Abstract
The three-dimensional X-ray crystal structure of the ‘rare cutting’ type II restriction endonuclease SgrAI bound to cognate DNA is presented. SgrAI forms a dimer bound to one duplex of DNA. Two Ca2+ bind in the enzyme active site, with one ion at the interface between the protein and DNA, and the second bound distal from the DNA. These sites are differentially occupied by Mn2+, with strong binding at the protein–DNA interface, but only partial occupancy of the distal site. The DNA remains uncleaved in the structures from crystals grown in the presence of either divalent cation. The structure of the dimer of SgrAI is similar to those of Cfr10I, Bse634I and NgoMIV, however no tetrameric structure of SgrAI is observed. DNA contacts to the central CCGG base pairs of the SgrAI canonical target sequence (CR|CCGGYG, | marks the site of cleavage) are found to be very similar to those in the NgoMIV/DNA structure (target sequence G|CCGGC). Specificity at the degenerate YR base pairs of the SgrAI sequence may occur via indirect readout using DNA distortion. Recognition of the outer GC base pairs occurs through a single contact to the G from an arginine side chain located in a region unique to SgrAI.
INTRODUCTION
Sequence-specific endonucleases capable of cleaving longer recognition sequences are highly sought for use in genomic work, as longer sequences occur less frequently and allow the manipulation of larger DNA fragments. Typical type II restriction endonucleases (REs) cleave 6 bp target sequences, however, enzymes capable of cleaving longer sequences of 8 bp have also been identified. The DNA bound structures of only two such enzymes, NotI (1) and SfiI (2), have been reported to date. The ‘rare cutting’ RE SgrAI cleaves an 8 bp recognition (or cognate) sequence, CR|CCGGYG (|denotes cut site) (3), and exhibits unusual biochemical properties including faster cleavage of plasmids bearing two sites over those with only one (4,5), and the cleavage of secondary sites (CR|CCGGY(A,C,T) and CR|CCGGGG) upon cleavage at cognate sites (6). The molecular mechanism of this allostery and specificity modulation is unknown.
Type II REs typically bind and recognize palindromic sequences as dimers (7,8), but the unusual biochemical properties exhibited by SgrAI suggest the formation of a higher order oligomer such as a tetramer. At low enzyme concentrations SgrAI cleaves plasmids bearing one or two sites at equal rates, however higher concentrations of enzyme result in the faster cleavage of the two site plasmid (5). The stimulation of DNA cleavage activity occurs through three-dimensional space as shown by the concerted cleavage of SgrAI cognate sequences on plasmids each bearing a single site but connected by concatenation (9). Cleavage at cognate sequences in plasmids can also be stimulated by the addition of oligonucleotides containing the cognate sequence, either intact or mimicking the cleavage products of SgrAI (6). Analytical ultracentrifugation shows that SgrAI exists as a dimer in the absence of DNA, but forms high molecular mass aggregates in the presence of a 20 bp DNA containing its recognition site (5). The cleavage kinetics also suggest that DNA binding stimulates the formation of an activated conformation, which is presumably a tetramer or higher molecular weight species, as the DNA cleavage velocity shows sigmoidal dependence on DNA concentration (10).
DNA cleavage at cognate sequences by SgrAI stimulates cleavage rates at other cognate sites and at secondary sites as well. The cleavage at secondary sites is 200-fold reduced relative to cognate (6), but this difference is reduced to only 10-fold when the secondary sites are adjacent to DNA ends simulating the products from cognate DNA cleavage (6). In addition, the secondary sites are cleaved 15-fold more slowly relative to cognate when present in a plasmid concatenated to one containing the cognate sequence, and 2.5-fold more slowly when present in the same plasmid (9).
The target site of SgrAI, CRCCGGYG, is similar to those of two 6 bp cleaving REs with known three-dimensional structures, Cfr10I (RCCGGY) and NgoMIV (GCCGGC), that are known to form tetramers. However, although both Cfr10I (11) and NgoMIV (12) cleave two sites in a concerted manner, neither exhibit the modulation of specificity found in the cleavage activity of SgrAI. Here we present the structure of SgrAI bound to DNA and Ca2+ and Mn2+ and compare it to the structures of the related enzymes, Cfr10I and NgoMIV, in order to investigate the mechanism of allosteric modulation of activity and specificity by SgrAI.
MATERIALS AND METHODS
Protein purification
SgrAI restriction endonuclease was expressed in Escherichia coli strain ER2566, from plasmid pET21a_SgrA1R, in the presence of the MspI methyltransferase expressed from the plasmid pBAKMspIM (New England Biolabs). Cells were grown at 37°C in LB medium supplemented with 50 μg/ml ampicillin. Cells were induced to overexpress at an OD600 of 0.5–0.6 with 1 mM isopropyl β-d-thiogalactopyranoside, and incubated for further 4 h, then harvested by centrifugation. Cells were resuspended in 20 mM potassium phosphate buffer (pH 7.0), 50 mM NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol, 50 μM PMSF and 100 μM benzamide, then lysed on ice by sonication. The lysate was centrifuged at 9979g for 1 h and the supernatant was centrifuged again at 125 171g for 1 h. The lysate was loaded onto a Heparin FF (Pharmacia) column and washed with 20 mM potassium phosphate buffer (pH 7.0), 50 mM NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol. Protein was eluted with 20 mM potassium phosphate buffer (pH 7.0), 1 M NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol. The pooled fractions containing SgrAI protein were dialyzed against 40 mM potassium phosphate buffer (pH 6.9), 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol, loaded onto an SP Sepharose FF (Pharmacia) column, and eluted with 40 mM potassium phosphate buffer (pH 6.9), 1 M NaCl, 0.1 mM EDTA, 5% v/v glycerol. The pooled fractions containing SgrAI protein were dialyzed against 10 mM Tris–HCl (pH 8.0), 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol, loaded onto a Q FF (Pharmacia) column, and eluted with 10 mM Tris–HCl (pH 8.0), 1 M NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol. The pooled fractions containing SgrAI protein were loaded directly onto a Heparin FF (Pharmacia) column, and the column was washed with 10 mM Tris–HCl (pH 8.0), 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol. SgrAI was eluted with 10 mM Tris–HCl (pH 8.0), 1 M NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 5% v/v glycerol. SgrAI activity was followed throughout the purification using activity assays and visualization of purity via SDS–PAGE and Coomassie blue staining.
DNA preparation
All oligonucleotides were made synthetically (W. M. Keck Foundation Biotechnology Resource Center, Yale University, New Haven, CT) and purified using C18 reverse phase HPLC (13). The concentration of each single strand was then measured spectrophotometrically, with extinction coefficients calculated from standard values for the nucleotides (14). All oligonucleotides are self complementary (typically 100–400 mM each) and were annealed by heating to 90°C for 10 min, followed by slow-cooling to 4°C over 4–5 h in an Eppendorf thermocycler. The sequences of the DNA duplexes used for cocrystallization are:
Non-cognate 17-2 (G–G mismatch in bold):
5′- AAGTCGACCGGTGGACT-3′
3′- TCAGGTGGCCACCTGAA-5′
Cognate 18:
5′- GAGTCCACCGGTGGACTC-3′
3′- CTCAGGTGGCCACCTGAG-5′
Crystallization
Crystals were prepared with SgrAI and the two duplex oligonucleotides described earlier in the presence of 5–10 mM CaCl2 or 10 mM MnCl2. The crystallization conditions included 1.5–3.0 μl of the protein:DNA mixture with 1.0–1.5 μl of the precipitating solution [25 to 21% PEG 4000, 0.1 M imidazole buffer (pH 6.5) or 0.1 M HEPES buffer (pH 7.5), 0.15–0.20 M NaCl] per drop, and placed over 1 ml of the precipitating solution. The SgrAI concentration varied between 10 and 30 mg/ml in 20 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM DTT, 1 mM EDTA, and was mixed with up to an equal volume of DNA (in sterile deionized water) to give a 1 : 2 molar ratio of SgrAI dimer:DNA duplex. Crystals grow overnight to 1 week at 17°C. The crystal used for dataset S208 was grown in the presence of 1 mM Cu(OAc)2, and that for data set S215 was soaked in a solution containing 1 mM HgI4K2. Crystals used for datasets S30 and S37 were soaked in stabilization solution [25% PEG 4000, 0.1 M HEPES (pH 7.5), 0.3 M NaCl] with 1 mM K2PtCl6 or Hg(SCN)2, respectively, overnight at 17°C. The crystals were then exchanged into a cryoprotection solution [25% PEG 4000, 0.1 M imidazole (pH 6.5) or HEPES buffer (pH 7.5), 0.3 M NaCl, and 30% glycerol] and flash-frozen in liquid nitrogen. X-ray diffraction was measured using synchrotron radiation at the Stanford Synchrotron Light Source (SSRL) BL7-1 (dataset S31), BL1-5 (dataset S30), BL9-1 (dataset S37), BL9-2 (datasets S215, S208), and the Advanced Light Source (Berkeley, CA) BL 5.0.3 (dataset S332). Data collection was performed while maintaining the crystal at 100 K. Image processing and data reduction were performed with MOSFLM (15) and SCALA (16) (datasets S30, S215), HKL2000 (HKL Research, Inc.) (datasets S31, S37, S332) or CrystalClear (Rigaku Americas Corporation) (dataset S208).
Structure solution and refinement
Heavy-atom sites were identified in two derivative datasets using RSPS (17) (Table 1). The MIR phases were combined with molecular replacement (MR) phases prior to density modification, including 2-fold averaging with DM, to produce an interpretable map. The MR solution was obtained in a two-step process. First, a solution was obtained in the C2221 crystal form (Form 3) using PHASER (18,19) and a 219-residue search model based on Cfr10I (PDB entry 1CFR). Electron density encompassing a crystallographic dimer from the C2221 crystal form was used as the search model for the P21 crystal form (Form 1), using PHASER. The model was built using RESOLVE (20), BUCCANEER (21) and O (22). This initial model was refined and modified using the Form 1 (S31) data, and several iterations of refinement with REFMAC5 (23) and model building with the software O (22). The Form 2 structure was determined by molecular replacement and PHASER (18,19), and refined using CNS (24), PHENIX (25), and the model-building program XtalView (26). The Mn2+ bound structure was refined with the Form 2 model, and CNS (24), PHENIX (25), and the model-building program XtalView (26). Coordinates have been deposited in the Protein Data Bank (PDB IDs 3DPG, 3DVO, and 3DW9, for SgrAI with noncognate DNA and Ca2+ (dataset S31), SgrAI with cognate DNA and Ca2+ (dataset S208), and SgrAI with cognate DNA and Mn2+ (dataset S332), respectively.)
Table 1.
Heavy-atom phasing statistics
| Hg sites (data set S37) | Pt sites (data set S30) | |
|---|---|---|
| Number of sites | 2 | 4 |
| Resolution | 2.45 Å | 2.50 Å |
| R-Cullis (acentric/centric) | 0.91/0.80 | 0.96/0.90 |
| Phasing power (acentric/centric) | 0.77/0.63 | 0.43/0.37 |
| FOM | acentric 0.24, centric 0.45, all 0.25 | |
Solvent accessible surface area calculation
The amount of solvent accessible surface area (SASA) buried in the dimer–dimer interface of the Cfr10I, NgoMIV and modeled SgrAI tetramers was calculated using the program AREAIMOL in the CCP4 suite of programs (27), which uses the algorithm of Lee and Richards (28). The SASA was calculated for the whole tetramer, as well as for each dimer independent of the other. The amount of buried SASA was calculated as the difference between the sum of the two dimers with that of the tetramer.
RESULTS
Overall structure
The structure of SgrAI bound to DNA has been determined in two crystal forms, Forms 1 and 2. Form 1 has been refined to 1.91 Å with Rcryst of 18.3% and Rfree of 22.6%, while Form 2 has been determined to a resolution of 1.89 Å with Rcryst of 20.7% and Rfree of 26.7% (Table 2). An additional crystal form, Form 3, was used in the structure solution, but was not refined due to the limiting resolution (2.98 Å). Forms 2 and 3 contain cognate DNA with the cognate sequence CACCGGTG (Cognate 18), while Form 1 contains DNA with G–G mismatches in one side of the recognition sequence (Non-cognate 17-2). The forms differ in the contents of the crystallographic asymmetric units; Form 1 contains one dimer and one duplex of DNA, Form 2 contains two dimers and two duplexes of DNA and Form 3 contains one monomer and one strand of DNA in the respective asymmetric units.
Table 2.
Diffraction data and structure refinement statistics
| Form 1 | K2PtCl6 | Hg(SCN)2 | Form 2 | Form 3 | Mn2+ | |
|---|---|---|---|---|---|---|
| Code | S31 scrai | S30 | S37 | S208 | S215 | S332_Mn |
| BeamLine | SSRL BL7-1 | SSRL BL 1-5 | SSRL BL 9-1 | SSRL BL9-2 | SSRL BL9-2 | ALS 5.0.3 |
| Processing software | MOSFLM/SCALA | MOSFLM/SCALA | HKL2000 | CRYSTALCLEAR | MOSFLM/SCALA | HKL2000 |
| DNA | Noncognate 17-2 | Noncognate 17-2 | Noncognate 17-2 | Cognate 18 | Cognate 18 | Cognate 18 |
| Space group | P21 | P21 | P21 | P21 | C2221 | P21 |
| Cell | 69.73Å, 78.62Å, 86.83Å, β = 109.76° | 70.04Å, 79.38Å, 86.69Å, β = 109.71° | 69.68Å, 78.30Å, 86.91Å, β = 109.82° | 59.14Å, 119.25Å, 123.80Å, β = 95.60° | 50.05Å, 369.47Å, 122.87Å | 58.26 Å, 117.10 Å, 63.70 Å, β = 112.96° |
| Resolution | 1.91 Å | 2.99 Å | 2.24 Å | 1.89 Å | 2.98 Å | 2.2 Å |
| Total observations | 157 240 | 62 540 | 91 302 | 429 421 | 101 213 | 120 959 |
| Unique observations | 66 598 | 18 210 | 39 972 | 126 082 | 25 938 | 35 535 |
| Complete (%) | 96.1 (66.8) | 99.7 (99.9) | 94.2 (95.1) | 92.8 (81.0) | 99.3 (98.2) | 88.8 (57.4) |
| I/sigma | 6.8 (1.5) | 12.6 (3.0) | 16.3 (1.9) | 6.5 (202) | 3.9 (2.2) | 16 (2.0) |
| Multiplicity | 2.4 (2.1) | 3.4 | 2.3 (2.3) | 3.4 (3.2) | 3.9 (4.0) | 3.4 (2.2) |
| Rmergea (%) | 7.1 (48.3) | 9.3 (31.7) | 7.3 (51.1) | 12.0 (53.2) | 23.7 (56.9) | 7.4 (32.2) |
| Rcrystb (%) | 18.3 | 20.7 | 17.4 | |||
| Rfreec (%) | 22.6 | 26.7 | 24.0 | |||
| Overall B factor (Å2, Wilson plot) | 23.8 | 25.1 | 33.7 | |||
| RMSD-bonds | 0.006 | 0.023 | 0.019 | |||
| RMSD-angles | 1.06 | 2.1 | 1.9 | |||
| Numbers of waters | 470 | 776 | 320 | |||
| Number of metal ions | 4 Ca2+ | 8 Ca2+ | 4 Mn2+ |
aRmerge = ∑hkl(|<Ihkl>−Ihkl|)/(∑hkl Ihkl) where <Ihkl> is the average intensity over symmetry related and equivalent reflections and Ihkl is the observed intensity for reflection hkl.
bRcyst = ∑hkl(||Fobs|−|Fcalc||)/(∑hkl|Fobs|) where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitude for reflection hkl. The sum is carried out over the 95–97% of the observed reflections which are used in refinement.
cRfree refers to the R factor for the test reflection set (3–5% of the total observed) which was excluded from refinement.
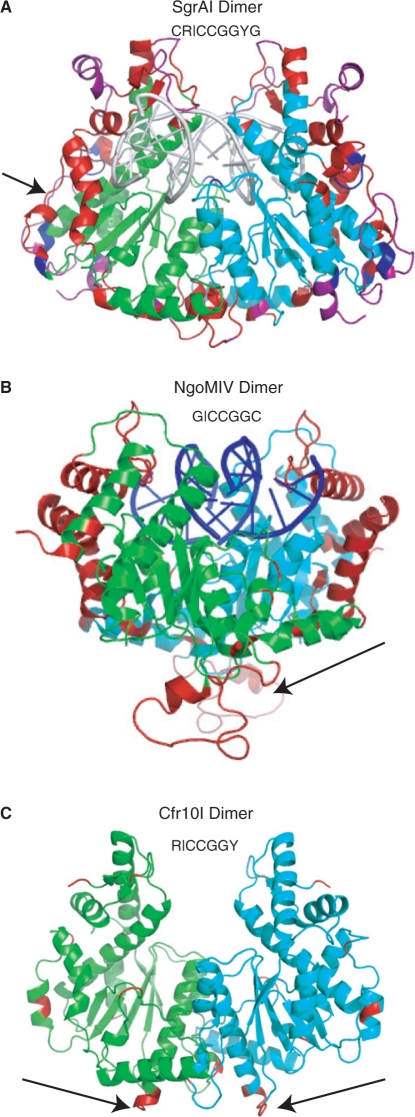
All refined structures of SgrAI bound to DNA show the same global conformation (Figure 1A). In all forms, SgrAI (recognition sequence CR|CCGGYG) forms a dimer very similar to those of NgoMIV (recognition sequence G|CCGGC) (12), Cfr10I (recognition sequence R|CCGGY) (11) and Bse634I (recognition sequence R|CCGGY) (29). (As Bse634I is 31% identical, and 54% similar, in sequence to Cfr10I, structural comparisons were made only with Cfr10I.) The DALI (30) server was used to align the structures of SgrAI with NgoMIV and Cfr10I, shown in Figure S1 as underlined regions of the sequences. In Figure 1A, portions of SgrAI that do not align with NgoMIV or Cfr10I are colored in red or blue, respectively, and those that align with neither are colored in purple. The segments of NgoMIV and Cfr10I that do not align with SgrAI are colored red in Figure 1B and C, respectively. NgoMIV and Cfr10I form tetramers at the face of the dimer opposite the DNA binding face (arrows, Figures 1B–C, 2A and 3A). Segments of these proteins involved in tetramer formation are not conserved in SgrAI (red, Figure 1B and C) and no tetrameric structures like those of NgoMIV or Cfr10I are found in any of the SgrAI crystal forms.
Figure 1.
Dimer structures of SgrAI, NgoMIV and Cfr10I. (A) SgrAI dimer with two subunits in green and cyan. Regions that do not align with NgoMIV, Cfr10I or either, are shown in red, blue and purple, respectively. Arrow indicates the segment corresponding to the loops of NgoMIV that form the dimer–dimer interface. DNA shown as a cartoon in white. (B) NgoMIV dimer with two subunits in green and cyan. Regions that do not align with SgrAI are shown in red. The arrow indicates loops that form the dimer–dimer interface. Bound DNA is shown as a cartoon in blue. (C) Cfr10I dimer with two subunits in green and cyan. Regions that do not align with SgrAI are shown in red. The arrows indicate the extensions of the alpha helices involved in dimer–dimer contacts.
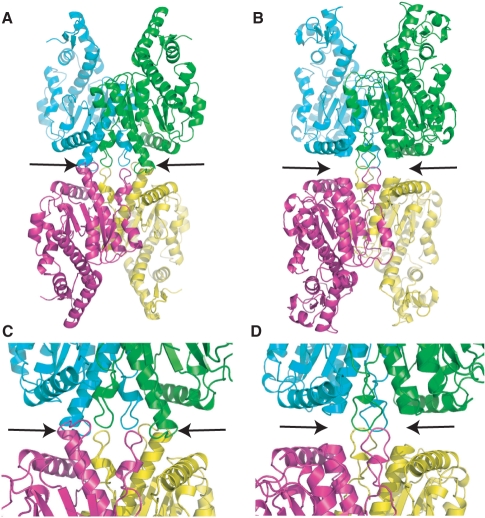
Figure 2.
Cfr10I Tetramer. (A) Tetramer of Cfr10I with individual subunits shown in cyan, green, pink and yellow. Arrows indicate dimer–dimer contacts formed by helical extensions identified in Figure 1C. (B) Model of a SgrAI tetramer where individual subunits of SgrAI are superimposed onto individual subunits of Cfr10I in the Cfr10I tetramer. Alignments were made using the alignment provided by DALI (30), shown in Figure S1. (C) Close-up of the dimer–dimer interface of the Cfr10I tetramer. Arrows as in A. (D) Close-up of the dimer–dimer interface of the SgrAI tetramer model shown in B. Arrows as in B.
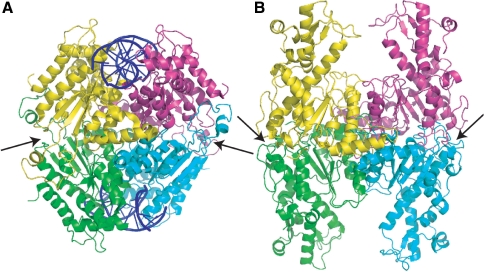
Figure 3.
NgoMIV tetramer. (A) Tetramer of NgoMIV with individual subunits shown in cyan, green, pink and yellow. Arrows indicate dimer–dimer interface formed by the loop extensions identified in Figure 1B. (B) Model of SgrAI tetramer where individual subunits of SgrAI are superimposed onto individual subunits of NgoMIV in the NgoMIV tetramer. Alignments were made using the alignment provided by DALI (30), shown in Figure S1.
Modeling was performed to assess the possibility that SgrAI could form tetrameric complexes like those of Cfr10I and NgoMIV. Figure 2 shows the tetrameric interface of Cfr10I (Figure 2A and C) and the model of SgrAI (Figure 2B and D) after each individual subunit of SgrAI was aligned onto each Cfr10I subunit. It is clear that the additional sequences in Cfr10I (arrows, Figures 1C, 2A and C) serve the purpose of stabilizing this interface. The extensions are absent in SgrAI (Figure 1A), and therefore cannot form the same interface (Figure 2B and D). The amount of solvent accessible surface area (SASA) buried in the dimer–dimer interfaces of the Cfr10I tetramer and the modeled SgrAI tetramer is 4172 and 1027 Å2, respectively. Figure 3 shows the tetrameric interface of NgoMIV (Figure 3A) and the model of SgrAI (Figure 3B) after superposition of each subunit of SgrAI onto those of NgoMIV. Rather than the lack of contacts seen for the Cfr10I-type interface (Figure 2B and D), the interface is overlapped in the SgrAI tetramer model (arrows, Figure 3B). The amount of buried SASA in the NgoMIV tetrameric interface and that of the SgrAI-modeled tetramer is 12 918 and 10 937 Å2, respectively. However, over 3000 close contacts or steric overlaps are found in the SgrAI tetramer model. Clearly, if SgrAI forms a tetramer at this surface, it is different from those of Cfr10I and NgoMIV, which are dissimilar from each other as well.
Protein–DNA contacts
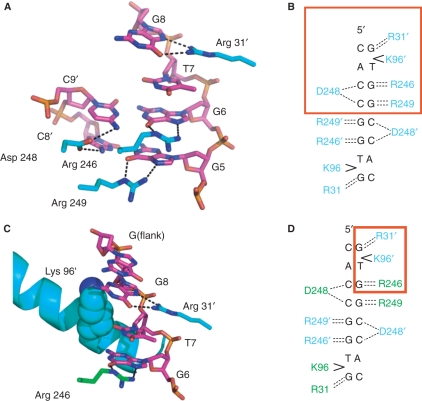
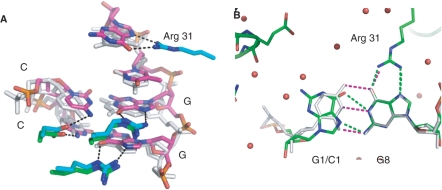
Contacts between SgrAI and the base edges of one-half site of the bound DNA from the Form 1 structure are shown in Figure 4A and C, and schematically in Figure 4B and D. The contacts to the central CCGG bases are identical to those found in the NgoMIV/DNA structure (recognition sequence G|CCGGC, Figure 5A) and utilize the common segment RSDR in both proteins (found as RPDR in Cfr10I). No direct contacts are made to the TA base pair, where the SgrAI specificity is YR, however an unusual DNA distortion is made by Lys 96 (Figure 4C). The side chain of Lys 96 contacts the outer G of the SgrAI recognition sequence CRCCGGYG from the minor groove and unstacks this base from the T that just precedes it. This distortion could be contributing to sequence specificity. The outer G is directly contacted by Arg 31; both Arg 31 and Lys 96 are from a different subunit in the dimer than the residues contacting the GG nucleotides that just precede the T (as well as their base pairing C nucleotides).
Figure 4.
SgrAI–DNA contacts. (A) Contacts to the base edges in the major groove of half of the 2-fold symmetric DNA recognition sequence by SgrAI. (B) Schematic of the SgrAI–DNA contacts with the boxed region indicating the contacts shown in A. (C) Close-up of the Lys 96–DNA contacts in the minor groove. (D) Schematic of the SgrAI–DNA contacts with the boxed region indicating the contacts shown in C.
Figure 5.
Comparisons of DNA and metal ion binding. (A) Superposition of one-half site of the recognition sequences of SgrAI (pink, blue and red) and NgoMIV (white) and side chains of contacting residues (SgrAI in cyan, blue, red, NgoMIV in green, blue, red). (B) Superposition of SgrAI bound to cognate DNA (Form 2, white) on SgrAI bound to DNA containing a G–G mismatch at the outer base pairs of the SgrAI recognition sequence (Form 1, color) at the site of the mismatch. The G is contacted by Arg 31, however, no contacts are made to the base pairing partner. The non-contacted G (G1) adopts the syn conformation at the glyosidic bond. Hydrogen bonds are shown as purple dashes (Form 2 with cognate DNA) and green dashes (Form 1, mismatched non-cognate DNA).
The G:G mismatch does not perturb the protein–DNA interface since no contacts are made to the C of CRCCGGYG, which is replaced by a G in the mismatch of Form 1. This mismatched and non-cognate G (GRCCGGYG) takes on a syn orientation, allowing hydrogen bonding to the opposite G, and minimally perturbing the duplex conformation (Figure 5B).
Active site structure
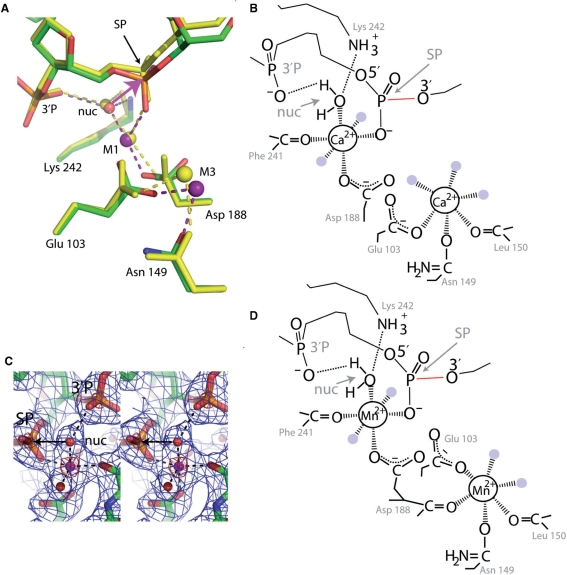
All three crystal form structures (Forms 1–3) of SgrAI bound to DNA were determined from crystals grown in the presence of Ca2+, and that of Form 2 has also been determined from crystals grown in the presence of Mn2+. The structures with Ca2+ (yellow, Figure 6A) show two Ca2+ bound in each active site, near the phosphate of the scissile phosphodiester bond (SP, Figure 6A) in sites identified as M1 and M3 (Figure 6A). A schematic describing the interactions around the Ca2+ ions is shown in Figure 6B, however the ligations around the M3 Ca2+ are variable in the different active sites and crystal forms, with ligation by both oxygens of Glu 103 in some, and also differing in the number of ligated water molecules. The structure with Mn2+ has been refined to 2.2 Å with Rcryst of 17.4% and Rfree of 24.0% (Table 1). The M1 site is strongly occupied by Mn2+ (purple sphere at M1, Figure 6A), identified by very strong electron density (simulated annealing omit 2Fo−Fc electron density, blue, 1σ, red, 8σ, Figure 6C), and strong peaks in the anomalous difference map (11σ and 7σ in each active site). A second Mn2+ in each active site is identified by a weaker but substantial anomalous difference signal in the M3 site (3σ), and refinement indicates partial occupancy (0.3–0.4 in the two active sites) (purple sphere at M3, Figure 6A). A schematic describing interactions around the Mn2+ ions is shown in Figure 6D. The DNA is uncleaved in both Ca2+ and Mn2+ bound structures, and the conformations of the protein and DNA are very similar (Figure 6A).
Figure 6.
Metal ion binding. (A) Superposition of Form 2 structures with Ca2+ (yellow, Ca2+ shown as yellow sphere) and Mn2+ (green, red, orange, blue, Mn2+shown as purple spheres) using the alpha carbon atoms of residues 103, 149, 188 and 242. SP, phosphate of the scissile phosphodiester bond, 3′P, phosphate 3′ of SP, nuc, water molecule positioned for in-line attack on the SP, M1 and M2, metal ion binding sites, pink arrow, direction of nucleophilic attack, red line, scissile bond. (B) Schematic of the interactions around the two Ca2+ of the Form 2 Ca2+ bound structure of SgrAI, shown in Figure 6A. Dashed lines indicate ligation to the Ca2+ ions. Some water molecules ligated to the Ca2+ are shown as light blue circles. (C) Stereo diagram of the electron density around an M1 Mn2+ ion in the Form 2 Mn2+ bound structure of SgrAI. Simulated annealing omit 2Fo−Fc electron density shown in blue (1σ) and red (8σ). SP, 3′P, nuc defined as in Figure 6A, arrow, direction of nucleophilic attack. The Mn2+ is shown as a pink sphere and ligated water molecules as red spheres. Ligations to the Mn2+ are shown as dashed lines. (D) Schematic of the interactions around the Mn2+ ions of the Form 2 Mn2+ bound structure of SgrAI, shown in Figure 6A. Dashed lines indicate ligation to the Mn2+ ions. Water molecules ligated to the Mn2+ ions are shown as light blue circles.
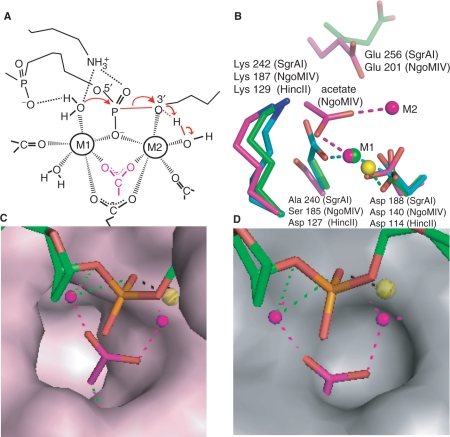
The type II REs contain an active site motif, PDX9–18 (E/D)YK, where X represents any amino acid type, and Y represents a hydrophobic residue (8). The D and (E/D) residues of the motif have been found to ligate the divalent cation(s) in RE crystal structures, and as proposed in the two-metal-ion mechanism (Figure 7A). The PDX9–18(E/D)YK motif differs in Cfr10I, where the (E/D) residue is replaced with serine. Experimental data suggest that the (E/D) residue in Cfr10I derives instead from a different residue, namely Glu 204, making the motif PDX55KX13E (31). Glu 204 of Cfr10I is conserved in both NgoMIV (Glu 201) and SgrAI (Glu 256), however, does not directly ligate either of the two Mg2+ found in the NgoMIV structure (closest approach, 4.3 Å), nor the Ca2+ or Mn2+ of the SgrAI structures (closest approach, 12 Å, Figure 7B). Residue Glu 103 of SgrAI, also conserved in Cfr10I (Glu 71) and NgoMIV (Glu 70), ligates the M3 ion in the SgrAI structures, however, Glu 70 of NgoMIV does not directly ligate either bound Mg2+ ion. One or both of these acidic residues may serve to replace the (D/E) residue of the motif by interacting with the M2 metal ion through its water ligands rather than through direct ligation. As in Cfr10I, both SgrAI and NgoMIV are also missing the (E/D) of PDX9–18(E/D)YK, having alanine or serine at this position, respectively. Interestingly, an acetate molecule (pink and red, Figure 7B) is found in the structure of NgoMIV in approximately the same location as the side chain of the missing (E/D) residue (HincII Asp 127 shown in cyan, red, Figure 7B), and ligates both M1 and M2 Mg2+. However, acetate is not required for DNA cleavage by NgoMIV (12) or SgrAI (Figure S3), and was not located in any of the SgrAI structures. A surface representation of NgoMIV reveals a pocket where the acetate binds (Figure 7C). This pocket is shallower in SgrAI (Figure 7D), and blocks the position occupied by acetate in NgoMIV. Therefore, acetate appears unlikely to bind SgrAI in its current conformation.
Figure 7.
Comparison of NgoMIV and SgrAI active sites. (A) Model of the two-metal-ion mechanism of DNA cleavage. Red arrows indicate changes in bonding. Scissile P-O3′ bond shown as red line. Group in pink is an acetate molecule in the structure of NgoMIV. (B) Superposition of the active sites of HincII (cyan, blue, red, yellow), SgrAI (green, blue, red) and NgoMIV (magenta, blue, red) using the alpha carbon atoms of the residues shown (with the exception of Glu 256 and Glu 201). The positions of the bound metal ions are shown as spheres at M1 and M2. Interactions between metal ions and ligands are shown as dashed lines. Glu 201 and Glu 256 do not directly ligate metal ions. NgoMIV contains an acetate at roughly the same position as Ser 185, which is Asp in the typical RE motif PDX9–18(E/D)YK. (C) Superposition used in Figure 7B with the solvent accessible surface area of NgoMIV residues shown in pink, and the DNA of the SgrAI Form 2 Mn2+ bound structure shown in green, red and orange. The Mn2+ ion is shown as a yellow sphere, and the Mg2+ ions of the NgoMIV structure shown as pink spheres. The acetate molecule that ligates to both Mg2+ in the NgoMIV structure is shown in pink and red. The acetate fits neatly into a deep pocket in the NgoMIV structure. (D) As in Figure 7C, with the surface of the SgrAI protein shown in grey. The deep pocket found in the NgoMIV surface is absent in SgrAI, and the acetate molecule overlaps with residues of SgrAI forming the surface.
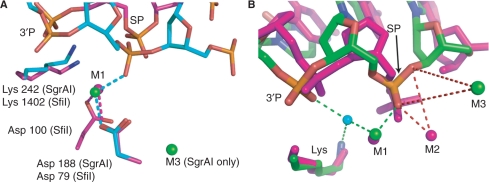
The enzyme SfiI shares some properties with SgrAI; it is also a ‘rare cutter’ (cut site GGCCNNNN|NGGCC) and biochemical data suggest the formation of a tetramer. SfiI requires the binding of two recognition sites for cleavage, and cleavage occurs in a concerted manner (32). The structure of SfiI bound to DNA has been determined (2) and shows a tetramer where each active site binds a single Ca2+ bound in what we term the M1 site (SfiI shown in magenta, red, blue, SgrAI shown in cyan, red, blue, Figure 8A). The positions of the bound DNA relative to the active sites differ in the two structures (Figure 8A).
Figure 8.
Comparison of SgrAI active site with SfiI and NgoMIV. (A) Superposition of active sites of SgrAI (cyan, blue, red, orange, green) and SfiI (magenta, blue, red, orange). Metal ions shown as spheres. (B) Superposition using the alpha carbon atoms of Lys 242, Glu 103 and Asp 188 of SgrAI Form 2 with Ca2+ onto Lys 187, Glu 70 and Asp 140 of NgoMIV, respectively. NgoMIV shown in magenta. Pink spheres indicate Mg2+ ions. SgrAI is shown in green, blue, orange and red. Green spheres indicate Ca2+ ions. Blue sphere indicates water molecule positioned for in-line attack. SP, phosphate of the scissile phosphodiester bond, 3′P, phosphate 3′ to the SP. M1 and M2 refer to the two-metal-ion binding sites. Green dashed lines indicate interactions at distances consistent with metal ion–ligand interactions or hydrogen bonds. Red dashes indicate distances too long for metal ion–ligand interactions to occur. Interactions predicted to occur in the two-metal-ion mechanism to the SP and O3′ of the SgrAI DNA by a metal ion at either M2 site are too distant to occur.
DISCUSSION
DNA cleavage and analytical ultracentrifuge data indicate that SgrAI exists predominantly as a dimer in solution, but forms a higher order oligomer upon binding to DNA containing the SgrAI recognition sequence, CR|CCGGYG (4–6,9,10). In addition, DNA cleavage by SgrAI is faster on plasmids containing two copies of this site than on plasmids bearing only a single site (4). The acceleration occurs at both the cognate recognition sequence, and at secondary sites: CR|CCGGY(A,T,C) and CR|CCGGGG. A plausible mechanism for the activation involves the formation of a tetramer of SgrAI, with two DNA-binding sites, where DNA cleavage at one binding site can be communicated to the other. Tetrameric complexes with two DNA-binding sites have been observed in the structures of the relatives NgoMIV (12) and Cfr10I (11), which also cleave plasmids bearing two recognition sites faster than those bearing only one. However, neither NgoMIV nor Cfr10I show the alteration of DNA specificity exhibited by SgrAI.
Structures of SgrAI bound to cognate (CACCGGTG) and non-cognate (GACCGGTG) DNA with Ca2+ or Mn2+ have been determined. In all structures, SgrAI forms a dimer very similar to those of Cfr10I and NgoMIV (Figure 1). Alignments of the structures show that SgrAI is more similar to Cfr10I, having some small deletions and several insertions relative to Cfr10I (blue, Figure 1A, red, Figure 1C, Figure S1). NgoMIV and Cfr10I form tetramers in the crystal structures, with the tetrameric interface at the end of the dimer opposite to that of the DNA-binding site (arrows, Figures 1B–C, 2A and 3A). However, the tetrameric interfaces of NgoMIV and Cfr10I are very different. The NgoMIV dimer–dimer interface is extensive, burying over 12 000 Å2 of solvent accessible surface area. NgoMIV also uses a segment to form a loop which binds to the surface of the second dimer in the tetramer (arrows, Figures 1B and 3A). This segment is folded back onto the body of the dimer in SgrAI (arrow, Figure 1A) and Cfr10I. A model of a SgrAI tetramer based on that of NgoMIV shows many steric overlaps and close contacts (Figure 3B). The tetramer of Cfr10I is stabilized by residues at the end of an alpha helix (arrow, Figures 1C, 2A and C), which are absent in SgrAI (Figures 1A, 2B and D). The dimer–dimer interface of Cfr10I buries a little over 4000 Å2, however, a model of a similar interface in SgrAI buries only 1000 Å2. Clearly, the tetrameric interface of SgrAI must be unique from those found in NgoMIV and Cfr10I.
No crystal structure of Cfr10I bound to DNA has been determined to date, however, that of NgoMIV bound to DNA is known (12). The DNA contacts by NgoMIV are very similar to those by SgrAI at the core CCGG sequence (Figure 5A). The contacts made by NgoMIV to the G–C base pairs just outside of the CCGG sequence are absent in SgrAI, instead, SgrAI may specify the R–Y base pairs using indirect readout (Figure 4C). Lys 96 approaches the DNA from the minor groove and may stabilize the conformation of the DNA where the outer G of the SgrAI recognition sequence CRCCGGYG (Gua 8) is unstacked from the Y (Thy 7). Unstacking of YR steps is one of the more common types of DNA distortions used in the indirect readout of DNA sequences (33,34). The outer C–G base pair, unique to the SgrAI recognition sequence (CRCCGGYG), is recognized by Arg 31 to the G (Figure 4A), and no contacts are made to the outer C of CRCCGGYG. Interestingly, the structure formed by residues 1–31 in SgrAI is absent in both Cfr10I and NgoMIV (Figure 1, Figure S1).
SgrAI is only the third ‘rare cutting’ RE structure determined in the presence of bound DNA, the others being NotI (1) and SfiI (2). In addition, the structure of SdaI (35) has been determined without bound DNA. The DNA bound structures show a common theme, namely that the outer two base pairs of the 8 bp recognition sequence are contacted only singly, while the inner base pairs are fully saturated with interactions at both bases, just as is typically seen in 6 bp cutting REs. The comparison of DNA recognition by SgrAI and the 6 bp cutting NgoMIV shows that the recognition of the common core base pairs (CCGG) is identical in the two enzymes, and the additional base pair at the beginning and end of the SgrAI site is recognized only at the G of the CG base pair (by Arg 31). The fact that only the G of the outer CG base pairs is contacted accounts for the ability of SgrAI to bind the GG mismatch at this position (Figure 5B).
Several models for the catalytic mechanism of DNA cleavage by REs have been proposed (36,37) which are also similar to those proposed for other classes of DNA nucleases (38). The two-metal-ion mechanism shown in Figure 7A can be used as a framework for understanding the variations in the proposed mechanisms. Mechanistic differences include the number of metal ions, where 1, 2 and 3 have been proposed. Other differences include the absence or presence of ligations of the metal ions to the phosphate of the scissile phosphodiester bond (SP, Figure 7A) and to the O3′ atom, the leaving group of the reaction. Interactions with the lysine and 3′ phosphate (3′P, Figure 7A) may also be absent, and some REs contain a different amino acid in the place of the lysine. In the two–metal-ion mechanism shown in Figure 7A, both metal ions, M1 and M2, ligate the phosphate of the scissile phosphodiester bond (SP, Figure 7A) to position and electrostatically stabilize the transition state. The M1 ion ligates and orients a water molecule activating it for an attack on the phosphorus by stabilizing its deprotonated state hydroxide, which is more nucleophilic than water. The neighboring 3′ phosphate is often within hydrogen bonding distance to the nucleophilic water and may act as the proton acceptor. The M2 ion also ligates the O3′ leaving group and stabilizes the negative charge that forms on this atom after cleavage of the P-O3′ bond (red line, Figure 7A), and an M2 bound water molecule donates a proton to the O3′. However, direct ligation of the O3′ is not observed in all RE structures. The function of the lysine is not known, but it may serve to orient and/or activate the nucleophile, or in electrostatic stabilization of the transition state. In NgoMIV, this lysine (Lys 187) forms a salt bridge with the cleaved phosphate, but in SgrAI it is too distant for hydrogen bonding to SP (4.7 Å in the Mn2+ bound structure), or for strong hydrogen bonding to the nucleophilic water (3.3 Å in the Mn2+ bound structure).
The in vivo ion cofactor of SgrAI is thought to be Mg2+, however, Ca2+ has been shown to inhibit DNA cleavage and enhance binding affinity to DNA in several Mg2+-dependent endonucleases (39,40). In addition, Ca2+ will often bind in the active site at the protein–DNA interface close to known Mg2+ binding sites, although it can show binding behavior unique from that of Mg2+(40–42). The structure of SgrAI bound to DNA and Ca2+ shows two ions per active site, one in the M1 site, but the second is distal from the DNA (Figure 6A and C), which does not make the expected contacts to the DNA (Figure 7A), and we call this site the M3 site. As expected, the DNA is uncleaved in the Ca2+ bound structure (Figure 6A).
The divalent cation Mn2+ will often confer activity to Mg2+-dependent enzymes (43–46). The greater number of electrons of Mn2+ allows for better identification in the electron density relative to Mg2+, which has the same number of electrons as water molecules. The structure of SgrAI with Mn2+ shows strong occupancy in the M1 site (purple sphere, Figure 6B). A second, partially occupied Mn2+ is found in the M3 site. The M1 ion, whether Ca2+ or Mn2+, ligates a water molecule (nuc, Figure 6A) positioned for in-line attack of the phosphorus and displacement of the O3′ atom with water-P distance of 3.4 Å and a water-P-O3′ angle of 173° in the Mn2+ bound structure. This water molecule is also hydrogen bonded to the 3′phosphate (3′P, Figure 5A), an interaction that may be catalytic (47). Surprisingly, the DNA is uncleaved (red, green, blue, Figure 5A), and occupies a similar position in all of the SgrAI structures. The absence of an M2 ion may be responsible for the lack of DNA cleavage, if DNA cleavage of SgrAI utilizes the two-metal-ion mechanism (Figure 7A). Two Mg2+ ions are found in the structure of NgoMIV bound to DNA, and NgoMIV is proposed to utilize a two-metal-ion mechanism (12).
A superposition of the structures of NgoMIV and SgrAI, using the alpha carbon atoms of conserved active site residues, is shown in Figure 8B. The M1 Ca2+ of SgrAI (green sphere, M1, Figure 8B) is very close (0.6 Å) to the M1 Mg2+ of NgoMIV (magenta sphere, M1, Figure 8B). The DNA is cleaved in the NgoMIV structure indicating that the enzyme has carried out the DNA cleavage reaction. The DNA is uncleaved in the SgrAI structures, although Mn2+ is known to confer catalytic activity to the enzyme (Figure S2). In both the SgrAI and NgoMIV structures, the M1 ion is positioned as predicted by the two-metal-ion mechanism, ligated to both an oxygen of the phosphate of the scissile phosphodiester bond (SP, Figure 8B), and a water molecule positioned for in-line attack on the phosphorus of the SP. No M2 ion is found in either Mn2+ or Ca2+ bound structures of SgrAI, and that found in the M3 site is distal from the DNA, indicating that the M2 site predicted by the two-metal-ion mechanism is not available in the SgrAI structure for binding by either divalent cation. Interestingly, a third, distal, metal ion binding site is also found in EcoRV (48), and movement of metal ions between sites has been suggested to occur during the DNA cleavage reaction (49). In addition, even the M2 ion found in NgoMIV is incorrectly positioned in SgrAI for the two-metal-ion mechanism of DNA cleavage shown in Figure 7A. Using the superposition of the SgrAI and NgoMIV structures, the M2 Mg2+ of the NgoMIV structure is found to be too distant from the SgrAI DNA to ligate the phosphate of the scissile phosphodiester bond or the O3′ (2.8 and 4.3 Å, respectively, red dashes, Figure 8B). It is possible that the M2 Mg2+ of NgoMIV is shifted as a result of DNA cleavage, and would occupy a more optimal position for the cleavage reaction prior to cleavage. Alternatively, the DNA may not be positioned optimally in the SgrAI structure, which may be the cause for the lack of DNA cleavage in the Mn2+ bound structure. An altered position of the DNA could decrease the affinity of the second ion for the M2 site.
The biochemical data suggest that SgrAI can exist in at least two conformations, with one possessing an inherently greater DNA cleavage activity than the other. The observed stimulation of DNA cleavage activity could be accomplished by shifting the equilibrium from the low to the high activity form, possibly through formation of a higher order oligomer favoring the high activity conformation. The rate of DNA cleavage may be controlled by the positioning of groups in the active site, where the optimal alignment results in faster DNA cleavage kinetics. Analysis of the active sites of SgrAI shows very similar placement of all groups, including the DNA, in the various crystal structures (Figure 6A), indicating that only a single conformation of the enzyme has been determined. Given that SgrAI should favor a low activity conformation prior to activation, that activation appears to require binding of two copies of the target sequence by presumably a tetramer of SgrAI, and the possibility that the uncleaved DNA is mispositioned in the structures of SgrAI, we conclude that these structures represent the low activity conformation of SgrAI.
In summary, we present the three-dimensional crystal structure of SgrAI bound to DNA in three crystal forms. In all forms, SgrAI forms a dimer very similar to NgoMIV, and especially Cfr10I, but in none does SgrAI form a tetramer. The first and last base pair of the 8 bp recognition sequence is recognized by one amino acid contact to one base of the base pair, as in other ‘rare cutting’ restriction endonucleases, perhaps as a result of recent evolutionary changes (1). The conformation within the active site is very similar in all SgrAI structures, and has been characterized with Ca2+ and Mn2+. Two Ca2+ or Mn2+ bind per active site, however only one is in a position predicted by the two-metal-ion mechanism, the M1 position. The DNA is not cleaved, indicating that the structure is of an inactive form. Inactivity could be due to the positioning of the DNA and/or the lack of binding the second metal ion, in the M2 position. The structure of SgrAI with cleaved DNA will be necessary to fully understand the allosteric mechanism employed by this enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (NIH R01 GM066805 to N.C.H.). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Portions of this work were carried out at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Funding to pay the Open Access publication charges for this article was provided by NIH 5R01GM066805.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lambert AR, Sussman D, Shen B, Maunus R, Nix J, Samuelson J, Xu SY, Stoddard BL. Structures of the rare-cutting restriction endonuclease NotI reveal a unique metal binding fold involved in DNA binding. Structure. 2008;16:558–569. doi: 10.1016/j.str.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. EMBO J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tautz N, Kaluza K, Frey B, Jarsch M, Schmitz GG, Kessler C. SgrAI, a novel class-II restriction endonuclease from Streptomyces griseus recognizing the octanucleotide sequence 5′-CR/CCGGYG-3′ [corrected] Nucleic Acids Res. 1990;18:3087. doi: 10.1093/nar/18.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilcock DT, Daniels LE, Bath AJ, Halford SE. Reactions of type II restriction endonucleases with 8-base pair recognition sites. J. Biol. Chem. 1999;274:36379–36386. doi: 10.1074/jbc.274.51.36379. [DOI] [PubMed] [Google Scholar]
- 5.Daniels LE, Wood KM, Scott DJ, Halford SE. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J. Mol. Biol. 2003;327:579–591. doi: 10.1016/s0022-2836(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 6.Bitinaite J, Schildkraut I. Self-generated DNA termini relax the specificity of SgrAI restriction endonuclease. Proc. Natl Acad. Sci. USA. 2002;99:1164–1169. doi: 10.1073/pnas.022346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and function. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 8.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood KM, Daniels LE, Halford SE. Long-range communications between DNA sites by the dimeric restriction endonuclease SgrAI. J. Mol. Biol. 2005;350:240–253. doi: 10.1016/j.jmb.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 10.Hingorani-Varma K, Bitinaite J. Kinetic analysis of the coordinated interaction of SgrAI restriction endonuclease with different DNA targets. J. Biol. Chem. 2003;278:40392–40399. doi: 10.1074/jbc.M304603200. [DOI] [PubMed] [Google Scholar]
- 11.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 12.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal AK. Crystallization of DNA binding proteins with oligodeoxynucleotides. Methods. 1990;1:83–90. doi: 10.1016/S1046-2023(05)80150-1. [DOI] [PubMed] [Google Scholar]
- 14.Cavaluzzi,M.J. & Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 16.Evans PR. Scaling and assessment of data quality. Acta Crysr. 2006;D62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 17.Knight SD. RSPS version 4.0: a semi-interactive vector-search program for solving heavy-atom derivatives. Acta Crystallogr. D Biol. Crystallogr. 2000;56:42–47. doi: 10.1107/s0907444999012913. [DOI] [PubMed] [Google Scholar]
- 18.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 19.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView/Xfit–a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 29.Grazulis S, Deibert M, Rimseliene R, Skirgaila R, Sasnauskas G, Lagunavicius A, Repin V, Urbanke C, Huber R, Siksnys V. Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res. 2002;30:876–885. doi: 10.1093/nar/30.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skirgaila R, Grazulis S, Bozic D, Huber R, Siksnys V. Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues. J. Mol. Biol. 1998;279:473–481. doi: 10.1006/jmbi.1998.1803. [DOI] [PubMed] [Google Scholar]
- 32.Wentzell LM, Nobbs TJ, Halford SE. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 33.Neugebauerova S, Kypr J. Invariant and variable base stacking geometries in B-DNA and A-DNA. J. Biomol. Struct. Dyn. 2000;18:73–81. doi: 10.1080/07391102.2000.10506648. [DOI] [PubMed] [Google Scholar]
- 34.Ulyanov NB, Zhurkin VB. Sequence-dependent anisotropic flexibility of B-DNA. A conformational study. J. Biomol. Struct. Dyn. 1984;2:361–385. doi: 10.1080/07391102.1984.10507573. [DOI] [PubMed] [Google Scholar]
- 35.Tamulaitiene G, Jakubauskas A, Urbanke C, Huber R, Grazulis S, Siksnys V. The crystal structure of the rare-cutting restriction enzyme SdaI reveals unexpected domain architecture. Structure. 2006;14:1389–1400. doi: 10.1016/j.str.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Dupureur C. An integrated look at metallonuclease mechanism. Curr. Chem. Biol. 2008;2:159–173. [Google Scholar]
- 37.Galburt EA, Stoddard BL. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 38.Horton NC. In: Protein-Nucleic Acid Interactions Structural Biology. Rice PA, Correll CC, editors. The Royal Society of Chemistry: Cambridge, UK; 2008. pp. 333–363. [Google Scholar]
- 39.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 40.Etzkorn C, Horton NC. Ca2+ binding in the active site of HincII: implications for the catalytic mechanism. Biochemistry. 2004;43:13256–13270. doi: 10.1021/bi0490082. [DOI] [PubMed] [Google Scholar]
- 41.Etzkorn C, Horton NC. Mechanistic insights from the structures of HincII bound to cognate DNA cleaved from addition of Mg2+ and Mn2+ J. Mol. Biol. 2004;343:833–849. doi: 10.1016/j.jmb.2004.08.082. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Sam MD, Perona JJ. Mn2+-dependent catalysis by restriction enzymes: pre-steady-state analysis of EcoRV endonuclease reveals burst kinetics and the origins of reduced ativity. J. Am. Chem. Soc. 1999;121:1444–1447. [Google Scholar]
- 44.Baldwin GS, Sessions RB, Erskine SG, Halford SE. DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J. Mol. Biol. 1999;288:87–103. doi: 10.1006/jmbi.1999.2672. [DOI] [PubMed] [Google Scholar]
- 45.Vermote CL, Vipond IB, Halford SE. EcoRV restriction endonuclease: communication between DNA recognition and catalysis. Biochemistry. 1992;31:6089–6097. doi: 10.1021/bi00141a019. [DOI] [PubMed] [Google Scholar]
- 46.Parry D, Moon SA, Liu HH, Heslop P, Connolly BA. DNA recognition by the EcoRV restriction endonuclease probed using base analogues. J. Mol. Biol. 2003;331:1005–1016. doi: 10.1016/s0022-2836(03)00861-1. [DOI] [PubMed] [Google Scholar]
- 47.Jeltsch A, Alves J, Wolfes H, Maass G, Pingoud A. Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA. 1993;90:8499–8503. doi: 10.1073/pnas.90.18.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horton NC, Newberry KJ, Perona JJ. Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases. Proc. Natl Acad. Sci. USA. 1998;95:13489–13494. doi: 10.1073/pnas.95.23.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton NC, Perona JJ. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.