could be detected in the immunoprecipitates of the control rabbit IgG. These results, taken together, suggest that GS32 interacts preferentially with a syntaxin 6–containing complex.
Figure 12.
Preferential interaction of GS32 with immobilized GST–syntaxin 6. (A) Golgi extract (100 μg) was incubated with beads coupled with 8 μg of GST–syntaxins 3, 4, 5, and 6. After extensive washing, the beads were analyzed by immunoblot to detect the amount of GS32 retained by the beads. (B) One-half of the amount (4 μg) of GST–syntaxins 3, 4, 5, and 6 was analyzed by SDS-PAGE and stained with Coomassie blue.
Figure 13.
Antibodies against GS32 but not GS28 coimmunoprecipitate syntaxin 6 from Golgi extracts. Golgi extracts were immunoprecipitated with antibodies against GS32, GS28 or with control rabbit IgG. The immunoprecipitates (lanes 1, 2, and 3) and postimmunoprecipitate supernatants (lanes 4, 5, and 6) were resolved by SDS-PAGE and processed for immunoblot to detect GS32 (A), syntaxin 5 (B), and syntaxin 6 (C). As shown, syntaxin 6 but not syntaxin 5 was selectively coimmunoprecipitated by GS32-specific antibodies. Lanes 1–3 show the beads, whereas lanes 4–6 show 20% of the postimmunoprecipitated supernatants.
DISCUSSION
Our results suggest that GS32 is a SNAP-25–related SNARE localized to the Golgi apparatus. Like SNAP-25 and SNAP-23, there are two regions of the polypeptide with a high probability of forming coiled-coil domains in GS32 (the N-terminal coiled coil spans amino acid residues 50–112, and the C-terminal coiled coil spans amino acid residues 195–257). It has been proposed that protein–protein interaction is mediated by coiled-coil domains in vesicle docking and fusion. Therefore, the conserved coiled-coil domains of GS32, SNAP-23, and SNAP-25 as well as the syntaxins were compared and phylogenetically studied. The sequence alignment and phylogenetic analysis clearly establish that GS32 is a SNAP-25/23–related protein but not a member of the syntaxin family. Furthermore, like SNAP-25/23 but unlike the syntaxins, GS32 does not contain a C-terminal hydrophobic membrane anchor. Previously, the existence of a similar EST clone (accession number, AA150357) encoding GS32 was also noticed by Bock and Scheller (1997). However, they have classified this protein as a member of the syntaxin family and named it syntaxin 9. Our results suggest that the name syntaxin 9 is inappropriate for this protein, and we would like to propose the name GS32 for this protein.
The existence of GS32 in the 20-S SNARE complex suggests that GS32 is a SNARE, and because SNAP-25 is a t-SNARE that is preferentially associated with the target membrane for which the respective docking and fusion events are occurring, it may be reasonable to speculate that GS32 is also a t-SNARE. Distinct from other t-SNAREs, SNAP–25 lacks a C-terminal anchor transmembrane domain (Oyler et al., 1989). The membrane binding of SNAP-25 requires a posttranslational modification of its cysteine residues near the center of the primary amino acid sequence (Hess et al., 1992; Veit et al., 1996; Gonzalo and Linder, 1998). However, in the case of GS32, no cysteine residue is present in the primary amino acid sequence. Therefore, the association of GS32 with the Golgi membranes must be by other means. One of the possibilities is that GS32 may bind stably to an integral membrane protein as a receptor. Another possibility is that GS32 interacts with another protein in the cytosol that harbors membrane-binding capability and therefore is targeted to the membrane. How the specific association of GS32 with membranes is regulated remains to be investigated. Phosphorylation could be one way of regulating the specific association of soluble proteins with membranes. This has been shown at least in the case of the vesicle-docking protein p115 (Sohda et al., 1998). During interphase, two forms of p115 were detected in the cells; the phosphorylated form was detected exclusively in the cytosol, whereas the unphosphorylated form was associated with Golgi membrane. In GS32, there are 26 serine and 8 tyrosine residues present in the primary amino acid sequence, and PROSITES search revealed several potential phosphorylation sites in GS32 (our unpublished observations). Further work is required to test this hypothesis.
Although the functional aspects of GS32 remain to be established, its Golgi association and establishment as a novel SNARE suggest that GS32 may be involved in a docking and/or fusion event of the Golgi apparatus. SNAP-25 and SNAP-23 interact with several surface syntaxins; Golgi association of GS32 suggests that it may interact with syntaxins associated with the Golgi apparatus. By the use of an in vitro interaction assay, it was found that GS32 in the Golgi extract could interact preferentially with Golgi syntaxin 6. Furthermore, coimmunoprecipitation of syntaxin 6 by antibodies against GS32 demonstrates that GS32 and syntaxin 6 exist in a protein complex in the Golgi extract. Syntaxin 6 is a SNARE that localized to the trans-Golgi network (TGN) with significant amino acid sequence homology to syntaxin 3, Pep12, and SNAP-25 (Bock et al., 1996). In the TGN, syntaxin 6 was found on noncoated and on AP-1– and/or clathrin-coated membranes as well as on clear vesicles in the vicinity of endosomes (Bock et al., 1997). Although GS32 is demonstrated here to interact with syntaxin 6 in a syntaxin 6–containing complex, colocalization between GS32 and syntaxin 6 labeling at the Golgi apparatus is not perfect (our unpublished observations). In addition, under the BFA condition, GS32 was shown to be redistributed to the ER-like structures (Figures 6B and 7B), but syntaxin 6 was concentrated at the MTOC (Bock et al., 1996; our unpublished observations) consistent with its TGN localization. However, interestingly, some of the syntaxin 6 is also observed to redistribute to the ER-like structures, although to a much lesser extent (our unpublished observations). The redistribution of some of the syntaxin 6 to the ER under the BFA condition indicates that some syntaxin 6 could also be localized to the Golgi apparatus and therefore interact with GS32. Thus, it is likely that syntaxin 6 is present in two different protein complexes (SNARE complex) that are confined to the TGN and Golgi, respectively. Alternatively, other than interacting with syntaxin 6, GS32 may also interact with other proteins of the Golgi apparatus that are redistributed to the ER by BFA. Additional studies are needed to test these possibilities.
SNAP-25 has been documented to interact with neuronal members of the syntaxin family (Söllner et al., 1993; Chapman et al., 1994). Here, we have demonstrated such an interaction between GS32, a SNAP-25–like protein, and syntaxin 6, although we do not yet know whether this represents a direct binding. During the revision of our manuscript, it has been shown that Vam7p is a SNAP-25–like protein interacting with Vam3p, a vacuolar syntaxin-like protein involved in vacuolar protein trafficking (Sato et al., 1998; Ungermann and Wickner, 1998). This study, together with our present one, clearly establishes that SNAP-25–like proteins may also function in intracellular trafficking events.
The molecular, cell biological, and biochemical characterization of GS32 and the demonstration of its interaction with syntaxin 6 will provide a novel avenue for studying the functional and mechanistic aspects of these two proteins in the Golgi apparatus.
ACKNOWLEDGMENTS
We thank James E. Rothman for the generous gift of plasmids for producing HisX6–α-SNAP and HisX6-NSF, Richard H. Scheller for the cDNA clones of the syntaxins, Anje Habermann for technical assistance, Dr. Tommy Nilsson for the SA:48 cells, Dr. Paramjit Singh for the control GST–RB recombinant protein, members of Dr. Hong’s laboratory for critical reading of the manuscript, and Dr. Y.H. Tan for his continuous support. This work was funded by the Institute of Molecular and Cell Biology (to W.H.).
REFERENCES
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related mammalian proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani RJ, Bae HR, Bock JB, Chao DS, Doung YC, Prekeris R, Yoo JS, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- Araki S, Tamori Y, Kawanishi M, Shinoda H, Masugi J, Mori H, Niki T, Okazawa H, Kubota T, Kasuga M. Inhibition of binding of SNAP-23 to syntaxin 4 by Munc18c. Biochem Biophys Res Commun. 1997;234:257–262. doi: 10.1006/bbrc.1997.6560. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HRB. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HRB. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark IC, Wilson MC. Human cDNA clones encoding two different isoforms of the nerve terminal protein SNAP-25. Gene. 1994;139:291–292. doi: 10.1016/0378-1119(94)90773-0. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of luminal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 function in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Lin RC, Scheller RH. A new syntaxin family member implicated in targeting of intracellular transport vesicle. J Biol Chem. 1996;271:17961–17965. doi: 10.1074/jbc.271.30.17961. [DOI] [PubMed] [Google Scholar]
- Bock JB, Scheller RH. A fusion of new ideas. Nature. 1997;387:133–135. doi: 10.1038/387133a0. [DOI] [PubMed] [Google Scholar]
- Boivin D, Beliveau R. Subcellular distribution and membrane association of Rho-related small GTP-binding proteins in kidney cortex. Am J Physiol. 1995;269:F180–F189. doi: 10.1152/ajprenal.1995.269.2.F180. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Chapman ER, An S, Barton N, Jahn R. SNAP-25, a t-SNARE which binds to both syntaxin and synaptobrevin via domains that may form coiled coils. J Biol Chem. 1994;269:27429–27432. [PubMed] [Google Scholar]
- Colley KJ, Lee EU, Adler B, Browne JK, Paulson JC. Conversion of a Golgi apparatus sialyltransferase to a secretory protein by replacement of the NH2-terminal signal anchor with a signal peptide. J Biol Chem. 1989;264:17619–17622. [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. A receptor for selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Darsow T, Burd CG, Emr SD. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Matteson J, Balch WE. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J Biol Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink LA, Trimble WS, Scheller RH. Two vesicle-associated membrane protein genes are differentially expressed in the rat CNS. J Biol Chem. 1989;264:11061–11064. [PubMed] [Google Scholar]
- Fernandez CJ, Warren G. In vitro synthesis of sulfated glycosaminoglycans coupled to intercompartmental Golgi transport. J Biol Chem. 1998;273:19030–19039. doi: 10.1074/jbc.273.30.19030. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Gaisano HY, Ghai M, Malkus PN, Sheu L, Bouquillon A, Bennett MK, Trimble WS. Distinct cellular localizations of syntaxin family of proteins in rat pancreatic acinar cells. Mol Biol Cell. 1996;7:2019–2027. doi: 10.1091/mbc.7.12.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T, Chilcote T, Mundigl O, Binz T, Niemann H, De Camilli P. Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J Cell Biol. 1994;125:1015–1024. doi: 10.1083/jcb.125.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T, Zahraoui A, Vaidyanathan VV, Raposo G, Tian JM, Karin M, Niemann H, Louvard D. A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol Biol Cell. 1998;9:1437–1448. doi: 10.1091/mbc.9.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Linder ME. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G. Fine Structure Immunocytochemistry. Berlin: Springer-Verlag; 1993. Cryo and replica techniques for immunolabelling; pp. 137–203. [Google Scholar]
- Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson M, Goud B, Soling H, Tang BL, Wong SH, Hong W. Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HRB. SEC5 encodes a 39-kDa integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:147–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hess DT, Slater TM, Wilson MC, Skene JHP. The 25 kDa synaptosomal-associated protein SNAP-25 is the major methionine-rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J Neurosci. 1992;12:4634–4641. doi: 10.1523/JNEUROSCI.12-12-04634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HRB. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- Lian JP, Ferro-Novick S. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell. 1993;73:735–745. doi: 10.1016/0092-8674(93)90253-m. [DOI] [PubMed] [Google Scholar]
- Louvard D, Reggio H, Warren G. Antibodies to the Golgi complex and the rough endoplasmic reticulum. J Cell Biol. 1982;92:92–107. doi: 10.1083/jcb.92.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock C, Lucocq J. Quantitative immunoelectron microscopy reveals α2,6-sialyltransferase is concentrated in the central cisternae of rat hepatocyte Golgi apparatus. Eur J Cell Biol. 1998;76:18–24. doi: 10.1016/s0171-9335(98)80013-7. [DOI] [PubMed] [Google Scholar]
- Low SH, Chapin SJ, Weimbs T, Komuves L, Bennett MK, Mostov KE. Differential localization of syntaxin isoforms in polarized Madin-Darby canine kidney cells. Mol Biol Cell. 1996;7:2007–2018. doi: 10.1091/mbc.7.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SL, Wong SH, Hong W. The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J Cell Sci. 1996;109:209–220. doi: 10.1242/jcs.109.1.209. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol Biol Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshansky V, Fleser A, Noel J, Bourgoin S, Vinay P. Isolation of heavy endosomes from dog proximal tubules in suspension. J Membr Biol. 1996;153:59–73. doi: 10.1007/s002329900110. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Sudhof TC. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- McNew JA, Sogaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Sollner TH. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- Mu F, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo J, Tock EPC, Toh B. EEA1, an early endosome-associated protein: EEA1 is a conserved α-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Newman AP, Groesch ME, Ferro-Novick S. Bos1p, a membrane protein required for ER to Golgi transport in yeast, copurifies with carrier vesicles and with Bet1p and the ER membrane. EMBO J. 1992;11:3609–3617. doi: 10.1002/j.1460-2075.1992.tb05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: a coat of many colors. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Okazaki I, Himeno M, Ezaki J, Ishikawa T, Kato K. Purification and characterization of an 85 kDa sialoglycoprotein in rat liver lysosomal membranes. J Biochem (Tokyo) 1992;111:763–769. doi: 10.1093/oxfordjournals.jbchem.a123833. [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte H-H, Schmitt HD, Gallwitz D. The yeast SLY genes products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom. FE, Wilson MC. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V, Virbasius J, lane WS, Toh B, Shpetner HS, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XR, Yao XB, Chow DC, Forte JG, Bennett MK. Association of syntaxin 3 and vesicle-associated membrane protein (VAMP) with H+/K+-ATPase-containing tubulovesicles in gastric parietal cells. Mol Biol Cell. 1997;8:399–407. doi: 10.1091/mbc.8.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in nonneuronal tissue. J Biol Chem. 1996;271:300–303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanism of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1989. [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1532. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Scheller RH. Membrane trafficking in the presynaptic nerve terminal. Neuron. 1995;14:893–897. doi: 10.1016/0896-6273(95)90328-3. [DOI] [PubMed] [Google Scholar]
- Shim J, Newman AP, Ferro-Novick S. The BOS1 gene encodes an essential 27-kDa putative membrane protein that is required for vesicular transport from the ER to the Golgi complex in yeast. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sögaard M, Tani K, Ye RB, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yano A, Takami N, Ikehara Y. Phosphorylation of the vesicle docking protein p115 regulates its association with the Golgi membrane. J Biol Chem. 1998;273:5385–5388. doi: 10.1074/jbc.273.9.5385. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh B, D’Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Stones S, Sacher M, Mao Y, Carr C, Lyons P, Quinn AM, Ferro-Novick S. Bet1p activates the v-SNARE Bos1p. Mol Biol Cell. 1997;8:1175–1181. doi: 10.1091/mbc.8.7.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam VN, Krijnse-Locker J, Tang BL, Ericsson M, Yusoff ARbM, Griffiths G, Hong W. Monoclonal antibody HFD9 identifies a novel 28 kDa integral membrane protein on the cis-Golgi. J Cell Sci. 1995;108:2405–2414. doi: 10.1242/jcs.108.6.2405. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Loh E, Hong W. N-Ethylmaleimide-sensitive factor (NSF) and α-soluble NSF attachment proteins (SNAP) mediate dissociation of GS28-syntaxin 5 Golgi SNAP receptors (SNARE) complex. J Biol Chem. 1997;272:25441–25444. doi: 10.1074/jbc.272.41.25441. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Peter F, Philip R, Wong SH, Hong W. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science. 1996;272:1161–1163. doi: 10.1126/science.272.5265.1161. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Yusoff ARbM, Wong SH, Lim GB, Chew M, Hong W. Biochemical fractionations and characterization of proteins from Golgi-enriched membranes. J Biol Chem. 1992;267:12016–12021. [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Südhof TC, Baumert M, Perin MS, Jahn R. A synaptic vesicle membrane protein is conserved from mammals to Drosophila. Neuron. 1989;2:1475–1481. doi: 10.1016/0896-6273(89)90193-1. [DOI] [PubMed] [Google Scholar]
- Tang BL, Low DY, Tan AE, Hong W. Syntaxin 10: a member of the syntaxin family localized to the trans-Golgi network. Biochem Biophys Res Commun. 1998;242:345–350. doi: 10.1006/bbrc.1997.7966. [DOI] [PubMed] [Google Scholar]
- Trimble WS, Cowan DM, Scheller RH. VAMP-1: a synaptic vesicle associated integral membrane protein. Proc Natl Acad Sci USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Wickner W. Vamp 7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 1998;17:3269–3276. doi: 10.1093/emboj/17.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Sollner TH, Rothman JE. Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 1996;385:119–123. doi: 10.1016/0014-5793(96)00362-6. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimbs T, Mostov K, Low SH, Hofmann K. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Kubalek EW. SNAPs and NSF: general members of the fusion apparatus. Trends Cell Biol. 1995;5:64–69. doi: 10.1016/s0962-8924(00)88948-5. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Whiteheart SW, Wiedmann M, Brunner M, Rothman JE. A multisubunit particle implicated in membrane fusion. J Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PPC, Daneman N, Volchuk A, Lassam N, Wilson MC, Klip A, Trimble WS. Tissue distribution of SNAP-23 and its subcellular localization in 3T3–L1 cells. Biochem Biophys Res Commun. 1997;230:64–68. doi: 10.1006/bbrc.1996.5884. [DOI] [PubMed] [Google Scholar]
- Wong SH, Low SH, Hong W. The 17-residue transmembrane domain of β-galactoside α-2,6-sialyltransferase is sufficient for Golgi retention. J Cell Biol. 1992;117:245–258. doi: 10.1083/jcb.117.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Xu Y, Zhang T, Hong W. Syntaxin 7, a novel syntaxin member associated with the early endosomal compartment. J Biol Chem. 1998a;273:375–380. doi: 10.1074/jbc.273.1.375. [DOI] [PubMed] [Google Scholar]
- Wong SH, Zhang T, Xu Y, Subramaniam VN, Griffiths G, Hong W. Endobrevin, a novel synaptobrevin/VAMP-like protein preferentially associated with the early endosome. Mol Biol Cell. 1998b;9:1549–1563. doi: 10.1091/mbc.9.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wong SH, Zhang T, Subramaniam VN, Hong W. GS15, a 15-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) homologous to rbet1. J Biol Chem. 1997;272:20162–20166. doi: 10.1074/jbc.272.32.20162. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Subramaniam VN, Wong SH, Tang BL, Parton RG, Rea S, James DE, Hong W. A novel synaptobrevin/VAMP homologous protein (VAMP5) is increased during in vitro myogenesis and present in the plasma membrane. Mol Biol Cell. 1998;9:2423–2437. doi: 10.1091/mbc.9.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wong SH, Tang BL, Xu Y, Peter F, Subramaniam VN, Hong W. The mammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in vesicular transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1997;139:1157–1168. doi: 10.1083/jcb.139.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]