Abstract
We have previously described the transcriptional changes that occur in the hippocampal CA1 field of aged rats following a Morris Water Maze (MWM) training paradigm. In this report we proceed with the analysis of the dentate region from the same animals. Animals were first identified as age learning-impaired or age-superior learners when compared to young rats based on their performance in the MWM. Messenger RNA was isolated from the dentate gyrus of each animal to interrogate Affymetrix RAE 230A rat genome microarrays. Microarray profiling identified 1129 genes that were differentially expressed between aged and young rats as a result of aging, and independent of their behavioral training (p<0.005). We applied Ingenuity Pathway Analysis (IPA) algorithms to identify the significant biological processes underlying age-related changes in the dentate gyrus. The most significant functions, as calculated by IPA, included cell movement, cell growth and proliferation, nervous system development and function, cellular assembly and organization, cell morphology and cell death. These significant processes are consistent with age-related changes in neurogenesis, and the neurogenic markers were generally found to be downregulated in senescent animals. In addition, statistical analysis of the different experimental groups of aged animals recognized 85 genes (p<0.005) that were different in the dentate gyrus of aged rats that had learned the MWM when compared to learning impaired and a number of controls for stress, exercise and non-spatial learning. The list of learning-related genes expressed in the dentate adds to the set of genes we previously described in the CA1 region. This long list of genes constitutes a starting tool to elucidating the molecular pathways involved in learning and memory formation.
Keywords: aging, hippocampus, learning and memory, Morris water maze, nervous system, gene expression, learning impaired, superior learner, dentate gyrus, microarray, class prediction, pathway analysis, neurogenesis
Introduction
The process of aging results in cognitive decline both in humans and animals. Memory deficits associated with aging have been reported in a wide variety of animal models. Numerous studies have been carried out in rats, where it has been demonstrated that age-related deficits show large variability between individuals (Gage, Dunnett et al. 1984; deToledo-Morrell, Geinisman et al. 1988; Aitken and Meaney 1989; Markowska, Stone et al. 1989; Clark, Magnusson et al. 1992; Gallagher and Nicolle 1993). Some rats maintain their cognitive abilities and can perform as well as young animals in spatial learning tasks, whereas others show cognitive impairments and perform poorly on these learning paradigms. Rats can be separated into learning impaired (AI) or superior learners (SL) based on their spatial learning performance (Gallagher, Burwell et al. 1993; Schulz, Huston et al. 2002), and have been used to study the mild cognitive impairments associated with aging. We have hypothesized that differential changes in gene expression, particularly in the hippocampus, account for some of the age-related changes in learning ability. In order to test this hypothesis, we have performed a microarray analysis of genes that are differentially expressed in rats that show age-related cognitive impairment in the Morris water maze (MWM) compared to age-matched rats that were less impaired in this spatial learning task.
In a previous experiment we categorized aged rats as learning-impaired (AI) and superior learners (SL) based on their behavior in the MWM and examined the genome-wide transcriptional changes that occurred in the CA1 region (Burger, Cecilia Lopez et al. 2007). Here we describe the outcome of the analysis of transcriptional profiles of genes in the dentate gyrus that were differentially expressed between aged animals that had learned the MWM and those who where learning-impaired and a number of controls who did not have a chance to learn the paradigm. In addition, we describe the expression patterns between the young and the aged dentate gyrus. We used pathway analysis in order to find relevant biological functions for the large number of genes identified.
2. Methods
2.1. Animals
Fisher-344 male rats, 3 and 24 month old, were obtained from the National Institutes of Aging colony (NIA, Washington DC). Rats were housed (2 per cage) in a 12:12 light-dark cycle with ad libitum access to food and water. Behavioral testing occurred during the light phase of the cycle. Procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee and were in accordance with guidelines established by the US Public Health Policy on Humane Care and Use of Laboratory Animals. The animals used in this experiment are the same animals reported previously in (Burger, Cecilia Lopez et al. 2007).
2.2. Behavioral Design
The water maze was composed of a black tank 2 m in diameter filled with water, no dye added. The lucite hidden platform was 12 cm2 situated 3cm under water. Because the tank is black, the transparent Lucite platform cannot be seen. Distance traveled to the platform was measured with a San Diego Instruments video tracking system and SDI chromotrack software. The temperature of the water was measured every day and was 28° C. The animals were habituated for two days in a water maze with a visible platform. This training also served as a test for visual acuity, since aged Fischer 344 rats are prone to develop retinal degeneration {Markowska, 1990 #176}. The animals were tested for two days (four trials per day) using a visible platform to make sure no animals were included in the analysis who were blind or not motivated to escape the water. Animals that could not see the platform were removed from the study. We found only one such animal, which was removed from the study (Burger, Cecilia Lopez et al. 2007).
The animals were then trained in sets of 4 trials per day for 8 days (32 trials total) with a two day break after day three. In each trial, the rat was introduced into the tank with his head pointing towards the tank wall from one of four randomly chosen entry points (N, S, E, and W). The order of the entry points was randomly varied each day. The length of the swimming path to find an escape (path length) was recorded. If the animal did not find the platform after 90 sec, he was placed on the platform for 15 seconds. A probe trial was carried out at the end of trial 32 by removing the platform and allowing the animal to swim in the maze for 60 seconds. The wall annulus of the MWM is quite large, taking up 88.5% of the total area of the maze. Thus, even well trained animals must swim some large relative distance in the wall annulus in order to reach the platform.
For one group of animals, the platform was visible and was changed to different positions during trials (animal groups are shown in Table 1). An additional control group was placed in the maze and swam without a platform present for the average amount of time that the hidden platform animals spent to find the platform on each individual day. Finally, a group of age-matched animals remained in their cages for the entirety of the experiment.
Table 1. Experimental groups used for the microarray analysis.
A total of 80 chips (1 per rat) were included in the analysis for the aged versus young comparison (figure 2). For the learning-related supervised analysis the intermediate learners were removed to highlight the differences between SL and AI, leaving 66 chips for the comparison of SL vs CTL (figure 4).
| Age | Hidden platform | Visible platform | Yoked control | Cage control |
|---|---|---|---|---|
| 3 months | 9 | 5 | 6 | 7 |
| 24 months | 14 Unimpaired | 5 | 7 | 3 |
| 14 Intermediate | ||||
| 10 Impaired | ||||
2.2.1. Statistical analysis and definition of aged- superior learner animals
Animals were identified as aged-superior learners when their hidden platform acquisition was similar to that of young rats as determined by their acquisition curve using distance to find the platform and time spent in the target quadrant in the probe trial. Aged learning-impaired animals were identified as those who did not improve at the end of the hidden platform training period and showed no quadrant bias in the probe trial. A group of intermediate learners (table I. n= 14) were removed from the experiment to maximize gene expression differences between SL and AI for the microarray experiment. This group of animals was still used for the aged versus young comparison. The MWM acquisition data were analyzed using repeated measures ANOVA. For the probe trial percent distance data, one-way ANOVA was performed. Significant main-effects were followed by Scheffe'post-hoc test for contrasts between groups. In the results section, we report post hoc values only when there was a highly significant main effect. We do not report each for various water maze variables main effect because these groups were artificially chosen and this is not a true unbiased experiment.
It is important to note that we originally started with 100 animals, but due to failure to amplify some of the RNAs using the Affymetrix Protocol II, or in some cases, failure of the amplified RNA to hybridize to the chip, a number of samples could not be analyzed (see Table I for total number of animals). Therefore, because the dataset in this study is not the same as was used in the CA1, i.e. there were some differences in the animals included in this dentate experiment compared to the previously reported CA1 experiment, we have re-analyzed the behavior for the set of animals used in this dentate study (Burger, Cecilia Lopez et al. 2007).
2.3. Tissue Dissection and RNA Isolation
Animals were sacrificed 24 hours after the last behavioral trial and the dentate gyri were isolated using a micro-dissection knife (Electron Microscopy Sciences, Hatfield, PA) as previously described (Burger, Cecilia Lopez et al. 2007) and the tissue was placed in 100 µl RNAlater. RNA was isolated using the Qiagen RNeasy protocol (Qiagen, Valencia, CA). The tissue was homogenized in 400 µl of RLT buffer and 1% mercaptoethanol, and treated with 200 µg/ml proteinase K for 10 minutes at 55°C. The RNA was stored at −80°C until further use.
2.4. RNA amplification, cDNA Synthesis and Test RNA labeling
Two rounds of RNA amplification were performed following the Affymetrix Small Sample Target Labeling version II protocol (with minor modification) using 200 ng of total RNA from the dentate of each animal. After the second round of amplification, in vitro transcription was carried out using the Enzo Bioarray RNA transcript labeling kit (Enzo, Farmingdale, NY). Hybridization and washes were performed using Affymetrix protocol Euk GE-WS2v4.
2.5. Statistical Analysis of Affymetrix Microarray Gene Expression
2.5.1. Normalization Modeling and Expression filter
The Affymetrix data including young and aged animals was normalized and modeled using DChip version 1.3 (Li and Hung Wong 2001), as previously described (Burger, Cecilia Lopez et al. 2007). Unsupervised analysis was performed using probe sets whose hybridization signal intensities varied the most across the data set. Probe sets with a coefficient of variation greater than 0.5 were identified and subjected to hierarchal cluster analysis using DChip clustering algorithms (Li and Hung Wong 2001).
2.5.2. Supervised learning, discrimination analysis, and cross-validation
The DChip expression matrix was filtered to remove probe sets that were never detected above background on any array in the analysis. From the 15,923 probe sets present in the Affymetrix 230A chip, this filter removed 4,332 probe sets. The resulting expression matrix contained 11,591 probe sets that were detected above background on at least one array using the Affymetrix detection call algorithms. This expression matrix was imported to BRB Array Tools (v3.4 Beta 2; http://linus.nci.nih.gov/BRB-ArrayTools.html) for class prediction analysis. BRB array tools was used to identify genes that differentiated among the treatment classes: aged vs. young, for the first class prediction analysis (p<0.005); or superior learners vs. controls (p<0.005) for the aged group class prediction. The ability of gene identification to predict treatment class was assessed by using Leave-one-out–cross validation, using a nearest-neighbor prediction model. This analysis identified 1129 aged-associated genes and 85 learning-associated genes.
To fulfill MIAME standards (Brazma, Hingamp et al. 2001), Affymetrix DAT and CEL. files, as well as the TXT file that results from the former two, as well as the Dchip expression matrix have been deposited at the Gene Expression Omnibus website (GEO: http://www.ncbi.nlm.nih.gov/geo/ Accession series record number: GSE4821).
2.6. Pathway analysis
For the 85 learning-associated genes, we used a combination of bioinformatics software that included E! Ensemble, Protonet, Pandora, and Pubmed and Pubmatrix searches (Becker, Hosack et al. 2003). We also used http://bind.ca for protein-protein interaction information. Using this approach (Velardo, Burger et al. 2004; Burger, Cecilia Lopez et al. 2007) we found information on 50 genes (Table 3 and supplementary Table 3); the other 35 transcripts were expressed sequence tags (EST).
Table 3. SL vs CTL gene annotations.
Genes were classified based on their putative role in nervous system function. Unidentified ESTs were kept in a separate group. The values for the geometrical means of signal intensity (geom. means of intensity) and the assigned p values were calculated using BRB (see Methods). Blue indicates downregulation, red indicates that expression is higher relative to the other class. Class 1: SL, Superior Learners. Class 2: CTL, aged impaired, cage, visible and yoked controls.
% CV, percent cross-validation support. A value of 100 means that the probe set was significant each time an array was left out of the data set during the leave-one-out-cross validation study. If the value is 0 it means that that particular probe set was only identified as significant when all of the arrays were present in the data set.
| Molecular Function/Cellular Component | Gene Name | Gene symbol | Gene ID | Geom mean of intensities: AU | Geom mean of intensities CTL | Parametric p-value | % CV support |
|---|---|---|---|---|---|---|---|
| Protein modification | |||||||
| Alpha-mannosidase-like protein mRNA, 3' UTR | 1372533 at | 251.1 | 316.6 | 0.000715 | 100 | ||
| proteaseome (prosome, macropain) 28 subunit, 3 (predicted) | Psme3 predicted | 1388582 at | 377.2 | 454.1 | 0.000768 | 100 | |
| ubiquitin associated domain containing 1 | Ubadc1 | 1388387 at | 341.5 | 438.1 | 0.000987 | 100 | |
| leucine rich repeat containing 41 | Lrrc41 | 1372784 at | 716.5 | 882.2 | 0.00478 | 38 | |
| ring finger protein 25 (predicted) | Rnf25 predicted | 1372088 at | 660.4 | 806.9 | 0.002096 | 97 | |
| similar to glucosamine-phosphate N-acetyltransferase 1 | GNPNAT | 1375530_at | 222.8 | 271.3 | 0.000807 | 100 | |
| Protease, serine, 25 (predicted) | PRSS25, htra2 | 1367478 at | 980.3 | 1154.6 | 0.003924 | 59 | |
| Mitogen activated protein kinase kinase kinase 7 (predicted) | Map3k7 | 1399075 at | 502.6 | 642.9 | 0.00346 | 74 | |
| v-raf oncogene homolog 1 (murine sarcoma 3611 virus) | Araf1 | 1388305 at | 317.6 | 440.9 | 0.000152 | 100 | |
| PX domain containing serine/threonine kinase | Pxk | 1390422 at | 174.7 | 217.2 | 0.001049 | 100 | |
| calcium/calmodulin-dependent protein kinase kinase 1, alpha | Camkk1 | 1368156_at | 2712.5 | 3383 | 0.003017 | 87 | |
| Similar to mitogen activated protein Kinase associated protein 1 | MAPKAP1 | 1377007_at | 282.4 | 489.6 | 0.003639 | 64 | |
| Phosphatase and actin regulator 2 | Phactr2 | 1392105_at | 97.8 | 118.6 | 0.004672 | 44 | |
| PTK2 protein tyrosine kinase 2 | Ptk2 | 1387875 at | 424.6 | 527.5 | 0.001078 | 100 | |
| Transcribed locus, strongly similar to protein phosphatase 1, regulatory | PPP1R1C | 1372625_at | 1164 | 832.4 | 0.001311 | 100 | |
| Transcription/Translation | |||||||
| cAMP responsive element binding protein-like 2 (predicted) | Crebl2 | 1373618_at | 433.4 | 524.8 | 0.001388 | 100 | |
| Transcribed locus; thrapi (thyroid hormone receptor associated protein 1) | Thrap1 | 1386156_at | 59.2 | 84.2 | 0.001651 | 97 | |
| similar to interleukin enhancer binding factor 2 | ILF2 | 1371582_at | 1173.7 | 1472 | 0.004394 | 46 | |
| similar to DNA polymerase epsilon p17 subunit (DNA polymerase epsilon | POLE3 | 1388833 at | 513.8 | 613.6 | 0.000695 | 100 | |
| chromatin accessibility complex 1 (predicted) | Chrac1_predicted | 1388943_at | 271.8 | 317.4 | 0.004511 | 44 | |
| high mobility group box 1 | Hmgb1 | 1368042_a_at | 2251 | 2828.7 | 0.002422 | 92 | |
| translin-associated factor X | Tsnax | 1398823 at | 257.3 | 359.8 | 0.003994 | 44 | |
| WD repeat domain 3 (predicted) | Wdr3 predicted | 1374793 at | 237.7 | 311.6 | 0.003787 | 69 | |
| Similar to ankyrin repeat domain protein 17 isoform b (Ankrd17) | Ankrd17 b | 1399008 at | 518.2 | 619.7 | 0.002595 | 90 | |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 (predicted) | Ddx42_predicted | 1376596_at | 394.4 | 475.4 | 0.004412 | 41 | |
| eukaryotic translation initiation factor 2, subunit 3 | EIF2S3 | 1398936 at | 294 | 423.9 | 0.00034 | 100 | |
| neurotransmission | |||||||
| SEC6-like 1 (S. cerevisiae) Sec6l1 | EXO3 | 1389624_s_at | 537.5 | 625.1 | 0.001243 | 100 | |
| tocopherol (alpha) transfer protein | Ttpa | 1369435 at | 51.5 | 66.4 | 0.003635 | 72 | |
| choline kinase alpha | Chka | 1368692 a at | 155.1 | 180.7 | 0.0026 | 90 | |
| tyrosine hydroxylase | Th | 13734432_a_at | 97.8 | 119.1 | 0.003209 | 87 | |
| DnaJ (Hsp40) homolog, subfamily C, member 14 | DNAJC14 | 1373402_at | 152.6 | 179.2 | 0.001953 | 95 | |
| phosducin-like | Pdcl | 1387499_a at | 126.1 | 158.7 | 0.002668 | 87 | |
| reticulon 3 | Rtn3 | 1368807_at | 280.6 | 337.7 | 0.002983 | 87 | |
| neurofibromin 2 (bilateral acoustic neuroma) | NF2 | 1372502_at | 987.1 | 1327.8 | 0.001684 | 97 | |
| CLIP associated protein | Clasp1_predicted | 1375685 at | 494 | 631.1 | 3.40E-05 | 100 | |
| Rad and gem related GTP binding protein 2 | Rem2 | 1374035_at | 474.7 | 338 | 0.002704 | 90 | |
| chromogranin A | Chga | 1387235 at | 1015.9 | 742 | 0.002257 | 95 | |
| Iipid metabolism | |||||||
| peroxisomal biogenesis factor 3 | Pex3 | 1368526_at | 369.9 | 444.1 | 0.00096 | 100 | |
| hydroxysteroid (17-beta) dehydrogenase 4 | Hsd17b4 | 1367672 at | 448.1 | 564.1 | 0.001009 | 100 | |
| UDP-glucuronosyltransferase 8 | Ugt8 | 1368858_at | 167.8 | 209.1 | 0.004106 | 51 | |
| G0/G1 switch gene 2 (predicted) | G0s2_predicted | 1388395 at | 263.4 | 335.6 | 0.003386 | 90 | |
| Transcribed locus, strongly similar to NP_112224.1 SET binding factor 2 | SBF2 | 1375171 at | 30.6 | 43.6 | 0.004257 | 49 | |
| Low density lipoprotein-related protein 12 (predicted) | LRP12 | 1374108 at | 332.2 | 442.7 | 0.003416 | 79 | |
| phospholipase A2, activating protein | Plaa | 1368668 at | 145 | 191.2 | 0.001631 | 97 | |
| Cell Surface/ Cell Transduction | |||||||
| TPA regulated locus (predicted) | CXCL12 | 1373392_at | 375.5 | 504.7 | 5.60E-05 | 100 | |
| CD9 antigen | Cd9 | 1371499 at | 197.6 | 274.6 | 0.002284 | 95 | |
| wingless-related MMTV integration site 6 (predicted) | Wnt6_predicted | 1376063_at | 63.3 | 82.6 | 0.000855 | 100 | |
| Solute carrier family 9 (sodium/hydrogen exchanger), isoform 3 regulator 2 | Slc9a3r2 | 1388831 at | 553.1 | 378.1 | 0.002645 | 92 | |
| Mitochondria | |||||||
| nicotinamide nucleotide adenylyltransferase 3 (predicted) | Nmnat3_predicted | 1374025_at | 117.9 | 150.4 | 0.004718 | 44 | |
| mitochondrial ribosomal protein L50 (predicted) | Mrpl50_predicted | 1376585_at | 211.3 | 260.2 | 0.001482 | 97 | |
| mitochondrial translation optimization 1 homolog (S. cerevisiae) | Mto1_predicted | 1377493_at | 290.2 | 342.6 | 0.003489 | 69 | |
| EST | |||||||
| 1388524_at | 623.8 | 773.4 | 0.000605 | 100 | |||
| 1372714_at | 408 | 512.5 | 0.000794 | 100 | |||
| 1371522_at | 447.3 | 543.5 | 0.001065 | 100 | |||
| 1390622_at | 90.9 | 110 | 0.001292 | 100 | |||
| 1372550_at | 105.1 | 134 | 0.00134 | 100 | |||
| 1373634_at | 278.6 | 368.4 | 0.001438 | 100 | |||
| 1390467_at | 445.6 | 542.7 | 0.001532 | 100 | |||
| 1376071_at | 240.8 | 302.3 | 0.001664 | 97 | |||
| 1390449_at | 425.5 | 502.5 | 0.001759 | 100 | |||
| 1376627_at | 328.8 | 436 | 0.00185 | 100 | |||
| 1371773_at | 972.6 | 1247.1 | 0.001934 | 95 | |||
| 1374233_at | 703.2 | 831.1 | 0.002042 | 100 | |||
| 1388662_at | 240.7 | 287.7 | 0.002054 | 97 | |||
| 1376822_at | 40.4 | 58.1 | 0.002135 | 92 | |||
| 1374498_at | 208.6 | 254 | 0.00242 | 92 | |||
| 1389562_at | 595.1 | 793.3 | 0.002535 | 97 | |||
| 1373846_at | 201.9 | 252.3 | 0.002777 | 92 | |||
| 1374260_at | 183.7 | 236.1 | 0.002974 | 90 | |||
| 1390458_at | 84.5 | 108.3 | 0.003182 | 90 | |||
| 1376318_at | 336.7 | 407.3 | 0.00345 | 79 | |||
| 1373285_at | 652 | 746.6 | 0.00366 | 69 | |||
| 1389411_at | 398 | 526.6 | 0.003812 | 56 | |||
| 1373859_at | 163.6 | 199.5 | 0.003913 | 54 | |||
| 1383406_at | 216 | 268 | 0.004066 | 51 | |||
| 1389417_at | 298.3 | 354.5 | 0.004088 | 56 | |||
| 1389000_at | 486.7 | 582.7 | 0.004141 | 54 | |||
| 1390385_at | 533.7 | 647.4 | 0.004357 | 51 | |||
| 1374001_at | 355.8 | 458.1 | 0.004522 | 44 | |||
| 1376288_at | 338 | 410.2 | 0.004524 | 36 | |||
| 1377149_at | 655.7 | 768.2 | 0.004593 | 38 | |||
| 1375952_at | 513.9 | 595.5 | 0.004933 | 31 | |||
| 1373520_at | 959.3 | 1145.3 | 0.002418 | 92 | |||
| 1389004_at | 368 | 291.5 | 0.002752 | 92 | |||
| 1390167_at | 1683.1 | 1184.7 | 0.002425 | 90 |
In addition, the functional analysis algorithm from Ingenuity Pathway Analysis (IPA; Ingenuity® Systems, www.ingenuity.com) was used to identify the biological functions and/or diseases that were most significant to the data set. Genes from the dataset that met the p value cutoff of 0.005 and were associated with biological functions and/or diseases in the Ingenuity Pathways Knowledge Base were considered for the analysis. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. A dataset containing gene identifiers and their corresponding fold change values and the p values from the BRB class prediction results for each gene were uploaded as an Excel spreadsheet into IPA. The 85 genes that were found significant were uploaded for the dentate analysis of SL versus control comparison and fold change was calculated as SL/controls, from the geometrical mean intensities (supplementary Table 2 and supplementary Table 4)
IPA was also used to investigate the functions in the aged and young comparison. Fold change was calculated as Aged/Young.
3. Results and Discussion
3.1 Aged Rats can be separated into learning impaired and superior learners based on their performance in the Morris Water Maze Paradigm
The MWM paradigm was utilized to segregate aged rats into superior learners and learning-impaired rats as we have previously described (Burger, Cecilia Lopez et al. 2007). The RNA from each characterized animal was used to interrogate Affymetrix chips. Because gene expression changes have been observed in the hippocampus due to factors such as stress, exercise, and aging (Goyns, Charlton et al. 1998; Wei, Zhang et al. 1999; Lee, Weindruch et al. 2000; Jiang, Tsien et al. 2001; Tong, Shen et al. 2001; Cotman and Berchtold 2002; Vaynman, Ying et al. 2003; Alfonso, Pollevick et al. 2004; Lu, Pan et al. 2004; Lukiw 2004; Nagata, Takahashi et al. 2004; Sawada, Morinobu et al. 2004), it was imperative to include a number of control groups in the array experiment (Burger, Cecilia Lopez et al. 2007). A visual version of the MWM task was performed in one group of animals to control for the motor aspect of this training paradigm and the procedure of finding a platform. A separate group of animals were time-yoked controls that swam in the absence of a platform to partially control for stress and to control for the act of swimming. The final group was untreated and remained in their cages for the duration of the training to control for gene expression patterns simply related to senescence in the brain (Goyns, Charlton et al. 1998; Wei, Zhang et al. 1999; Lee, Weindruch et al. 2000; Jiang, Tsien et al. 2001; Lu, Pan et al. 2004).
In order to insure that all rats in the MWM groups were not blind and were motivated to escape the pool, a two day visible platform paradigm was undertaken where the visible platform was placed in the middle of the maze (Fig. 1A). Only one animal was not able to swim to the visible platform and was excluded from the study (Burger, Cecilia Lopez et al. 2007). As additional evidence that the aged animals did not have visual deficits, we must point out that all of our “visible platform controls” were able to find the platform every single time. Because the platform was randomly moved around the platform annulus for each visible trial and every animal included in this study swam to the platform regardless of the platform placement, we must conclude that these animals were not blind.
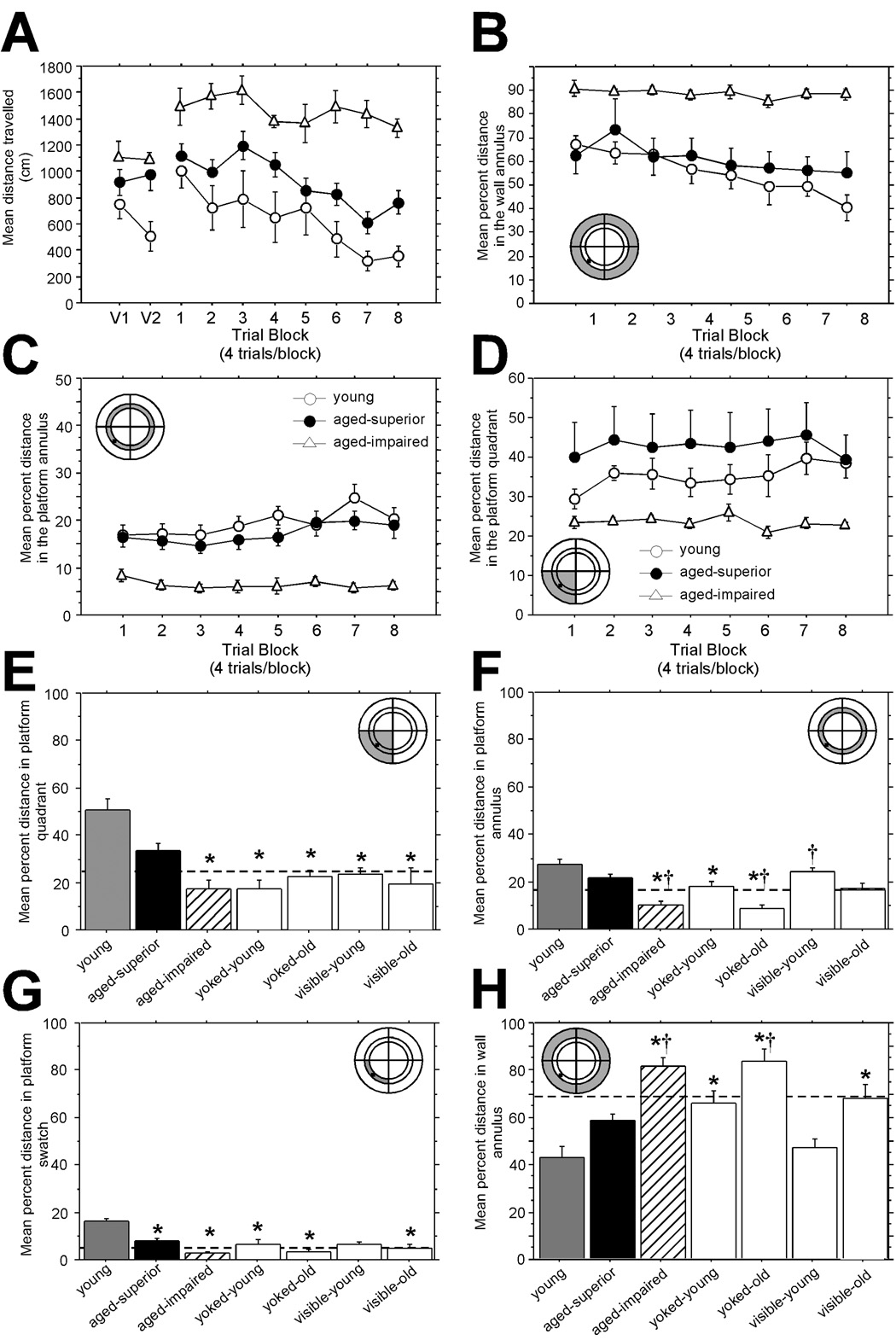
Figure 1. A schematic representing the experimental design of the MWM testing depicting the conceptual basis for each control group is presented in a previous publication (Burger, Cecilia Lopez et al. 2007).
(A) Visible platform and acquisition curve. The distance traveled by the young animals decreased significantly over the eight days (open circles). Similarly, aged SL animals (closed circles) learned the task but not quite to the efficiency of the young animals. On the other hand, AI animals (open triangles) did not reduce their path length and rarely found the platform. For all individual statistical contrasts, see the text. (B) Path length percentage in the wall annulus is used to evaluate thigmotaxic behavior. Both the young and the aged SL animals swam less over time in the wall annulus while the AI animals essentially only swam near the wall over the entire training period. (C) Path length percentage in the platform annulus. While the young animals searched the most of any group in the platform annulus, both young and the SL animals searched more in the platform annulus than the AI animals. (D) Path length percentage in the platform quadrant. The young animals searched more of their time in the platform quadrant throughout the training period than either aged group. The aged SL animals increased their searching of the platform quadrant over time but the aged impaired animals never altered their search strategy. Maze schematics in the corner of panels A–D: the shaded areas depict the location of the maze pertinent to that panel.
(E–H) Evaluation of search strategy from the probe trial. (E) Proportion of swimming in the platform quadrant. The dashed line indicates the proportion of the MWM taken up by this quadrant (25%). Asterisks indicate a significant difference from young animals. Young and aged superior learners used a spatial learning strategy as shown by the increased amount of path length in the platform quadrant. AI rats searched an identical amount in the platform quadrant as animals that did not learn the spatial task. (F) Proportion of searching in the platform annulus. The dashed line indicates the percent of the MWM area occupied by the platform annulus (17%). The asterisks indicate significant differences from young animals and daggers (†) indicate significant differences from aged SL. The SL animals searched the platform annulus as much as the young animals while the AI animals rarely swam in the platform annulus. (G) Proportion of searching in the portion of the platform annulus that is in the platform quadrant. This measure is intended to serve as a more sensitive indicator of whether the animals are using a true spatial strategy by restricting their searching to a small portion of the MWM surrounding the platform (termed, the platform swatch, dashed line indicates 4.5% of the MWM area) versus using a response strategy circling around the platform annulus. Asterisks indicate significant differences from the young group and daggers indicate significant differences from the aged SL animals. A similar pattern was found when evaluating platform entries (data not shown). (H) Proportion of path length in the wall annulus. This analysis was intended to evaluate the animals’ level of thigmotaxia. The dashed line indicates the area of the MWM covered by the wall annulus (67%). Asterisks indicate significant differences from the young animals and daggers indicate a significant difference from aged SL animals. With the exception of the young animals trained on the visible platform, all the other groups displayed similar thigmotaxia to the AI animals whereas both the young and the aged SL animals searched less in the wall quadrant. Maze schematics in the corner of panels E–H: the shaded areas depict the location of the maze pertinent to that panel.
Another factor that had to be considered was the fact that old rats are prone to swim slower than young rats and therefore path length to find the platform instead of latency to the platform was used as the dependent variable (Gallagher and Nicolle 1993). Both acquisition and probe trial measures were examined in order to separate aged SL from AI animals (Fig. 1). As reported previously (Gage, Dunnett et al. 1984; Markowska, Stone et al. 1989; Clark, Magnusson et al. 1992), aged impaired rats generally needed longer swim paths to find the visible platform compared to young rats. However, while the superior learners (SL) displayed longer swim paths to the visible platform than young rats, this increased distance did not reach statistical significance [effect of group F(2,30) = 6.3, p = 0.005, post-hoc, young vs. SL, p = 0.06]. In addition, the two aged groups, SL and AI, performed identically in the visible portion of the training (post hoc, p > 0.43). The experimental and control groups that were to be exposed to the MWM were equalized based on their visible platform performance prior to separation into their respective treatment groups (data not shown for control groups).
We investigated each groups’ learning strategies during the acquisition phase of the MWM. Initially, all animals display thigmotaxia which is a natural defensive reaction to a novel stressful situation. The SL rats decreased their distance to find the hidden platform over time (Fig. 1A) in the MWM (effect of trials for SL group F[7,91] = 5.8, p <0.0001). In addition, while the SL rats did not find the hidden platform as efficiently as the young rats (post hoc, p = 0.03), the SL rats did reduce their search distance at the same rate as the young animals over the 8 days of training (SL vs. young × trials interaction F[7,147] = 1.6, p > 0.60). In contrast, AI animals performed significantly poorer than both young and SL animals (post hoc, p < 0.0001 and p < 0.0002, respectively). Indeed, the AI animals showed no significant reduction of swim path over the training period (effect of trials for the AI group F[7,63]) = 1.7, p > 0.1). The SL and young animals were already equivalent in distance to find the platform during the first day of training (post hoc, p > 0.9). We hypothesized that the young and SL rats had learned the procedural portion of the MWM, i.e, that escape is possible, while the AI rats apparently did not benefit from visible platform training. (Schulz, Huston et al. 2002).
The SL and young rats suppressed their thigmotaxia to a greater extent than the AI rats, spending significantly less distance searching in the wall annulus (Fig. 1B; post hoc, young vs. SL, p = 0.8, young vs. AI, p = 0.0025 and SL vs. AI, p = 0.005). The AI rats never suppressed their thigmotaxic searching despite being placed on the hidden platform after every trial. This perseveration of thigmotaxis in AI rats has been previously reported (Schulz, Huston et al. 2002; Burger, Cecilia Lopez et al. 2007). Stress-induced thigmotaxia in the AI animals might have prevented them from adopting a spatial search strategy.
The young rats and the SL group swam greater distances in the platform annulus (Fig. 1C, 17% of the MWM, young vs. SL post hoc, p > 0.50) than the AI rats (post hoc, both hidden platform groups vs AI, p < 0.0001, respectively). Likewise, the young and SL animals restricted their swimming to the platform quadrant during acquisition more over time than AI animals (Fig. 1D, post hoc, p < 0.005, respectively) as demonstrated by the percent distance swam in the platform quadrant (25% of the MWM).
At the end of the last training trial (trial 32), a probe trial was carried out by removing the hidden platform (Fig. 1 E–H). This test of spatial learning is particularly important since all the control groups can be interrogated about their spatial knowledge during this trial. The young rats searched for the platform using a spatial strategy as evidenced by the majority of their search pattern being located in the platform quadrant (Fig. 1E, dotted line = percent of the maze taken up by the quadrant). The SL rats searched the platform quadrant at an intermediate rate in that their path length in the platform quadrant was not different from either the young rats (p < 0.07) or the AI rats (p < 0.09). On the other hand, AI rats did not show a preference for the target quadrant during the probe trial, and this behavior was similar to that of the control rats that did not have an opportunity to learn the spatial task (yoked and visible controls, p > 0.96 in all contrasts).
A second indicator of spatial strategy is the percent distance traveled in the platform annulus (Fig. 1F). The young rats also restricted their search strategy relatively more than the the AI rats (post hoc, p < 0.0001). However, in contrast to the quadrant analysis, the aged SL rats searched the platform annulus as much as the young rats (post hoc, p > 0.49) and significantly more than the AI rats (post hoc, p = 0.0003).
To determine more strictly if the rats were using a true spatial strategy to search for the platform in the probe trial, we evaluated the percent path length in the portion of the platform annulus that is in the platform quadrant (Fig. 1G). This small swatch of the MWM takes up 4.5% percent of the total area of the maze. The young rats limited their search strategy to the platform swatch significantly more than the aged SL (post hoc, p < 0.04 ), and the AI (post hoc, p < 0.0001 ). The swatch analysis did not reveal any differences between the aged SL rats and the AI rats (p = 0.28).
The AI animals displayed a highly thigmotaxic swim pattern in the probe trial (Fig. 1H). Swim distance in the wall annulus (68% of the MWM area) clearly demonstrated that the young animals and the aged SL rats suppressed their natural tendency to swim next to the wall compared to the AI rats (post hoc, p ≤ 0.0009 in both contrasts).
The overall probe data shows that the young animals adopted a spatial strategy and the aged SL also searched the pool in a pattern that is consistent with spatial learning and was clearly superior to the search strategy of the AI animals. However, the aged SL rats appeared to rely on a more annulus-based search strategy to find the hidden platform compared to the young rats as revealed in the probe trial. Thus, while some of the behavioral details are different compared to the population of animals analyzed previously in (Burger, Cecilia Lopez et al. 2007), the overall strategy of the young vs. SL vs. AI remained essentially unchanged.
3.2 Supervised Analysis of Differential Expression Genes Associated with Aging in the Hippocampus Reveals Changes in Over a Thousand Transcripts
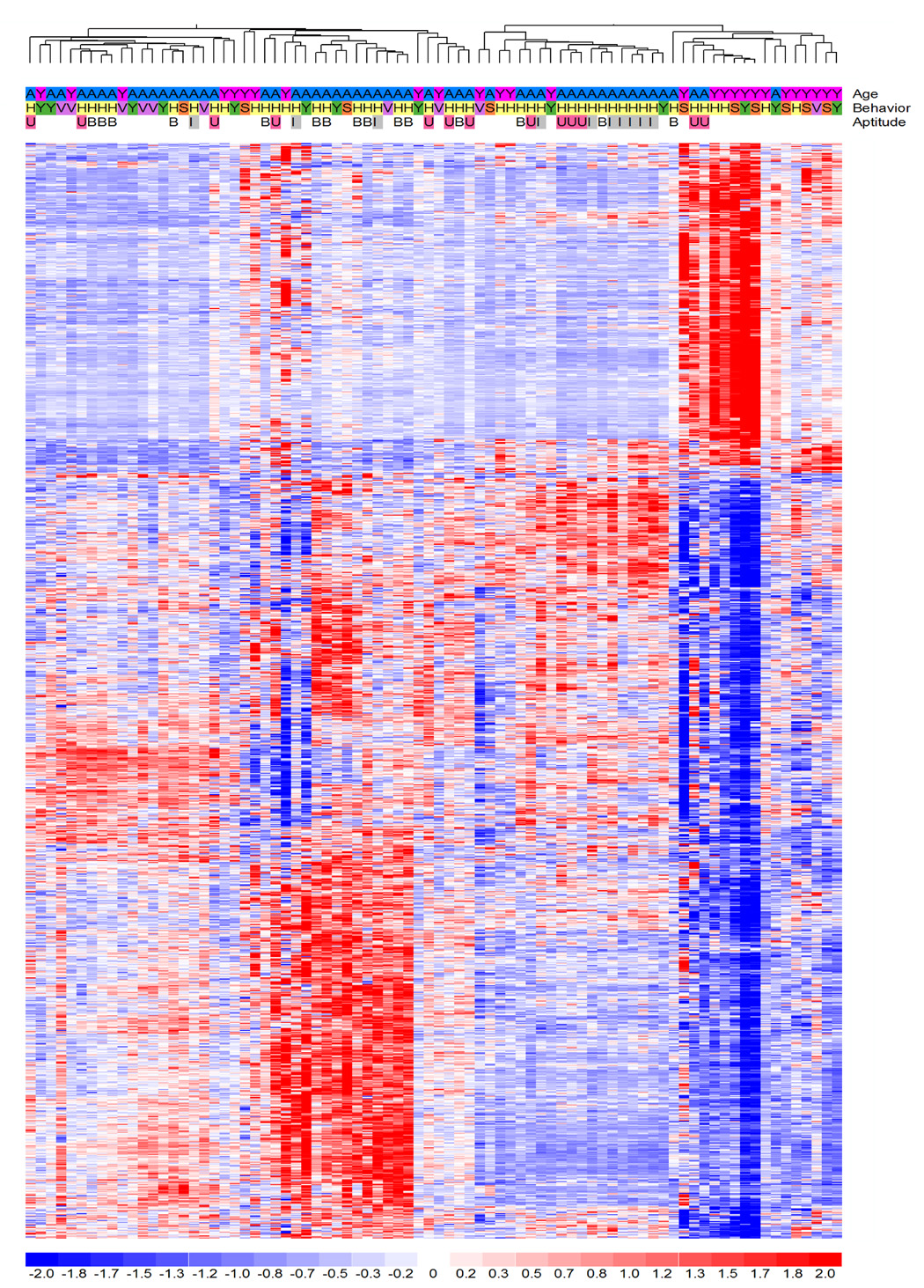
Supervised analysis of the data set was performed to identify expression profiles that separated young and aged animals (Fig. 2), and aged-superior learning animals from the rest of the groups (Figure 4). Analysis of aged and young animals at a significance threshold of p < 0.005 revealed significant changes in gene expression of 1129 genes between the two groups (Supplementary Table 1). We typically use a more stringent statistical significance level of p<0.001 to minimize the number of false positive, but for this study we wanted to have a larger number of genes available for pathway analysis to interpret our results (1129 genes at p<0.005 versus 457 at p<0.001). In addition, we performed leave-one-out-cross-validation analysis using BRB array tools to verify whether probe sets significant at p<0.005 differentiated between the groups of aged and young rats. The gene expression classifier correctly identified the array left out 69% of the time. This is significantly higher than the 50% expected by chance alone (p<0.017; Supplementary Table 1).
Figure 2. Expression Profiles of genes whose expression changed significantly between aged and young rats in the dentate gyrus.
Significant (p<0.005) probe sets (1129) were subjected to hierarchical clustering. Different genes are represented by the horizontal rows of pixels, while experimental animals are represented by the vertical rows. The red signal indicates genes whose expression is increasing under the given condition, blue indicates those that are decreasing with a maximum luminescence at two standard deviations. The length of the branch between any two replicates or groups on the dendrogram is a representation of the overall similarity of each global gene expression dataset. Abreviations: A: aged; Y: young; HID I: hidden platform impaired. HID U: hidden platform unimpaired (superior learners); VIS: visible platform control; SIT: cage control; YOKE: yoked control.
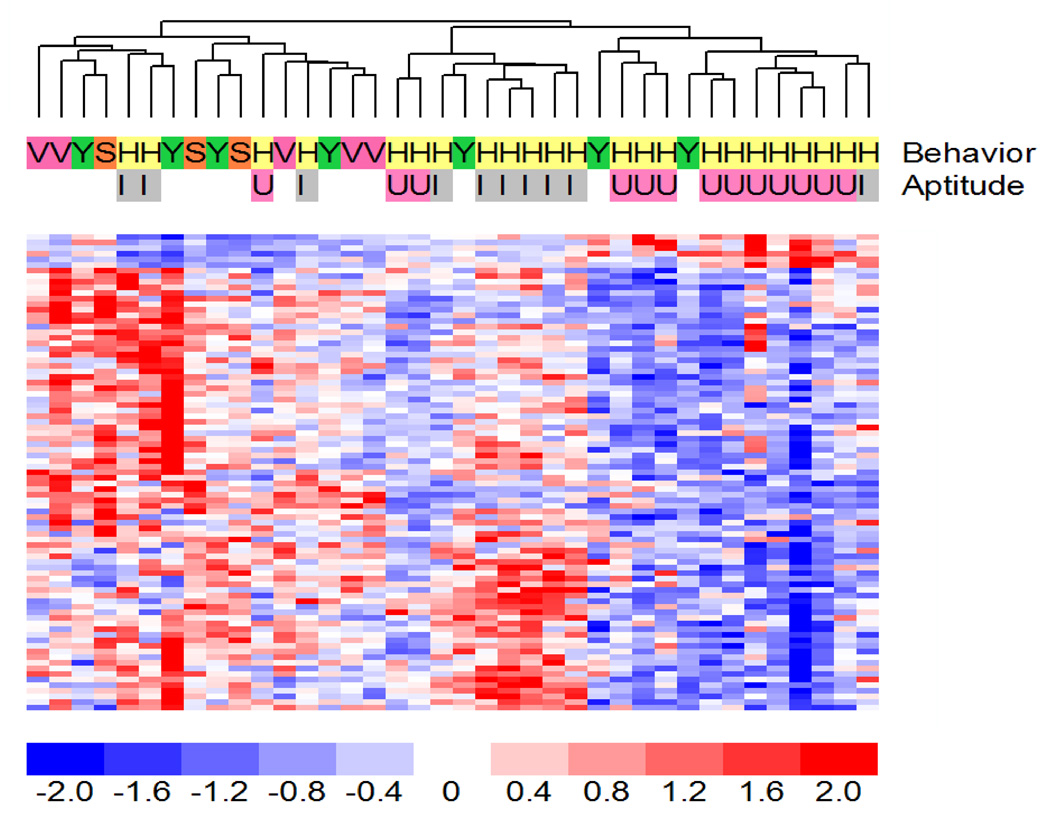
Figure 4.
Gene expression patterns of genes whose expression varied between the different experimental groups of aged rats in the dentate. The dendogram shows the hierarchical cluster patterns of the 85 genes identified as significant at the p < 0.005 significance threshold between the aged superior learners and aged impaired and control animals. Gene expression is displayed using a red-blue color scale as in figure 2.
3.3. Pathway Analysis of Transcriptional Differences Determined by Aging in the Dentate
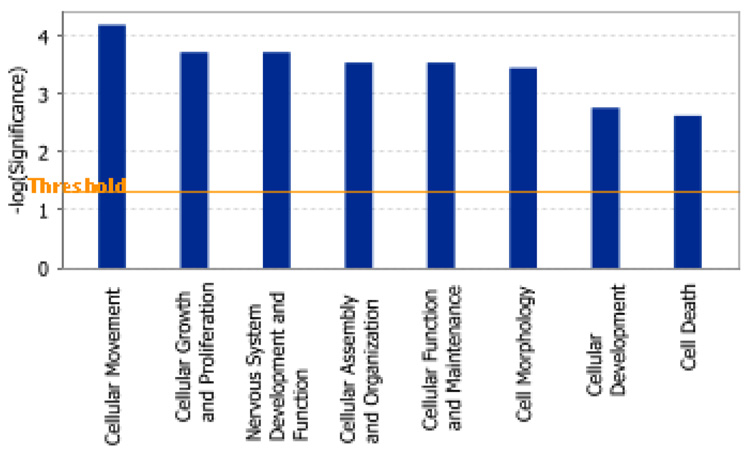
We used Ingenuity Pathway Analysis (IPA) algorithms and database to identify known pathways induced by aging. For this purpose, the 1129 differentially expressed genes were subjected to IPA. Of these genes, 427 genes were identified in the knowledge bank (i.e. focus genes eligible for generating networks; see materials and methods and supplementary data for details on IPA algorithms).
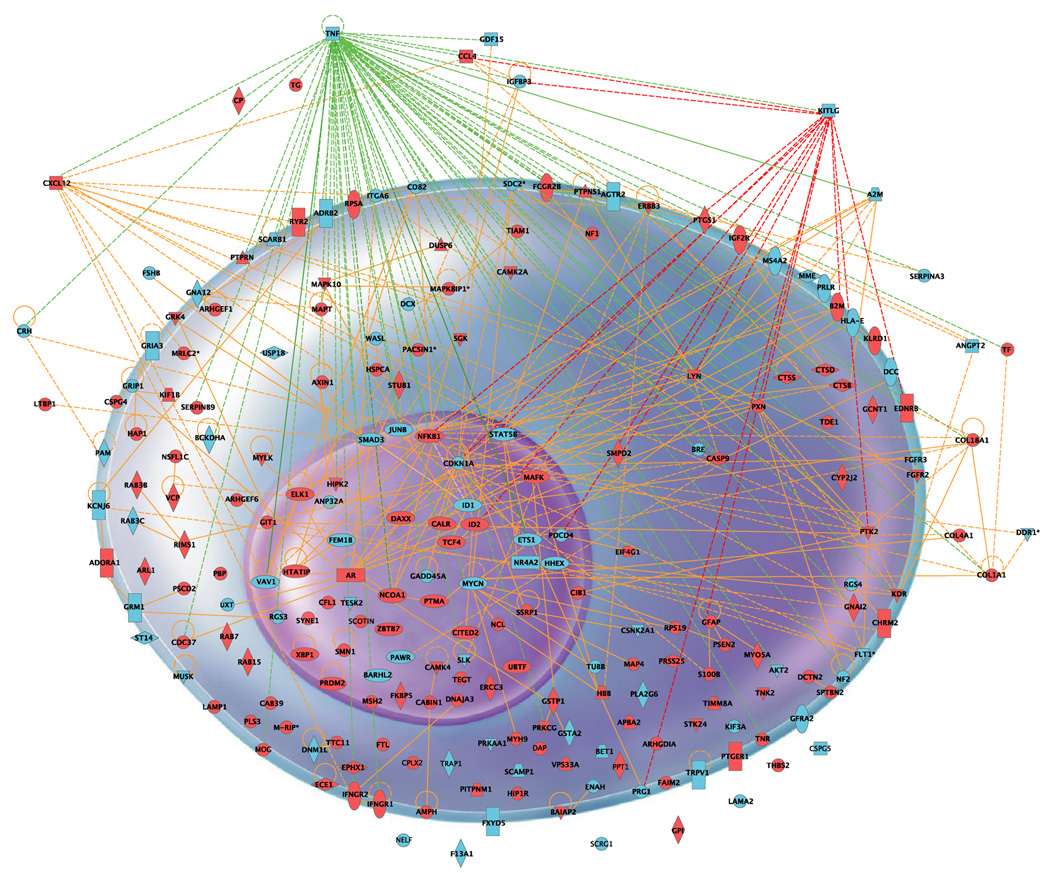
The networks illustrate functional relationships between gene products based on known interactions in the literature. Each network is composed of biological functions and canonical pathways. Canonical pathways are defined as known metabolic or signaling pathways. Ingenuity analysis revealed Cell Movement, Cell Growth and Proliferation, and Nervous System Development and Function as the three top functions influenced by age in the dentate (Figure 3a and supplementary table 3). These were followed by other functions such as Cellular Assembly and Organization, Cell Morphology, Cell Development and Cell Death. We find these particular functions relevant to the process of neurogenesis in the hippocampus. To limit the focus of this paper, we will only briefly discuss the different functions and a few key genes.
Figure 3. Pathway Analysis of the biological processes underlying age-related changes in the dentate.
(A) Bar chart representing eight top functional categories identified by IPA as significant for the genes that were differentially expressed in the aged dentate. Functional categories are represented in the × axis. The y axis designates the significance score (negative log of p value, see materials and methods for details). The horizontal line indicates the significance threshold.
(B) Age Related changes in expression of genes involved in Neurogenesis. 220 genes were involved in the IPA functions shown in fig3a are shown with their respective interrelationships in the cell. Genes that showed statistically significant increase in expression with aging are depicted in red. Those for which expression is decreased are shown in blue.
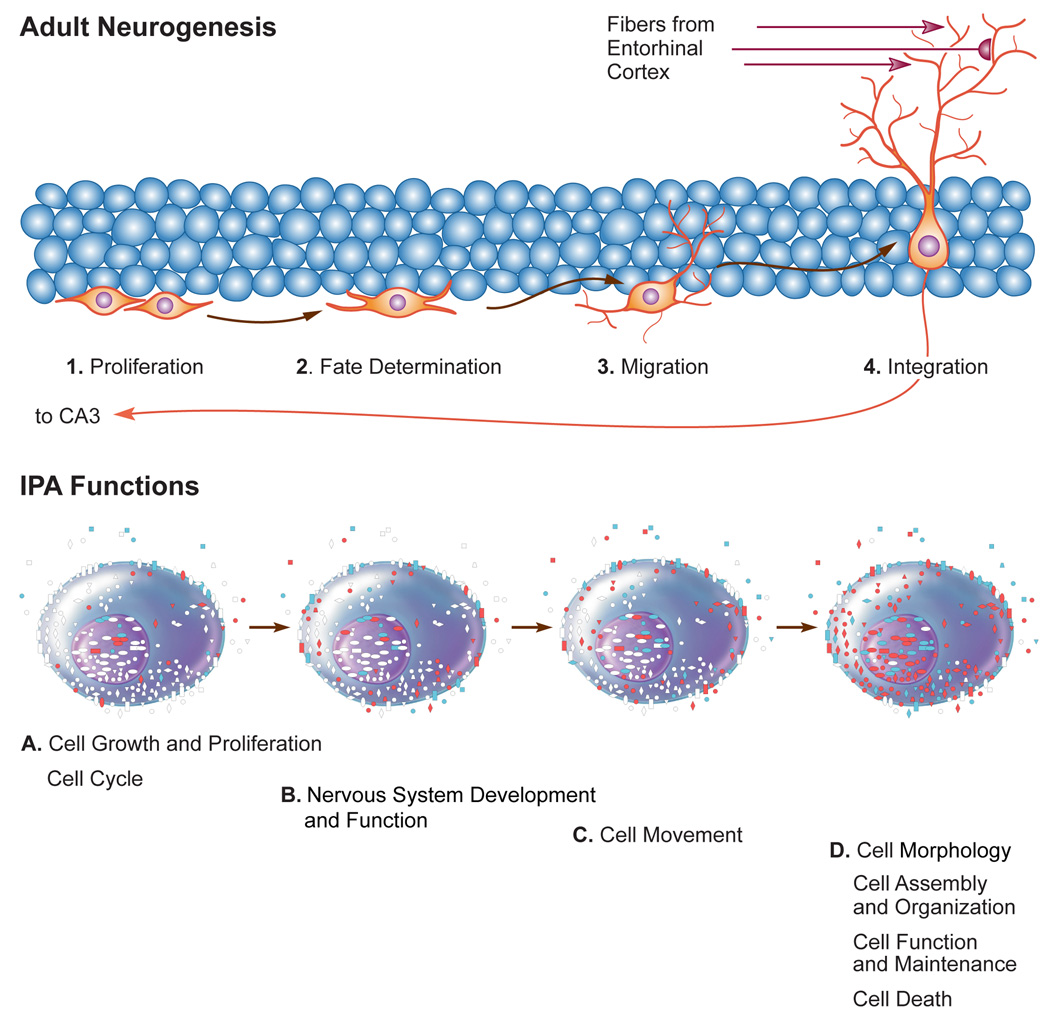
(C) Diagrammatic representation of the parallel between the cellular stages of neurogenesis and the molecular functions identified by pathway analysis that changed in the senescent dentate gyrus. The top of the image represents the dentate granule cell layer and the different stages associated with adult neurogenesis in the dentate. At the bottom, the Ingenuity Pathway Analysis functions representing the molecular signature of each of the stages in neurogenesis. Each cell shows which of the genes shown in fig 3b represent a given IPA function. Genes that were not part of a given function are depicted in white, while those who were upregulated are shown in red; downregulated genes are shown in blue.
3.3.1. Cell Movement; Cellular Growth and Proliferation; and Cell Death
Neurogenesis involves cellular processes that include cell proliferation, fate determination, migration of precursor cells, and neuronal differentiation. Therefore the functions we identified, including Cell Growth and Proliferation, Cell Cycle, Cell Migration, Cell Death, and Cell Differentiation seem relevant to these aspects of neurogenesis. 85 genes were associated with Cell Movement (Figure 3a and supplementary Table 3). This category includes cell migration, invasion, chemotaxis, transmigration and cell movement. Cell migration is important in the context of neurogenesis in the dentate gyrus where differences in the rate of migrating cells have been reported as a result of aging (Kuhn, Dickinson-Anson et al. 1996; Heine, Maslam et al. 2004; Rao, Hattiangady et al. 2005). Not surprisingly, in this group we found doublecortin (DCX), which regulates migration of neurons and is a known marker for newly generated neurons in the dentate (Rao and Shetty 2004). DCX was downregulated in aged rats.
It also is worth noting two genes in this group that appear to interact with a large number of genes in the network (see Figure 3b and supplementary Table 3), namely tumor necrosis factor-alpha (TNF) and Kit Ligand (KITL). TNF belongs to the cytokine family and regulates a wide spectrum of biological processes including cell proliferation, differentiation, apoptosis, lipid metabolism, and neuroprotection. A role for TNF in learning and memory has been reported (Aloe, Properzi et al. 1999; Golan, Levav et al. 2004) and TNF is found to interact with a large number of focus genes in the study (Figure 3b, green lines).
KITL encodes the ligand of the tyrosine-kinase receptor encoded by KIT proto-oncogene receptor. This ligand is a pleiotropic factor that acts in neural cell development, and plays a role in cell migration and apoptosis. KITL is expressed in the dentate gyrus and its expression is restricted to neurons (Wong and Licinio 1994). Animals that lack the receptor show impairments in spatial learning and synaptic potentiation (Katafuchi, Li et al. 2000). Animals mutant for KITL also are deficient in hippocampal learning (Motro, Wojtowicz et al. 1996).
The next category was Cellular Growth and Proliferation, which includes growth, proliferation, expansion and differentiation of cells and is also pertinent to the possible formation of new cells in this area of the hippocampus. 37 genes were associated with this function. Not surprisingly, in the Cell Cycle function (supplementary Table 3) we found thirty genes involved in cell cycle progression indicating the activity of dividing cells in this region.
Interestingly, a few well-known markers of the distinct differentiation stages of dentate neurogenesis were downregulated with age, namely S100b which is involved in the proliferation of astrocytes and it is used as a marker for early progenitors. Also, collapsing-response mediated protein (DPYSL5), which interacts with DPYSL2 another widely used developmental-stage marker (better known as TOAD 64), is known to be expressed in immature neurons. HTR2A is another well-characterized gene that was downregulated in the aged dentate. Loss of this serotonin receptor has been reported with age and serotonin is known to affect production of new neurons (Reynolds, Jansson et al. 2006).
TUBB (beta tubulin) was downregulated and interacts with TUBB3 (Tuj1) which is also an early mature neuronal marker (Miller, Naus et al. 1987). Downregulation of all these genes seems to indicate a decrease in neuronal development in this region associated with aging. It is important to point out that even though astrocytes serve as neuronal precursors (Doetsch, Caille et al. 1999; Laywell, Rakic et al. 2000; Noctor, Flint et al. 2001; Seri, Garcia-Verdugo et al. 2001; Noctor, Flint et al. 2002), there was only differential expression of GFAP (it was actually upregulated), but not other astrocytic markers such as vimentin, musashi, 3-PGDH, tenascin-C, Mash1 (a transcription factor that maintains precursor state), or nestin (Johnson, Zimmerman et al. 1992; Steindler and Laywell 2003; Seri, Garcia-Verdugo et al. 2004). Only S100b which is involved in the proliferation of horizontal astrocytes was downregulated, as mentioned before. This could indicate that the rate limiting step in the decrease in neurogenesis with senescence is not the proliferation of radial astrocytes but other proliferative precursor cells that mark the transition between glia-like states and neuronal differentiation (as suggested by the fact that markers for later fate determintation such as DCX, TUBB, DPYSL5 and HTR2A were downregulated (Encinas, Vaahtokari et al. 2006; Steiner, Klempin et al. 2006)).
We would like to emphasize that we dissected the entire dentate gyrus, which represents a heterogeneous group of cell types, therefore we cannot discriminate which cell types are differentially expressed. We could be looking at changes in any type of dividing cells in this area, and some of these progenitor cells could give rise to glia and not new neurons.
Cell Death was the group that contained the largest number of associated genes, 134 in total (Supplementary Table 3). Of those, 120 genes were found associated with apoptosis which plays a key role in nervous system development, and which is an integral aspect of neurogenesis and aging (Heine, Maslam et al. 2004). The genes that were found to be involved specifically in the apoptosis of neurons were CASP9, CCL4, CTSB, CXCL12, ERBB3, HIPK2, ID2, KCNJ6, MAPK10, MAPK8IP1, MAPT, MYCN, NR4A2, PAWR, PTPNS1, SMN1, SMPD2, and TNF.
3.3.2. Nervous System Development and Function; Cellular Development
Sixty nine genes were catalogued as part of Nervous System Development and Function. Of these, the largest subdivision included 26 genes specifically associated with neurogenesis (Table 2 and Supplementary Table 2). Twelve genes were found to be involved in migration of neurons. In addition six were found as implicated in migration of neuroglia.
Table 2. List of genes found to be related to Nervous System Development and Function.
Process annotation is the description found by IPA that describes the cellular process of a particular gene. The significance is calculated by the IPA software as described in materials and methods. The genes are represented by their gene symbol.
| Process Annotation | Significance | Genes |
|---|---|---|
| quantity of nervous tissue cell lines | 1.88E-04 | COL18A1, SMPD2, TNF |
| quantity of synaptic vesicles | 5.53E-03 | KIF1B, MYO5A, RIMS1 |
| migration of Schwann cells | 1.57E-03 | CSPG4, KDR, LAMA2, NF1, TIAM1 |
| migration of neurons | 4.68E-03 | BARHL2, CXCL12, DCC, DCTN2, DCX, EDNRB, ITGA6, LAMA2, MAPT, MRLC2, PTK2, TIAM1 |
| migration of neuroglia | 1.55E-02 | CSPG4, KDR, LAMA2, NF1, TIAM1, TNR |
| binding of microqlia | 3.30E-03 | ITGA6, TNF, TNR |
| differentiation of oligodendrocytes | 4.61 E-03 | ERBB3, FGFR3, ID2, TG, TNF |
| differentiation of neuroglia | 4.63E-02 | ERBB3, FGFR3, ID2, NF1, TG, TNF |
| exocytosis of synaptic vesicles | 5.06E-03 | APBA2, CPLX1, CPLX2, RIMS1 |
| chemotaxis of nervous tissue cell lines | 5.53E-03 | CXCL12, PTPNS1, SMAD3 |
| long term depression | 8.77E-03 | B2M, CAMK4, CAMK2A, GRIA3, GRM1, MYO5A |
| long term depression of synapse | 1.83E-02 | GRM1, MYO5A |
| transport of synaptic vesicles | 1.05E-02 | AMPH, APBA2, CPLX1, CPLX2, RIMS1 |
| extension of neuritis | 1.39E-02 | BARHL2, CAB39, CDKN1A, ELK1, LAMA2, MUSK, MYH9, PITPNM1 |
| elongation of neuritis | 1.40E-02 | AGTR2, CAMK2A, JUNB, MAPT |
| plasticity of synapse | 1.40E-02 | NCDN, PPT1, RIMS1, S100B |
| short-term memory of mice | 1.83E-02 | CAMK2A, TNF |
| survival of retinal ganglion cells | 1.83E-02 | CXCL12, TNF |
| learning of rats | 2.93E-02 | JUNB, SGK |
| long-term memory | 3.49E-02 | CAMK2A, CRH, NFKB1 |
| neurogenesis | 4.56E-02 | ADORA1, ADRB2, APBA2, BARHL2, CHRM2, CNTN3, CSPG5, CXCL12, DCC, EDNRB, GFAP, GFRA2, GRIP1, HAP1, ID1, LAMA2, MAFK, MYCN, NELF, NR4A2, PPT1, PSCD2, SCRG1, TIMM8A, TNR, TRPV1 |
Cellular Development includes genes involved in development, maturation, differentiation and morphogenesis. The largest number of genes corresponded to development of cells (59 associated genes), and included PTK2, a gene that encodes a cytoplasmic protein tyrosine kinase which is found concentrated in the focal adhesions between cells. PTK2 is a member of the FAK (focal adhesion kinase) subfamily of protein tyrosine kinases. Activation of this gene by neural peptides or by molecules in the extracellular matrix could be an essential early step in cell growth. This multifunctional gene appeared in several of the functional groups we identified such as cell movement, cellular growth and proliferation, cell morphology and cell death. It has been found to interact with NMDA receptors and to be necessary for long-term potentiation in the dentate (Yang, Ma et al. 2003).
3.3.3. Cellular Assembly and Organization; Cellular Function and Maintenance; Cell Morphology
The functional groups comprising Cellular Assembly and Organization, Cellular Function and Maintenance, and Cell Morphology, bring together genes engaged in the establishment and maintenance of cell shape. Cellular Assembly, Organization and Maintenance functional groupings include genes whose products are involved with the formation and organization of cytoskeleton, formation of filaments, contact growth inhibition, dynamics of synaptic vesicles, extension of plasma membrane projections, exocytosis, endocytosis, and elongation of neurites.
Cell Morphology involve morphogenesis (29 genes), cell transformation (28 genes), spreading, cell change (30 genes), and polarity genes. All these categories are relevant to the stages of the post-mitotic mature neuron in acquiring its final morphology, and extending dendrites and axons through the granule cell layer (Zhao, Teng et al. 2006).
3.3.4 Neurogenesis as the prominent biological process associated with aging in the dentate
The dentate gyrus has been the focus of recent interest due to the discovery of ongoing neurogenesis in adults (Altman and Das 1965; Cameron, Woolley et al. 1993; Kuhn, Dickinson-Anson et al. 1996). Moreover, it has been suggested that neurogenesis in this region of the hippocampus plays a role in learning and memory (Barnea and Nottebohm 1996; Gould, Beylin et al. 1999; Shors, Miesegaes et al. 2001), although reports are conflicting (Leuner, Gould et al. 2006). For example, one study reported that learning enhances neurogenesis in young animals. (Gould, Beylin et al. 1999; Shors, Miesegaes et al. 2001). On the other hand, even though cell genesis in the hippocampus appears to decline with age (Seki and Arai 1995; Kuhn, Dickinson-Anson et al. 1996; Lemaire, Koehl et al. 2000; Lichtenwalner, Forbes et al. 2001), this neurogenesis doesn’t appear to correlate with learning ability in aged rats (Bizon and Gallagher 2003; Merrill, Karim et al. 2003).
Neurogenesis in the hippocampal dentate gyrus involves several cellular processes that begin with the proliferation of progenitor cells in the subgranular zone. These cells grow, differentiate and mature into adult neurons as they migrate through the granule cell layer. The differentiated neurons must then extend axons and dendrites. Finally, they must establish electrophysiological properties to form functional connections (van Praag, Schinder et al. 2002; Seri, Garcia-Verdugo et al. 2004). Superimposed on these neuro-developmental stages, a balance between neurogenesis and cell death must occur to keep a continuous rate of cell turnover. This developmental program is likely to be maintained by the orchestrated expression of a set of genes that govern the development and timing of neurogenesis, as well as maintain the equilibrium between neurogenesis and cell death. A number of genes have been identified as driving these processes, and markers for the different fate determining stages have been described, but the complete molecular pathway has not been elucidated.
In conclusion, based on the number and statistical significance of genes that were found to be affected by aging, the most prominent appeared to be involved in processes that involve cell division, cell death and apoptosis, migration of cells, and differentiation, all of which are consistent with changes in the different stages of neurogenesis. These changes at the molecular level agree with studies at the cellular level that report changes in rate of migration, differentiation and neurogenesis with aging (Seki and Arai 1995; Kuhn, Dickinson-Anson et al. 1996; Lichtenwalner, Forbes et al. 2001; Heine, Maslam et al. 2004). This study shows that the dentate gyrus is a very dynamic region and that aging results in changes in this activity. We note that using Ingenuity Pathway Analysis we found relationships for only 427 genes out of the 1129 identified by the microarray analysis. Of those 1129 probe sets, approximately 250 corresponded to EST sequences, and approximately 400 were genes that using IPA could not find any relationships with other genes. It is remarkable that of the 427 genes identified by the IPKB, 52% percent (220 genes) were involved in the functions described above (Figure 3b). It is also important to note that this is an under-representation of the neurogenic events going on since not all the genes involved in neurogenesis are known, and half of the genes that were differentially expressed have unknown identities.
In looking at the functions so far described (namely cell movement, growth, proliferation and death; nervous system development and function; cellular development, assembly and organization; cellular function maintenance and morphology), and the number of genes differentially transcribed between the young and aged rats, one can easily picture the putative progression from stem cell proliferation, apoptosis, migration, differentiation, and final establishment of shape with the definition of axons and dendrites. This would be followed by cell death, and the cycle would start again (Figure 3c). This study is a contribution of interest to those studying aging, neurogenesis and learning and memory since we have provided a long list of candidate genes that are likely involved in these biological processes.
The age-related genes we described in the dentate are different than those we found in the CA1 region except for an overlap of 59 genes (from a total of 431 probe sets identified in the CA1 at p<0.005, and the 1129 probe sets identified in the dentate). The functions containing the majority of the common genes were related to cell morphology, cell transformation and cell death (Burger, unpublished data). The functions found in the CA1 field were different than for the dentate (Burger, unpublished data) and no genes were found to be involved in neurogenesis. We must stress the fact that this is only a qualitative comparison, since for the CA1 we used the U34 rat chip (which contains approximately 1/3 of the rat genome) whereas for this one we used the second generation RAE 230A (approximately 2/3 of the rat genome). In addition, different RNA amplification techniques were used for the different studies (Burger, Cecilia Lopez et al. 2007).
Finally, there are many other interesting molecular functions in the list of genes identified, such as lipid metabolism, amino acid metabolism, cell signaling, behavior, neurological disease, but for the sake of conciseness we narrowed the discussion of the biological processes down to the most salient and significant one, i.e. neurogenesis (Fig 3b and c). The reader can find other facets of aging that have not been described here by looking at the genes in this long list of significant changes that occur during aging that are available as supplementary data (Supplementary Table 1 and 2).
3.4. Supervised Analysis of Learning-associated Genes in Aged Rats
We were interested in identifying the transcriptional profiles related to learning in aged animals that distinguished between superior learners and learning-impaired rats. We have previously described the changes in transcription between these two groups of animals for the CA1 region. In that study we distinguished two classes of aged animals. Similarly, in this study we also separated the experimental animals into two classes. To subtract contributions from stress (yoked group), motor activity (visible platform group) and other (cage group), all these groups were considered as one single class for the analysis. In this group we also included the animals that were impaired in the MWM, and we referred to them together as the control class (CTL class). The other experimental group was composed of the superior learning animals (SL class).
The rationale for collapsing the control groups and the AI group as one class for the supervised analysis is based on the following argument: we wanted to look at the gene differences that were a result of learning, not those patterns of expression due to steady-state or constitutive differences that were present between the two groups: SL vs AI. Since the control groups (yoked, visible, caged) had not been segregated into SL or AI prior to the experiment, these groups must certainly represent a mixture of both superior learners and impaired animals. Therefore, any gene expression patterns that might have present in SL animals in a constitutive manner, would have also been present in the control groups. A consequence of this control group design is that the genes we identified are likely to include genes that are directly causal for spatial learning.
To minimize false positives, we used at least three true biological replicates for each condition (see Table 1) and performed the t-test at a stringency of p<0.005 to assess the reproducibility of this model system. At a significance threshold of p<0.005, we identified 85 probe sets with significant differences in hybridization signal intensity (Fig 4, Table 3, and Supplementary Table 3). Leave-one-out-cross validation studies were used to determine if probe sets significant at p < 0.005 were capable of truly distinguishing between the groups. Leave-one-out validation studies indicated that the genes were true 79% of the time (the 1-nearest neighbors classifier had a p-value of 0.003).
3.5. Pathway Analysis of Transcriptional Profiles Determined by MWM Training in Aged rats
In order to classify these learning-associated genes into functional groups a series of bioinformatics tools were used (see Methods). A putative classification of the genes according to function has been summarized in Table 3 and supplementary Table 4. The tentative classification involves cytoskeletal genes, genes involved in regulation of transcription and translation, posttranslational modification including kinases and phosphatases, growth factors, and ten genes whose function could be correlated with neurotransmission. A smaller number of genes were found involved in lipid metabolism and mitochondrial function. The rest of the genes were uncharacterized EST. Only six genes were upregulated in the SL animals. This is striking compared to the differences between young and aged rats where about half of the genes are upregulated and half are downregulated. We do not understand the meaning of this result, but in the CA1 region we also found three fourths of the genes downregulated in SL animals compared to the controls.
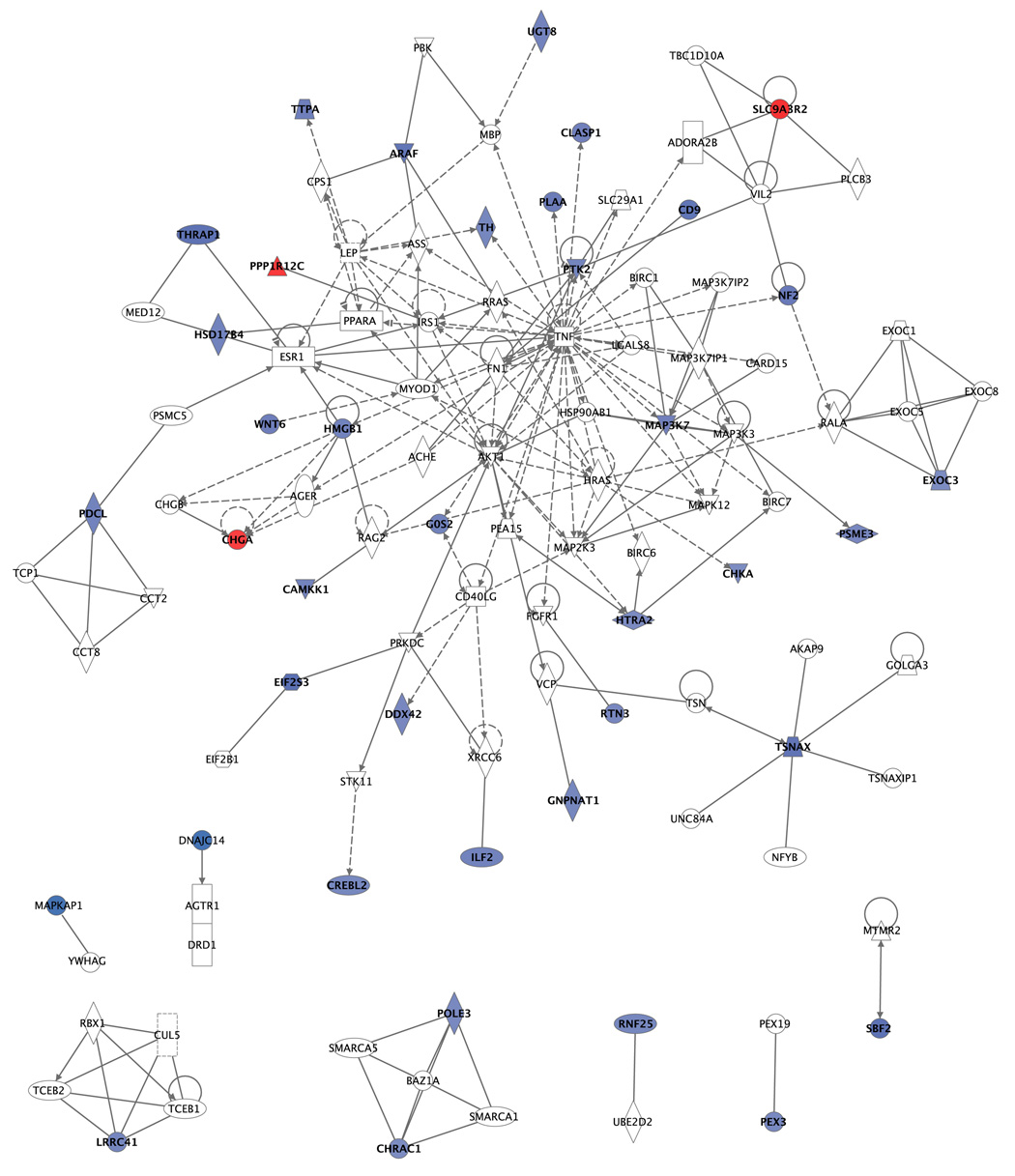
We then investigated the relationships between these genes. IPA analysis revealed that 37 of the genes could be used to generate networks (see supplementary information on IPA for details). Figure 5 illustrates the types of interactions that occur between the genes identified in the study, and also illustrates that there are a lot of missing links (genes in white are genes that did not significantly change but were used to connect the significant genes). Functions included Cellular Assembly and Organization, Cell Morphology, Cell Cycle, Cell Movement, DNA Replication, Recombination and Repair. Except for the first function which contained 10 genes (HMGB1, CLASP1, CD9, CHRAC1, POLE3, PTK2, TH, CHGA, HTRA2, PEX3), the others were only populated by five or less focus genes (Data not shown).
Figure 5. Network representation of the genes identified by IPA in the SL versus CTL comparison (tentative figure).
37 genes were used to generate networks that include interactions between the genes in the network. Genes that are colored in blue were downregulated in the SL compared to controls. Genes in red were upregulated in SL. Genes in white color were not significantly changed in the analysis. They were found by IPA as the “missing links” to network the significant genes in the study.
There were nineteen transcripts that overlapped between the aged versus young and the SL versus CTL groups (Table 4). Namely, ten EST, and nine known genes: PXK, EXO3, CAMKK1; MAPKAP1, PTK2; HTRA2, CREBL2, E1F2B3 and NF2. Interestingly, all these genes were upregulated in aged rats and were downregulated in SL (i.e. SL animals have “young” levels of expression of these genes). The list is too short (and some of the genes are uncharacterized) to make any connections between these genes. Based on the large number of genes that were regulated by aging (1129 genes), the question that emerges is how do the aged superior learners compensate for these changes to manage to learn the task almost as well as young animals? Possibly the aged-related genes do not have a role in learning and memory in spite of their apparent role in neurogenesis. Some reports argue that neurogenesis is not necessary for learning in the aged rodent (Bizon and Gallagher 2003) but that indeed it might be detrimental (Bizon and Gallagher 2005), whereas it might be important in the young animal (Kempermann and Gage 2002). If so, are the 85 genes that are differentially expressed between SL and the AI sufficient to result in the learning deficits observed in the impaired rats? We must point out that the list of learning genes is incomplete due to the stringency of the statistical analysis which removes false negatives at the expense of false positives. So probably the list is longer and other genes are involved in the learning deficits associated with aging.
Table 4. Nineteen transcripts overlap in the two class prediction analyses.
The transcripts that overlap between the aged versus young and SL versus controls.
| Probe set | Gene Symbol |
|---|---|
| 1398936_at | E1F2B3 |
| 1389411_at | Transcribed locus |
| 1377149_at | Transcribed locus |
| 1367478_at | HTRA2/PRSS25 |
| 1376318_at | Transcribed locus |
| 1372502_at | NF2 |
| 1374233_at | Similar to RIKEN cDNA 1810055E12 |
| 1376627_at | Transcribed locus |
| 1377007_at | MAPKAP1 |
| 1389624_s_at | EXO3 |
| 1390422_at | PXK |
| 1388524_at | Transcribed locus |
| 1389417_at | Transcribed locus |
| 1387875_at | PTK2 |
| 1390467_at | Transcribed locus |
| 1371522_at | Transcribed locus (Similar to mKIAA0945 protein) |
| 1373618_at | CREBI2 |
| 1368156_at | CAMKK1 |
| 1375952_at | Transcribed locus |
Finally, the list of learning-related genes expressed in the dentate represent a different set of genes than those we previously described in the CA1 region (Burger, Cecilia Lopez et al. 2007). We must reiterate that because of the technical differences in the analysis of the two hippocampal regions, sampling might have been incomplete and we might have only identified in each case a small number of the genes that are truly different between CA1 and dentate. Still, these results are suggestive of differential age-related molecular changes in these two distinct anatomical regions. This is not surprising since distinct anatomical hippocampal boundaries have been shown to reflect differential gene expression
3. 6. Are these transcripts differentially expressed as a result of learning?
It is important to stress that we designed the supervised statistical analysis to look at differences in gene expression after learning. Different groups of controls were included to rule out other sources of gene expression unrelated to learning the spatial task. In addition, the different groups of control animals were included with the AI animals in a single class in order to discard any differences in gene expression between AI and superior learners that might have existed a priori (see section 3.4 for detailed explanation). If indeed a constitutive set of genes existed that might have conferred the superior learners the ability to learn the paradigm, we should have found expression profiles in the control groups that matched the SL group. To test this possibility, we performed supervised analysis using different classes in which each experimental group was placed in a separate class (i.e., cage controls, yoked, visible, SL, and AI). We also tried controls (yoked, visible and cage controls as one class) vs. SL and AI. Finally, we also tried just SL vs. AI alone. None of these alternative comparisons produced statistically significant differences (using a significance threshold of p < .001 or p < .005) in gene expression. We found differences between the different groups at a lower statistical significance threshold of p <.01, but this is not an adequate level of statistical confidence for analysis of microarrays.
Statistically signifcant differences emerged only when we did the controls plus impaired versus SL. Thus, the fact that no significant transcriptional patterns were found in the different comparisons suggests that the list of differentially expressed mRNAs identifed in this study are likely due to MWM training and not to pre-existing constitutive expression of genes that improved learning in aged SL animals.
Even though stress associated with the MWM can produce dramatic effects on cognition, neurochemistry and gene expression, no significant hippocampal stress-related genes were identified in this study (Seligman ME 1967; Sapolsky, Krey et al. 1984; Mendelson and McEwen 1991; Amat J 1998; Deak T 1999; Grillo, Piroli et al. 2003; Alfonso, Pollevick et al. 2004; Liu, Bertram et al. 2004; Nacher, Pham et al. 2004; Piroli, Grillo et al. 2004; Reagan, Rosell et al. 2004; Huang, Jen et al. 2005; Kohen, Kirov et al. 2005; MacPherson, Dinkel et al. 2005; Mitsukawa, Mombereau et al. 2005; Wiedenmayer, Magarinos et al. 2005; Marini, Pozzato et al. 2006). Still, differential response to stress, other sensorimotor differences, or motivational differences between AI and aged SL animals (as opposed to learning differences) cannot be ruled out. Individual animals might respond to the stress of the MWM differently. Indeed, AI animals showed much longer path lengths on the early trials than SL and young animals (Fig. 1A). AI animals also displayed thigmotaxis (Fig. 1B). This stress response in the learning impaired animals might have prevented them from adopting a spatial search strategy, and therefore, prevented spatial learning. In fact, the results show that AI animals did not adopt a spatial search strategy during training or in the probe trial (Fig 1). So, whether the transcriptional profiles are a cause or a result of the behavioral differences between the groups of animals will have to be investigated in experiments where the time course of expression of these putative genes is analyzed.
3.7. What is the role that these age- and learning-related changes play in the hippocampal formation?
We have designed a MWM experiment that, together with microarray analysis, has potentially identified some of the players involved in learning and memory in the aged dentate gyrus. It was particularly difficult to find the biological role that these 85 transcripts might play in learning and memory in the hippocampus for two reasons. First, many pieces of the puzzle are missing due to the large number of EST, the stringency of the statistical analysis that resulted in false negatives, and the lack of knowledge about how these genes are related to each other. Second, pathway analysis works when there are a large number of gene differences to populate the pathway. The first step following gene discovery requires the demonstration that these genes are functionally implicated in learning and memory. This task will be possible using genetic techniques such as transgenic animals or viral gene delivery to alter the normal function of these candidates, since we have a relatively short list of genes to screen. Therefore it will be crucial to validate the role of each individual gene in learning and memory formation, to then begin piecing together individual pathways involving the genes in this array experiment. Neuronal-specific genes represent one third of the genome, and functional genomic approaches used in the characterization of these genes will facilitate understanding how the nervous system operates.
Supplementary Material
Supplementary Table 1. Statistical validation of the microarray data for the age comparison. The file contains the output data from BRB listing the 1129 genes identified as significant in the aged versus young class comparison (p< 0.005). The first part of the file shows the results of the statistical validation. The next table in the file shows the significant genes with their corresponding parametric p value, geometrical mean of expression intensity for each class, and a cross-validation % support (%CV). The Genebank accession numbers for each gene are hyperlinked to Netaffix. The gene symbols are hyperlinked to Entrez.
Supplementary Table 2. Genes in Top 15 networks Generated from IPA. The gene symbols are colored in red or blue, to indicate that the gene was upregulated or downregulated in the aged group when compared to young.
Supplementary Table 3. Same as Table 3, but with annotations and hyperlinks to Netaffix. List of 85 genes identified by BRB as significantly different between superior learners and controls including learning impaired. Column Y indicates the 28 rat probe sets that were identified by IPA.
Acknowledgements
Support for this work was provided by an Evelyn F. McKnight Brain Research grant to CB, and by an NIH grant NS PO136302 to NM and RJM. We would like to acknowledge the rat subjects who contributed to this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Aitken DH, Meaney MJ. Temporally graded, age-related impairments in spatial memory in the rat. Neurobiol Aging. 1989;10(3):273–276. doi: 10.1016/0197-4580(89)90062-6. [DOI] [PubMed] [Google Scholar]
- Alfonso J, Pollevick GD, et al. Identification of genes regulated by chronic psychosocial stress and antidepressant treatment in the hippocampus. Eur J Neurosci. 2004;19(3):659–666. doi: 10.1111/j.1460-9568.2004.03178.x. [DOI] [PubMed] [Google Scholar]
- Amat J, M-A P, Watkins LR, Maier SF. Escapable and inescapable stress differentially and selectively alter extracellular levels of 5-HT in the ventral hippocampus and dorsal periaqueductal gray of the rat. brain research. 1998;797(1):12–22. doi: 10.1016/s0006-8993(98)00368-0. [DOI] [PubMed] [Google Scholar]
- Aloe L, Properzi F, et al. Learning abilities, NGF and BDNF brain levels in two lines of TNF-alpha transgenic mice, one characterized by neurological disorders, the other phenotypically normal. Brain Res. 1999;840(1–2):125–137. doi: 10.1016/s0006-8993(99)01748-5. [DOI] [PubMed] [Google Scholar]
- Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124(3):319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- Barnea A, Nottebohm F. Recruitment and replacement of hippocampal neurons in young and adult chickadees: an addition to the theory of hippocampal learning. Proc Natl Acad Sci U S A. 1996;93(2):714–718. doi: 10.1073/pnas.93.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Hosack DA, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003;4(1):61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon JL, Gallagher M. Production of new cells in the rat dentate gyrus over the lifespan: relation to cognitive decline. Eur J Neurosci. 2003;18(1):215–219. doi: 10.1046/j.1460-9568.2003.02733.x. [DOI] [PubMed] [Google Scholar]
- Bizon JL, Gallagher M. More is less: neurogenesis and age-related cognitive decline in Long-Evans rats. Sci Aging Knowledge Environ. 2005;2005(7):re2. doi: 10.1126/sageke.2005.7.re2. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Burger C, Cecilia Lopez M, et al. Changes in transcription within the CA1 field of the hippocampus are associated with age-related spatial learning impairments. Neurobiol Learn Mem. 2007;87(1):21–41. doi: 10.1016/j.nlm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Cameron HA, Woolley CS, et al. Differentiation of newly born neurons and glia in the dentate gyrus of the adult rat. Neuroscience. 1993;56(2):337–344. doi: 10.1016/0306-4522(93)90335-d. [DOI] [PubMed] [Google Scholar]
- Clark AS, Magnusson KR, et al. In vitro autoradiography of hippocampal excitatory amino acid binding in aged Fischer 344 rats: relationship to performance on the Morris water maze. Behav Neurosci. 1992;106(2):324–335. doi: 10.1037//0735-7044.106.2.324. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC. Exercise: a behavioral intervention to enhance brain health and plasticity. Trends Neurosci. 2002;25(6):295–301. doi: 10.1016/s0166-2236(02)02143-4. [DOI] [PubMed] [Google Scholar]
- Deak TNK, Cotter CS, Fleshner M, Watkins LR, Maier SF, Spencer RL. Long-term changes in mineralocorticoid and glucocorticoid receptor occupancy following exposure to an acute stressor. Brain Res. 1999;847(2):211–220. doi: 10.1016/s0006-8993(99)02050-8. [DOI] [PubMed] [Google Scholar]
- deToledo-Morrell L, Geinisman Y, et al. Age-dependent alterations in hippocampal synaptic plasticity: relation to memory disorders. Neurobiol Aging. 1988;9(5–6):581–590. doi: 10.1016/s0197-4580(88)80117-9. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, et al. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Vaahtokari A, et al. Fluoxetine targets early progenitor cells in the adult brain. Proc Natl Acad Sci U S A. 2006;103(21):8233–8238. doi: 10.1073/pnas.0601992103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Dunnett SB, et al. Spatial learning and motor deficits in aged rats. Neurobiol Aging. 1984;5(1):43–48. doi: 10.1016/0197-4580(84)90084-8. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Burwell R, et al. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107(4):618–626. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Nicolle MM. Animal models of normal aging: relationship between cognitive decline and markers in hippocampal circuitry. Behav Brain Res. 1993;57(2):155–162. doi: 10.1016/0166-4328(93)90131-9. [DOI] [PubMed] [Google Scholar]
- Golan H, Levav T, et al. Involvement of tumor necrosis factor alpha in hippocampal development and function. Cereb Cortex. 2004;14(1):97–105. doi: 10.1093/cercor/bhg108. [DOI] [PubMed] [Google Scholar]
- Gould E, Beylin A, et al. Learning enhances adult neurogenesis in the hippocampal formation. Nat Neurosci. 1999;2(3):260–265. doi: 10.1038/6365. [DOI] [PubMed] [Google Scholar]
- Goyns MH, Charlton MA, et al. Differential display analysis of gene expression indicates that age-related changes are restricted to a small cohort of genes. Mech Ageing Dev. 1998;101(1–2):73–90. doi: 10.1016/s0047-6374(97)00166-8. [DOI] [PubMed] [Google Scholar]
- Grillo CA, Piroli GG, et al. Region specific increases in oxidative stress and superoxide dismutase in the hippocampus of diabetic rats subjected to stress. Neuroscience. 2003;121(1):133–140. doi: 10.1016/s0306-4522(03)00343-9. [DOI] [PubMed] [Google Scholar]
- Heine VM, Maslam S, et al. Prominent decline of newborn cell proliferation, differentiation, and apoptosis in the aging dentate gyrus, in absence of an age-related hypothalamus-pituitary-adrenal axis activation. Neurobiol Aging. 2004;25(3):361–375. doi: 10.1016/S0197-4580(03)00090-3. [DOI] [PubMed] [Google Scholar]
- Huang AM, Jen CJ, et al. Compulsive exercise acutely upregulates rat hippocampal brain-derived neurotrophic factor. J Neural Transm. 2005 doi: 10.1007/s00702-005-0359-4. [DOI] [PubMed] [Google Scholar]
- Jiang CH, Tsien JZ, et al. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci U S A. 2001;98(4):1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Zimmerman K, et al. Induction and repression of mammalian achaetescute homologue (MASH) gene expression during neuronal differentiation of P19 embryonal carcinoma cells. Development. 1992;114(1):75–87. doi: 10.1242/dev.114.1.75. [DOI] [PubMed] [Google Scholar]
- Katafuchi T, Li AJ, et al. Impairment of spatial learning and hippocampal synaptic potentiation in c-kit mutant rats. Learn Mem. 2000;7(6):383–392. doi: 10.1101/lm.33900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Gage FH. Genetic determinants of adult hippocampal neurogenesis correlate with acquisition, but not probe trial performance, in the water maze task. Eur J Neurosci. 2002;16(1):129–136. doi: 10.1046/j.1460-9568.2002.02042.x. [DOI] [PubMed] [Google Scholar]
- Kohen R, Kirov S, et al. Gene expression profiling in the hippocampus of learned helpless and nonhelpless rats. Pharmacogenomics J. 2005;5(5):278–291. doi: 10.1038/sj.tpj.6500322. [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, et al. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16(6):2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laywell ED, Rakic P, et al. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc Natl Acad Sci U S A. 2000;97(25):13883–13888. doi: 10.1073/pnas.250471697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, et al. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25(3):294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lemaire V, Koehl M, et al. Prenatal stress produces learning deficits associated with an inhibition of neurogenesis in the hippocampus. Proc Natl Acad Sci U S A. 2000;97(20):11032–11037. doi: 10.1073/pnas.97.20.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner B, Gould E, et al. Is there a link between adult neurogenesis and learning? Hippocampus. 2006 doi: 10.1002/hipo.20153. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8) doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenwalner RJ, Forbes ME, et al. Intracerebroventricular infusion of insulin-like growth factor-I ameliorates the age-related decline in hippocampal neurogenesis. Neuroscience. 2001;107(4):603–613. doi: 10.1016/s0306-4522(01)00378-5. [DOI] [PubMed] [Google Scholar]
- Liu YF, Bertram K, et al. Stress induces activation of stress-activated kinases in the mouse brain. J Neurochem. 2004;89(4):1034–1043. doi: 10.1111/j.1471-4159.2004.02391.x. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Gene expression profiling in fetal, aged, and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29(6):1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- MacPherson A, Dinkel K, et al. Glucocorticoids worsen excitotoxin-induced expression of pro-inflammatory cytokines in hippocampal cultures. Exp Neurol. 2005;194(2):376–383. doi: 10.1016/j.expneurol.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Marini F, Pozzato C, et al. Single exposure to social defeat increases corticotrophin-releasing factor and glucocorticoid receptor mRNA expression in rat hippocampus. Brain Res. 2006;1067(1):25–35. doi: 10.1016/j.brainres.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Markowska AL, Stone WS, et al. Individual differences in aging: behavioral and neurobiological correlates. Neurobiol Aging. 1989;10(1):31–43. doi: 10.1016/s0197-4580(89)80008-9. [DOI] [PubMed] [Google Scholar]
- Mendelson SD, McEwen BS. Autoradiographic analyses of the effects of restraint-induced stress on 5-HT1A, 5-HT1C and 5-HT2 receptors in the dorsal hippocampus of male and female rats. Neuroendocrinology. 1991;54(5):454–461. doi: 10.1159/000125951. [DOI] [PubMed] [Google Scholar]
- Merrill DA, Karim R, et al. Hippocampal cell genesis does not correlate with spatial learning ability in aged rats. J Comp Neurol. 2003;459(2):201–217. doi: 10.1002/cne.10616. [DOI] [PubMed] [Google Scholar]
- Miller FD, Naus CC, et al. Isotypes of alpha-tubulin are differentially regulated during neuronal maturation. J Cell Biol. 1987;105(6 Pt 2):3065–3073. doi: 10.1083/jcb.105.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsukawa K, Mombereau C, et al. Metabotropic Glutamate Receptor Subtype 7 Ablation Causes Dysregulation of the HPA Axis and Increases Hippocampal BDNF Protein Levels: Implications for Stress-Related Psychiatric Disorders. Neuropsychopharmacology. 2005 doi: 10.1038/sj.npp.1300926. [DOI] [PubMed] [Google Scholar]
- Motro B, Wojtowicz JM, et al. Steel mutant mice are deficient in hippocampal learning but not long-term potentiation. Proc Natl Acad Sci U S A. 1996;93(5):1808–1813. doi: 10.1073/pnas.93.5.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Takahashi Y, et al. Profiling of genes associated with transcriptional responses in mouse hippocampus after transient forebrain ischemia using high-density oligonucleotide DNA array. Brain Res Mol Brain Res. 2004;121(1–2):1–11. doi: 10.1016/j.molbrainres.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Nacher J, Pham K, et al. Chronic restraint stress and chronic corticosterone treatment modulate differentially the expression of molecules related to structural plasticity in the adult rat piriform cortex. Neuroscience. 2004;126(2):503–509. doi: 10.1016/j.neuroscience.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, et al. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, et al. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22(8):3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroli GG, Grillo CA, et al. Biphasic effects of stress upon GLUT8 glucose transporter expression and trafficking in the diabetic rat hippocampus. Brain Res. 2004;1006(1):28–35. doi: 10.1016/j.brainres.2004.01.044. [DOI] [PubMed] [Google Scholar]
- Rao MS, Hattiangady B, et al. Newly born cells in the ageing dentate gyrus display normal migration, survival and neuronal fate choice but endure retarded early maturation. Eur J Neurosci. 2005;21(2):464–476. doi: 10.1111/j.1460-9568.2005.03853.x. [DOI] [PubMed] [Google Scholar]
- Rao MS, Shetty AK. Efficacy of doublecortin as a marker to analyse the absolute number and dendritic growth of newly generated neurons in the adult dentate gyrus. Eur J Neurosci. 2004;19(2):234–246. doi: 10.1111/j.0953-816x.2003.03123.x. [DOI] [PubMed] [Google Scholar]
- Reagan LP, Rosell DR, et al. Chronic restraint stress up-regulates GLT-1 mRNA and protein expression in the rat hippocampus: reversal by tianeptine. Proc Natl Acad Sci U S A. 2004;101(7):2179–2184. doi: 10.1073/pnas.0307294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CA, Jansson M, et al. Longitudinal change in memory performance associated with HTR2A polymorphism. Neurobiol Aging. 2006;27(1):150–154. doi: 10.1016/j.neurobiolaging.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Krey LC, et al. Stress down-regulates corticosterone receptors in a site-specific manner in the brain. Endocrinology. 1984;114(1):287–292. doi: 10.1210/endo-114-1-287. [DOI] [PubMed] [Google Scholar]
- Sawada T, Morinobu S, et al. Reduction in levels of amphiphysin 1 mRNA in the hippocampus of aged rats subjected to repeated variable stress. Neuroscience. 2004;126(2):461–466. doi: 10.1016/j.neuroscience.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Seligman ME, M S. Failure to escape traumatic shock. J Exp Psychol. 1967;74(1):1–9. doi: 10.1037/h0024514. [DOI] [PubMed] [Google Scholar]
- Schulz D, Huston JP, et al. Water maze performance, exploratory activity, inhibitory avoidance and hippocampal plasticity in aged superior and inferior learners. Eur J Neurosci. 1967;16(11):2175–2185. doi: 10.1046/j.1460-9568.2002.02282.x. [DOI] [PubMed] [Google Scholar]
- Seki T, Arai Y. Age-related production of new granule cells in the adult dentate gyrus. Neuroreport. 1995;6(18):2479–2782. doi: 10.1097/00001756-199512150-00010. [DOI] [PubMed] [Google Scholar]
- Seri B, Garcia-Verdugo JM, et al. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J Comp Neurol. 2004;478(4):359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- Seri B, Garcia-Verdugo JM, et al. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21(18):7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors TJ, Miesegaes G, et al. Neurogenesis in the adult is involved in the formation of trace memories. Nature. 2001;410(6826):372–376. doi: 10.1038/35066584. [DOI] [PubMed] [Google Scholar]
- Steindler DA, Laywell ED, et al. Astrocytes as stem cells: nomenclature, phenotype, and translation. Glia. 2003;43(1):62–69. doi: 10.1002/glia.10242. [DOI] [PubMed] [Google Scholar]
- Steiner B, Klempin F, et al. Type-2 cells as link between glial and neuronal lineage in adult hippocampal neurogenesis. Glia. 2006;54(8):805–814. doi: 10.1002/glia.20407. [DOI] [PubMed] [Google Scholar]
- Tong L, Shen H, et al. Effects of exercise on gene-expression profile in the rat hippocampus. Neurobiol Dis. 2001;8(6):1046–1056. doi: 10.1006/nbdi.2001.0427. [DOI] [PubMed] [Google Scholar]
- van Praag H, Schinder AF, et al. Functional neurogenesis in the adult hippocampus. Nature. 2002;415(6875):1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, et al. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience. 2003;122(3):647–657. doi: 10.1016/j.neuroscience.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Velardo MJ, Burger C, et al. Patterns of gene expression reveal a temporally orchestrated wound healing response in the injured spinal cord. J Neurosci. 2004;24(39):8562–8576. doi: 10.1523/JNEUROSCI.3316-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Zhang Y, et al. Differential display and cloning of the hippocampal gene mRNas in senescence accelerated mouse. Neurosci Lett. 1999;275(1):17–20. doi: 10.1016/s0304-3940(99)00717-x. [DOI] [PubMed] [Google Scholar]
- Wiedenmayer CP, Magarinos AM, et al. Age-specific threats induce CRF expression in the paraventricular nucleus of the hypothalamus and hippocampus of young rats. Horm Behav. 2005;47(2):139–150. doi: 10.1016/j.yhbeh.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Wong ML, Licinio J. Localization of stem cell factor mRNA in adult rat hippocampus. Neuroimmunomodulation. 1994;1(3):181–187. doi: 10.1159/000097159. [DOI] [PubMed] [Google Scholar]
- Yang YC, Ma YL, et al. Focal adhesion kinase is required, but not sufficient, for the induction of long-term potentiation in dentate gyrus neurons in vivo. J Neurosci. 2003;23(10):4072–4080. doi: 10.1523/JNEUROSCI.23-10-04072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Teng EM, et al. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci. 2006;26(1):3–11. doi: 10.1523/JNEUROSCI.3648-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Lein ES, et al. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J Comp Neurol. 2001;441(3):187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Statistical validation of the microarray data for the age comparison. The file contains the output data from BRB listing the 1129 genes identified as significant in the aged versus young class comparison (p< 0.005). The first part of the file shows the results of the statistical validation. The next table in the file shows the significant genes with their corresponding parametric p value, geometrical mean of expression intensity for each class, and a cross-validation % support (%CV). The Genebank accession numbers for each gene are hyperlinked to Netaffix. The gene symbols are hyperlinked to Entrez.
Supplementary Table 2. Genes in Top 15 networks Generated from IPA. The gene symbols are colored in red or blue, to indicate that the gene was upregulated or downregulated in the aged group when compared to young.
Supplementary Table 3. Same as Table 3, but with annotations and hyperlinks to Netaffix. List of 85 genes identified by BRB as significantly different between superior learners and controls including learning impaired. Column Y indicates the 28 rat probe sets that were identified by IPA.