Abstract
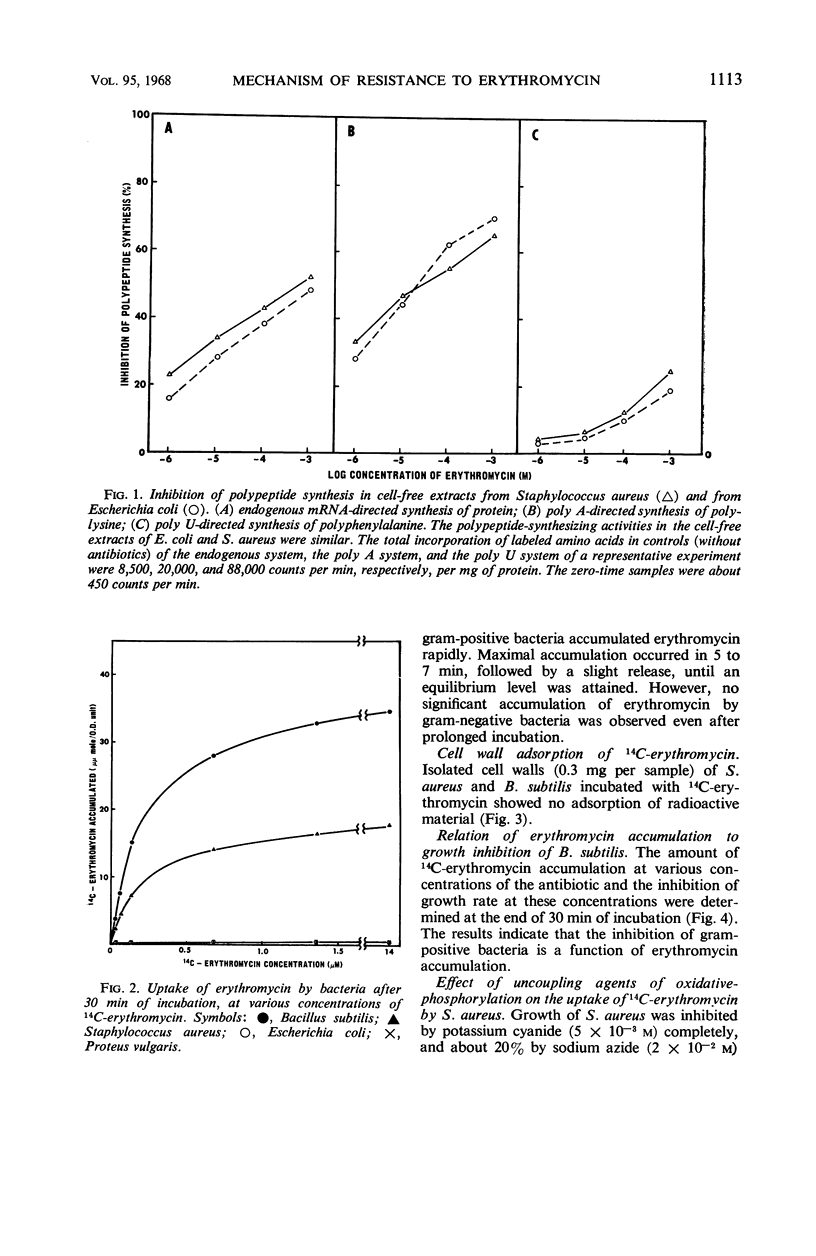
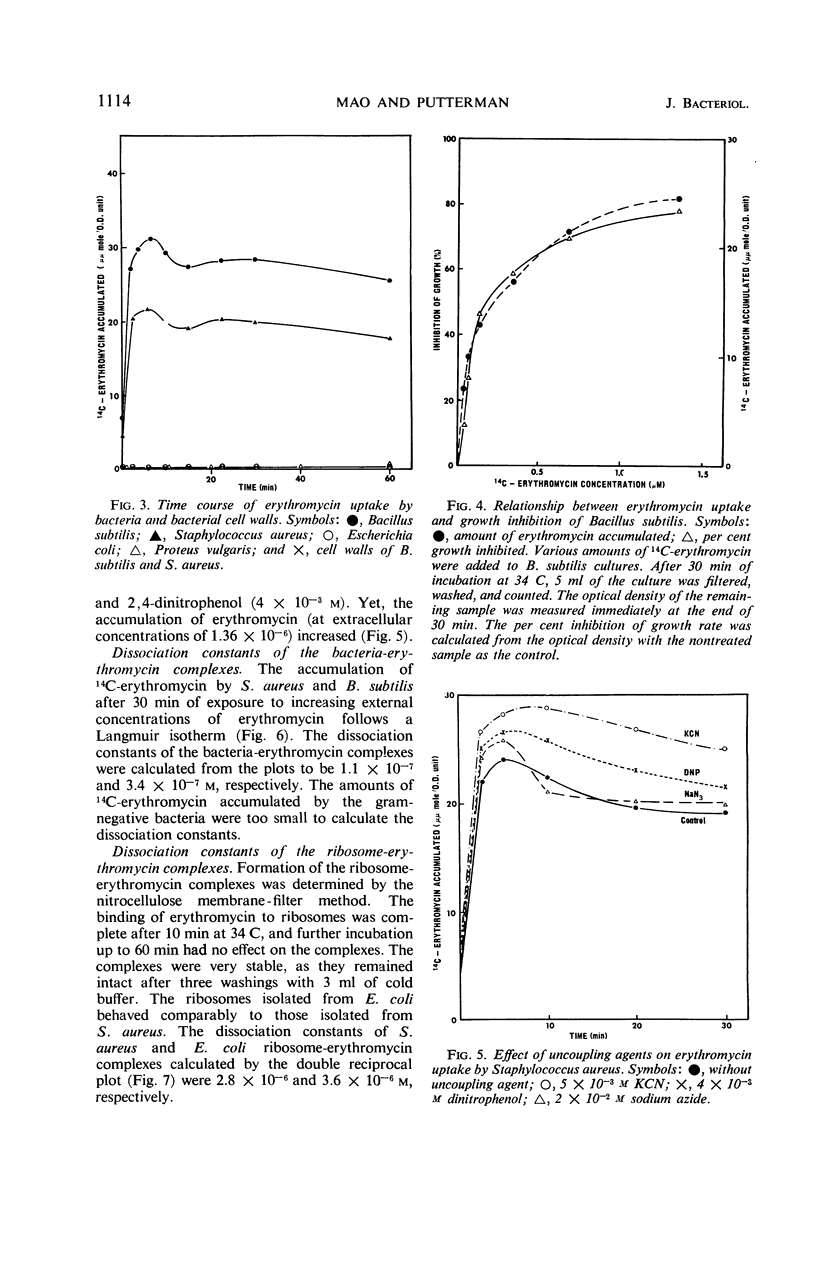
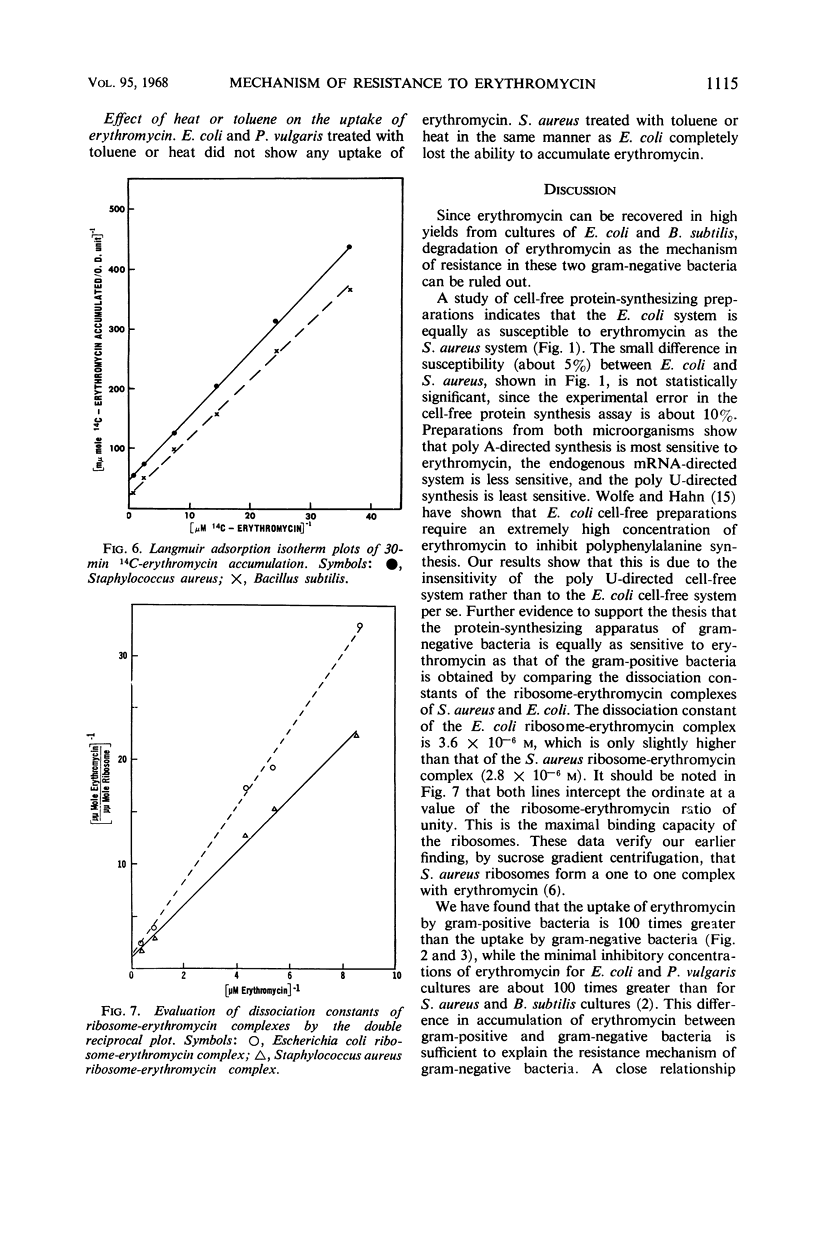
Erythromycin was recovered in high yield after incubation with gram-negative bacteria. The cell-free protein-synthesizing preparation from gram-negative bacteria is equally as susceptible to the antibiotic as is that from gram-positive bacteria. Thus, neither destruction of erythromycin nor the absence of the step susceptible to the antibiotic plays an important role in the resistance mechanism of gram-negative bacteria. A 100-fold difference in accumulation of erythromycin between gram-positive and gram-negative bacteria was observed. This alone explains the resistance of gram-negative bacteria to erythromycin. Furthermore, data showed that the inhibition of growth is closely related to the accumulation of erythromycin. The concentration of intracellular erythromycin in gram-positive bacteria was found to be 44- to 90-fold greater than that of the extracellular medium. However, the antibiotic did not accumulate on the cell walls, nor was the accumulation energy-dependent. It is proposed that it takes place by the binding of erythromycin to the bacterial ribosomes, forming a very stable complex. The dissociation constants of erythromycin-Staphylococcus aureus complex and erythromycin-Bacillus subtilis complex were determined to be 1.1 × 10−7 and 3.4 × 11−7m, respectively.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- HAIGHT T. H., FINLAND M. The antibacterial action of erythromycin. Proc Soc Exp Biol Med. 1952 Oct;81(1):175–183. doi: 10.3181/00379727-81-19815. [DOI] [PubMed] [Google Scholar]
- HURWITZ C., ROSANO C. L. Accumulation of label from C14-streptomycin by Escherichia coli. J Bacteriol. 1962 Jun;83:1193–1201. doi: 10.1128/jb.83.6.1193-1201.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAGAN B. M., ZOLLA S., BUSSER R., LIEPNIEKS S. SENSITIVITY OF COCCAL AND L FORMS OF STAPHYLOCOCCUS AUREUS TO FIVE ANTIBIOTICS. J Bacteriol. 1964 Sep;88:630–632. doi: 10.1128/jb.88.3.630-632.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C. Protein synthesis in a cell-free extract from Staphylococcus aureus. J Bacteriol. 1967 Jul;94(1):80–86. doi: 10.1128/jb.94.1.80-86.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Tardrew P. L. Demethylation of erythromycins by rabbit tissues in vitro. Biochem Pharmacol. 1965 Jul;14(7):1049–1058. doi: 10.1016/0006-2952(65)90033-x. [DOI] [PubMed] [Google Scholar]
- Mao J. C. The stoichiometry of erythromycin binding to ribosomal particles of Staphylococcus aureus. Biochem Pharmacol. 1967 Dec;16(12):2441–2443. doi: 10.1016/0006-2952(67)90232-8. [DOI] [PubMed] [Google Scholar]
- POLLOCK M. R. Drug resistance and mechanisms for its development. Br Med Bull. 1960 Jan;16:16–22. doi: 10.1093/oxfordjournals.bmb.a069785. [DOI] [PubMed] [Google Scholar]
- TAUBENECK U. Susceptibility of Proteus mirabilis and its stable L-forms to erythromycin and other macrolides. Nature. 1962 Oct 13;196:195–196. doi: 10.1038/196195b0. [DOI] [PubMed] [Google Scholar]
- TAUBMAN S. B., SO A. G., YOUNG F. E., DAVIE E. W., CORCORAN J. W. EFFECT OF ERYTHROMYCIN ON PROTEIN BIOSYNTHESIS IN BACILLUS SUBTILIS. Antimicrob Agents Chemother (Bethesda) 1963;161:395–401. [PubMed] [Google Scholar]
- Tanaka K., Teraoka H. Binding of erythromycin to Escherichia coli ribosomes. Biochim Biophys Acta. 1966 Jan 18;114(1):204–206. doi: 10.1016/0005-2787(66)90272-3. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Teraoka H., Nagira T., Tamaki M. Formation of C14-erythromycin-ribosome complex. J Biochem. 1966 Jun;59(6):632–634. doi: 10.1093/oxfordjournals.jbchem.a128355. [DOI] [PubMed] [Google Scholar]
- Taubman S. B., Jones N. R., Young F. E., Corcoran J. W. Sensitivity and resistance to erythromycin in Bacillus subtilis 168: the ribosomal binding of erythromycin and chloramphenicol. Biochim Biophys Acta. 1966 Aug 17;123(2):438–440. doi: 10.1016/0005-2787(66)90301-7. [DOI] [PubMed] [Google Scholar]
- WOLFE A. D., HAHN F. E. ERYTHROMYCIN: MODE OF ACTION. Science. 1964 Mar 27;143(3613):1445–1446. doi: 10.1126/science.143.3613.1445. [DOI] [PubMed] [Google Scholar]


