Abstract
A closed-loop control system drives progression of the coupled stalked and swarmer cell cycles of the bacterium Caulobacter crescentus in a near-mechanical step-like fashion. The cell-cycle control has a cyclical genetic circuit composed of four regulatory proteins with tight coupling to processive chromosome replication and cell division subsystems. We report a hybrid simulation of the coupled cell-cycle control system, including asymmetric cell division and responses to external starvation signals, that replicates mRNA and protein concentration patterns and is consistent with observed mutant phenotypes. An asynchronous sequential digital circuit model equivalent to the validated simulation model was created. Formal model-checking analysis of the digital circuit showed that the cell-cycle control is robust to intrinsic stochastic variations in reaction rates and nutrient supply, and that it reliably stops and restarts to accommodate nutrient starvation. Model checking also showed that mechanisms involving methylation-state changes in regulatory promoter regions during DNA replication increase the robustness of the cell-cycle control. The hybrid cell-cycle simulation implementation is inherently extensible and provides a promising approach for development of whole-cell behavioral models that can replicate the observed functionality of the cell and its responses to changing environmental conditions.
Keywords: Caulobacter, model, regulatory circuit, hybrid system, symbolic model checking
The identification of regulatory pathways controlling the cell cycle of the bacterium Caulobacter crescentus has progressed to the point that an experimentally based system-level characterization of the cell-cycle control circuit is available. Aspects of the cyclical circuit that drives progression of the C. crescentus cell cycle are described in recent papers (1–5). Recent findings have identified novel mechanisms providing signals that tightly integrate the biochemical and genetic components of the core cell-cycle circuit with the 3D topology of the cell (4, 6, 7) and DNA replication (8).
Many of the regulatory mechanisms and pathways in the C. crescentus cell-cycle circuit operate very rapidly so that they approximate discrete switching elements, whereas others, e.g., the transcriptional regulatory networks, operate relatively slowly. This suggested use of hybrid control analysis methods (9–13) to simulate this cell-cycle control system and formal analysis methods to analyze its properties. Hybrid control systems, by definition, include both continuous and discrete regulatory mechanisms (9). We constructed a hybrid control system simulation model and derived an equivalent asynchronous discrete circuit model from the simulation model. The analysis showed that the cell-cycle control is robust and that DNA methylation-based cell-cycle regulatory mechanisms (8) enhance robustness of the cell cycle.
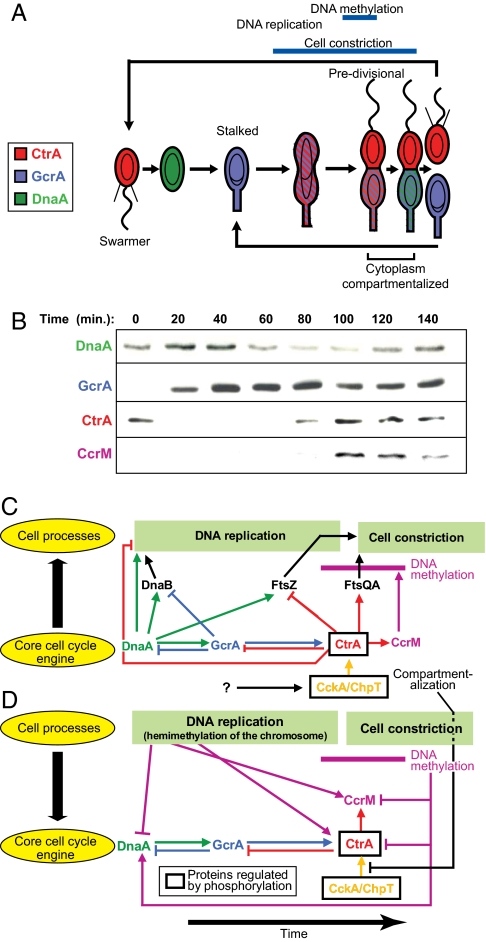
C. crescentus always divides asymmetrically, producing morphologically distinct daughters (Fig. 1A), so it is a model system not only for bacterial cell-cycle control but also for asymmetric cell division. The daughter cells have identical genotypes but different morphology and cell fates. The swarmer daughter cell has a single polar flagellum, polar chemotaxis receptors, and polar pili, and it cannot initiate DNA replication until after a period of motility when the swarmer differentiates into a stalked cell. Swarmer-to-stalked cell differentiation involves loss of the flagellum and polar chemotaxis receptors, retraction of the pili, construction of a polar stalk, and initiation of DNA replication. In contrast, the stalked daughter cell initiates chromosome replication after cytoplasmic compartmentalization during cytokinesis. This compartmentalization event occurs ≈20 min before completion of cell division after chromosome decatenation (Fig. 1A) (6). Compartmentalization initiates divergent genetic programs in each compartment, so that the next cell cycle effectively begins in each compartment well before cell division (4, 6, 7).
Fig. 1.
Genetic circuit that drives cell-cycle progression. (A) Schematic of the C. crescentus cell cycle showing changes in master regulatory protein concentrations that control activation of numerous modular functions that implement the cell cycle. Predivisional cells are compartmentalized ≈20 min before cell separation (6). (B) Western blots showing concentrations of the master regulatory proteins during the cell cycle (8, 26). (C) Schematic of protein interactions that create the cyclical core engine and the regulatory connections from the core engine to the processive DNA replication and cell constriction functions. Events are positioned to indicate their approximate timing. (D) Signals returning from the controlled subsystems synchronize the core engine with the state of cell-cycle progression. The interval of DNA methylation is indicated in C and D by the horizontal purple bar. All of the pathways indicated in C and D are included in the cell-cycle control model.
The C. crescentus cell-cycle control system is hierarchical, distributed, asynchronous, and parallel. The control circuit comprises phosphosignaling pathways, genetic regulatory logic, dynamical control of regulatory protein localization, activation, and stability, and two processive subsystems, chromosome replication and cytokinesis. Parallelism arises from activation of multiple modular cell-cycle functions that operate at the same time. Four master regulator proteins (DnaA, GcrA, CtrA, and CcrM), organized as a cyclical genetic circuit, serially activate many subsystems to implement the cell cycle (Fig. 1 B–D and supporting information (SI) Fig. S1) (3). Fig. 1B shows the changing concentrations of these regulatory proteins that activate or repress controlled subsystems. Fig. 1C shows principal signal pathways from the core circuit to the DNA replication and cell constriction subsystems. Figs. 1D and Fig. S1 show feedback signal pathways from these two controlled subsystems to the core circuit to tie the rate of progression of the core circuit to the progression of the cell cycle. The cell-cycle control system is synchronized tightly with the progression of chromosome replication and cytokinesis by both genetic and nongenetic mechanisms (8, 14). The rates of progress of the parallel independent reaction cascades are inherently unpredictable, but some pathways must be completed in a particular order. Synchronization is accomplished by several mechanisms: (i) tying subsystem activation time to cyclical regulatory protein concentrations and activation levels, (ii) tying gene expression levels to intermediate events in the processive subsystems [e.g., promoter hemimethylation during DNA replication (8)], and (iii) checkpoint signaling (e.g., delaying completion of cell constriction until after DNA decatenation).
In the next section, we describe the hybrid control simulation model and its validation by comparison with experimental observations. Then we describe use of model checking to analyze robustness of the cell-cycle control and control circuit features that contribute to robustness.
Cell-Cycle Hybrid Control System Simulation Model.
Several descriptions of the aspects of C. crescentus control system circuitry are available (1, 3, 5). A stalked cell-cycle subcircuit model has been reported (15), but with no analysis of robustness or modeling of the control of CtrA phosphorylation. We developed a scalable simulation of control of the coupled swarmer and stalked cell cycles with emphasis on compatibility with formal analysis. We wanted to predict the progress of the regulatory machinery into either compartment of the predivisional cell to enable comparative analysis of regulatory events within the respective compartments. These events that establish asymmetry are difficult to observe experimentally.
Sources for the signaling in the cell-cycle control system, the regulatory logic in promoter regions, and protein stability models are in Table S1 in SI Appendix. The protein components of the genetic circuit include the four master regulator proteins (DnaA, GcrA, CtrA, and CcrM) that comprise the core cyclical circuit and DnaB, FtsZ, and FtsQA (Fig. S2B). DnaB, FtsZ, and FtsQA are components of pathways that connect the core engine with DNA replication and cytokinesis (Fig. 1 C and D). This is a parsimonious model of the cell-cycle control circuitry. For example, DnaB is only one of the proteins in the replication complex whose synthesis is activated by DnaA (16). FtsQA represents two proteins, FtsQ and FtsA (whose genes are in an operon), required for initiation of cell constriction. The simulation includes phenomenological models of the progress of chromosome replication and cell constriction. These two subsystems determine the timing of the changes in the methylation state of the dnaA, ctrA, and ccrM promoter regions and of cell compartmentalization (8, 17).
Cytoplasmic compartmentalization is essentially instantaneous (6). The promoter methylation state changes when the replication fork traverses promoters that involve this mechanism (8) are also instantaneous, as is the remethylation reaction. Positive autoregulation of the ctrA P2 promoter by CtrA∼P leads to rapid synthesis of CtrA (Fig. S1). Methylation state dependence of the ctrA P1 promoter tends to delay ctrA activation until the gene is replicated, providing additional assurance that CtrA is synthesized rapidly and at the right time. CtrA∼P is cleared from the cell by simultaneous activation of proteolysis and inhibition of its activation so that the elimination of CtrA∼P is also rapid. The rapid creation and destruction of CtrA∼P shortens average switching times of downstream genes and other processes by activated CtrA∼P because of accelerated traversal of the “threshold” regions for these reactions (18).
Each of these events triggers discrete changes in the network of gene transcription, phosphosignaling, and protein-level reactions that control the cell cycle. Additional description of pathways is in SI Appendix. In modeling the system, we used mechanistic models where the reaction mechanisms are known, for example, the promoter control mechanisms of the master regulator genes in the core engine (Fig. S1). In the case of the CckA/ChpT phosphosignaling pathway (Fig. 1 C and D and Fig. S1), mechanisms of the pathway are not completely identified, but the function of the pathway and the timing of its operation within the cell cycle are well characterized (14). Similarly, the time from onset to completion of DNA replication and the time from initial cell constriction to compartmentalization are known (19). In these cases, we used phenomenological, but functionally accurate, models with correct cell-cycle timing. Reactions that occur essentially instantaneously were modeled as discrete switching events. The resulting hybrid control simulation enabled analysis of the C. crescentus cell-cycle control as an integrated system.
The simulation model is constructed by using the Matlab tools, Simulink and Stateflow, that are widely used for control system analysis (see SI Appendix). Innovative aspects of this simulation model are: (i) a top-down hierarchical simulation architecture that mimics the design of the cell's control system (Figs. S2–S4) designed for future incorporation of additional regulatory subsystems (e.g., control of polar organelle development, a mechanistic model of the CckA-related phosphosignaling pathway, plus nutrient and stress signal/response systems); (ii) realistic treatment of the hybrid control aspects of the system; and (iii) simulation of molecular-level differentiation within the nascent daughter cell compartments from the instant of compartmentalization. The complete model description is in SI Appendix.
Patterns of protein and mRNA concentrations vs. the simulation predictions shown in Fig. 2 and in SI Appendix agree within experimental error (recognizing loss of synchrony in the predivisional cell data). In silico analogs of mutant strains were simulated, and the resulting predictions were consistent with the mutant strain phenotypes (see below and SI Appendix). The consistency of the simulation results with these experimental observations validated the hybrid control simulation of the in vivo cell-cycle control system. The simulations were performed by using nominal kinetic parameter values (Table S3 in SI Appendix) characteristic of cells in log growth phase. Subsequent analysis (described below) of the robustness of the cell-cycle control then showed that the cell cycle will operate correctly even if there is wide variation in kinetics of reaction rates in different pathways.
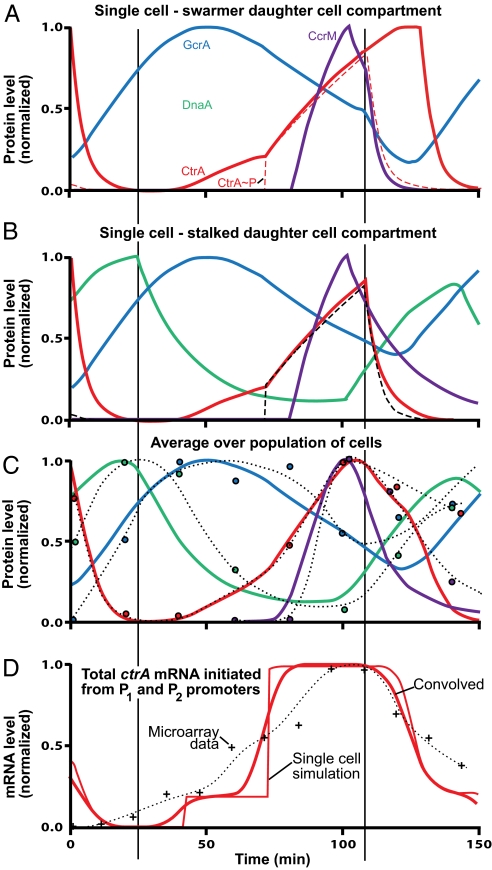
Fig. 2.
Simulation of protein levels (normalized) during cell-cycle progression. A and B show predicted (normalized) levels of the master regulatory proteins (Fig. 1) tracked into the swarmer and stalked cell compartments, respectively, at the single-cell level. After inner membrane compartmentalization at ≈117 min, protein concentration levels diverge in the stalked and swarmer daughter cell compartments. (C) Circles: observed protein levels in synchronized cells (quantified Western blots, Fig. 1B). (The dotted lines are continuous approximations of the experimental levels.) Curves: simulated protein levels made comparable with experimental observations by averaging results in A and B and convolving with a Gaussian distribution to approximate random variation around an average in different cell's progression through the cell cycle. The errors in the experimental values are approximately ±10% of the peak value. Loss of synchrony degrades experimental data in predivisional cell. (D) +, observed ctrA mRNA levels from Affymetrix microarray assays (20).
The progression of protein and mRNA concentrations can be tracked within the simulation into either of the nascent daughter cell compartments. Fig. 2A shows predicted normalized patterns of CtrA, CtrA∼P, GcrA, DnaA, and CcrM concentrations as a function of time in the cell cycle from the instant of cell separation through cytoplasmic compartmentalization into the stalked daughter cell compartment until cell division. Fig. 2B follows the protein concentrations into the swarmer daughter cell compartment. Experimental observations from synchronized populations (e.g., from Western blots and microarray assays) are always averages over many cells, and measurements of samples taken late in the synchronized cell cycle always include signals from both the nascent swarmer and stalked daughter cell compartments and some cells from the next generation. To compare the single-cell predictions of protein and mRNA patterns from the simulation to experimental observations in synchronized cell populations we (i) averaged the predictions from the swarmer and stalked daughter cell branches of the simulation (Fig. 2 A and B) and (ii) convolved the result with a Gaussian distribution with a 5-min standard deviation to approximate dispersions in cell-cycle stage among cells in experimental samples. Fig. 2C shows the resulting protein level predictions (the curved lines) along with normalized experimental results quantified from Western blots (circles).
The temporal profiles of ctrA mRNA levels in Fig. 2D are predicted by a Hill function approximation for gene activity (averaged and convolved as for protein levels in Fig. 2C). Experimental results from Affymetrix microarray assays of time samples from synchronized cell populations (20) are shown for comparison. For additional comparisons of predicted temporal patterns of mRNA levels to experimental levels for all genes in the model, see SI Appendix.
In Silico Analysis of Mutant Phenotypes.
We created variants of the simulation corresponding to four mutant strains that have been characterized in vivo and compared the in silico mutant simulations with the in vivo phenotypes. Table S4 in SI Appendix shows the four strains and the changes to the wild-type model to create the mutant simulation. In each case, the simulation model predicted, (i) the concentration profile of each protein in the model in single cells as a function of cell-cycle time when followed into either the swarmer or the stalked compartment of the predivisional cell, (ii) whether the cell can progress through each stage of the cell cycle, and (iii) whether DNA replication and cytokinesis occur normally. The simulation predictions were consistent with the in vivo phenotypes.
Robustness of Cell-Cycle Control.
Because (i) both the connectivity of the cell-cycle circuit design and the nodal logic is strongly experimentally based, (ii) the simulation predicts key protein and mRNA temporal concentration patterns, and (iii) in silico predictions of mutant strain phenotypes are consistent with experimental observations, we concluded that the control circuit design in the simulation was essentially correct. Simulation with nominal parameter values, however, is not sufficient to demonstrate robustness, that is, “invariance of phenotypes in the face of perturbation” (21). Commonly, robustness of biological circuits is analyzed by assessing the range of parameter variation within which the circuit functions satisfactorily by checking performance with random choices of parameter values. We performed a limited parameter sensitivity analysis on a subset of the parameters in the simulation, and the results suggested that the operation of the cell-cycle control is robust. Then we used the validated simulation model to identify an equivalent asynchronous sequential digital circuit model and used formal model checking software for further analysis.
Fig. 3A illustrates the rationale underlying our approach to identifying the discrete abstraction to the hybrid simulation model and application of the model checking methodology for robustness analysis. The figure shows eight events in an interval centered on the time of daughter cell separation that will occur in a stalked cell cycle (Fig. 1A and Fig. S1) and affect operation of the control circuit. The events occur at times labeled by the Tis. T1, for example, is the time of cytoplasmic compartmentalization that interrupts the CckA/ChpT phosphosignal and precipitates elimination of CtrA∼P. The specific timing of events and the intervals between the events will depend on the kinetic parameters of the system, whether in vivo or in silico. (Because of stochastic variation in reaction rates from cell to cell in a population, actual reaction rates in different cells will vary, and consequently cells will show dispersion in the rate of progression through the cell cycle.) Thus, the rate of production of any protein X will depend on the kinetic parameters of its transcription and translation, and the time for X to activate a dependent process will depend on the kinetics of its binding at a target site that determine the concentration range for action. The rate of production of X will also depend on availability of substrates (e.g., amino acids) that will vary with environmental nutrient levels. There is a minimum average cell generation time under optimal high nutrient conditions. Under either carbon or nitrogen starvation, the cell cycle comes to a controlled stop (see below), suggesting there is also a maximum to the sustainable generation time. Thus, a defining element of the robustness of the C. crescentus cell-cycle control is its ability to achieve reliably correct ordering of cell-cycle functions between these extremes of cell growth rate. Our method for exploring this is to consider variation of the event times (i.e., the Tis in Fig. 3A) directly, rather than individual variation of the many kinetic parameters that determine the times. This approach to robustness analysis is more computationally efficient and complete than using a Monte Carlo method to explore the parameter space.
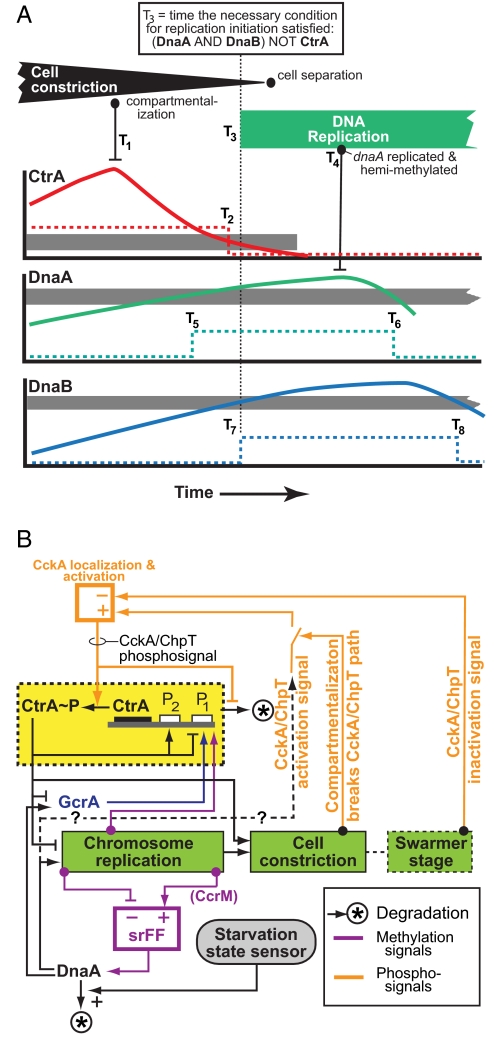
Fig. 3.
Logic circuit of Caulobacter cell cycle control system. (A) Stalked cell-cycle events affecting control circuit operation in the interval around cell separation. Graphs show protein concentration patterns. Gray bands: “threshold” ranges relating to initiation of DNA replication. [CtrA] above the band represses initiation; below, it does not. The dotted CtrA line illustrates the binary signal abstraction used in the robustness analysis. Thus, T2 approximates the time when [CtrA] transitions from repressing to not-repressing initiation. Other Tis are times of other events. The robustness analysis examined the effect of all patterns of Ti orderings on cell-cycle control. (B) Logical diagram of cell-cycle control system operations. Green rectangles: Serially ordered processive subsystems. Yellow box: cyclical synthesis and destruction of CtrA∼P that is a central element of the circuit. Orange pathways: CckA/ChpT phosphosignal pathways differentially controlled in swarmer and stalked cell compartments that play an essential role in both enabling resynthesis of CtrA∼P when the signal is active and triggering CtrA∼P destruction when it is inactive. Purple: DNA methylation-state based control of DnaA synthesis that creates a logical component functionally similar to a electrical set-reset flipflop (srFF) circuit (8). Carbon or nitrogen starvation halts cell-cycle progression by accelerating DnaA proteolysis (27).
For example, consider the particular situation shown in Fig. 3A, in which the condition for activation of DNA replication involving the CtrA, DnaA, and DnaB proteins is satisfied at T3. After a delay, the dnaA gene is duplicated at T4. Consequently, its promoter region becomes hemimethylated and the rate of dnaA transcription is greatly reduced (8). Although there are essential ordering relationships between some timing events, for example: T2 > T1, T4 > T3, T6 > T5, T6 > T4, and T8 > T7, the lengths of the various interevent intervals are determined by the specific reaction rates that occur by chance in each cell under the prevailing environmental conditions.
To facilitate exploration of the effects of timing variations, we simplify the protein concentration profiles as shown in Fig. 3A so that they are either “low” (under the thresholds of downstream sites of action) or “high” (above the thresholds of downstream sites of action), because other than time and direction of traversing the threshold region, details of the temporal profile of these variables are not significant to the operation of the circuit. This yields a discrete logic abstraction of the signaling and the time points that identify the timing of the transitions between the two states (e.g., T5 and T6 in Fig. 3A). Other events can also be characterized as binary signals or finite state machines with discrete stages. For example, in Fig. 3A, the cell changes from not compartmentalized to compartmentalized at T1, and the dnaA promoter changes from methylated to hemimethylated at T4. This simplification of the cell-cycle control circuit by using discrete signal levels with discrete transition timing produces an abstraction of the biological circuit equivalent to an asynchronous sequential digital circuit, a network of logic elements and simple-state machines with variable delays. Within this abstraction, it is necessary only to examine each distinctive ordering of events to determine whether the circuit will function correctly. Following electrical circuit analysis procedures for similar problems, we use a software model checking tool to search the immense space of all feasible orderings of the Tis for cases where the circuit might fail. This approach using a symbolic model checker (22) is used to check for correct operation of concurrent systems such as electrical circuits and network protocols; we use it to check the cell-cycle control circuit, which is also a concurrent system. The model checker we used, NuSMV (22, 23), takes as input the logic description of the C. crescentus cell-cycle model (created from the validated simulation model as described above) and a specification of the limitations on ordering of transitions that must apply for a viable cell. The program checks all feasible orderings of the transition times and notifies the user of any event timing “hazards” where the specification would be violated.
A challenge in any method for assessing biological robustness is defining requirements for successful operation of the modeled system. For the C. crescentus cell-cycle control, the key functionality of the control circuit is to ensure that actions that affect completion of the cell cycle, cell growth, chromosome replication, asymmetric cell division, and so forth, repeatedly occur in the proper order. This includes ordering of expression of the seven proteins in the model (Fig. S1), switching of the CckA/ChpT phosphosignaling pathway, and activation of DNA replication followed by cell constriction, compartmentalization into nascent swarmer and stalked cell compartments, and cell division. Thus, we required that for all feasible orderings of the event Tis, the model system would successfully produce an unbounded succession of Stalked→DNA-replication→Compartmentalization→(Swarmer or Stalked) states as occurs in the tree of descendent daughter cells of an initial swarmer cell. This criterion is written in computational tree logic to cover all possible paths in the state space (24). For the subset of cell-cycle subsystems we model and for the asynchronous logic abstraction of the model, satisfaction of this requirement is evidence that the cell-cycle control is robust. A hazard detected by the model checking can be a biological possibility that has acceptably low probability of occurrence or it may represent a feature of the biological system that prevents the hazard from occurring, but is missing in the simulation model.
Model-Checking Results.
The model-checking program exhaustively checked the enormous number of alternative possible Ti combinations, and only two potential hazards were identified. This suggests that the overall design of the cell-cycle control circuit has been optimized by evolutionary selection to operate over a wide range of nutrient conditions and to be resistant to stochastic variations in time to complete various subsystem operations or signaling pathways. Further, it is evidence that conclusions relating to operation of the cell-cycle control based on the simulation model do not depend on the exact parameter values for the simulation. The two hazards identified both relate to repressive feedback signals (GcrA repression of DnaB expression and CtrA repression of FtsZ expression; see Fig. S1). In both of these cases, the hazard identified is that anomalously slow synthesis of the protein coupled with anomalously fast synthesis of the repressive feedback signal might block cell-cycle progression. Both cases appear to have relatively low probability of occurring, and there also may be undiscovered aspects of the regulation of both DnaB and FtsZ that eliminate the hazard.
We also examined the incremental contribution of the methylation-based control of the ctrA P1 promoter to robustness of the cell-cycle control. When we simulated an in silico mutant where full methylation does not repress the ctrA P1 promoter, the cell cycle operated correctly; however, applying the model checking program to this in silico mutant identified an additional hazard: in this mutant, CtrA could to be synthesized too early, leading to premature repression of FtsZ and thus failure of cytokinesis. This suggests that, although this methylation-based mechanism is not essential, it contributes to fitness of the organism by assuring correct operation of the cell cycle in the fraction of cells where stochastic variation produces anomalously early expression of CtrA.
The DNA methylation-based control of dnaA transcription also contributes to robustness of cell-cycle control by reducing its expression during DNA replication, so that the likelihood of reinitiation of replication is reduced. However, the methylation-based repression of dnaA is not complete, so that some expression remains (8). Model checking analysis of an in silico mutant without this remaining basal expression identified another hazard: in a situation where DNA replication is unusually slow, GcrA could be degraded before the ctrA P1 promoter becomes hemimethylated. Then, CtrA would not reaccumulate in predivisional cells to activate the synthesis of FtsQA, and cell constriction would not occur. The low-level synthesis of DnaA by the dnaA basal expression, however, can restart GcrA synthesis and, in turn, CtrA synthesis to rescue the cell.
Halting and Restarting the Cell Cycle.
The DnaA protein is strategically located in the circuit to simultaneously activate DNA replication and the GcrA-CtrA-CcrM pathway at the beginning of each cell cycle (Fig. 1C and Fig. S1) (16, 25, 26). Elimination of DnaA halts the cell cycle (25), and control of DnaA stability is a mechanism used to halt the C. crescentus cell cycle in response to stress (27). When C. crescentus cells are starved for carbon or nitrogen, DnaA is rapidly proteolyzed, and relief of the starvation rapidly restabilizes DnaA (27). We used model checking to assess whether this DnaA stability-dependent method of halting and restarting the cell cycle is robust by adding an additional switch in the model to modulate DnaA stability. This analysis showed that the cell-cycle stops and restarts correctly regardless of when DnaA stability changes during the cell cycle.
Discussion
A small number of spatially and temporally controlled regulatory proteins create a core cell-cycle engine that drives progression of the C. crescentus cell cycle. The cell-cycle control system is hierarchically organized, and it activates functional modules just when needed. The cascaded cell-cycle reaction pathways proceed in a near mechanical step-like fashion. The engine and modular cell-cycle subfunctions are synchronized by mechanisms that connect the timing of gene expression and the phosphorylation of CtrA to progression of cell-cycle functions. There are checkpoint signals between modules that further assure proper ordering of functions. Top-level regulatory proteins are redundantly regulated at the levels of transcription, stability, and activity. We considered operation of a bacterial cell cycle from a hybrid control system engineering perspective to ask whether the cell-cycle control is robust, and whether stopping and restarting the cell cycle in response to nutritional stress is robust. Our analysis shows the control system design will operate correctly over a wide range of cell generation times as necessary to adapt to nutrient availability, and, in periods of starvation, to completely stop cell-cycle progression. Furthermore, we showed through analysis by model checking that the recently discovered “methylation ratchet” (8) is a mechanism that has evolved to enhance robustness of cell-cycle control by preventing deleterious effects of stochastic variation in the circuit timing.
Fig. 3B shows a logical diagram of the C. crescentus cell-cycle control circuit operation. This view of the circuit shows that the alternating CtrA∼P signal (Fig. 3B, yellow box) is central to achieving serial activation of the DNA replication, cell constriction and the swarmer stage in one daughter cell (Fig. 3B, green boxes). Separate feedback signals synchronized with different points in the swarmer and stalked cell-cycle switch the CckA/ChpT phosphosignaling pathway (Fig. 3B, orange) to control activation and deactivation of autoregulated CtrA expression and the CtrA phosphorylation state. DnaA is essential for initiation of DNA replication (25), and it is redundantly regulated to ensure that C. crescentus has one, and only one round, of replication per cell cycle. As with CtrA, regulation of the time when active DnaA is available is coupled tightly to cell-cycle progression, particularly by the repression of dnaA transcription by the hemimethylation state of its promoter following the passing of the replication fork early in DNA replication until it is remethylated as replication nears completion (Fig. 3B, purple) (8).
Our hybrid cell-cycle control system simulation architecture is inherently extensible. A top-down incremental model development paradigm, building on the approach used here, is a promising avenue for development of whole-cell behavioral models that can replicate the observed functionality of the cell and its response to a host of environmental conditions. These comprehensive cell behavioral models (and, eventually, models of linked systems of cells) will have far more parameters than the C. crescentus cell-cycle control system examined here, so that assessment of robustness by conventional parameter variation analysis will be computationally limited. In contrast, robustness analysis using the methodology applied here is computationally efficient and scalable to systems that are far more complex.
Supplementary Material
Acknowledgments.
H.H.M., M.H., L.S., and X.S. were partially supported by Department of Energy, Office of Science Grant DE-FG02-05ER64136. L.S. was partially supported by National Institutes of Health Grants GM51426 and GM32506. M.H. and D.D. were partially supported by a Stanford Computer Science Department seed grant. D.D. was also partially supported by National Institutes of Health Grant 5U56CA112973-02. J.C. was supported by a Stanford Dean's Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805258105/DCSupplemental.
References
- 1.Laub MT, Shapiro L, McAdams HH. Systems biology of Caulobacter. Annu Rev Genet. 2007;41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- 2.Chen JC, Stephens C. Bacterial cell cycle: Completing the circuit. Curr Biol. 2007;17:R203–R206. doi: 10.1016/j.cub.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 3.McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 4.McGrath PT, Viollier P, McAdams HH. Setting the pace: Mechanisms tying Caulobacter cell-cycle progression to macroscopic cellular events. Curr Opin Microbiol. 2004;7:192–197. doi: 10.1016/j.mib.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Biondi EG, et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 3006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 6.Judd EM, et al. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alur R, et al. Modeling and analyzing biomolecular networks. Comput Sci Eng. 2002;4:20–31. [Google Scholar]
- 10.Ghosh R, Tomlin C. Symbolic reachable set computation of piecewise affine hybrid automata and its application to biological modelling: Delta-Notch protein signalling. Syst Biol. 2004;1:170–183. doi: 10.1049/sb:20045019. [DOI] [PubMed] [Google Scholar]
- 11.Ye P, et al. Hybrid automata as a unifying framework for modeling excitable cells. Conf Proc IEEE Eng Med Biol Soc. 2006;1:4151–4154. doi: 10.1109/IEMBS.2006.259294. [DOI] [PubMed] [Google Scholar]
- 12.Batt G, et al. Validation of qualitative models of genetic regulatory networks by model checking: Analysis of the nutritional stress response in Escherichia coli. Bioinformatics. 2005;21(Suppl 1):i19–28. doi: 10.1093/bioinformatics/bti1048. [DOI] [PubMed] [Google Scholar]
- 13.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Brazhnik P, Sobral B, Tyson JJ. A quantitative study of the division cycle of Caulobacter crescentus stalked cells. PLoS Comput Biol. 2008;4:e9. doi: 10.1371/journal.pcbi.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 17.Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 2002;21:4969–4977. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keiler KC, Shapiro L. TmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGrath PT, et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 21.de Visser JA, et al. Perspective: Evolution and detection of genetic robustness. Evol Int J Org Evol. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 22.Clarke EM, Emerson EA, Sistla AP. Automatic verification of finite-state concurrent systems using temporal logic specifications. ACM Trans Program Lang Syst. 1986;8:244–263. [Google Scholar]
- 23.Cimatti A, et al. International Conference on Computer-Aided Verification (CAV 2002) Denmark: Copenhagen; 2002. pp. 359–364. [Google Scholar]
- 24.Emerson EA, Halpern JY. Decision procedures and expressiveness in the temporal logic of branching time. J Comput Syst Sci. 1985;30:1–24. [Google Scholar]
- 25.Gorbatyuk B, Marczynski GT. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol. 2001;40:485–497. doi: 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- 26.Collier J, Murray SR, Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 2006;25:346–356. doi: 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.