Abstract
In addition to pathways that regulate flowering in response to environmental signals such as photoperiod or cold temperatures (vernalization), flowering time is also regulated by light quality. In many species, far-red (FR) light is known to accelerate flowering. This is environmentally significant because leaves absorb more red light than FR light; thus, plants growing under a canopy experience light that is enriched in FR light. In this article, we have explored the promotion of flowering by FR-enriched light (FREL) in Arabidopsis (Arabidopsis thaliana). Previous work has shown that the floral promoter CONSTANS (CO) plays a critical role in day-length perception and exhibits complex regulation; CO mRNA is regulated by the circadian clock and CO protein is stabilized by light and degraded in darkness. We find that plants grown under FREL contain higher levels of CO mRNA in the early part of the day than plants under white light. Furthermore, transgenic plants expressing CO under the control of a constitutive promoter accumulate higher levels of CO protein under FREL, indicating that FREL can increase CO protein levels independently of transcription. Consistent with the model that FREL promotes flowering through CO, mutants for co or gigantea, which are required for CO transcript accumulation, are relatively insensitive to FREL. Because the red:FR ratios used in these experiments are in the range of what plants would experience under a canopy, these results indicate that the regulation of CO by light quality likely plays a key role in the regulation of flowering time in natural environments.
As sessile organisms, plants do not have the option of migrating from a suboptimal environment to a more favorable one. Thus, plants have evolved mechanisms that allow them to alter their growth and development in response to environmental signals, thereby increasing the likelihood of survival and reproductive success. One well-studied example of how the external environment can regulate plant development is the transition from vegetative to reproductive development. In Arabidopsis (Arabidopsis thaliana), flowering is promoted by a group of genes referred to as floral integrators. These genes, including FLOWERING LOCUS T (FT), the FT homolog TWIN SISTER OF FT (TSF), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1)/AGAMOUS-LIKE20, all act as strong promoters of flowering (Kardailsky et al., 1999; Kobayashi et al., 1999; Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Pathways that regulate flowering time in response to environmental stimuli, such as cold or daylength, do so, in large part, by promoting or repressing the expression of these floral integrators. Thus, signals from multiple environment-sensing pathways are integrated at the levels of FT, TSF, and SOC1 expression (Putterill et al., 2004).
In many plant species, flowering is promoted by prolonged exposure to cold temperatures, such as plants in temperate climates would experience in winter. This promotion of flowering is known as vernalization (Chouard, 1960). Winter-annual accessions of Arabidopsis occur naturally and are late flowering unless vernalized. This vernalization-responsive block to flowering is created by two genes, FLOWERING LOCUS C (FLC), a MADS-domain-containing transcription factor that acts as a repressor of FT, TSF, and SOC1 (Michaels et al., 2005; Moon et al., 2005), and FRIGIDA (FRI), which is required for high levels of FLC expression (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). Vernalization, in turn, promotes flowering through an epigenetic shut off of FLC expression that is mediated by repressive histone modifications at the FLC locus (Bastow et al., 2004; Sung and Amasino, 2004). In contrast to winter annuals, most rapid-cycling accessions of Arabidopsis contain naturally occurring loss-of-function mutations in FRI and therefore have low levels of FLC expression and are early flowering even in the absence of vernalization (Johanson et al., 2000). Forward-genetic screens conducted in rapid-cycling backgrounds have identified a group of genes, collectively referred to as the autonomous floral-promotion pathway, that act to constitutively repress FLC expression (Koornneef et al., 1991; Michaels and Amasino, 1999; Sheldon et al., 1999). The phenotype of loss-of-function autonomous-pathway mutants is similar to that of FRI-containing winter annuals; autonomous-pathway mutants are late flowering due to elevated levels of FLC (Michaels and Amasino, 1999, 2001) and this late-flowering phenotype is eliminated by vernalization (Koornneef et al., 1991). It is important to note that, although vernalization removes the block to flowering created by FLC, vernalization alone is not sufficient to induce rapid flowering. Early flowering in Arabidopsis also requires the activation of the floral integrators by inductive daylengths.
In both winter-annual and rapid-cycling Arabidopsis, flowering occurs more rapidly in long days than in short days. The floral promoter CONSTANS (CO) is a key component in the promotion of flowering by long days. CO is a B-box-containing protein that acts to promote the expression of FT, TSF, and SOC1 and is regulated at both the mRNA and protein levels (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000; Suarez-Lopez et al., 2001; Hepworth et al., 2002; Valverde et al., 2004). CO transcription is regulated by the circadian clock such that peak expression occurs late in the day under long days, but after dark in short days (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002; Valverde et al., 2004). This circadian expression of CO mRNA is dependent upon GIGANTEA (GI) because CO transcript levels are greatly reduced in gi mutants (Suarez-Lopez et al., 2001). CO protein is stabilized by white, blue, or far-red (FR) light by PHYTOCHROME A (PHYA) and CRYPTOCHROMEs (CRY1 and CRY2), but is degraded under red (R) light by PHYB or in darkness (Valverde et al., 2004). Because CO transcription is only coincident with light under long days, CO protein accumulation and subsequent activation of floral integrators provide a long-day specific flowering signal.
In addition to the duration of the light period (daylength), plants also perceive the quality of light (i.e. wavelength). Because leaves absorb more R light than FR light, plants growing under a canopy experience lower R:FR ratios than plants growing in full sun. Low R:FR ratios are perceived by the phytochrome family of photoreceptors and induce a range of responses including stem and petiole elongation, hyponastic leaves, reduced branching, and early flowering (Smith, 1995). The effect of FR-enriched light (FREL) on flowering time has been investigated in wild-type Arabidopsis, as well as late-flowering lines containing mutations in autonomous-pathway or photoperiod-pathway genes (Martinez-Zapater and Somerville, 1990; Eskins, 1992; Bagnall, 1993; Lee and Amasino, 1995; Cerdan and Chory, 2003). The molecular details of how FREL promotes flowering are not well understood, but low R:FR ratios are known to increase expression of FT (Cerdan and Chory, 2003; Halliday et al., 2003). Interestingly, a correlation has been observed between vernalization responsiveness and the promotion of flowering by FREL (Bagnall, 1993). FRI-containing winter annuals and autonomous-pathway mutants, which are late flowering due to high levels of FLC, show a strong early-flowering phenotype when vernalized or grown under FREL. In contrast, vernalization and FREL are much less effective in promoting flowering in late-flowering photoperiod-pathway mutants, such as gi or co, in which FLC levels are not elevated.
Although the reason for the correlation between the effects of vernalization and FREL is unclear, at least three explanations seem plausible. Because FLC is required for the late-flowering phenotype of FRI and autonomous-pathway mutants, but not for that of photoperiod-pathway mutants (Michaels and Amasino, 2001), it is possible that, like vernalization, FREL represses FLC expression. Alternatively, FREL may promote flowering through activation of the photoperiod pathway, in which case photoperiod-pathway mutants would be predicted to have an attenuated response to FREL. Finally, FREL may promote flowering independently of both FLC and the photoperiod pathways, possibly through pathways that are responsible for the induction of other aspects of the shade-avoidance response. In this article, we provide evidence that the effect of FREL on flowering time is independent of FLC and genetically separable from other aspects of the shade-avoidance response (Vandenbussche et al., 2005). Furthermore, we show that FREL results in elevated levels of both CO mRNA and protein. Together, these data support a model in which low R:FR ratios promote flowering through activation of the photoperiod pathway.
RESULTS
FREL Promotes Flowering in FRI and Autonomous-Pathway Mutants Independently of FLC
A variety of light sources have been utilized to add supplemental FR light, including specialized fluorescent tubes (Eskins, 1992), incandescent bulbs (Bagnall, 1993), and, more recently, FR light-emitting diodes (LEDs; Salter et al., 2003). LEDs have several advantages over other sources of FR light, including negligible heat output and more precise control over light quality (wavelength). For our experiments, plants were grown under cool-white fluorescent lights alone (white-light [WL] conditions) or supplemented with FR LEDs (Fig. 1A). The R:FR ratio (660 nm:730 nm) was 6.0 in WL and 0.4 under FREL. A R:FR ratio of 0.4 is in the range of what plants might experience growing under a canopy (Smith, 1982). Previous work using incandescent bulbs as a source of supplemental FR light has shown a positive correlation between vernalization responsiveness and FR responsiveness in late-flowering mutants in the Landsberg erecta (Ler) background (Bagnall, 1993). Autonomous-pathway mutants, whose phenotype can be eliminated by vernalization, showed the greatest reduction in flowering time under FREL, whereas photoperiod pathway mutants are less responsive to both FREL and vernalization (Bagnall, 1993). Given this result, one possible explanation for the responsiveness of autonomous-pathway mutants to FREL is that, like vernalization, FREL promotes flowering through a reduction in FLC expression.
Figure 1.
Effect of FREL on flowering time. A, Spectra of WL provided by cool-white fluorescent lamps (black line) or FREL (gray line) supplied by WL supplemented with FR LEDs. Light intensity is given in μmol m−2 s−1. The absorption peaks for the Pr and Pfr forms of phytochrome are indicated with vertical broken lines. B, Bars represent the total number of leaves formed prior to flowering under long days (rosette + cauline). White and black bars indicate plants grown under WL and FREL, respectively. Gray bars indicate the percentage of reduction in leaf number between WL and FREL. Error bars indicate 1 sd.
Because Ler contains a weak allele of FLC due to the insertion of a transposon in the first intron (Gazzani et al., 2003; Michaels et al., 2003), we investigated the effect of FREL on flowering in the Columbia (Col) background, which contains a more typical strong allele of FLC. When grown under FREL, autonomous-pathway mutants (fca, fld, flk, fve, or ld) or FRI-Col flowered significantly earlier than when grown under WL (Figs. 1B and 2, A–D); all lines showed at least a 30% reduction in total leaf number when grown under FREL (Fig. 1B). In addition to flowering early, plants grown under FREL also exhibited other phenotypes associated with low R:FR ratios, including increased petiole elongation (Fig. 2E) and elevated expression of genes, such as PHYTOCHROME-INTERACTING FACTOR3 (PIF3)-LIKE1 (PIL1; Fig. 2F), whose expression is induced by low R:FR ratios (Makino et al., 2002; Salter et al., 2003). To determine whether FLC levels are affected by FREL, we examined FLC mRNA levels under WL or FREL. In all backgrounds tested, FLC mRNA levels were similar under both light conditions (Fig. 2F). Similarly, no significant change in expression was detected in an FLC∷GUS-containing line grown under WL or FREL (Fig. 3, A and B). These results suggest that the regulation of FLC mRNA levels does not play a major role in the acceleration of flowering by FREL. As a further test of this hypothesis, we examined the flowering time of a line containing a constitutively expressed 35S∷FLC construct. Consistent with the result that FLC levels are not reduced under FREL, 35S∷FLC flowered earlier under FREL than under WL (Fig. 1B).
Figure 2.
Effect of FREL on FRI and late-flowering mutants. A to D, Col (A and B) and FRI-Col (C and D) plants grown under WL (A and C) or FREL (B and D). E, Leaves 5 to 9 taken from FRI-Col plants. In each pair of leaves shown, the leaf on the left was grown under WL and the leaf on the right was grown under FREL. F, Semiquantitative RT-PCR analysis of gene expression under WL and FREL. RNA was isolated from 14-d-old seedlings 4 h after dawn. UBQ was used as a control for loading. Numbers indicate the fold change in gene expression in response to FREL. All plants (A–F) were grown under long days. [See online article for color version of this figure.]
Figure 3.
Effect of FREL on photoperiod-pathway mutants. A to D, FLC∷GUS (A and B) and FT∷GUS (C and D) expression in 10-d-old seedlings grown under WL (A and C) or FREL (B and D). E and F, CO∷GUS expression in the first true leaves of 15-d-old seedlings grown under WL (E) or shifted to FREL for 8 h (F). G, Bars represent the total number of leaves formed prior to flowering (rosette + cauline). White and black bars indicate plants grown under WL and FREL, respectively. Gray bars indicate the percentage of reduction in leaf number between WL and FREL. Error bars indicate 1 sd. H to K, gi (H and I) and co (J and K) mutants grown under WL (H and J) or FREL (I and K). All plants (A–K) were grown under long days.
gi and co Mutants Show an Attenuated Response to FREL
An alternative explanation for the promotion of flowering by FREL in FRI and autonomous-pathway mutants is that the repression of flowering conferred by FLC is overcome through the activation of an alternative floral-promotion pathway. To examine this possibility, we determined the flowering time of mutants in the photoperiod pathway. Interestingly, the photoperiod-pathway mutants gi and co showed little difference in flowering time when grown under WL or FREL (Fig. 3, G–K). The relative insensitivity of gi and co mutants to FREL suggests that FR light may accelerate flowering through the photoperiod pathway. If this were the case, then a photoperiod-pathway mutation would be predicted to block or reduce the response of FRI or autonomous-pathway mutants to FREL. To test this hypothesis, a FRI co line (Michaels and Amasino, 2001) was grown under WL and FREL. When grown under FREL, FRI-Col flowered with 57% fewer leaves than under WL (Fig. 1B). In contrast, FRI co showed only a 20% reduction in leaf number under FREL (Fig. 3G). Thus, an active photoperiod pathway is required for the full effect of FREL in a FRI-containing background.
CO is known to promote flowering in long days by activating the expression of the floral promoters FT, TSF, and SOC1 (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000). Consistent with FREL promoting flowering through the photoperiod pathway, the transcript levels of FT, TSF, and SOC1 are elevated in FRI or ld plants grown under FREL, but not in co mutants (Fig. 2F). The induction of FT by FREL was also evident in plants containing an FT∷GUS fusion (Fig. 3, C and D). Taken together, these data are consistent with a model in which FREL acts through GI and CO to activate FT, TSF, and SOC1 expression.
FT, TSF, and SOC1 Act Redundantly to Promote Flowering in Response to FREL and CO
Given the fact that FREL results in increased expression of floral integrators, we investigated the effect of FREL on ft, tsf, and soc1 mutants. All three mutants flowered significantly earlier under FREL, forming 35% to 44% fewer leaves than when grown under WL (Fig. 3G). Given that mutations in co render plants almost completely insensitive to FREL (Fig. 3G), it is interesting that mutations in the downstream targets of CO do not prevent early flowering in response to FR light. One explanation for why single mutants in the floral integrators remain sensitive to FREL is that they act redundantly. In support of this model, FT and TSF are homologous genes that have been shown to have redundant functions (Michaels et al., 2005; Yamaguchi et al., 2005). In addition, evidence suggests that CO directly regulates both FT and SOC1 (Samach et al., 2000; Hepworth et al., 2002). To investigate the cumulative roles of FT, TSF, and SOC1 on the promotion of flowering in response to low R:FR ratios, we examined the response of double and triple mutants to FREL. ft and tsf single mutants each formed approximately 40% fewer leaves under FREL; the ft tsf double mutant, however, showed a much weaker response to FREL, flowering with only a 15% reduction in leaf number (Fig. 4A). Because CO is thought to directly regulate FT and SOC1, we examined the flowering time of an ft soc1 double mutant. Interestingly, the ft soc1 double mutant showed a relatively strong response to FREL, flowering with 30% fewer leaves. The observation that the ft tsf is less sensitive to FREL than the ft soc1 suggests that TSF may play a significant role in promoting flowering in response to FREL in ft soc1 plants. If so, then an ft tsf soc1 triple mutant would be predicted to be much less sensitive to FREL than the ft soc1 double. Indeed, this is the case; the ft tsf soc1 triple mutant was less sensitive to FREL than ft soc1 (Fig. 4A) and flowered with only 7% fewer leaves under FREL. Thus, FT, TSF, and SOC1 function redundantly to promote flowering in response to FREL.
Figure 4.
Redundant roles of FT, TSF, and SOC1 in the promotion of flowering in response to FREL and CO. Bars represent the total number of leaves formed prior to flowering (rosette + cauline). A, White and black bars indicate plants grown under WL and FREL, respectively. Gray bars indicate the percentage of reduction in leaf number between WL and FREL. B, Flowering time of T1 plants transformed with a 35S∷CO construct. Horizontal lines indicate the flowering time of the untransformed parental lines. Plants were grown under WL. C, Flowering time of ft tsf soc1 triple mutants and T2 lines transformed with a 35S∷CO. Plants were grown under WL. All plants (A–C) were grown under long days. Error bars indicate 1 sd (A and C).
Like FREL, CO also promotes the expression of the floral integrators. Previous work has shown that mutations in ft or soc1 can partially suppress the early-flowering phenotype of CO overexpression (Onouchi et al., 2000; Samach et al., 2000; Yoo et al., 2005). Therefore, we investigated whether FT, TSF, and SOC1 play redundant roles in promoting flowering in response to CO. Overexpression of CO causes a strong early-flowering phenotype regardless of photoperiod (Simon et al., 1996); therefore, we transformed single, double, and triple mutants with a construct containing CO under control of the 35S cauliflower mosaic virus promoter and examined the flowering time of T1 plants. To test the effectiveness of our construct, 35S∷CO was transformed into FRI-Col. Most T1 plants flowered early, with many plants flowering with fewer than 10 leaves compared to approximately 75 leaves for untransformed controls (Fig. 4B). The effect of CO overexpression in various ft, tsf, and soc1 mutant backgrounds was largely similar to that of FREL (Fig. 4, A and B). In tsf single mutants, 35S∷CO was very effective at accelerating flowering. CO overexpression in ft, ft tsf, or ft soc1 backgrounds also showed a strong early-flowering phenotype. The earliest flowering T1 plants in these backgrounds, however, were later than those obtained from the transformation of tsf or FRI with 35S∷CO. As with FREL, the ft tsf soc1 triple mutant showed the smallest change in flowering time in response to 35S∷CO, with the earliest T1 plants flowering with greater than 55 leaves. To ensure that the early flowering observed in the transformed triple mutant was not due to stress associated with herbicide selection, seed was collected from three early-flowering T1 lines and flowering time was determined in the T2 generation without herbicide selection. Similar to the T1 results, all three lines flowered slightly earlier than the untransformed parent (Fig. 4C). Thus, FT, TSF, and SOC1 act redundantly to promote flowering in response to both FREL and CO.
FREL Increases CO Transcript Levels
The results above suggest that the promotion of flowering in FRI and autonomous-pathway mutants by FREL is due to increased expression of floral integrators, whose expression is normally repressed by high levels of FLC in these backgrounds. The requirement for CO and GI in the promotion of flowering by FREL suggests that the photoperiod pathway is involved in this activation. To determine where in the photoperiod pathway FREL acts, we first examined the expression of genes associated with the circadian clock. In plants shifted from WL to FREL for 1, 2, 4, or 6 h, ZEITLUPE (ZTL), LOV KELCH PROTEIN2 (LKP2), EARLY FLOWERING3 (ELF3), FLAVIN-BINDING KELCH-REPEAT F-BOX (FKF1), and PIF3 (Zhou et al., 2007) showed no significant change in expression (Fig. 5A). This suggests that FREL may act downstream of the circadian clock to promote flowering. GI, which acts as an output of the circadian clock and regulates CO transcription (David et al., 2006), also showed no change in expression under WL or FREL (Fig. 5A). CO, however, showed significant increases in transcript levels after transfer to FREL (Fig. 5, A and B). This increase in CO transcription was also observed using a construct containing GUS fused to the CO promoter (Fig. 3, E and F). Consistent with the increase in CO expression, FT, and to a lesser extent SOC1, also showed increased expression (Fig. 5A). These results indicate that FREL promotes flowering, at least in part, through increased expression of CO mRNA.
Figure 5.
Regulation of CO mRNA by FREL. A, Col plants were grown in long days under WL for 14 d. Six hours after dawn on day 15, plants were either maintained in WL or shifted to FREL. After the indicated number of hours, plants were harvested and subjected to semiquantitative RT-PCR analysis. UBQ was used as a control for loading. Numbers indicate the fold change in gene expression in response to FREL. B, Quantitative RT-PCR analysis of CO expression. Col and gi plants were grown in long days under WL for 14 d. Six hours after dawn on day 15, plants were either maintained in WL or shifted to FREL. After the indicated number of hours, plants were harvested and subjected to quantitative RT-PCR analysis. The expression of CO was determined relative to ACTIN. Diamonds and squares indicate expression in Col under WL and FREL, respectively. Circles and crosses represent expression in gi under WL and FREL, respectively. Error bars indicate the se of the mean of three biological replicates. C, Quantitative RT-PCR analysis of CO and FT expression in 8-d-old Col plants grown in long-day conditions under WL (black line/diamonds) or FREL (gray line/squares). Plants were grown under WL or FREL from germination. Error bars indicate the se of the mean of three biological replicates. The horizontal bar indicates periods of light (white) and dark (black).
Mutations in either gi or co lead to insensitivity to FREL (Fig. 3G). Previous work has shown that mutations in gi prevent the accumulation of CO transcript (Suarez-Lopez et al., 2001). Thus, a possible explanation for the insensitivity of gi mutants is that the CO transcript cannot be up-regulated by FREL in a gi mutant background. To test this hypothesis we examined the levels of CO mRNA in a gi mutant grown under WL or shifted to FREL. Under WL, CO transcript levels were much lower than that observed in Col (Fig. 5B). Moreover, no increase in CO transcript was observed in the gi mutant background after shifting to FREL. Thus, the inability of gi mutant plants to up-regulate CO mRNA levels in response to FREL may explain the similar FR-insensitive phenotypes of co and gi mutants.
In addition to the experiments described above in which plants are shifted to FREL, we also investigated the expression of CO and FT in plants grown continuously under WL or FREL. CO transcription is regulated by the circadian clock, with mRNA levels increasing late in the day (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002). Therefore, it is possible that the increased levels of CO observed after shifting to FREL may be due to a shift in the phase of CO expression, such that CO is expressed at higher levels earlier in the day under FREL than under WL. To investigate this possibility, we determined the expression of CO during a 24-h time course in plants grown continuously under WL or FREL. CO expression was significantly higher under FREL early in the day (near dawn; Fig. 5C). Interestingly, however, CO levels were similar under WL and FREL for the remainder of the day (Fig. 5C). Consistent with the expression of CO, FT levels were also significantly higher under FREL only in the early part of the day (Fig. 5C). Thus, growth under FREL does not appear to shift the phase of CO expression, but rather causes increased CO expression during the early part of the day.
FREL Increases CO Protein Levels
If FREL acts to promote flowering solely by increasing the level of CO transcription, one would predict that constitutive expression of CO would lead to insensitivity to low R:FR ratios. To test this hypothesis, 35S∷CO was transformed into a co mutant. A transformed line was chosen, which flowered similarly to wild-type Col (Fig. 6A). When grown under WL and FREL, the 35S∷CO line showed approximately the same acceleration of flowering as seen in Col (Fig. 6A). Therefore, plants constitutively expressing CO remain sensitive to FREL.
Figure 6.
Regulation of CO protein by FREL. A, C, and D, Flowering time of plants of the indicated genotypes under WL (white bars) and FREL (black bars). Gray bars indicate the percentage of reduction in leaf number between WL and FREL. Error bars indicate 1 sd. Plants were grown under long days (A and D) or short days (C). B, Semiquantitative RT-PCR (top two images) and western-blot analysis (bottom three images) of a 35S∷GFP∷CO transgenic line. UBQ was used as a loading control for RT-PCR. A cross-reacting band (*) and a Coomassie-stained gel are shown as loading controls for western-blot analysis.
The results above indicate that, in addition to increasing CO mRNA levels, FREL also accelerates flowering through mechanisms that are downstream of CO transcription. One possible explanation is that CO protein may accumulate to higher levels under FREL than under WL alone. This model is supported by previous work demonstrating that CO protein accumulates to higher levels under FR light than in darkness (Valverde et al., 2004). Although these data indicate that FR light is able to stabilize CO protein, it should be noted that CO accumulated to similar levels under WL or FR light (supplied by a mixture of incandescent bulbs and FR LEDs) in these experiments (Valverde et al., 2004). It remains possible, however, that WL supplemented with FR light (i.e. FREL) may be more effective at stabilizing CO than WL alone.
To determine whether CO protein levels might also play a role in accelerating flowering under FREL, we investigated the levels of CO protein under WL and FREL. A 35S∷GFP∷CO construct was created and transformed into Col. The resulting line was early flowering, indicating that the fusion protein was functional. Plants were grown under WL for 17 d and then were either transferred to FREL or maintained in WL for 2 d. Reverse transcription (RT)-PCR analysis indicated that CO mRNA is indeed overexpressed and that the level of CO transcript is unaffected by light quality (Fig. 6B). CO protein levels were determined by western blot using an anti-GFP antibody to detect the GFP∷CO fusion protein in nuclear protein extracts. In contrast to 35S∷GFP∷CO mRNA levels, the level of CO protein was 2.6-fold higher under FREL than under WL (Fig. 6B). Thus, CO protein does indeed accumulate to higher levels under FREL.
As a final experiment to test the model that FREL acts to promote flowering through CO protein accumulation, we investigated the effect of FREL on flowering under short days. Because of the circadian regulation of CO transcript, CO mRNA (and therefore CO protein) does not accumulate during the light period in short days (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002; Valverde et al., 2004). If this is the case, then FREL should have little effect on flowering time in short days because there is no CO protein to stabilize. This is indeed the case. Col plants flowered similarly under short days regardless of light quality (Fig. 6C), supporting the model that the enhanced stabilization of CO protein by low R:FR ratios is key to the promotion of flowering by FREL.
Taken together, these results indicate that CO transcript and protein levels play an important part in the regulation of flowering time by light quality. CO expression is known to be regulated by several photoreceptors. CO transcript is increased in phyB mutants and decreased in phyA mutants (Yanovsky and Kay, 2002; Cerdan and Chory, 2003) and, at the protein level, CO is stabilized by PHYA, CRY1, and CRY2 and is destabilized by PHYB (Valverde et al., 2004). Consistent with the model that multiple photoreceptors are involved in CO regulation and previous observations (Mockler et al., 2003), we find that loss-of-function mutants in individual photoreceptors maintain a significant response to FREL under our conditions (Fig. 6D).
Acceleration of Flowering by FREL Is Genetically Separable from Other Shade-Avoidance Responses
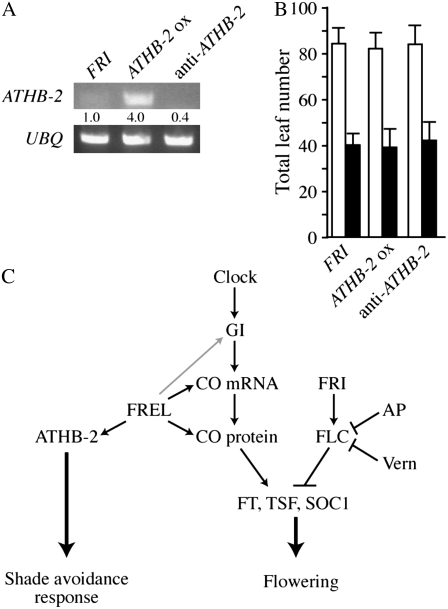
In many plant species, the low R:FR ratios experienced under a canopy induce a number of responses collectively referred to as the shade-avoidance response. These include increased apical dominance and stem elongation, diminished leaf expansion, and accelerated flowering (Smith, 1995). ATHB-2 is a homeodomain-Leu-zipper protein that is rapidly induced by low R:FR ratios (Carabelli et al., 1993). Plants overexpressing ATHB-2 have phenotypes indicative of the shade-avoidance response in the absence of FREL, including reduced cotyledon expansion and increased hypocotyl elongation (Steindler et al., 1999). Conversely, plants with reduced ATHB-2 expression have increased cotyledon expansion and decreased hypocotyl elongation (Steindler et al., 1999). Thus, ATHB-2/HAT4 is a key regulator of certain aspects of the shade-avoidance response (Ruberti et al., 1991; Schena and Davis, 1992). Given its role in the regulation of other shade-avoidance-associated phenotypes, we investigated the possibility that ATHB-2 might also be involved in the early-flowering phenotype of plants grown under low R:FR ratios. To determine whether altering ATHB-2 expression could mimic the early-flowering phenotype observed under FREL, overexpression (35S∷ATHB-2) and antisense (αATHB-2) constructs (Steindler et al., 1999) were transformed into a line containing FRI in the Col background (FRI-Col). The transgenic lines showed the previously described effects on hypocotyl elongation and cotyledon expansion (Steindler et al., 1999), indicating that the constructs were functional. RT-PCR was also performed to confirm the overexpression of ATHB-2 in the 35S∷ATHB-2 transgenic line and reduced expression in the αATHB-2-containing line (Fig. 7A). When grown under FREL, FRI-Col plants flowered significantly earlier than when grown under WL (Fig. 7B). Transgenic lines containing 35S∷ATHB-2 or αATHB-2, however, flowered similarly to the untransformed controls when grown under WL or FREL (Fig. 7B). Thus, although ATHB-2 expression is sufficient to induce other shade-avoidance phenotypes in the absence of low R:FR ratios, it is ineffective at promoting flowering. Thus, at the level of ATHB-2 expression, the effects of FREL on flowering time and other aspects of the shade-avoidance response are genetically separable.
Figure 7.
ATHB-2 expression levels do not affect flowering time. A, Semiquantitative RT-PCR analysis of ATHB-2 expression in 10-d-old seedlings. UBQ was used as a control for loading. Numbers indicated the fold change in gene expression relative to untransformed FRI plants. B, Bars represent the total number of leaves formed prior to flowering (rosette + cauline). White and black bars indicate plants grown under WL and FREL, respectively. Error bars indicate 1 sd. All plants (A and B) were grown under long days. C, Model for the promotion of flowering by FREL. Speculative interactions are depicted with gray arrows.
DISCUSSION
A number of laboratories have examined the effect of light quality on flowering time in Arabidopsis. In general, this work has shown that late-flowering FRI-containing lines or autonomous-pathway mutants are more responsive to FREL than photoperiod-pathway mutants (Martinez-Zapater and Somerville, 1990; Bagnall, 1993; Lee and Amasino, 1995). It should be noted, however, that some reports have shown that co mutants exhibit strong responses to FREL (Halliday et al., 1994; Devlin et al., 1996). Although the reasons for the discrepancies are not clear, differences in R:FR ratios, light sources, co alleles, and genetic backgrounds may play a role. Our results are consistent with those that have reported that photoperiod-pathway mutants are relatively insensitive to FREL (Martinez-Zapater and Somerville, 1990; Bagnall, 1993; Lee and Amasino, 1995) and can provide a molecular explanation for this observation. FREL leads to hyperactivation of the photoperiod pathway, thereby partially bypassing the block to flowering created by FLC (Fig. 7C). In autonomous-pathway mutants, high levels of FLC act to repress the expression of FT, TSF, and SOC1. Under FREL, however, CO mRNA and protein accumulate to higher levels and can increase the expression of the floral integrators. Consistent with this model, ft tsf soc1 triple mutants show greatly reduced sensitivity to FREL and CO overexpression. It is interesting to note, however, that triple mutants are not completely insensitive to FREL and CO (Fig. 4). This suggests that FREL and CO may promote flowering through FT/TSF/SOC1-independent mechanisms. Another possibility, however, is that our triple mutant does not completely eliminate the function of all three genes. Although the tsf and soc1 alleles used in this study are T-DNA insertional alleles and are likely to be nulls, the ft allele used (ft-1 backcrossed into Col) contains a missense mutation and may not completely eliminate protein function (Kardailsky et al., 1999; Kobayashi et al., 1999). That ft-1 is not a null is also supported by the observation that ft-10, a T-DNA allele, flowers later than ft-1 (Yoo et al., 2005).
We find that FREL acts to independently promote both the accumulation of CO mRNA and CO protein. Although previous studies have shown that CO mRNA levels are up-regulated by pure FR light (Tepperman et al., 2001) and that CO transcript levels are altered in phytochrome mutants (Suarez-Lopez et al., 2001; Tepperman et al., 2001; Yanovsky and Kay, 2002; Cerdan and Chory, 2003), we believe this to be the first demonstration of the effect of environmentally relevant changes in R:FR ratios on CO transcript levels in wild-type plants. It is particularly interesting that the up-regulation of CO mRNA in plants growing under FREL occurs primarily in the early part of the day. The molecular mechanism underlying this up-regulation of CO mRNA by FREL is not clear, but it may involve GI. gi mutants have very low levels of CO expression and are relatively insensitive to FREL (Suarez-Lopez et al., 2001; Fig. 5B). Furthermore, CO mRNA levels are not up-regulated by FREL in a gi mutant background. Thus, GI is required for proper expression of the CO transcript. Interestingly, GI protein is also regulated by light. GI protein is stabilized in light (white, red, or blue) and is degraded in darkness by the 26S proteasome (David et al., 2006). Unfortunately, the effect of FR light on GI protein accumulation has not yet been determined. An interesting possibility, however, is that GI protein may accumulate to higher levels or show high activity under FREL. Given that increased expression of GI has been shown to increase expression of CO mRNA (Mizoguchi et al., 2005), it seems reasonable to expect that any enhanced stabilization of GI protein by FREL would lead to increased CO transcription.
In addition to the up-regulation of CO mRNA levels, we have found that FREL also regulates CO protein levels. Because increased protein levels were observed using a GFP∷CO fusion protein driven by the constitutive 35S promoter, the increase in protein under FREL cannot be attributed to increased transcription. Light quality has previously been implicated in the accumulation of CO protein; however, no significant difference was observed in CO protein accumulation under WL or pure FR light (Valverde et al., 2004). Coupled with our results, this suggests that FREL (i.e. both WL and FR light) may be more effective at promoting CO protein accumulation than WL or FR light separately. The increased CO protein levels observed under FREL could be a result of increased translation and/or decreased degradation; however, reduced protein degradation may be more likely. The addition of proteasome inhibitors to nuclear protein extracts increases CO protein levels in both dark- and light-treated material (Valverde et al., 2004). This result suggests that a significant amount of proteasome-dependent degradation of CO takes place even in the light. It is possible, then, that WL and FR light (i.e. FREL) may have an additive stabilizing effect on CO.
The significant changes in CO mRNA and protein accumulation observed under FREL illustrate the sensitivity of plants to changes in light quality in the range normally experienced in the environment. In addition to accelerating flowering, FREL also induces other phenotypes associated with the shade-avoidance response. Here, we have been able to demonstrate that accelerated flowering in response to FREL is genetically separable from other phenotypes of the shade-avoidance response; overexpression of ATHB-2 is sufficient to induce hypocotyl elongation and reduced cotyledon expansion, but does not accelerate flowering. Thus, despite the fact that early flowering and other phenotypes associated with shade avoidance occur as a result of low R:FR ratios, they are controlled by separate outputs from the light-signaling mechanisms.
MATERIALS AND METHODS
Plant Material and Growth Conditions
FRI-Col (Lee et al., 1994b), fca-9 (Bezerra et al., 2004), fld-3 (He et al., 2003), fve-4 and FRI-co (Michaels and Amasino, 2001), ld-1 (Lee et al., 1994a), gi-2 (Park et al., 1999), ft-1 in Col, tsf, and soc1 (Michaels et al., 2005), co (SAIL24H04) (Kim and Michaels, 2006), and phyA-211, phyB-9, cry1-304, and cry2-1 (Mockler et al., 2003) have been described previously. flk (SALK_112850) was obtained from the Arabidopsis Biological Resource Center. The FLC overexpression line was created by transforming a 35S∷FLC construct into the flc-3 mutant. Plants were grown at 22°C under cool-white fluorescent light with a light intensity of 125 μmol m−2 s−1 (400–700 nm) with or without 10 μmol m−2 s−1 of supplemental FR provided by FR LEDs (L-D-735-H; Plasma Ireland Ltd.) at a density of 730 LED/m2. Light spectra were measured with a USB4000 spectrophotometer (Ocean Optics) using neutral density filters. Long and short days consisted of 16 h light/8 h dark and 8 h light/16 h dark, respectively.
Constructs
ATHB-2 overexpression and antisense constructs were kindly provided by I. Ruberti and have been described previously (Steindler et al., 1999). FLC∷GUS (Michaels et al., 2005) and FT∷GUS and CO∷GUS (Takada and Goto, 2003) constructs have been described previously. The 35S∷CO and GFP∷CO fusion constructs were created by cloning CO cDNAs into the pEarleyGate203 (Earley et al., 2006) and pEGAD (Cutler et al., 2000) vectors, respectively. 35S∷FLC was created by placing a genomic FLC clone under control of the 35S promoter in pPZP211 (Hajdukiewicz et al., 1994).
RNA Expression Analysis
Semiquantitative PCR (Michaels et al., 2004) and quantitative PCR (Mockler et al., 2004) were performed as described previously. Primers for semiquantitative RT-PCR were as follows: PIL1 (CAACGTAGGCAATCTCTCCTGGA and GCATGAACTTGTGTCTTCGCATC), GI (CTGTCTTTCTCCGTTGTTTCACTGT and TCATTCCGTTCTTCTCTGTTGTTGG), ZTL (GATGAAGAGGGAGGTCTTTTTCC and CCAAGAACAGGTCCAAGGTCAAT), ELF3 (TTCCTTCTCAGAGGTTTGGTGA and AGAGATTACAAAGCCACCTGAC), LKP2 (AGATGAAGTGGCGGAGGATGGAT and GCTCTCCGATTGGTAAAGCAGAA), SOC1 (CTGAGGCATACTAAGGATCG and GAACAAGGTAACCCAATGAA), FT (AGACGTTCTTGATCCGTTTA and GTAGATCTCAGCAAACTCGC), CO (AAACTCTTTCAGCTCCATGACCACTACT and CCATGGATGAAATGTATGCGTTATGGTTA), FKF1 (GTCTTCGAAGTCTTCACTGG and TTCCTCACACTCTCGTTCTT), PIF3 (GGGTTTGGGTTCAAAGAGAAGC and CGACGATCCACAAAACTGATCAGAAG), ATHB-2 (TCAAGGATCCATGATGTTCGAGAAAGACGATCTGGG and GTAAGAGCTCTTAGGACCTAGGACGAAGAGCGTCA), FLC (TTCTCCAAACGTCGCAACGGTCTC and GATTTGTCCAGCAGGTGACATCTC), and UBIQUITIN (UBQ; GATCTTTGCCGGAAAACAATTGGAGGATGGT and CGACTTGTCATTAGAAAGAAAGAGATAACAGG). For quantitative PCR, primers were as follows: CO (CATTAACCATAACGCATACATTTCATC and TCCGGCACAACACCAGTTT), FT (CAACCCTCACCTCCGAGAATAT and TGCCAAAGGTTGTTCCAGTTGT), and ACTIN2 (GCTGAGAGATTCAGATGCCCA and GTGGATTCCAGCAGCTTCCAT). All experiments were replicated at least three times with similar results.
Protein Expression Analysis
Plants were grown in WL under long-day conditions for 17 d. Plants were then either maintained under WL or transferred to FREL for an additional 2 d. All above-ground portions of the plants from WL and FREL were harvested at dusk. Crude preparation of nuclei was conducted using the CelLytic plant nuclei isolation and extraction kit (Sigma) according to the manufacturer's instructions. Proteins were extracted from the nuclei preparation, boiled in 4× SDS sample buffer, fractioned on a 10% SDS-PAGE minigel, and then blotted to polyvinylidene difluoride membrane. The resulting membrane was probed with anti-GFP antibody, reacted with goat anti-rabbit IgG conjugated with horseradish peroxidase, and visualized using ECL western-blotting substrate (Pierce).
Acknowledgments
We thank the C. Walczak lab for the gift of GFP antibody.
This work was supported by the National Science Foundation (grant no. IOB–0447583 to S.D.M.) and the National Institutes of Health (grant no. 1R01GM075060–01 to S.D.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Scott D. Michaels (michaels@indiana.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Bagnall DJ (1993) Light quality and vernalization interact in controlling late flowering in Arabidopsis ecotypes and mutants. Ann Bot (Lond) 71 75–83 [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 [DOI] [PubMed] [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM (2004) Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40 112–119 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24 591–599 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4 469–479 [DOI] [PubMed] [Google Scholar]
- Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423 881–885 [DOI] [PubMed] [Google Scholar]
- Chouard P (1960) Vernalization and its relations to dormancy. Annu Rev Plant Physiol 11 191–238 [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Armbruster U, Tama N, Putterill J (2006) Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 580 1193–1197 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Halliday KJ, Harberd NP, Whitelam GC (1996) The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: novel phytochromes control internode elongation and flowering time. Plant J 10 1127–1134 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Eskins K (1992) Light-quality effects on Arabidopsis development. Red, blue and far-red regulation of flowering and morphology. Physiol Plant 86 439–444 [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Koornneef M, Whitelam GC (1994) Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol 104 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33 875–885 [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751–1754 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim SY, Michaels SD (2006) SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133 4699–4707 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229 57–66 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM (1995) Effect of vernalization, photoperiod and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol 108 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994. a) Isolation of LUMINIDEPENDENS—a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM (1994. b) The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6 903–909 [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43 58–69 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Somerville CR (1990) Effect of light quality and vernalization of late-flowering mutants of Arabidopsis thaliana. Plant Physiol 92 770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S, Amasino R (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA 100 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, et al (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46 292–299 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Putterill J, Laurie R, Macknight R (2004) It's time to flower: the genetic control of flowering time. Bioessays 26 363–373 [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Lucchetti S, Morelli G (1991) A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J 10 1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426 680–683 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schena M, Davis RW (1992) HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA 89 3894–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS Box Gene. A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Igeno MI, Coupland G (1996) Activation of floral meristem identity genes in Arabidopsis. Nature 384 59–62 [DOI] [PubMed] [Google Scholar]
- Smith H (1982) Light quality, photoperception, and plant strategy. Annu Rev Plant Physiol 33 481–518 [Google Scholar]
- Smith H (1995) Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol 46 289–315 [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126 4235–4245 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar FF, Voesenek LA, Van Der Straeten D (2005) Reaching out of the shade. Curr Opin Plant Biol 8 462–468 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Sun X, Ni M (2007) Timing of photoperiodic flowering: light perception and circadian clock. J Integr Plant Biol 49 28–34 [Google Scholar]