Abstract
Thioredoxins (Trxs) constitute a family of small proteins in plants. This family has been extensively characterized in Arabidopsis (Arabidopsis thaliana), which contains six different Trx types: f, m, x, and y in chloroplasts, o in mitochondria, and h mainly in cytosol. A detailed study of this family in the model legume Medicago truncatula, realized here, has established the existence of two isoforms that do not belong to any of the types previously described. As no possible orthologs were further found in either rice (Oryza sativa) or poplar (Populus spp.), these novel isoforms may be specific for legumes. Nevertheless, on the basis of protein sequence and gene structure, they are both related to Trxs m and probably have evolved from Trxs m after the divergence of the higher plant families. They have redox potential values similar to those of the classical Trxs, and one of them can act as a substrate for the M. truncatula NADP-Trx reductase A. However, they differ from classical Trxs in that they possess an atypical putative catalytic site and lack disulfide reductase activity with insulin. Another important feature is the presence in both proteins of an N-terminal extension containing a putative signal peptide that targets them to the endoplasmic reticulum, as demonstrated by their transient expression in fusion with the green fluorescent protein in M. truncatula or Nicotiana benthamiana leaves. According to their pattern of expression, these novel isoforms function specifically in symbiotic interactions in legumes. They were therefore given the name of Trxs s, s for symbiosis.
Thioredoxins (Trxs) are small and ubiquitous proteins with two close and active redox-active Cys residues in a conserved WCG/PPC motif. In their dithiol form, they are powerful disulfide reductases (Holmgren, 1985) that play a posttranslational regulatory role in proteins involved in an ever-increasing number of cellular processes (Buchanan and Balmer, 2005). In plants, Trxs constitute a small protein family. Twenty-two genes have been detected in the fully sequenced Arabidopsis (Arabidopsis thaliana) genome, including two encoding monocysteinic isoforms (WCXXS) first described in this species (Meyer et al., 2002, 2005) but also present in poplar (Populus spp.; Gelhaye et al., 2003, 2004a). In contrast, the genomes of Escherichia coli, Saccharomyces cerevisiae, and Homo sapiens contain only two to three genes, all of which encode bicysteinic isoforms.
The plant Trx family is divided into six different types according to the primary structure and localization of its members: the f, m, x, and y types are in the chloroplasts (Buchanan, 1991; Mestres-Ortega and Meyer, 1999; Lemaire et al., 2003b), the o type is in the mitochondria (Laloi et al., 2001), and the h type (Johnson et al., 1987; Florencio et al., 1988; Rivera-Madrid et al., 1995) is mainly found in the cytosol and the phloem sap (Ishiwatari et al., 1995; Schobert et al., 1998), although members of this type have also recently been demonstrated to be targeted to mitochondria or the extracellular matrix (Gelhaye et al., 2004b; Juarez-Diaz et al., 2006). The h type, which has been further divided in three groups (Gelhaye et al., 2004a), is the largest type with 11 isoforms in Arabidopsis. Trxs h from groups I and II are reduced by NADP-Trx reductases (NTRs), whereas isoforms from group III (Gelhaye et al., 2003) are reduced by either glutathione (GSH) or glutaredoxins, a second family of disulfide reductases related to Trxs.
Despite the existence of numerous isoforms in each type and some redundancy in their functions, some plant Trx isoforms were found to be preferentially involved in interactions of plants with both pathogenic and symbiotic microorganisms. Indeed, a Trx x isoform in tomato (Solanum lycopersicum) plants was shown to render the interaction of tomato with Cladosporium fulvum compatible by decreasing the defense response through its interaction with the cf-9 protein (Rivas et al., 2004). Similarly, a Trx h isoform from Arabidopsis (AtTrx h5), first described to be induced by wounding, exposure to elicitors, and oxidative stress, was then demonstrated to confer sensitivity to victorin (Reichheld et al., 2002; Laloi et al., 2004; Sweat and Wolpert, 2007). Another Trx h isoform was also found to be required for the nodulation process in soybean (Glycine max; Lee et al., 2005).
The Trx family has been most intensively studied in the model species Arabidopsis. However, whether the phylogeny and function of Trxs in other plants are similar to those reported for Arabidopsis remains to be established. Here, we have taken advantage of the growing body of knowledge concerning the genes and genome of the model Medicago truncatula, a galegoid legume, to characterize the Trx family in leguminous plants. In contrast to Arabidopsis, legumes are characterized by symbiotic relationships with both nitrogen-fixing bacteria and mycorrhizal fungi. For that purpose, Trx sequences were searched in EST and genomic databases and compared to those of Arabidopsis. While M. truncatula contains members of each of the Trx types known to be present in Arabidopsis, it also contains two isoforms of a novel type. This novel type, which to our knowledge has not been described previously, contains both an atypical catalytic site and a signal peptide. Because these novel isoforms, described below, seem to be dedicated to symbiotic interactions in legumes, they were given the name of Trxs s, s for symbiosis.
RESULTS
All the Types of Trxs Described for Arabidopsis Are Present in M. truncatula
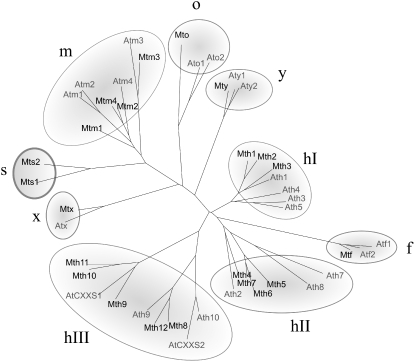
While the sequencing of M. truncatula genome is currently in progress (the release of the complete genome will only be effective by the end of the year 2008), large public EST databases are already available (MtGI, http//www.tigr.org/tdb/mtgi; MtDB, http://www.medicago.org). It is noteworthy that, at its last release (January 19, 2005), MtGI notably contained 226,923 ESTs in 36,878 unique sequences arising from various tissues of plants grown under either optimal or suboptimal conditions and in both the presence and the absence of symbiotic interactions. From these libraries, we were able to identify by homology searches about 80 sequences, tentative consensus (TC) or singletons, related to Trxs, among which 20 sequences of putative Trxs were identified (Table I). Eighteen encode complete open reading frames, while two (a Trx o and a Trx x) are only partially complete. Two additional Trx sequences were encountered in genomic data (Table I). All the Trx sequences were aligned with ClustalW and then compared to those of Arabidopsis in a phylogenetic tree (Fig. 1). All the types of Trxs previously described for Arabidopsis are present in M. truncatula. While some types are represented in M. truncatula by only a single member, others contain multiple isoforms: one Trx f, four Trxs m, one Trx x, one Trx y, one Trx o, and 12 Trxs h. However, M. truncatula also seems to have an additional type of Trx with two isoforms that forms a distinct cluster in the tree (see below).
Table I.
EST, TC, and BAC numbers corresponding to Trx sequences from M. truncatula Jemalong and the names they were given in this work
EST, TC, and BAC numbers were found in MtGI (http://www.tigr.org/tdb/mtgi). BAC, Bacterial artificial chromosome.
| Types | Here Referred to as: | EST and TC Nos. | BAC Nos. |
|---|---|---|---|
| H I | h1 | TC100552 | |
| h2 | TC101267 | AC160013_5 | |
| AC159535_22 | |||
| h3 | CR955005_24 | ||
| H II | h4 | TC93965 | AC151524_26 |
| h5 | TC104708 | CR954189_16 | |
| h6 | TC96215 | CR954189_24 | |
| h7 | TC104047 | CR954189_15 | |
| H III | h8 | TC95944 | AC152552_10 |
| h9 | TC101091 | AC169075_9 | |
| h10 | TC103046 | ||
| h11 | TC96360 | AC144502_13 | |
| h12 | AC174331_22 | ||
| AC151949_11 | |||
| F | f | TC94813 | AC186678_25 |
| M | m1 | TC93953 | |
| m2 | TC102114 | ||
| m3 | TC96118 | AC174368_22 | |
| AC175027_18 | |||
| m4 | TC99811 | ||
| O | o | BF644832 | |
| X | x | TC96299 | AC151824_7 |
| AC157490_16 | |||
| Y | y | TC102372 | AC125473_21 |
| AC126014_25 | |||
| S | s1 | TC102486 | |
| s2 | TC107795 |
Figure 1.
Darwin tree of M. truncatula and Arabidopsis Trx protein sequences. The protein sequences were deduced from nucleic sequences available for each species. The numbers of M. truncatula Trx sequences are indicated in Table I. The numbers of Arabidopsis Trx genes are (in parentheses): h1 (AT3G51030), h2 (AT5G39950), h3 (AT5G42980), h4 (AT1G19730), h5 (AT1G45145), h7 (AT1G59730), h8 (AT1G69880), h9 (AT3G08710), h10 (AT3G56420), CxxS1 (AT2G40790), CxxS2 (AT1G11530), f1 (AT3G02730), f2 (AT5G16400), m1 (AT1G03680), m2 (AT4G03520), m3 (AT2G15570), m4 (AT3G15360), o1 (AT2G35010), o2 (AT1G31020), x (AT1G50320), y1 (AT1G76760), and y2 (AT1G43560).
A Novel Type of Trx Exists in M. truncatula
As mentioned above, two sequences that do not seem to belong to the types of Trxs described so far were found in M. truncatula EST data banks. The deduced sequences of these two proteins share 53% identity and form a distinct cluster in the phylogenetic tree (Fig. 1) that is close to the cluster of isoforms of the m type, which explains their annotation as m type in the databases. As the corresponding genomic sequences are not yet known, further analysis required the cloning of the genes from M. truncatula. As in the case of Arabidopsis Trxs m, the gene sequences have only one intron that separates the regions encoding a putative transit peptide and the rest of the protein (Supplemental Fig. S1). We also cloned their coding regions (DQ121444, DQ121445). The deduced protein sequences are shown in Supplemental Figure S1 and Figure 2. Comparison with the sequence of Trx h2 (DQ121443), a Trx of h type also cloned in our study, reveals that both proteins have an N-terminal extension that may correspond to a transit peptide (Fig. 2). This extension, which is highly similar for the two isoforms (20 out of 27 first amino acids are identical), is nevertheless shorter than the chloroplast target peptides of Trxs m (Fig. 2) and does not contain the charge characteristics required for a chloroplast import. In contrast, according to the predictions of different computer programs (Predotar, SignalP, TargetP, Psort), it could correspond to a signal peptide that targets the proteins to the endoplasmic reticulum (ER). The potential cleavage site of the signal peptide in each isoform sequence is indicated in Supplemental Figure S1. To our knowledge, this is the first example of Trxs harboring a transit peptide of this nature have been encountered. In addition, the two isoforms contain an atypical putative catalytic site, LCSPC or WCGQNC, that differs from that of all previously described Trx types.
Figure 2.
Alignment of Trxs s with Trxs m and Trx h2 from M. truncatula. The protein sequences deduced from the coding regions of Trx s1 (DQ121444) and Trx s2 (DQ121445) were aligned with those of Trx h2 (DQ121443) and Trxs m. Identical and conserved amino acid residues appear in red and blue, respectively. Residues that are identical in several sequences but not all appear in green.
Because of the similarity of the two isoforms to Trxs m (at both the level of gene structure and protein sequence), these isoforms appeared to be bona fide members of the Trx family. Nevertheless, because of the presence in their sequences of both an atypical active site and a predicted signal peptide, they clearly define a novel Trx type. They were given the name of Trxs s for symbiosis, because the corresponding ESTs were present only in symbiotic root libraries of M. truncatula, interacting with Sinorhizobium meliloti (s1, eight ESTs; s2, eight ESTs) or Glomus species (s2, two ESTs). In addition, both isoforms were detected in a recent transcriptomic analysis of nodule development of M. truncatula during symbiosis with S. meliloti (El Yahyaoui et al., 2004) in which they were the sole Trxs that were overexpressed in this situation.
To determine whether Trxs s exist in other plant species, we examined all the nucleic sequences available in databases using the BLAST program. This search did not detect any possible orthologs, even when it was extended to the rice (Oryza sativa) and poplar genomic sequences or to ESTs of the legumes Lotus japonicus and soybean.
Overexpression of Trx s1 and Trx s2 in E. coli and Production of Specific Antibodies
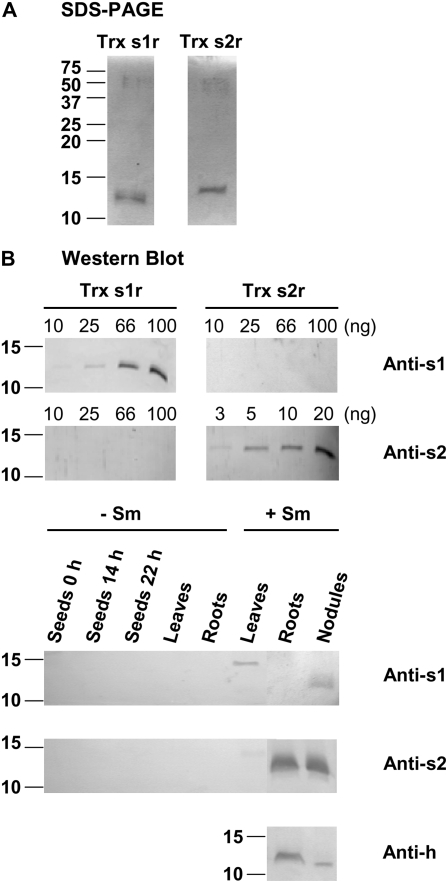
To investigate whether the newly discovered Trxs are functional and expressed proteins, cDNAs encoding the putative mature forms of Trxs s were cloned in pRSF2, a vector designed to produce recombinant proteins in fusion with an N-terminal His-tag. The proteins were overexpressed in E. coli and purified to an apparent homogeneity in two steps consisting of an initial anion-exchange followed by an affinity chromatography on nickel-chelating Sepharose. The His-tag was then cleaved and eliminated. When subjected to SDS-PAGE (Fig. 3A), recombinant Trx s1 (Trx s1r) and Trx s2 (Trx s2r) stained with Coomassie Blue appear as single bands with the expected molecular masses of about 13 and 14 kD, well in agreement with the theoretical masses of 13.099 and 13.998 kD calculated for the Trx s1r and Trx s2r without the transit peptide, respectively.
Figure 3.
Analyses of proteins by SDS-PAGE and western blot. For analyses of recombinant Trxs s, Trx s1r and Trx s2r were overexpressed in fusion with a His-tag in E. coli and purified. After the removal of the His-tag, proteins were resolved by 15% (w/v) SDS-PAGE and stained with Coomassie Blue (A) or transferred onto PVDF membranes (B). For SDS-PAGE analysis, 1 μg of each recombinant protein was loaded per lane. For western blotting, different quantities were loaded per lane ranging from 3 to 100 ng as indicated. For analyses of M. truncatula soluble proteins, they were extracted from whole seeds either dry (0 h) or imbibed for 14 h (before the radicle protrusion) or 22 h (after the radicle protrusion), leaves and roots from 20-d-old nonsymbiotic plants (−Sm), and leaves, roots, and functioning nodules from 20- to 25-d-old plants grown in symbiosis with S. meliloti (+Sm). They were resolved by 15% (w/v) SDS-PAGE and then transferred onto PVDF blots (B). Then 25 μg/lane was loaded for anti-s2 and anti-h blots and 50 μg/lane was loaded for anti-s1 blot. Blots were probed using the alkaline phosphatase assay with a 1:1,000 (v/v) dilution of antibodies raised against either a synthetic peptide derived from Trx s1 (Anti-s1) or the whole Trx s2r (Anti-s2). For anti-h blot, a mix of anti-h antibodies raised against P. sativum Trx h3 and Trx h4 was used at a 1:500 (v/v) dilution. Molecular masses of standard proteins are indicated in kilodaltons at the left. Data are representative of results obtained in three independent experiments.
Then, we used either a specific peptide, in the case of Trx s1, or the recombinant protein, in the case of Trx s2, to raise polyclonal antibodies in rabbit (see “Materials and Methods”). Each antibody was purified using either the corresponding peptide or the recombinant protein immobilized on Sepharose beads before use. The specificity of the purified antibodies was tested by western blotting using recombinant proteins. Figure 3B shows that both antibodies (at a 1/1,000 dilution) were able to react with the protein against which they were raised. Amounts per lane as low as 10 ng and 3 ng could be detected with the anti-s1 and the anti-s2, respectively. Figure 3B also shows that each antibody is specific for the isoform against which it was raised and does not cross-react with the other isoform. Because Trx h isoforms were reported to be abundant in plants, particularly in organs such as seeds, roots, and nodules, we also tested the reactivity of our antibodies against several M. truncatula Trx h recombinant isoforms of groups I, II, and III (h1, h2, h4, h5, h7, h8, and h9) separately produced in our laboratory (M. Renard and F. Montrichard, unpublished data). Anti-s antibodies did not react with any of these isoforms.
Trx s1r Is Reduced by NTR
As mentioned above, Trx s1 and Trx s2 have unusual putative catalytic sites when compared to the WCG/PPC sequence found in other Trxs. However, Trx s1 has a site (LCSPC) in which the two Cys, in positions with the same CXXC spacing found in other Trxs, are likely to be compatible with the formation of an intramolecular disulfide bond. Thus, this isoform may have a disulfide reductase function. In contrast, Trx s2, containing a putative catalytic site with three amino acids between the two Cys residues (WCGQNC), might not display the redox properties characteristic of classical Trxs.
To determine the possible functions of Trxs s, we performed enzymatic assays and compared the results with those obtained with a classical Trx, Trx h2 from M. truncatula. First, we assayed the disulfide reductase activity of Trx s isoforms using the test that is widely performed: the reduction of insulin. In this experiment, dithiothreitol (DTT) was used as a reducing agent for Trxs, and the electron transfer from DTT to insulin, resulting from the reaction sequence DTT → Trx→ insulin, was monitored. Neither of the two Trx s recombinant proteins was found to be active, even at the highest concentration tested (0.25 mg mL−1). In contrast, control experiments performed with Trx h2r produced insulin reduction at a rate comparable to those reported for many other Trxs (results not shown). It is important to point out that other Trxs, such as CDSP32 from potato (Solanum tuberosum; Broin et al., 2002), also do not catalyze insulin reduction despite the fact that these proteins display disulfide reductase activity with other substrates.
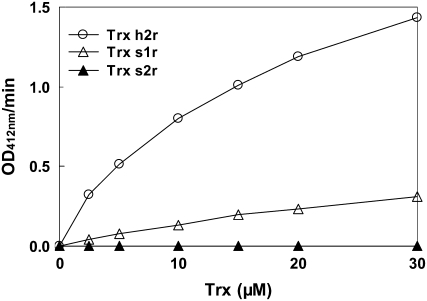
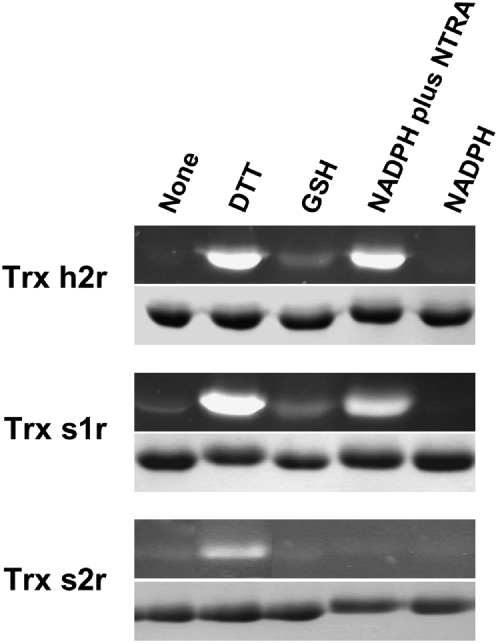
We also tested whether Trxs s could act as substrates for NTRs. M. truncatula NTRA (MtNTRA; Alkhalfioui et al., 2007) was chosen for these experiments, because it had previously been shown that NTRs of the A/B type from Arabidopsis were able to reduce Trxs of several types (Jacquot et al., 1994; Stein et al., 1995; Laloi et al., 2001) and were particularly effective with Trxs of the m type, the closest likely relatives of the s type. The results obtained, when dithiobisnitrobenzoate (DTNB) was used as a final electron acceptor (NADPH → NTRA → Trx s → DTNB), are shown in Figure 4. This figure shows that Trx s1r and Trx h2r, used as a control, could be reduced by NADPH in the presence of MtNTRA. In contrast, no reduction of DTNB was obtained with Trx s2r under these conditions. With Trx s1r, the reductase exhibits an apparent Km of 13.8 ± 2.1 μm with a kcat of 0.27 ± 0.02 s−1. The Km value obtained is similar to that calculated for Trx h2r (13.0 ± 1.1 μm) and is in the range of those determined for NTR and Trxs h and m from Arabidopsis and Chlamydomonas reinhardtii (Rivera-Madrid et al., 1995; Stein et al., 1995). However, the kcat value determined for Trx s1r is 5-fold lower than that measured for Trx h2r (1.47 ± 0.05 s−1), indicating that MtNTRA reduces Trx s1r less efficiently than Trx h2r. Of course, as Trx h2 may be a physiological substrate of the enzyme while Trx s1 might not be, the lower reduction efficiency of Trx s1r is not unexpected. Reduction of Trxs s in the presence of NADPH and MtNTRA was also assayed using the fluorescent probe monobromobimane (mBBr). For this purpose, recombinant Trxs s were incubated in the presence of MtNTRA plus NADPH for 15 min, after which any SH groups formed during this time were labeled with mBBr. Proteins were then resolved by SDS-PAGE, and the gel was observed under UV light. In parallel, the action of GSH and DTT was also tested. Figure 5 shows that both recombinant Trxs s and Trx h2r were almost fully oxidized in their native state. The incubation in the presence of DTT (lane DTT) has led to their reduction, although Trx s2r reduction could only be obtained when the concentration of this compound was increased to 20 mm. GSH was also able to reduce Trx s1r and Trx h2r (lane GSH), but to a lower extent than DTT. The figure further shows that MtNTRA in the presence of NADPH (lane NTRA plus NADPH) reduced Trx s1r and Trx h2r but not Trx s2r. As expected, in control experiments, NADPH alone (lane NADPH) was without effect on Trx s1r and Trx h2r. These results, showing that Trx s1r can act as a substrate for MtNTRA while Trx s2r cannot, are consistent with those described just above.
Figure 4.
Disulfide reductase activity of Trxs determined by the DTNB reduction assay. Different concentrations of Trxs were incubated in the presence of 1.6 μm MtNTRA, 0.2 mm NADPH, and 100 μm DTNB in 30 mm Tris-Cl, pH 8. DTNB reduction was followed at 412 nm.
Figure 5.
Reduction of Trx h2r, Trx s1r, and Trx s2r by NTR plus NADPH or other reducing agents. Trxs were incubated in the absence (None) or the presence of DTT (2.5 mm for Trx h2r and Trx s1r or 20 mm for Trx s2r), 5 mm GSH, 0.2 mm NADPH plus 1 μm MtNTRA, or 0.2 mm NADPH alone, during 15 min at RT, after which any SH groups formed during this time were labeled with 5 mm mBBr. Proteins were finally resolved by 15% (w/v) SDS-PAGE and observed under UV light (black segments) before staining with Coomassie Blue (gray segments).
The redox midpoint potentials (Em) of both recombinant Trx s isoforms were measured using redox posing with DTT redox buffers and mBBr detection of the reduced, dithiol state (Hirasawa et al., 1999). The titrations gave excellent fits to the Nernst equation for a two-electron redox couple and were independent of the redox equilibration times and total DTT concentrations used, as expected for equilibrium processes. At pH 7.0, Em values of −285 ± 10 and −315 ± 10 mV were determined for Trx s1r and Trx s2r, respectively. These values are in the range of those reported for Trxs of the m type (Hirasawa et al., 1999; Collin et al., 2003) and Trxs in general. These values are significantly lower than that for the GSSG/GSH couple (−245 mV at pH 7.0), which probably explains why they are only partially reduced by GSH but are fully reduced by the thermodynamically more potent reductant, DTT (Em = −330 mV at pH 7.0). The difference of 30 mV in the Em values of the two Trx s recombinant isoforms may also explain, at least in part, why a higher concentration of DTT is required to reduce Trx s2r. It should also be pointed out that the inability of MtNTRA to catalyze the reduction of Trx s2r by NADPH cannot be attributed to the fact that it has a more negative Em value than Trx s1r, as the NADP+/NADPH couple (Em = −320 mV at pH 7.0) is capable of reducing Trx s2r in a thermodynamically favorable reaction. Thus, the absence of Trx s2r reduction in this assay must arise instead from protein/protein recognition events between this NTR and Trx s2.
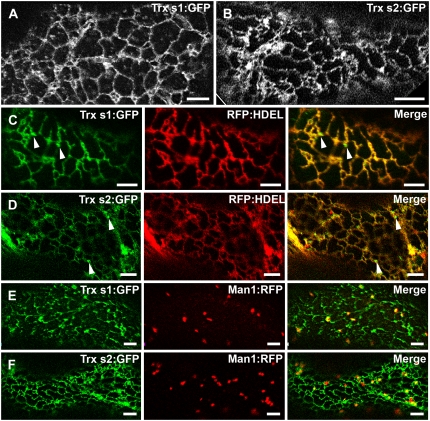
Trxs s Are Targeted to the ER
To determine whether the N-terminal sequence of Trxs s could act as a signal peptide that targets the proteins to the ER, as predicted from our bioinformatic analysis, transient 35S promoter-driven expression of Trx s1 and Trx s2, C terminally fused to GFP (35S:Trx s:GFP), was performed in M. truncatula and Nicotiana benthamiana. In agro-infiltrated M. truncatula leaf epidermal cells (Fig. 6A), Trx s1:GFP displayed a reticulated pattern very reminiscent to the ER (Yang et al., 2005; Thomas et al., 2008). The same pattern was observed in N. benthamiana (Supplemental Fig. S2). For unknown reasons, expression of Trx s2:GFP in M. truncatula leaf epidermal cells under identical experimental conditions as for Trx s1:GFP resulted in no detectable GFP fluorescence (data not shown). However, when expressed in N. benthamiana leaves, Trx s2:GFP displayed an ER-like pattern (Fig. 6B) very similar to the one observed for Trx s1:GFP (Fig. 6A; Supplemental Fig. S2). The ER localization of both Trx s1:GFP and Trx s2:GFP was confirmed upon coexpression with the modified red fluorescent protein (RFP):HDEL, an ER-targeted marker protein as described in Yang et al. (2005). Apart from some punctate structures (Fig. 6, C and D, arrowheads) that were visible with both Trx s1:GFP and Trx s2:GFP, nearly perfect colocalization between Trxs s:GFP and RFP:HDEL was observed, indicating that Trxs s accumulate in the ER. To determine whether the small punctuate structures could correspond to individual Golgi stacks, Trxs fused to GFP were coexpressed with Man1:RFP, a Golgi resident protein (Nebenführ et al., 1999). There, only very moderate colocalization, if any, was detected between the two proteins (Fig. 6, E and F), possibly as a consequence of the overexpression of the proteins due to transient expression. It is therefore concluded that Trxs s are not bona fide Golgi resident proteins.
Figure 6.
Visualization of Trxs s:GFP in M. truncatula and N. benthamiana leaf epidermal cells. A, ER-like fluorescence pattern resulting from Trx s1:GFP expression in M. truncatula. B, ER-like fluorescence pattern resulting from Trx s2:GFP expression in N. benthamiana. C and D, Coexpression of Trx s1:GFP and Trx s2:GFP, respectively, with the ER marker RFP:HDEL. In cells coexpressing Trxs s:GFP and RFP:HDEL, near perfect colocalization is observed. Punctuate structures in the ER vicinity are indicated by arrowheads. E and F, Coexpression of Trx s1:GFP and Trx s2:GFP, respectively, with the Golgi marker Man1:RFP. Only very moderate colocalization between Trxs s:GFP and Man1:RFP is observed. C and E, M. truncatula leaf epidermal cells. D and F, N. benthamiana leaf epidermal cells. Scale bars = 5 μm.
To make sure that the fluorescent proteins detected in leaves corresponded to Trxs fused to GFP and not to GFP alone, agro-infiltrated leaf samples were collected and analyzed by immunoblotting using either anti-GFP or anti-s antibodies. As anticipated, bands corresponding to the expected size (about 40 kD) for Trx s1 and Trx s2 fused to GFP were detected in all cases (Supplemental Fig. S3).
Trxs s Seem to Be Dedicated to Symbiosis
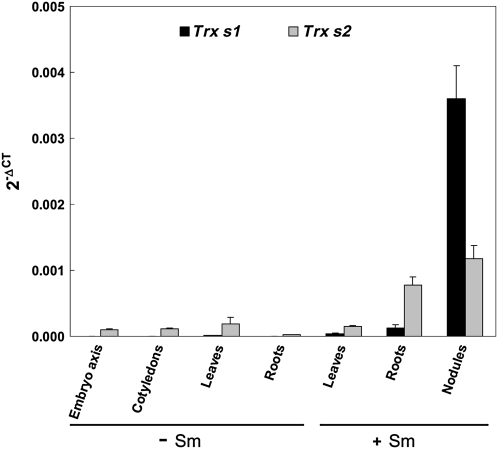
As mentioned above, ESTs and transcriptomic data suggest that expression of Trxs s seems to be linked, at least at the nucleic level, to symbiosis (El Yahyaoui et al., 2004). To provide additional evidence for this expression pattern, we performed an analysis of their expression by quantitative reverse transcription (RT)-PCR in different organs: seeds, leaves, and roots from nonsymbiotic plants and leaves, roots, and functioning nodules from plants grown in symbiosis with S. meliloti. The results obtained were expressed as 2−ΔCT, using mean values obtained with two constitutive genes, Msc27 and 18S RNA (Montrichard et al., 2003; Bouton et al., 2005). As expected Trx s1 and Trx s2 were found to be highly expressed in nodules, transcripts of Trx s1 being notably very abundant in these organs (Fig. 7). Both transcripts are also abundant in symbiotic roots, while they are present only at a basal level in leaves of symbiotic leaves and in roots and leaves from uninoculated plants. This basal level of Trx s gene expression is consistent with the fact that both of the Trx s sequences used in this study were cloned from nonsymbiotic roots by RT-PCR. When compared with the basal level found in roots from uninoculated plants, the respective abundance of the messengers of Trx s1 and Trx s2 are about 80- and 30-fold more abundant in symbiotic roots and 2,300- and 40-fold more abundant in nodules. Because the young radicle of germinating seeds is well known to prepare the plant for symbiosis, embryo axis and cotyledons dissected from dry and germinating seeds imbibed for 14 h (i.e. just before the radicle protrusion) or 22 h (after the radicle protrusion) were also analyzed. A basal expression of both genes was detected, independent of the part of the seeds considered, even in the radicle of seeds that had germinated (Fig. 7).
Figure 7.
Trx s1 and Trx s2 gene expression. Quantitative RT-PCR experiments were conducted using total RNAs extracted from different organs: embryo axis and cotyledons of germinated seeds (imbibed for 22 h), leaves, and roots from 20-d-old nonsymbiotic plants (−Sm), and leaves, roots, and functioning nodules from 20- to 25-d-old plants grown in symbiosis with S. meliloti (+Sm). Amplification of the sequences of two constitutive genes (18S RNA and Msc27) was followed using Sybr Green. Amplification of Trx s genes was monitored with fluorescent Taqman probes. The results obtained were then expressed as 2−ΔCT ± sd (ΔCT = CT of the gene of interest – mean of CTs of the constitutive genes). Data are representative of results obtained in three independent experiments.
To investigate whether the abundance of the corresponding proteins was correlated to the transcript accumulation during symbiosis, western-blot analyses were performed using protein extracts prepared from the same organs and the specific antibodies raised against Trx s1 and Trx s2 (Fig. 3B). For 50 μg of protein extracts loaded per lane, a faint band of approximately 13 kD was visible after 30 min to 1.5 h of staining using anti-s1 (1/1,000) in nodule extracts but not in other extracts examined (Fig. 3B). Thus, Trx s1 seems to be present only in nodules in a quantity that was estimated to be around ≤10 ng/50 μg protein by comparison with the results obtained with pure recombinant proteins. By contrast, for 25 μg of protein extracts loaded per lane, an intense band with a molecular mass of about 14 kD was observed in both lanes of symbiotic roots and nodules after only 1 to 2 min of staining using anti-s2 (1/1,000). Thus, Trx s2 is very abundant in these organs, in amounts >50 ng/25 μg protein. It is noted that, whatever the antibody used, a band with a mass of about 15 kD was also detected in leaf extracts from symbiotic plants. Although this band is not stained in lanes corresponding to leaf extracts from uninoculated plants on the blot shown in Figure 3B, this band was often stained in other blots not shown here. This band probably results from a nonspecific reaction of a protein present in leaf extracts with anti-s antibodies.
Interestingly, the molecular masses of the proteins detected in symbiotic extracts are lower than those calculated from the full-length sequences with the transit peptide, i.e. about 16 and 16.7 kD for Trx s1 and Trx s2, respectively. In fact, the masses are identical to those of the recombinant proteins alone (Fig. 3B). These results, consistent with the removal of the signal peptide in vivo, not only provide additional evidence that this novel type of Trx s is imported in the ER but also indicate that mature proteins may be soluble in this compartment, as the cleaved signal peptide is the only hydrophobic region seen in their sequences.
These results, showing that during symbiosis, Trx s1 is present in nodules while Trx s2 accumulates in both roots and nodules, correlate well with those obtained from quantitative RT-PCR and suggest a regulation of the expression of the genes of Trxs s at a transcriptional level.
One isoform of Trx h was reported to be abundant in symbiotic nodules of soybean (Lee et al., 2005). To see whether Trxs s are the sole Trxs accumulated in M. truncatula nodules or whether they are accumulated together with a Trx of h type as found in soybean, we performed western-blot analyses using a mix of anti-h antibodies raised against Pisum sativum Trx h3 and Trx h4 (Montrichard et al., 2003), antibodies that do not cross-react with recombinant Trxs s. We detected an intense band of protein with a molecular mass of 12 to 13 kD in both lanes of symbiotic roots and nodule extracts (Fig. 3B). Thus, it seems that both Trx s and Trx h isoforms are abundant in nodules of M. truncatula. As there is a slight difference in the apparent molecular mass of the bands of Trx h detected in roots and nodules, it is not yet clear whether they correspond to the same isoform.
DISCUSSION
We present here an overview of the Trx isoforms present in M. truncatula, highlighting that M. truncatula exhibits a novel type of Trx, with two different isoforms, in addition to all the types of Trxs previously described for Arabidopsis (Meyer et al., 2002, 2005). These two novel isoforms indeed belong to the Trx family. This classification is supported by their primary sequences, which are similar to those of Trxs, most notably to those of Trxs m (at both protein and gene levels) and by their redox potential values that are close to those of classical Trxs. Although neither of the two isoforms is able to reduce insulin, the fact that one of them can act as a substrate for MtNTRA provides additional support for their classification as members of the Trx family. However, because of the presence of an atypical catalytic site and of a signal peptide not previously found associated with Trxs, these two isoforms clearly belong to a novel type in this family. This is the first time, to our knowledge, that Trxs containing a signal peptide are described, although the presence of Trx h in the ER had been proposed previously (Marcus et al., 1991). Because the novel isoforms were found to be associated with symbiosis, they were given the name s for symbiosis: Trx s1 and Trx s2.
No orthologs were found in the genomes of Arabidopsis, rice, and poplar. Thus, it may be possible that Trxs s are specific to legume plants. However, no orthologs were found in ESTs from L. japonicus or soybean, two other model legumes, or P. sativum. The fact that no orthologs were detected in soybean is particularly surprising given that the soybean database from The Institute for Genomic Research contains 330,436 ESTs. This could be due to the relative low representation of ESTs from symbiotic roots in this database compared to M. truncatula. Alternatively, Trxs s could be unique to galegoid legumes that, like M. truncatula but unlike soybean, make indeterminate nodules. This suggests that a type of Trx could be specific to a plant family or class. Thus, the types f, m, x, y, o, and h that are common to Arabidopsis and M. truncatula and were also found to be present in cereals (Meyer et al., 2006) and the green alga C. reinhardtii (Lemaire et al., 2003a) are likely among the more ancient Trx types. In contrast, the s type that is present in M. truncatula but not in Arabidopsis, rice, or poplar may be a more recent type. The s type has probably evolved from the m type after the divergence of higher plant families. Later, a duplication event of the Trx s gene ancestor may have led to precursors of Trx s1 and Trx s2. Even later, an insertion in the nucleic sequence corresponding to Trx s2 may have occurred, leading to the appearance of a third amino acid in the catalytic site of Trx s2. The function of an isoform with this unique putative catalytic site remains to be elucidated.
In this study, we also demonstrated that this novel Trx type is associated with symbiosis. The regulation of expression of both genes during interaction of M. truncatula with S. meliloti occurs at the transcriptional level. However, Trx s isoforms show different patterns of accumulation either in nodules (Trx s1) or both roots and nodules (Trx s2), suggesting that they play different roles. Moreover, as suggested by the analysis of EST data, while Trx s1 may be specific of this symbiotic interaction, Trx s2 may also play a role in symbiosis with Glomus species. It is noted that high levels of reactive oxygen species are produced during the early step of the plant-symbiont interaction. Thus, in the case of Trx s2, one cannot rule out that it could be also functional in root tissue under other oxidative stress conditions.
We were also able to demonstrate, by transient expression of proteins in N. benthamiana or M. truncatula leaves, that Trxs s can be imported in the ER. This suggests that in symbiotic roots or/and nodules, Trxs s are primarily targeted to this compartment. However, their mechanism of retention in the ER is not known, because they do not harbor the C-terminal ER retention motif H/KDEL. It is noted that the signal peptide of Trxs s is highly conserved between the two isoforms, as is also the case for a family of small and Cys-rich proteins unique to galegoid legumes that are only expressed during nodulation (Mergaert et al., 2003). These Cys-rich proteins can be secreted thanks to a specific signal peptide peptidase that is also accumulated only during nodulation. Although Trxs s have a different signal peptide than proteins of this family, they may be secreted during symbiosis by a similar mechanism. The lack of the specific signal peptidase in plant leaves used for transient expression of Trxs s may have led to their accumulation and retention in the ER. It would be interesting to determine the localization of fusion proteins transiently expressed in symbiotic roots and nodules of M. truncatula, and experiments designed to provide this information are currently in progress.
This is not the first time that Trxs have been reported to be implicated in biotic interactions. Trxs as antioxidant agents are generally thought to be involved in scavenging of the reactive oxygen species that are produced at high levels under these conditions. It may be possible that Trxs s in M. truncatula symbiotic organs play a similar role. The Trxs s may also be involved in maintaining proteins that play important roles in symbiosis in the reduced state. For this reason, it would be interesting to identify their reductase and targets. However, Trxs s may also be involved more directly in the biotic interactions. Indeed, an important role of a Trx h in self-incompatibility has been reported in Brassica (Bower et al., 1996), and a Trx x isoform was shown to render the interaction of tomato with C. fulvum compatible by decreasing the defense response (Rivas et al., 2004; Nekrasov et al., 2006). Similarly, a Trx h isoform was demonstrated to confer sensitivity of Arabidopsis toward the toxin victorin (Sweat and Wolpert, 2007), while another Trx h isoform was found to be involved in the nodulation process of soybean, with plants deficient in this isoform having a reduced number of nodules (Lee et al., 2005). Thus, Trxs s may also play a role in the compatible interaction of M. truncatula with its symbionts. Interestingly, in the biotic interactions mentioned above, the disulfure reductase activity of the Trx isoforms was not required. This raises the possibility that the role of Trxs s in symbiosis may also not involve redox activity. This question, as well as other important aspects about the function of Trxs s in M. truncatula symbiosis, await investigation using mutants of Trxs s.
It is noteworthy that the Trxs of type s found in M. truncatula are clearly unique when compared with members of other types having a role in biotic interactions in that they are accumulated during symbiosis and are not synthesized under other situations. Nevertheless, Trxs s are not the sole isoforms present in symbiotic roots and nodules. They are accumulated together with one or more Trx h isoforms that may be related to the isoform reported to play a role in soybean nodulation (Lee et al., 2005). A key question for future research is then to determine the exact role of each type of Trx in symbiosis.
MATERIALS AND METHODS
Materials
Surface-sterilized seeds of Medicago truncatula (Paraggio and Jemalong A17) were imbibed on filter paper (Whatman no. 1) in a petri dish (9-cm diameter) soaked with 3.5 mL of distilled water and allowed to germinate at 20°C in the dark for 48 h. Some of the seedlings were then transferred to a mix of sand and vermiculite, and the other seedlings were transferred to soil. Plants were allowed to grow under a 16-h-light/8-h-dark photoperiod with regular watering. Symbiotic plants were obtained by inoculation of a part of 4-d-old seedlings transferred in sand with a fresh suspension of Sinorhizobium meliloti strain RCR2011 (300 μL of bacterial suspension with optical density at 600 nm of 0.1/plant). Leaves, roots, and functioning nodules (when present) were harvested after 20 to 25 d of growth. Seeds were obtained from plants that were allowed to develop in soil, generally about 3 months after the transfer of seedlings.
Methods
DNA Cloning and Overexpression of Recombinant Proteins
Genomic DNA was extracted from leaves of 13-d-old plants of M. truncatula Paraggio using the DNeasy plant mini kit (Qiagen). Sequences of Trx s isoforms were amplified by PCR with FideliTaq DNA polymerase (USB) using primers ORFsens and ORFanti during the course of cycles described in Table II.
Table II.
Primers and probes used in PCR experiments
The primers used for DNA cloning may be constituted of two distinct parts. The part of the sequence that appears in uppercase is specific, while the part that appears in lowercase was added to allow the insertion of the PCR product in the appropriate vector (pRSF2 or pDON207) according to the manufacturer's instructions (Novagen and Invitrogen, respectively). The genomic sequences of Trx s1 and Trx s2 were amplified using primers ORFsens and ORFanti, and leaf genomic DNA. The whole coding regions of Trx s1 and Trx s2 were amplified using the same primers, while the parts corresponding only to putative mature forms of the proteins were amplified with primers MATsens and ORFanti, using cDNAs reverse transcribed from root RNAs. After a preliminary denaturation step of 5 min at 94°C, DNA amplification was performed during five cycles (94°C × 1 min; annealing temperature of the specific part of the primers × 1 min; 72°C × 1 min) and 30 cycles (94°C × 1 min; 72°C × 1 min). Then, a final elongation step was realized for 10 min at 72°C. For quantitative RT-PCR, the amplification cycles in all cases consisted in a preliminary step of 5 min at 94°C, followed by 40 cycles of 94°C × 15 s and 60°C × 1 min.
| Primer Name | Sequence |
|---|---|
| Primers used for DNA cloning | |
| S1 ORFsens | gacgacgacaagATGACTACCGTCACAATAAC |
| S1 MATsens | gacgacgacaagATGCAACTCCTCAACGCTGC |
| S1 ORFanti | gaggagaagcccggtTCATAGGTTTTGTTCCATAC |
| S2 ORFsens | gacgacgacaagATGGCCACCGTCACATTA |
| S2 MATsens | gacgacgacaagATGACCGTACAACTCGAATCCT |
| S2 ORFanti | gaggagaagcccggtTCATATTGATAGTTGAATAAG |
| H2 ORFsens | gacgacgacaagATGGCAGCTGAAGAAGGA |
| H2 ORFanti | gaggagaagcccggtTCAAGCAGTAGCAACAGTT |
| AttB1s1sens | ggggacaagtttgtacaaaaaagcaggcttcgaaggagatagaaccATGACTACCGTCACAATA |
| AttB2s1anti | ggggaccactttgtacaagaaagctgggtctAGGTTTTGTTCCATACG |
| AttB1s2sens | ggggacaagtttgtacaaaaaagcaggcttcgaaggagatagaaccATGGCCACCGTCACATT |
| AttB2s2anti | ggggaccactttgtacaagaaagctgggtctATTGATAGTTGAATAAGTTC |
| Primers and probes used for quantitative RT-PCR | |
| S1TaqMsens | CGCTGCCACTCAAGATGCT |
| S1TaqManti | GAGTTAAGGACAAGGGAACCAAAA |
| S1TaqMprobe | FAM-CCAATGGAGTGGCTTATGTTACCGATGAAA-TAMRA |
| S2TaqMsens | CAGTGCCGCCGATGCT |
| S2TaqManti | GGGACGAAGGAACCAAAAGTT |
| S2TaqMprobe | FAM-CCGATGGAGTGGCTCCTGTTACTGATG-TAMRA |
| Qti-ARN18SFor | ACTGCGAAAGCATTTGCCAA |
| Qti-ARN18SRev | CGGCATCGTTTATGGTTGAGAC |
For cDNA cloning, total RNAs from roots of 20-d-old plants of M. truncatula Paraggio were extracted with the RNeasy plant kit (Qiagen) and reverse transcribed using the Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's instructions. Complete Trx coding regions or regions corresponding to the putative mature proteins were amplified by PCR using KOD HiFi DNA polymerase (Novagen) with primers (ORFsens or Matsens and ORFanti) and cycles indicated in Table II. The PCR products were resolved in 1.4% agarose gels, and the amplicons of expected size were excised from the gels, purified with the Qiaquick gel extraction kit (Qiagen), and inserted into the plasmid pRSF2 of the ligation-independent Ek/LIC cloning kit (Novagen). This vector was engineered to express target proteins fused to a N-terminal His-tag. Recombinant plasmids were introduced in Nova Blue Escherichia coli strain for multiplication. When appropriate, they were subsequently transferred into BL21 (pLysS) cells for the production of proteins. Protein purification on Q-Sepharose and on Ni2+-chelating Sepharose (Amersham) was carried out according to the manufacturer's instructions. The His-tag was cleaved and removed using the Enterokinase cleavage-capture kit (Novagen) according the manufacturer's instructions.
Quantitative RT-PCR
Total RNAs from different organs of M. truncatula Jemalong (dry and germinating seeds, leaves, roots, and nodules) were extracted and reverse transcribed as above for quantitative RT-PCR experiments. For that purpose, 0.2 to 2 μL of cDNAs obtained from 10 μg of total RNAs retro-transcribed in a total volume of 120 μL were used as templates using forward and reverse primers (Table II) in a total volume of 25 μL. Amplification of the sequence of the constitutive genes (18S RNA and Msc27) was followed using Sybr Green in the presence of 0.3 μm of each primer and 12.5 μL of 2× Sybr Green master mix (Applera). Amplification of the sequences of Trx s isoforms was followed using fluorescent Taqman probes in the presence of 1 μm of each primer and 2× Taqman master mix (Applera). The amplification cycles in all cases consisted of a preliminary step of 5 min at 94°C, followed by 40 cycles of 94°C × 15 s and 60°C × 1 min (ABI Prism 7000 SDS; Applied Biosystems). The results were expressed as 2−ΔCT (ΔCT = CT of gene of interest – mean of CTs of constitutive genes) ± sd (n = 6–18; three independent experiments).
Western-Blot Analysis
For soluble protein extraction, dry or germinating seeds, leaves, and roots from 20- to 25-d-old plants grown either under sterile conditions or within symbiosis interaction with S. meliloti, and functioning nodules were ground in 50 mm Tris-HCl, pH 7.8, 1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride (1–5 mL/g fresh weight). The resulting homogenates were centrifuged (50,000g for 20 min at 4°C), and the soluble proteins were stored at −20°C. For western-blot analysis, proteins (25–50 μg/lane) were resolved by 15% SDS-PAGE (Laemmli, 1970) and transferred onto polyvinylidene difluoride (PVDF) membranes as previously described (Duval et al., 2002). For the analysis of transient expression of Trxs fused to GFP in leaves of Nicotiana benthamiana or M. truncatula, proteins were extracted by grounding a disc (5-mm diameter) of a transformed leaf in the presence of 80 μL of Laemmli loading buffer ×2. Then, 10 to 30 μL of supernatant obtained after centrifugation (14,000 rpm; 1 h) was loaded per lane.
Two rabbit polyclonal antibodies were raised against a synthetic peptide (VDDNQLIPSKYGIKGIPNV) derived from Trx s1 and the recombinant Trx s2r (Davids Biotechnologie). Then they were respectively purified by affinity chromatography on the peptide and the recombinant protein bound to activated CNBr-Sepharose. Membranes were probed with 1:1,000 (v/v) dilutions of purified antibodies. Immunodetection was performed using the phosphatase alkaline assay in the presence of a mixture of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (Sigma).
Enzymatic Assays
Trx activity was measured using the insulin or DTNB reduction assays as previously described (Holmgren, 1979; Jacquot et al., 1995) using either DTT or NADPH plus recombinant MtNTRA (Alkhalfioui et al., 2007) as a reductant.
The ability of different reducing agents (DTT and GSH) or NADPH plus NTR to reduce Trxs was tested by incubating Trxs in the presence of these compounds followed by a treatment with mBBr, a fluorescent probe that labels SH. For that purpose, 100 μg of Trx in 20 mm KH2PO4 buffer, pH 7.2, was incubated in the absence or the presence of 2.5 or 20 mm DTT, 5 mm GSH, 0.2 mm NADPH alone or 0.2 mm NADPH plus 1 μm MtNTRA for 15 min, followed by addition of 5 mm mBBr. After an additional incubation period of 15 min, the proteins were precipitated by adding 5 volumes of acetone that had been prechilled at −20°C. After 2 h at −20°C, the proteins were resuspended in phosphate buffer and resolved in 15% SDS-PAGE (2 μg/lane). The fluorescent proteins were observed under UV light before the staining of proteins with Coomassie Blue.
Determination of Redox Potential
The midpoint redox potential of each Trx s was determined according to Hirasawa et al. (1999). Briefly, oxidation-reduction titrations were carried out using mBBr-labeling to monitor the dithiol/disulfide redox state of the Trxs after poising samples at defined ambient redox potentials (Eh) generated using different ratios of reduced to oxidized DTT in 100 mm Tricine-NaOH buffer, pH 7.0. The titrations were carried out at ambient temperature. The data was fitted to the Nernst Equation for a single two-electron redox couple as described previously (Hirasawa et al., 1999). Em values were independent of the redox equilibration time over a range from 1.5 to 3.0 h and independent of the total DTT concentration over a range from 1.0 to 2.5 mm.
Determination of Subcellular Localization
Transient expression of Trxs in fusion to GFP in M. truncatula or N. benthamiana leaves was used to determine the subcellular localization of Trxs s. For that purpose, constructs were made using Gateway cloning technology (Invitrogen) according to the manufacturer's instructions. The cDNA corresponding to the two Trx s isoforms were amplified by PCR using recombinant pRSF2 plasmids with “AttB” primers (Table II) that provided for the addition of attB recombination sites and then cloned into the pDON207 entry vector using the Gateway BP Clonase enzyme mix. The constructs were checked by DNA sequencing (MWG-Biotech). For expression of the recombinant Trxs fused to GFP, the cDNAs were transferred using the Gateway LR Clonase enzyme mix into the Destination vector pK7FWG2, allowing a constitutive transcription of the gene of interest under the control of a cauliflower mosaic virus 35 S promoter (35S:Trx:GFP). These constructs were checked by restriction digest analysis. Recombinant plasmids were then transferred in Agrobacterium tumefasciens (strains GV3101 or pMP90). Finally, positive clones grown in Luria-Bertani medium supplemented with spectinomycin were resuspended in water, and suspensions with optical density of 0.2 to 0.5 at 650 nm were used to infiltrate leaves of M. truncatula or N. benthamiana. Three to 4 d later, cortical regions of epidermal cells in discs of infiltrated leaves were observed by confocal microscopy.
Determination of Protein Concentration
Protein contents in extracts were determined according to Bradford (1976) using bovine serum albumin as a standard. The concentrations of recombinant Trxs s in purified fractions were determined using their theoretical absorption coefficients at 280 nm: 3,960 and 16,055 m−1 cm−1 for Trx s1 and Trx s2, respectively.
Accession Numbers
The nucleotide sequences of Trx h2, Trx s1, and Trx s2 from M. truncatula reported in this article have been submitted to GenBank under accession numbers DQ121443, DQ121444, and DQ121445, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic sequences and coding regions of Trxs s cloned from M. truncatula.
Supplemental Figure S2. ER-like fluorescence pattern resulting from Trx s1:GFP expression in N. benthamiana epidermal leaf cell.
Supplemental Figure S3. Detection on western blots of Trxs fused to GFP in leaves of N. benthamiana overexpressing s1-GFP (lane 1) and s2-GFP (lane 2) using antibodies raised against Trx s1 or Trx s2 (left) or GFP (right).
Supplementary Material
Acknowledgments
We thank R. Tsien for mRFP1 plasmid, A. Nebenführ for ManI:Tdtomato, and V. Gomord for RFP:HDEL, Institut de Biologie Moléculaire des Plantes, Strasbourg, France.
This work was supported by the “Contrat Etat Region des Pays de la Loire” (2000–2006), by the “Conseil Général de Maine et Loire” (fellowship to F.A.), and by the Robert A. Welch Foundation (grant no. D–0710 to D.B.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Françoise Montrichard (francoise.montrichard@univ-angers.fr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alkhalfioui F, Renard M, Montrichard F (2007) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58 969–978 [DOI] [PubMed] [Google Scholar]
- Bouton S, Viau L, Lelievre E, Limami AM (2005) A gene encoding a protein with a proline-rich domain (MtPPRD1), revealed by suppressive subtractive hybridization (SSH), is specifically expressed in the Medicago truncatula embryo axis during germination. J Exp Bot 56 825–832 [DOI] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR (1996) Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantity of protein utilizing the principle of protein dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Broin M, Cuine S, Eymery F, Rey P (2002) The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys 288 1–9 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278 23747–23752 [DOI] [PubMed] [Google Scholar]
- Duval F, Renard M, Jaquinod M, Biou V, Montrichard F, Macherel D (2002) Differential expression and functional analysis of three calmodulin isoforms in germinating pea (Pisum sativum L.) seeds. Plant J 32 481–493 [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florencio FJ, Yee BC, Johnson TC, Buchanan BB (1988) An NADP/thioredoxin system in leaves: purification and characterization of NADP-thioredoxin reductase and thioredoxin h from spinach. Arch Biochem Biophys 266 496–507 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Gérard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hiroshawa M, Knaff DB, et al (2004. b) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA 101 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555 443–448 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004. a) The thioredoxin h system of higher plants. Plant Physiol Biochem 42 265–271 [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schurmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38 5200–5205 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254 9627–9632 [PubMed] [Google Scholar]
- Holmgren A (1985) Thioredoxin. Annu Rev Biochem 54 237–271 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195 456–463 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Issakidis E, Decottignies P, Lemaire M, Miginiac-Maslow M (1995) Analysis and manipulation of target enzymes for thioredoxin control. Methods Enzymol 252 240–252 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Rivera-Madrid R, Marinho P, Kollarova M, Le Marechal P, Miginiac-Maslow M, Meyer Y (1994) Arabidopsis thaliana NAPHP thioredoxin reductase. cDNA characterization and expression of the recombinant protein in Escherichia coli. J Mol Biol 235 1357–1363 [DOI] [PubMed] [Google Scholar]
- Johnson TC, Cao RQ, Kung JE, Buchanan BB (1987) Thioredoxin and NADP-thioredoxin reductase from cultured carrot cells. Planta 171 321–331 [PubMed] [Google Scholar]
- Juarez-Diaz JA, McClure B, Vazquez-Santana S, Guevara-Garcia A, Leon-Mejia P, Marquez-Guzman J, Cruz-Garcia F (2006) A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNase in vitro. J Biol Chem 281 3418–3424 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA 98 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Shin KH, Kim YK, Suh JY, Gu YY, Kim MR, Hur YS, Son O, Kim JS, Song E, et al (2005) Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol 139 1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Collin V, Keryer E, Issakidis-Bourguet E, Lavergne D, Miginiac-Maslow M (2003. a) Chlamydomonas reinhardtii: a model organism for the study of the thioredoxin family. Plant Physiol Biochem 41 513–521 [Google Scholar]
- Lemaire SD, Collin V, Keryer E, Quesada A, Miginiac-Maslow M (2003. b) Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii. FEBS Lett 543 87–92 [DOI] [PubMed] [Google Scholar]
- Marcus F, Chamberlain SH, Chu C, Masiarz FR, Shin S, Yee BC, Buchanan BB (1991) Plant thioredoxin h: an animal-like thioredoxin occurring in multiple cell compartments. Arch Biochem Biophys 287 195–198 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestres-Ortega D, Meyer Y (1999) The Arabidopsis thaliana genome encodes at least four thioredoxins m and a new prokaryotic-like thioredoxin. Gene 240 307–316 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Riondet CCL, Abdelgawwad MR, Reichheld JP, Vignols F (2006) Evolution of redoxin genes in the green lineage. Photosynth Res 89 179–192 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Vignols F, Reichheld JP (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol 347 394–402 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval DF, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher L, Dunahay T, Frohlick J, Mazurkiewicz A, Meehl J, Staehelin L (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system1. Plant Physiol 121 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Ludwig AA, Jones JDG (2006) CITRX thioredoxin is a putative adaptor protein connecting Cf-9 and the ACIK1 protein kinase during the Cf-9/Avr9-induced defence response. FEBS Lett 580 4236–4241 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Mestres-Ortega D, Laloi C, Meyer Y (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol Biochem 40 685–690 [Google Scholar]
- Rivas S, Rougon-Cardoso A, Smoker M, Schauser L, Yoshioka H, Jones JD (2004) CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J 22 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivera-Madrid R, Mestres D, Marinho P, Jacquot JP, Decottignies P, Miginiac-Maslow M, Meyer Y (1995) Evidence for five divergent thioredoxin h sequences in Arabidopsis thaliana. Proc Natl Acad Sci USA 92 5620–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert C, Baker L, Szederkenyi J, Grossmann P, Komor E, Hayashi H, Chino M, Lucas WJ (1998) Identification of immunologically related proteins in sieve-tube exudate collected from monocotyledonous and dicotyledonous plants. Planta 206 245–252 [Google Scholar]
- Stein M, Jacquot JP, Jeannette E, Decottignies P, Hodges M, Lancelin JM, Mittard V, Schmitter JM, Miginiac-Maslow M (1995) Chlamydomonas reinhardtii thioredoxins: structure of the genes coding for the chloroplastic m and cytosolic h isoforms; expression in Escherichia coli of the recombinant proteins, purification and biochemical properties. Plant Mol Biol 28 487–503 [DOI] [PubMed] [Google Scholar]
- Sweat TA, Wolpert TJ (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Bayer E, Ritzenthaler C, Fernandez-Calvino L, Maule A (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17 1513–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.