Abstract
Streptomyces avermitilis, an industrial organism responsible for the production of the anthelminthic avermectins, harbors a 13.4-kb gene cluster containing 13 unidirectionally transcribed open-reading frames corresponding to the apparent biosynthetic operon for the sesquiterpene antibiotic pentalenolactone. The advanced intermediate pentalenolactone F, along with the shunt metabolite pentalenic acid, could be isolated from cultures of S. avermitilis, thereby establishing that the pentalenolactone biosynthetic pathway is functional in S. avermitilis. Deletion of the entire 13.4-kb cluster from S. avermitilis abolished formation of pentalenolactone metabolites, while transfer of the intact cluster to the pentalenolactone-nonproducer S. lividans 1326 resulted in production of pentalenic acid. Direct evidence for the biochemical function of the individual biosynthetic genes came from expression of the ptlA gene (SAV2998) in Escherichia coli. Assay of the resultant protein established that PtlA is a pentalenene synthase, catalyzing the cyclization of farnesyl diphosphate to pentalenene, the parent hydrocarbon of the pentalenolactone family of metabolites. The most upstream gene in the cluster, gap1 (SAV2990), was shown to correspond to the pentalenolactone resistance gene, based on expression in E. coli and demonstration that the resulting glyceralde-3-phosphate dehydrogenase, the normal target of pentalenolactone, was insensitive to the antibiotic. Futhermore, a second GAPDH isozyme (gap2, SAV6296) has been expressed in E. coli and shown to be inactivated by pentalenolactone.
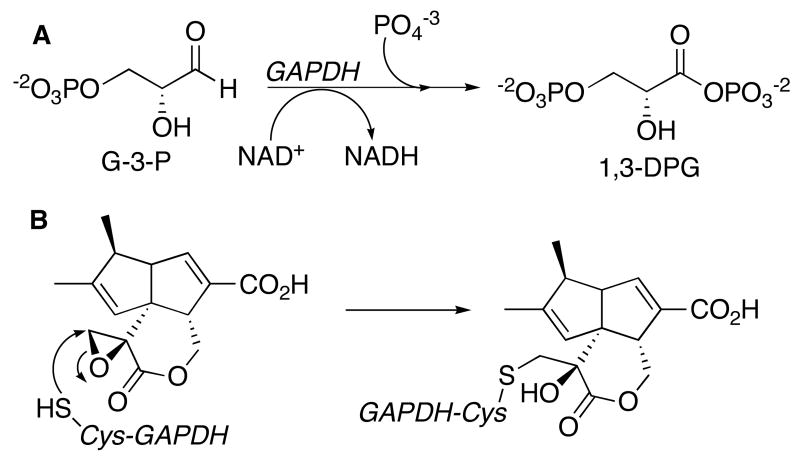
Pentalenolactone (1) (Scheme 1) is a sesquiterpenoid antibiotic, first isolated in 1957 from Streptomyces roseogriseus (1), and subsequently found in the extracts of numerous Streptomyces species, including Streptomyces UC5319 (2), S. arenae (3), S. chromofuscus, S. griseochromogenes, S. baarnensis, S. omiyaensis, S. albofaciens, and S. viridifaciens (4, 5). Pentalenolactone, which has been shown to be active against both Gram-positive and Gram-negative bacteria as well as a variety of fungi and protozoa (6), and to inhibit the replication of DNA viruses such as HSV-1 and HSV-2 (7). Pentalenolactone also inhibits vascular smooth muscle cell proliferation (8). The antibiotic selectively inhibits the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH)1 (9, 10). Incubation of rabbit muscle GAPDH with pentalenolactone results in time-dependent, irreversible inactivation of GAPDH as a result of alkylation of the active site Cys-149 residue, apparently by reaction with the electrophilic epoxide moiety of the antibiotic (Scheme 2) (11, 12). Self-resistance in the pentalenolactone-producer S. arenae is due to the presence of two GAPDH isozymes, an inducible, pentalenolactone-insensitive form and a constitutive, pentalenolactone-sensitive form (13-15).
Scheme 1.
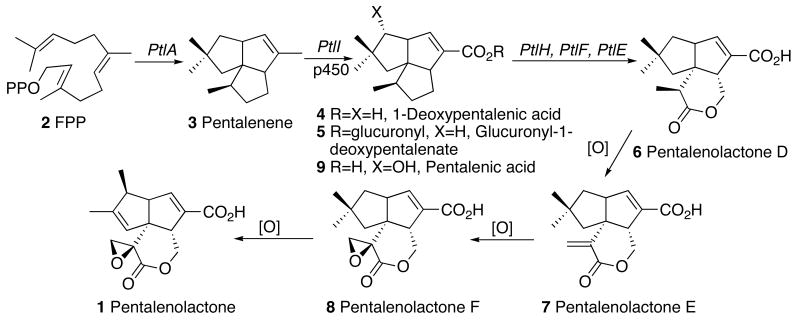
Biosynthesis of pentalenolactone (1) from farnesyl diphosphate (2, FPP), showing known metabolites and the proposed role of enzymes encoded by the S. avermitilis pentalenolactone gene cluster.
Scheme 2.
A. GAPDH-catalyzed conversion of D-glyceraldehyde-3-phosphate (G-3-P) to 1,3-diphosphoglycerate (1,3-DPG). B. Inactivation of GAPDH by pentalenolactone.
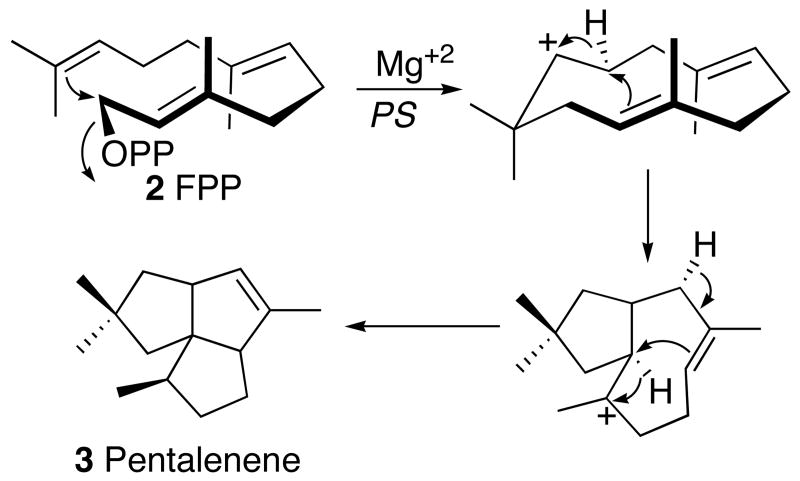
Pentalenolactone is derived from the 1-deoxyxylulose-5-phosphate/2-C-methylerythritol-4-phosphate pathway of isoprenoid biosynthesis (16).2 The committed step in pentalenolactone biosynthesis is the cyclization of farnesyl diphosphate (FPP, 2), the universal precursor of all sesquiterpenes, to pentalenene (3), the parent hydrocarbon of the pentalenolactone family of metabolites. Pentalenene synthase from Streptomyces UC5319 has been cloned and expressed in Escherichia coli (17) and its structure has been solved by X-ray crystallography (18). The enzyme, a monomer of MD 38002, requires only Mg2+ for activity. Pentalenene synthase contains two Mg2+-binding domains that are universally conserved across all known sesquiterpene synthases: an aspartate-rich DDLFD motif at amino acids (aa) 80-85, and an NSE triad in NDIASLEKE beginning at Asn-219. Extensive experiments with isotopically labeled FPP have firmly established a cyclization mechanism in which FPP (2), folded as shown, undergoes ionization and cyclization via an intermediate humulyl cation to pentalenene (3) (19, 20) (Scheme 3). Refeeding of labeled pentalenene (3) to cultures of Streptomyces UC5319 and analysis of the derived labeled pentalenolactone (1) has confirmed the intermediacy of pentalenene in the pentalenolactone biosynthetic pathway (20). A variety of plausible intermediates in the conversion of pentalenene to pentalenolactone have been isolated, including 1-deoxypentalenic acid (4) (as the derived glucuronyl ester 5) (4), pentalenolactone D (6) (21), pentalenolactone E (7) (22), and pentalenolactone F (8) (21, 23, 24), as well as pentalenic acid (9) (25), a demonstrated shunt metabolite of the main pentalenolactone biosynthetic pathway (20) (Scheme 1).
Scheme 3.
Mechanism of cyclization of FPP (2) to pentalenene (3) catalyzed by pentalenene synthase (PS).
Little is known about the organization of genes for entire terpenoid biosynthetic pathways. The intermediates of the biosynthesis of the fungal diterpene gibberellic acid have been intensively studied, and many of the relevant enzymes and genes have been characterized from both plant and fungal sources (26). Similarly many of the intermediates of trichothecane mycotoxin biosynthesis are known and a short biosynthetic gene cluster has been identified, although few of the open reading frames (ORFs) have been characterized biochemically (27). Croteau and his collaborators have systematically identified and characterized many of the genes responsible for the biosynthesis of the antitumor diterpene taxol, a formidable task that has been considerably complicated by the fact that the relevant structural genes are not clustered in Pacific yew (Taxus brevifolia) (28).
Streptomyces avermitilis (S. avermectinius is a junior homotypic synonym of S. avermitilis (29)) is the producer of the potent anthelminthic macrolide avermectin which is widely used in human and animal medicine. The complete genome sequence of S. avermitilis was determined in 2003, revealing the presence of 7,575 ORFs in the 9.03 Mb linear chromosome, with an average GC content of 70.7% (30, 31). Thirty gene clusters related to secondary metabolites were recognized, corresponding to nearly 7% of the genome. Among these gene clusters were at least six encoding putative terpenoid biosynthetic pathways. Centered at 3.75 Mb in the S. avermitilis chromosome is a ∼13.4-kb cluster containing 13 unidirectionally-transcribed ORFs, among which is the 1,011-bp ptlA (SAV2998), encoding a protein of 336 aa with 76% identity and 85% similarity to the pentalenene synthase of Streptomyces UC5319 (Figure 1 and Table 1; see Figure S1 in Supporting Information for sequence alignment). Just upstream of ptlA is ptlB (SAV2997), which encodes a typical farnesyl diphosphate synthase. Of the remaining 11 ORFs, 8 encode putative redox enzymes, including mono- or dioxygenases and a dehydrogenase, while ptlR (SAV3000) corresponds to a typical transcriptional regulator. Significantly, the most upstream gene in the cluster, gap1 (SAV2990), encodes a 335-aa protein with 88% identity and 92% similarity to the previously identified pentalenolactone-resistant GAPDH of S. arenae (See Figure S2, Supporting Information). Since many bacterial antibiotics are co-expressed with the corresponding genes for self-resistance, the 13 ORFs were proposed to make up an operon for the biosynthesis of pentalenolactone. We report below the results of combined molecular genetic and biochemical experiments that provide compelling evidence for the identity of the ptl-cluster.
Figure 1.
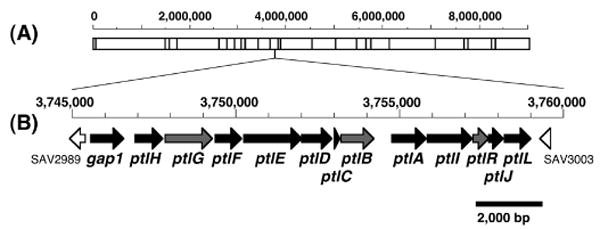
Gene cluster for pentalenolactone biosynthesis in S. avermitilis. AseI-physical map of a linear chromosome of S. avermitilis (A) and a region involving pentalenolactone biosynthesis (B). Gene names and definitions are given in Table 1.
Table 1.
Pentalenolactone (ptl) gene cluster of Streptomyces avermitilisa
| Gene | ID | Start nt | End nt | Definition | Best matchb |
|---|---|---|---|---|---|
| gap1 | SAV2990 | 3,745,497 | 3,746,501 | glyceraldehyde-3-phosphate dehydrogenase | ptlK, pentalenolactone resistance gene |
| ptlH | SAV2991 | 3,746,862 | 3,747,719 | putative hydroxylase | PF05721: Phytanoyl-CoA dioxygenase (PhyH) |
| ptlG | SAV2992 | 3,747,777 | 3,749,231 | putative transmembrane efflux protein | PF07690: Major Facilitator Superfamily |
| ptlF | SAV2993 | 3,749,307 | 3,750,119 | putative oxidoreductase | PF00106: short chain dehydrogenase |
| ptlE | SAV2994 | 3,750,177 | 3,751,961 | putative monooxygenase | PF00743: Flavin-binding monooxygenase-like |
| ptlD | SAV2995 | 3,751,962 | 3,752,882 | putative dioxygenase | PF02668: Taurine catabolism dioxygenase TauD, TfdA family |
| ptlC | SAV2996 | 3,752,934 | 3,753,110 | hypothetical protein | |
| ptlB | SAV2997 | 3,753,107 | 3,754,120 | polyprenyl diphosphate synthase | probable farnesyl diphosphate synthase (idsA2) |
| ptlA | SAV2998 | 3,754,725 | 3,755,735 | pentalenene synthase | PF03936: Terpene synthase family, metal binding domain |
| ptlI | SAV2999 | 3,755,792 | 3,757,141 | cytochrome P450 | CYP183A1 |
| ptlR | SAV3000 | 3,757,201 | 3,757,662 | putative AraC-family transcriptional regulator | PF00165: Bacterial regulatory helix-turn-helix proteins, AraC family |
| ptlJ | SAV3001 | 3,757,659 | 3,758,093 | putative lyase | PF00903: Glyoxalase/Bleomycin resistance protein, Dioxygenase superfamily |
| ptlL | SAV3002 | 3,758,163 | 3,758,933 | hypothetical protein | Nfa37730 of Nocardia farcinica IFM 10152 |
For detailed annotation and alignments, see the Streptomyces avermitilis Genome Project website: http://avermitilis.ls.kitasato-u.ac.jp/
PF Number is the NCBI PFAM reference.
Experimental Procedures
Materials
Streptomyces avermitilis cosmids CL_216_D07 and CL_214_C12, from the S. avermitilis genomic DNA library (http://avermitilis.ls.kitasato-u.ac.jp/) (30), were used to transform E. coli ElectroTenBlue (Stratagene. Oligonucleotide primers were purchased from Integrated DNA Technologies. S. arenae TÜ469 was obtained from the DMSZ Gmbh, Braunschweig, Germany. The pET28a expression vector was purchased from Stratagene. pET28e was prepared from pET28a by replacing the XbaI site with an EcoRI site. Restriction enzymes and ligases were purchased from Promega. Pentalenene synthase from Streptomyces UC5319 was purified from E. coli/pZW05 as previously described (17), omitting the DNAse treatment. Pentalenolactone was isolated from S. arenae TÜ469 (3) and converted to the benzylamine salt, PLBA (11). HiTrap Fast Flow columns were purchased from GE Healthcare. [1-3H]Farnesyl diphosphate triammonium salt (16.1 Ci/mmol), obtained from Perkin Elmer Life Sciences, Boston, was mixed with unlabeled FPP, prepared as previously described (32, 33), to a final specific activity of 44 μCi/μmol. All other chemicals and other buffer components were purchased from Sigma-Aldrich and were of the highest grade available.
Methods
Electrocompetent cells for electroporation were prepared from E. coli BL21(DE3) and E. coli BL21(DE3) RP Codon Plus (Stratagene) by standard methods. DNA sequencing of plasmid constructs was performed by the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource Laboratory (Yale University School of Medicine, New Haven CT using fluorescently labeled dideoxynucleotides. Polymerase chain reactions were carried out using Pfu Turbo polymerase (Stratagene) as recommended by the manufacturer. Qiaquick PCR purification kits, Qiaprep spin miniprep kits, and Ni-NTA resin were used as suggested by the supplier (Qiagen). The concentration of D-G-3-P was determined from the D/L mixture as suggested by Sigma. Standard molecular biological procedures were performed as described (34). Generation of Streptomyces exoconjugants and homologous recombination utilized standard methodology (35). Protein concentrations were determined by the method of Bradford (36) using the Bio-Rad kit. Steady-state kinetic and enzyme inactivation parameters were calculated by direct fitting to the Michaelis-Menten or other appropriate equations by nonlinear least squares regression using Kaleidagraph V 3.6 (Adelbeck Software, Reading, PA). UV spectra were recorded on a HP 8452A photodiode array spectrophotometer. GC-MS analysis of sesquiterpenes was carried out on a Hewlett-Packard (Agilent) GC-MSD Series 2. Liquid scintillation was measured with a Beckman LS6500 in 7 mL of Opti-Fluor.
Isolation of Pentalenolactone F
A mutant of S. avermitilis (ΔaveR ΔolmA5), blocked in the production of both avermectins and oligomycins, was used to examine the production of pentalenolactones. The spores of the mutant were transferred to a 250-mL flask containing 20 mL of vegetative medium (glucose (0.5 g), soy flour (1.5 g), and yeast extract (0.5 g) per 100 mL, pH 7.2) and allowed to grow while shaking at 30 °C for 2 days. A 1-mL portion was removed and used to inoculate 20 × 500-mL flasks containing 100 mL of production medium (soluble starch (40 g), soy flour (20 g), FeSO4•7H2O (0.5 g), K2HPO4 (1 g), and KCl (0.3 g) per liter, pH 6.5). After incubation while shaking at 28 °C for 4 days, the culture was filtered. The mycelium was washed with tap water and extracted with 500 mL of methanol. The concentrated supernatant was diluted with an equal volume of water and acidified with 2 N H2SO4 to pH 2.5. The products were then extracted twice with a half volume of chloroform. The organic fraction was concentrated under reduced pressure and subjected to flash silica gel chromatography (30:1::chloroform:methanol). Pentalenolactone-rich fractions were pooled and concentrated to an oil. This residue was dissolved in THF/diethyl ether (1:1) and treated with a solution of diazomethane in diethyl ether and 50% KOH for 10 min at 0 °C. After quenching by addition of ethereal acetic acid, the ether layer was washed with saturated bicarbonate solution. Methyl ester metabolites were further purified by preparative silica TLC and final purification was performed by ODS-HPLC using n-hexane/ethyl acetate (1:1), resulting in the isolation of 8 mg of pentalenolactone F methyl ester (8-Me).
Construction of a ptl-deletion mutant of S. avermitilis
Two DNA segments were amplified by PCR from cosmid CL_216_D07. A segment upstream of gap1 (nt 3,743,485 to 3,745,499) was amplified using the forward (5′-CTCGAGCAATTGCAACGCCTATCTGGACACCTCGAC-3′) and reverse (5′-CTCGAGACTAGTCATGGATCCCTCTCCTAGCGCTG-3′) primers to introduce MfeI and SpeI restriction sites (bold), respectively. The forward (5′-CTCGAGACTAGTCCTACCGGTCCGGGTCAGCCGTC-3′) and reverse (5′-CTCGAGCAATTGCTCGCCCTCGCCGAGTTCGTGAC-3′) primers were used for the amplification of a second segment downstream of ptlL (nt 3,758,936 to 3,760,758), with introduction of SpeI and MfeI sites (bold), respectively. The two amplified DNA segments were ligated together into EcoRI-cut pKU250 and transformed into E. coli DH5α. The resultant plasmid was digested with SpeI and the linearized plasmid was ligated with the SpeI-fragment of the aphII kanamycin resistance cassette. The plasmid pKU250::upstream-aphII-downstream construct in E. coli recA dcm/pUB307aph::Tn7 was introduced into S. avermitilis by conjugation. After exoconjugants were selected by vector marker (thiostreptone resistance), ptl-deletion mutants, generated by double homologous recombination upstream of gap1 and downstream of ptlL, were obtained by selection for thiostreptone-sensitivity and neomycin-resistance. The deletion was confirmed by PCR. Neither pentalenolactones nor pentalenic acid could be detected in cultures of the S. avermitilis ptl-deletion mutant.
Transfer of the ptl-cluster to S. lividans 1326
Cosmid CL_216_D07 was digested with AvrII and SnaBI and the 14.9-kb segment was purified by agarose gel electrophoresis and ligated with the XbaI/Ecl136II-cut φC31-based integrating vector carrying aphII as selectable marker. The resulting plasmid was used to transform E. coli S17.1 and then transferred into S. lividans 1326 by conjugation. The S. lividans::ptl exoconjugants, obtained by selection for neomycin resistance, produced ∼1 mg/L of pentalenic acid (9) along with traces of pentalenolactone metabolites. The insertion of the intact ptl gene cluster was verified by PCR.
Expression and Purification of S. avermitilis pentalenene synthase
The ptlA gene was amplified by PCR from template DNA from cosmid CL_216_D07 using the forward (5′-GCGCGCCATATGGCAATGCCCCAGGACGTCG-3′) and reverse (5′-GGCCGGAAGCTTCGGCTACAGCGTGCTGCC-3′) primers to introduce NdeI and HindIII restriction sites (bold) flanking the normal start and stop codons, respectively. The resulting amplicon and the pET28a vector were digested separately with NdeI and HindIII and purified using a PCR purification kit before ligation with T4 DNA ligase and transformation of E. coli ElectroTenBlue cells. The resultant pET28a/SAV2998 plasmid was then used to transform E. coli BL21(DE3) which was grown at 37 °C in LB media (1 L) supplemented with kanamycin (80 mg/L) to an OD600 ∼ 0.6, then induced by the addition of IPTG (0.5 mM). After a further 5 h at 30 °C, the cells were harvested by centrifugation. The cell pellet was resuspended in lysis buffer (30 mL, 20 mM imidazole, 500 mM NaCl, 50 mM Tris, 5 mM MgCl2, 2.7 mM β-mercaptoethanol, 10% glycerol (v/v), pH 8.0) and disrupted by sonication. After removal of the cellular debris by centrifugation (20 min, 30,000g, 4 °C), the recombinant protein was purified by HiTrap Ni affinity column chromatography using a linear gradient of imidazole (10-400 mM) in lysis buffer. Fractions were analyzed by SDS-PAGE and those containing the desired protein were concentrated by ultrafiltration (10 kDa NMWL) (9.4 mg protein). N-Terminal His6-tag-PtlA eluted at 90-118 mM imidazole.
His6-tag pentalenene synthase from Streptomyces UC5319
DNA from plasmid pZW05, harboring the pentalenene synthase gene of Streptomyces UC5319 (17), was amplified by PCR using the forward (5′-CCGCGCGCCATATGCCCCAGGACGTCGACTTCC-3′) and reverse (5′-GGCCGGAAGCTTACTAGTCAATTGCTAGTGGGCGTGCTGCCGAG-3′) primers. The first ten PCR cycles employed an annealing temperature of 60 °C while the next 25 cycles used an annealing temperature of 71 °C. The resulting amplicon and pET28e vector were digested separately with NdeI and HindIII, then purified by PCR purification kit before ligation with T4 DNA ligase overnight at 4 °C and transformation of E. coli XL1-Blue. The resultant pET28e/PS2HIS plasmid was used to transform E. coli BL21(DE3). The protein was expressed in LB media (1 L) supplemented with kanamycin (80 mg/mL). The cells were shaken at 37 °C to an OD600 of 0.6, at which point they were induced by the addition of IPTG (1 mM) and further incubated at 25 °C for 6 h. Recombinant protein was isolated and purified as above by Ni-affinity chromatography (10-500 mM imidazole in lysis buffer). The fractions containing pentalenene synthase (0.6 mg) which eluted between 97 and 118 mM imidazole were pooled and concentrated by ultrafiltration (10 kDa NMWL).
GC-MS analysis of pentalenene synthase extracts
The formation of pentalenene was verified by GC-MS analysis, using the protocol previously described to analyze the pentane-soluble, hydrocarbon product of the incubation of FPP with pentalenene synthase (37). GC-MS was performed using a chiral 25 m × 0.25 mm hydrodex-β-6-TBDM column (Macherey-Nagel) with a temperature ramp of 45 °C to 180 °C at 2 °C/min and an inlet temperature of 150 °C. Under these conditions pentalenene eluted as a single peak at Rt 45.13 min. An authentic sample of pentalenene was generated by parallel incubations with recombinant Streptomyces UC5319 pentalenene synthase. Pentalenene m/z (observed) 204.36 Da; m/z (calculated for C15H24) 204.35.
Pentalenene synthase assay
Assays were performed as previously described, using [1-3H]FPP (17). Assays with S. avermitilis pentalenene synthase and Streptomyces UC5319 His6-tag-pentalenene synthase used 15-80 μM FPP. The pentane extracts were passed through a 2-cm silica gel Pasteur pipette column before quantitation by liquid scintillation counting. The steady-state kinetic parameters, kcat and Km, were calculated by fitting the liquid scintillation data to the Michaelis-Menten equation.
Preparation of pentalenolactone-resistant GAPDH (Gap1)
S. avermitilis gap1 DNA was amplified by PCR from the cosmid CL_216_D07. The forward and reverse primers, (5′-CCGCGCGCCATATGACTGTTCGTGTCGGCATCAATGGC-3′) and (5′-GGCCGGAAGCTTACTAGTCAATTGTCAGAGGGTGTCGCCGACCAGC-3′), were used to introduce NdeI and HindIII sites (bold), respectively, flanking the coding region of the gene. The resulting amplicon and pET28a vector were digested separately with NdeI and HindIII and the digests were purified by a PCR purification kit before ligation with T4 DNA ligase overnight at 4 °C and transformation of E. coli XL-1 Blue. The resultant plasmid, pET28/SAV2990 was used to transform E. coli BL21(DE3) RP Codon Plus. The culture was grown in LB medium (1 L) supplemented with kanamycin (50 mg/L) at 37 °C to an OD600 of 0.5, then induced with IPTG (0.9 mM) and incubated for an additional 3 h at 37 °C. The cell pellet after centrifugation was resuspended in lysis buffer (20 mL, 50 mM Tris pH 8.0, 5 mM MgCl2, 10% glycerol (v/v), 2.7 mM β-mercaptoethanol, 5 mM imidazole, 300 mM NaCl, 1.5 mM benzamidine, 2 μg/mL pepstatin, 2 μg/mL leupeptin, 0.16 mM NAD+), and disrupted with a French press at 10,000 psi. Cellular debris was removed by centrifugation, and purification was carried out using Ni-NTA resin. GAPDH-containing fractions (0.7 mg protein, eluting at 40 mM imidazole) were pooled, concentrated by ultrafiltration (10 kDa NMWL) and exchanged into storage buffer (20 mM Tris pH 8.5, 2.7 mM β-mercaptoethanol, 20% glycerol (v/v), 20 mM NaCl, 0.16 mM NAD+) via PD-10 column (Amersham).
Preparation of pentalenolactone-sensitive GAPDH (Gap2)
S. avermitilis gap2 was amplified by PCR from the cosmid CL_214_C12, using the forward and reverse primers, 5′-GCGCGCCATATGACGATCCGCGTAGGCATCAAC-3′ and 5′-GGCCGGAAGCTTTTAGAGCTGGTTGCCGACGAAGAC-3′, respectively, to introduce NdeI and HindIII sites (bold) flanking the coding region. After ligation into pET28a, the Gap2 protein was expressed in E. coli BL21(DE3)/pET28a/SAV6296 and purified by Ni-affinity chromatography using same protocols used for S. avermitilis Gap1. The desired protein (4.2 mg) eluted at 60 mM imidazole.
GAPDH assay
GAPDH activity was assayed at room temperature, with substitution of arsenate for inorganic phosphate, as previously described (11, 38, 39). Under these conditions, the initially generated product 1-arseno-3-phosphoglycerate undergoes spontaneous hydrolysis to 3-phosphoglycerate, thereby avoiding interfering product inhibition by 1,3-diphosphoglycerate. To the assay buffer (480 μL, 133 mM Tris-HCl pH 8.75, 0.2 mM EDTA, 17 mM Na2HAsO4•7H2O, 3 mM DTT, 0.4 mM NAD+ and 0.098 μM Gap1 or 1.2 mM NAD+ and 0.067 μM Gap2) was added a 20-μL solution of D-G-3-P at concentrations of 25.5 μM to 956 μM. The formation of NADH was monitored by UV at 340 nm. The initial rates of each reaction were calculated using the UV-Visible ChemStation software (HP) and the data were fit to the Michaelis-Menten equation to determine kcat and Km.
Time-dependent inactivation of GAPDH
Recombinant Gap2 (2.1 μM) was pre-incubated with a range of fixed concentrations of PLBA (0, 74.5, 149, and 224 μM) in 500 μL of buffer containing Tris-HCl (133 mM, pH 8.75), EDTA (0.2 mM), Na2HAsO4•7H2O (17 mM), DTT (3 mM), NAD+ (0.61 mM) at 23 °C. Aliquots of 20 μL were withdrawn from the incubation mixtures and diluted 25-fold into GAPDH assay buffer at periodic intervals, and the residual activity was assayed at 23 °C. The enzymatic reactions were initiated by adding solutions of G-3-P (10 μL) to achieve an initial D-G-3-P concentration of 0.29 mM. The assays were performed in triplicate and the initial rates were measured as described above. The maximum rate of inactivation, kinact, and the PLBA inhibition constant, KI, were calculated by direct fitting of the observed rates of inactivation kobs, at individual concentrations of PLBA [I], to the standard equation for active-site directed irreversible inactivation, kobs = kinact[[I]/(KI +[I])] (40).
Analogous experiments were also carried out with recombinant Gap1 (4.55 μM) in the presence of PLBA (0, 24.4, 61, 183, and 610 μM). A pre-incubation temperature of 4 °C was used to compensate for slight thermal instability of the protein. Aliquots of 20 μL were withdrawn at periodic intervals (0.5 min, 1.5 min, 3 min, 5 min and 8 min) and diluted in 480 μL of the assay buffer and assayed in triplicate for residual GAPDH activity at 23 °C. Under these conditions there was no detectable decrease in GAPDH activity even after 8-min pre-incubation in the presence of 610 μM PLBA.
Results
Isolation of pentalenolactone F and characterization of a ptl-deletion mutant
Although pentalenolactones occur widely in Streptomyces, there have been no prior reports of their isolation from S. avermitilis. A 4-L culture of S. avermitilis (ΔaveR ΔolmA5), blocked in the formation of both avermectins and oligomycins, was grown for 4 days at 28 °C. Extraction of the harvested mycelium with methanol followed by chromatographic fractionation of the methylated extract gave 8 mg of purified pentalenolactone F methyl ester (8-Me) whose 1H NMR, IR, and mass spectra were identical in all respects with those previously reported for 8-Me (24). The microorganism also produced the shunt metabolite, pentalenic acid (9). Pentalenolactone (1) itself was not detected in the organic extract of S. avermitilis. An engineered S. avermitilis ptl-deletion mutant, lacking the 13.4-kb segment containing the entire ptl cluster (gap1 to ptlL; SAV2990-3002), produced neither pentalenolactone F (8) nor pentalenic acid (9). In a complementary experiment, when a 14.9-kb AvrII-SnaBI-segment (nt 3,744,874 to 3,759,745) containing the ptl cluster was introduced by conjugation into S. lividans 1326, which normally does not produce pentalenolactones, the resultant exoconjugants were found to produce ∼1 mg/L of pentalenic acid (9), accompanied by traces of pentalenolactone metabolites.
S. avermitilis pentalenene synthase
Using DNA from S. avermitilis cosmid CL_216_D07 as template (30), the 1011-bp presumptive coding region for pentalenene synthase was amplified by PCR while introducing NdeI and HindIII restriction sites at the 5′- and 3′-termini, respectively of the ORF. The amplified DNA was ligated into the expression vector pET28a. The resultant plasmid, pET28a/SAV2998 was purified from E. coli ElectroTenBlue and used to transform the T7 RNA polymerase-based expression host E. coli BL21(DE3). The resulting soluble protein was subjected to Ni2+-affinity purification. The recovered recombinant SAV2998 (9.4 mg from 1 L of culture) was judged to be >90% pure by SDS-PAGE. Incubation of the purified SAV2998 protein with 80 μM FPP and analysis of the pentane-soluble extract by capillary chiral GC-MS, revealed the formation of a single sesquiterpene product identical to pentalenene by direct comparison with an authentic sample, thereby confirming the predicted pentalenene synthase activity of PtlA (SAV2998) (see Supporting Information, Figure S3).
The steady-state kinetic parameters were determined for S. avermitilis pentalenene synthase as well as for N-terminal His6-tag-pentalenene synthase from Streptomyces UC5319 using [1-3H]FPP as substrate (Table 2). By direct comparison, the kcat for the S. avermitilis enzyme of 0.065 s-1 was ∼6-fold lower than the value of 0.40 s-1 determined for the Streptomyces UC5319 His6-tag synthase, while the Km values for FPP for each enzyme were very similar (36 μM compared to 29 μM). The addition of the N-terminal His-tag has little apparent effect on the kcat when compared to the native recombinant Streptomyces UC5319 pentalenene synthase (kcat 0.3 s-1), but results in an marked 100-fold increase in the Km for FPP, compared to a Km of 0.3 μM for the native cyclase (37).
Table 2.
Steady-state kinetic parameters for pentalenene synthase (PS) from S. avermitilis and Streptomyces UC5319
| Protein | kcat (s-1) | Km (μM) | kcat/Km (M-1s-1) |
|---|---|---|---|
| PtlA (SAV2998) | 0.065±002 | 36.0±3.6 | 1.8 × 103 |
| His-Tag-PS | 0.40±0.02 | 28.6±3.6 | 1.4 × 104 |
| PSa | 0.3 | 0.3 | 1.0 × 106 |
Data from Ref. 37.
Pentalenolactone-insensitive GAPDH
The gap1 gene (SAV2990), harbored within cosmid CL_216_D07, was amplified by PCR, ligated as an NdeI-HindIII fragment into pET28e, and expressed in E. coli BL21(DE3) RP Codon Plus. The resultant protein, carrying an N-terminal His6-tag was purified by Ni2+-affinity chromatography to yield 0.7 mg of Gap1 protein per L of culture. The NAD+-dependent GAPDH activity was confirmed by standard arsenate-decoupled assay (11, 38, 39), giving a kcat for D-glyceraldehyde-3-phosphate (G-3-P) of 33 s-1 and a Km of 0.33 mM. To assess the inhibitory effect of pentalenolactone, Gap1 (4.5 μM) was preincubated with concentrations up to 0.6 mM of the benzylamine salt of pentalenolactone (PLBA) (11). Aliquots were periodically withdrawn after 0.5 to 8 min, diluted 25-fold into arsenate assay buffer and monitored for GAPDH activity. Exposure of Gap1 to 0.6 mM pentalenolactone for as long as 8 min at 4 °C had no detectable effect on the measured GAPDH activity. The Gap1 protein is thus insensitive to pentalenolactone.
Pentalenolactone-sensitive GAPDH
S. avermitilis harbors a second apparent GAPDH gene, gap2 (SAV6296), located between nt 7559711 and nt 7560718 of the S. avermitilis chromosome and assigned to PFAM PF02800 (GAPDH). The gap2 gene was amplified by PCR from S. avermitilis cosmid CL_214_C12 and the derived NdeI-HindIII fragment was inserted into the corresponding cloning sites of pET28a. The resultant plasmid, pET28a/SAV6296, was used to transform E. coli BL21(DE3). IPTG-induced expression and Ni2+-affinity purification gave 4.2 mg of N-terminal His6-tag-Gap2 whose GAPDH activity was confirmed, giving a kcat 165 s-1 and Km for D-G-3-P of 0.33 mM. Incubation of S. avermitilis Gap2 at 23 °C with increasing concentrations of PLBA up to 225 μM resulted in time-dependent, irreversible inactivation. Semilog plots of the fraction of remaining GAPDH activity as a function of PLBA concentration showed the expected pseudo-first order inactivation (11, 40) (Figure 5). The rate of inactivation by pentalenolactone was saturable, with a maximum rate of inactivation, kinact, of 5.3 ± 1.6 min-1 (t1/2 = 0.13 min) and a KI for PLBA of 1.3 ± 0.4 mM.
Figure 5.
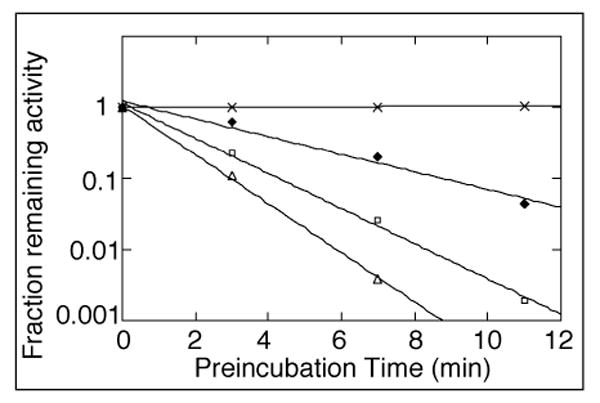
Time-dependent inactivation of S. avermitilis Gap2 by pentalenolactone. Semilog plot of the inactivation of Gap2 (2.1 μM) as a function of preincubation time and varied PLBA concentration: 0 (X), 74.5 (closed diamonds), 149 (open squares), and 223.5 μM (open triangles). Aliquots of 20 μL were withdrawn at periodic intervals and assayed for residual GAPDH activity in a total volume of 500 μL of assay buffer, as described in Methods. Each data point represents the mean of triplicate values.
Discussion
The increasing availability of complete microbial genome sequences has recently opened up the possibility of the top-down characterization of the molecular genetics and biochemistry of entire biosynthetic pathways whose genes are frequently organized in discrete clusters. As an essential first step in the elucidation of the enzymology and genetics of pentalenolactone biosynthesis in S. avermitilis, we have established that ptlA (SAV2998), one of 13 ORFs within a 13.4-kb cluster, encodes a 336-aa protein that catalyzes the cyclization of FPP to pentalenene (3), the well-established sesquiterpene hydrocarbon precursor of the pentalenolactone family of metabolites. The pentalenolactone biosynthetic pathway is functional in S. avermitilis, as confirmed by the isolation of both pentalenolactone F (8) and the shunt metabolite pentalenic acid (9). Deletion of the entire ptl-cluster from S. avermitilis abolished the production of pentalenolactone metabolites, while transfer of this same cluster to the naïve host, S. lividans 1326, endowed the transformants with the ability to generate pentalenic acid (9), accompanied by trace amounts of pentalenolactones. We have also demonstrated that gap1, located at the 5′-end of the biosynthetic gene cluster, corresponds to the essential locus for self-resistance to the antibiotic pentalenolactone, which acts as an active-site directed, irreversible inhibitor of the glycolytic enzyme GAPDH. Recombinant Gap1 retained full activity even after exposure to 0.6 mM pentalenolactone for 8 min at 4 °C, conditions under which rabbit GAPDH is completely inactivated (11). Gap1 shows considerable sequence identity to the homologous, pentalenolactone-insensitive GAPDH that confers resistance to pentalenolactone to S. arenae (15). Similar to S. arenae, S. avermitilis also harbors a second, pentalenolactone-sensitive, isoform of GAPDH. Interestingly, although the maximum rate of inactivation of Gap2 by PLBA is 2-3 times that previously observed for rabbit muscle GAPDH (kinact 5.3 min-1 compared to 1.3-3.3 min-1) (11), S. avermitilis Gap2 is considerably less sensitive to pentalenolactone than is the rabbit enzyme, exhibiting a KI of 1.3 mM that is nearly 100-fold higher than that for rabbit GAPDH (KI of 6-12 μM). In preliminary experiments on expression of gap1 and gap2 in S. avermitilis, gap2 was constitutively expressed during mycelial growth while expression of gap1 from the ptl gene cluster started after at least 15 h of growth. Thus prior to antibiotic production, Gap2 is utilized for the glycolytic pathway in S. avermitilis.
Within the ptl-cluster, there are 8 ORFs that appear to encode redox enzymes, as deduced from extensive sequence comparisons (Table 1). Cytochrome p450s are known to be able to mediate the three-step oxidative conversion of methyl groups to the corresponding carboxylic acids (41). It is therefore likely that CYP183A1, encoded by ptlI (SAV2999) (42), catalyzes the conversion of pentalenene (3) to 1-deoxypentalenic acid (4) (Scheme 1). Indeed we have already obtained preliminary experimental evidence in support of this hypothesis (Cane, D. E. & Quaderer, R., unpublished). PtlH (SAV2991), which resembles a non-heme iron, α-ketoglutarate-dependent hydroxylase, can oxidize 1-deoxypentalenic acid (4), with the resultant product undergoing oxidation to the corresponding ketone mediated by the dehydrogenase PtlF (SAV2993). Intriguingly, the putative monooxygenase, PtlE (SAV2994), shows a strong resemblance (49% identity and 62% similarity over 591 aa) to the monooxygenase of Rhodococcus ruber (Genbank AY052630.1) that catalyzes the Baeyer-Villiger-like oxidation of cyclodecanone to lauric acid. The latter three enzymes could therefore convert 1-deoxypentalenic acid (4) in three steps to the known metabolite pentalenolactone D (6). From pentalenolactone D, formation of pentalenolactones E (7) and F (8) would involve straightforward enzymatic desaturation adjacent to the lactonic carbonyl and epoxidation, possibly mediated by some combination of PtlD (SAV2995) and PtlJ (SAV3001). Formation of pentalenolactone (1) itself would require oxidative rearrangement of pentalenolactone F (8) (21). These and related possibilities are currently under active study.
Supplementary Material
Supporting Information Available. Sequence alignments for S. avermitilis PtlA (SAV2998) and for Gap1 (SAV2990) and Gap2 (SAV6296), and GC-MS identification of pentalenene. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Dr. Tun-Li Shen for carrying out the GC-MS analysis and Xiaofei He for the preparation of FPP
Footnotes
This work was supported by U.S. National Institutes of Health grant GM30301 to D.E.C., by Grant of the 21st Century COE Program, Ministry of Education, Culture, Sports, Science and Technology, Japan to H.I and S.O, and by Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science No. 17510168 to H.I.
Abbreviations: aa, amino acid; DTT, 1,4-dithiothreitol; farnesyl diphosphate, FPP; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; G-3-P, glyceraldehyde-3-phosphate; IPTG, isopropyl-β-D-thiogalactopyranoside; NAD+, β-nicotinamide adenine dinucleotide; NMWL, nominal molecular weight limit; PS, pentalenene synthase; PLBA, pentalenolactone benzylamine salt.peptide synthetase; PKS, polyketide synthase; ptl, pentalenolactone; TCEP, tris(2-carboxyethyl)phosphine.
Arigoni, D., Schwarz, M. & Eppacher, S. cited in Cane, D. E. (1999) in Comprehensive Natural Products Chemistry. Isoprenoids Including Carotenoids and Steroids, ed. Cane, D. E. (Elsevier, Oxford), Vol. 2, pp. 168.
References
- 1.Koe BK, Sobin BA, Celmer WD. PA 132, a New Antibiotic. I. Isolation and Chemical Properties. Antibiot Annu. 1957:672–675. [PubMed] [Google Scholar]
- 2.Martin DG, Slomp G, Mizsak S, Duchamp DJ, Chidester CG. The Structure and Absolute Configuration of Pentalenolactone (PA 132) Tetrahedron Lett. 1970:4901–4904. doi: 10.1016/s0040-4039(00)99739-9. [DOI] [PubMed] [Google Scholar]
- 3.Keller-Schierlein W, Lemke J, Nyfeler R, Zähner H. Stoffwechelprodukte von Mikroorganismen. 105. Arenaemycin E, D, und C. Arch Mikrobiol. 1972;84:301–316. [PubMed] [Google Scholar]
- 4.Takahashi S, Takeuchi M, Arai M, Seto H, Otake N. Studies on the Biosynthesis of Pentalenolactone. V. Isolation of Deoxypentalenylglucuron. J Antibiot. 1983;36:226–228. doi: 10.7164/antibiotics.36.226. [DOI] [PubMed] [Google Scholar]
- 5.Okazaki T, Enokita R, Torikata A, Inukai M, Takeuchi M, Takahashi S, Arai M. Studies on Actinomycetes Producing Pentalenolactone and Its New Related Compounds. Ann Rep Sankyo Res Lab. 1979;31:94–103. [Google Scholar]
- 6.English AR, McBride TJ, Lynch JE. PA 132, A New Antibiotic. II. In Vitro and in Vivo Studies. Antibiot Annu. 1957:682–687. [PubMed] [Google Scholar]
- 7.Nakagawa A, Tomoda H, Hao MV, Okano K, Iwai Y, Omura S. Antiviral Activities of Pentalenolactones. J Antibiot. 1985;38:1114–1115. doi: 10.7164/antibiotics.38.1114. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda M, Fukuda A, Takagi M, Morita M, Shimada Y. Inhibitory effect of pentalenolactone on vascular smooth muscle cell proliferation. Eur J Pharm. 2001;411:45–53. doi: 10.1016/s0014-2999(00)00894-3. [DOI] [PubMed] [Google Scholar]
- 9.Duszenko M, Balla H, Mecke D. Specific Inactivation of Glucose Metabolism from Eucaryotic Cells by Pentalenolactone. Biochim Biophys Acta. 1982;714:344–350. doi: 10.1016/0304-4165(82)90343-9. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann S, Neeff J, Heer U, Mecke D. Arenaemycin (Pentalenolactone): A Specific Inhibitor of Glycolysis. FEBS Lett. 1978;93:339–342. doi: 10.1016/0014-5793(78)81135-1. [DOI] [PubMed] [Google Scholar]
- 11.Cane DE, Sohng JK. Inhibition of Glyceraldehyde-3-Phosphate Dehydrogenase by Pentalenolactone: Kinetic and Mechanistic Studies. Arch Biochem Biophys. 1989;270:50–61. doi: 10.1016/0003-9861(89)90006-4. [DOI] [PubMed] [Google Scholar]
- 12.Cane DE, Sohng JK. Inhibition of Glyceraldehyde-3-phosphate Dehydrogenase by Pentalenolactone. 2. Identification of the Site of Alkylation by Tetrahydropentalenolactone. Biochemistry. 1994;33:6524–6530. doi: 10.1021/bi00187a020. [DOI] [PubMed] [Google Scholar]
- 13.Maurer KH, Mecke D. Regulation of Enzymes Involved in the Biosynthesis of the Sesquiterpene Antibiotic Pentalenolactone in Streptomyces arenae. J Antibiot. 1986;39:266–271. doi: 10.7164/antibiotics.39.266. [DOI] [PubMed] [Google Scholar]
- 14.Maurer KH, Pfeiffer F, Zehender H, Mecke D. Characterization of Two Glyceraldehyde-3-phosphate Dehydrogenase Isoenzymes from the Pentalenolactone Producer Streptomyces arenae. J Bacteriol. 1983;153:930–936. doi: 10.1128/jb.153.2.930-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frohlich KU, Kannwischer R, Rudiger M, Mecke D. Pentalenolactone-insensitive glyceraldehyde-3-phosphate dehydrogenase from Streptomyces arenae is closely related to GAPDH from thermostable eubacteria and plant chloroplasts. Arch Microbiol. 1996;165:179–186. doi: 10.1007/BF01692859. [DOI] [PubMed] [Google Scholar]
- 16.Cane DE, Rossi T, Tillman AM, Pachlatko JP. Stereochemical Studies of Isoprenoid Biosynthesis. Biosynthesis of Pentalenolactone from [UL-13C6]-Glucose and [6-2H2]-Glucose. J Am Chem Soc. 1981;103:1838–1843. [Google Scholar]
- 17.Cane DE, Sohng JK, Lamberson CR, Rudnicki SM, Wu Z, Lloyd MD, Oliver JS, Hubbard BR. Pentalenene Synthase. Purification, Molecular Cloning, Sequencing and High-Level Expression in Escherichia coli of a Terpenoid Cyclase from Streptomyces UC5319. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 18.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 19.Cane DE, Abell C, Tillman AM. Pentalenene Biosynthesis and the Enzymatic Cyclization of Farnesyl Pyrophosphate. Proof that the Cyclization is Catalyzed by a Single Enzyme. Bioorg Chem. 1984;12:312–328. [Google Scholar]
- 20.Cane DE, Oliver JS, Harrison PHM, Abell C, Hubbard BR, Kane CT, Lattman R. The Biosynthesis of Pentalenene and Pentalenolactone. J Am Chem Soc. 1990;112:4513–4524. [Google Scholar]
- 21.Cane DE, Sohng JK, Williard PG. Isolation and Structure Determination of Pentalenolactones A, B, D, and F. J Org Chem. 1992;57:844–852. [Google Scholar]
- 22.Cane DE, Rossi T. The Isolation and Structural Elucidation of Pentalenolactone E. Tetrahedron Lett. 1979:2973–2974. [Google Scholar]
- 23.Williard PG, Sohng JK, Cane DE. The x-ray crystal structure of pentalenolactone F methyl ester (epi-pentalenolactone F) J Antibiot. 1988;41:130–133. doi: 10.7164/antibiotics.41.130. [DOI] [PubMed] [Google Scholar]
- 24.Tillman AM, Cane DE. Pentalenolactone F, A New Metabolite Isolated from Streptomyces. Isolation and Structure Elucidation. J Antibiot. 1983;36:170–172. doi: 10.7164/antibiotics.36.170. [DOI] [PubMed] [Google Scholar]
- 25.Seto H, Sasaki T, Uzawa J, Takeuchi S, Yonehara H. Studies on the Biosynthesis of Pentalenolactone. Part II. Isolation of Pentalenic Acid and Pentalenolactone H. Tetrahedron Lett. 1978:4411–4412. [Google Scholar]
- 26.Tudzynski B. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl Microbiol Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 27.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet Biol. 2001;32:121–133. doi: 10.1006/fgbi.2001.1256. [DOI] [PubMed] [Google Scholar]
- 28.DeJong JM, L Y, Bollon AP, Jennewein S, Williams D, Croteau RB. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi Y, Matsumoto A, Seino A, Ueno J, Iwai Y, Omura S. Streptomyces avermectinius sp. nov., an avermectin-producing strain. Int J Syst Evol Microbiol. 2002;52:2163–2168. doi: 10.1099/00207713-52-6-2163. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 31.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cane DE, Ha HJ. Trichodiene Biosynthesis and the Role of Nerolidyl Pyrophosphate in the Enzymatic Cyclization of Farnesyl Pyrophosphate. J Am Chem Soc. 1988;110:6865–6870. [Google Scholar]
- 33.Cane DE, Yang G. Trichodiene Synthase. Stereochemical Studies of the Cryptic Allylic Diphosphate Isomerase Activity Using an Anomalous Substrate. J Org Chem. 1994;59:5794–5798. [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Second. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 35.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 36.Bradford M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantitites of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 37.Seemann M, Zhai G, de Kraker JW, Paschall CM, Christianson DW, Cane DE. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 38.Orsi BA, Cleland WW. Inhibition and Kinetic Mechanism of Rabbit Muscle Glyceraldehyde-3-phosphate Dehydrogenase. Biochemistry. 1972;11:102–109. doi: 10.1021/bi00751a018. [DOI] [PubMed] [Google Scholar]
- 39.Duggleby RG, Dennis DT. Nicotinamide Adenine Dinucleotide-specific Glyceraldehyde-3-phosphate Dehydrogenase from Pisum sativum. J Biol Chem. 1974;249:167–174. [PubMed] [Google Scholar]
- 40.Kitz R, Wilson IB. Esters of Methanosulfonic Acid as Irreversible Inhibitors of Acetylcholinesterase. J Biol Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 41.Pikuleva IA, Babiker A, Waterman MR, Bjorkhem I. Activities of recombinant human cytochrome P450c27 (CYP27) which produce intermediates of alternative bile acid biosynthetic pathways. J Biol Chem. 1998;273:18153–18160. doi: 10.1074/jbc.273.29.18153. [DOI] [PubMed] [Google Scholar]
- 42.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2) Biochem Biophys Res Commun. 2003;307:610–619. doi: 10.1016/s0006-291x(03)01231-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available. Sequence alignments for S. avermitilis PtlA (SAV2998) and for Gap1 (SAV2990) and Gap2 (SAV6296), and GC-MS identification of pentalenene. This material is available free of charge via the Internet at http://pubs.acs.org.