Abstract
The PspGI restriction–modification system recognizes the sequence CCWGG. R.PspGI cuts DNA before the first C in the cognate sequence and M.PspGI is thought to methylate N4 of one of the cytosines in the sequence. M.PspGI enhances fluorescence of 2-aminopurine in DNA if it replaces the second C in the sequence, while R.PspGI enhances fluorescence when the fluorophore replaces adenine in the central base pair. This strongly suggests that the methyltransferase flips the second C in the recognition sequence, while the endonuclease flips both bases in the central base pair out of the duplex. M.PspGI is the first N4-cytosine MTase for which biochemical evidence for base flipping has been presented. It is also the first type IIP methyltransferase whose catalytic activity is strongly stimulated by divalent metal ions. However, divalent metal ions are not required for its base-flipping activity. In contrast, these ions are required for both base flipping and catalysis by the endonuclease. The two enzymes have similar temperature profiles for base flipping and optimal flipping occurs at temperatures substantially below the growth temperature of the source organism for PspGI and for the catalytic activity of endonuclease. We discuss the implications of these results for DNA binding by these enzymes and their evolutionary origin.
INTRODUCTION
PspGI restriction–modification (R–M) enzymes are two of the most heat stable R–M enzymes to have been described. The PspGI R–M system was found in Pyrococcus species strain GI-H, which has an optimal growth temperature of 85°C and the PspGI endonuclease (R.PspGI) has a half life of 2 h at 95°C (1). The PspGI R–M enzymes recognize the sequence CCWGG where W is A or T. The endonuclease (REase), R.PspGI, is an isoschizomer of EcoRII that cleaves DNA before the first cytosine in its recognition sequence (1). The methyltransferase (MTase), M.PspGI, is believed to methylate the N4 position of cytosine. This conclusion is based in part on sequence analysis that shows that M.PspGI is highly similar to M.MvaI, a known N4-cytosine methyltransferase and contains the SPPY sequence motif which is unique to the N4-cytosine methyltransferases (1–4). Additionally, it has been argued that if hyperthermophiles are to methylate cytosines in their DNA to protect it from endogenous restriction endonucleases, then they are likely to methylate the exocyclic nitrogen rather than carbon-5. This is because 5-methylcytosine (5mC) has a high potential for deaminating to thymine causing C to T mutations (5). N4-methylcytosine does not present such a mutagenic hazard. It is not known whether M.PspGI methylates the first or the second cytosine in its recognition sequence, but R.PspGI is inhibited by methylation of the inner cytosine on carbon-5 by Escherichia coli Dcm (1) suggesting that this cytosine may also be the site of methylation by M.PspGI. Together these data suggest that M.PspGI methylates the second cytosine in its recognition sequence at the N4 position.
R.PspGI is thought to flip both bases in the central base pair of its recognition sequence out of the double helix. We originally suggested this based largely on genetic evidence (6). We found that R.PspGI protects its cognate sequence, CCWGG, against deamination of cytosines to uracil at high temperatures under conditions where the enzyme bound to DNA without cutting (6). In contrast, the enzyme or its catalytically inactive mutant, D138A, increased the rate of deamination of the third C in the sequence CCCGG by a factor of 14. We interpreted these data to mean that flipping of this base out of the duplex makes it more susceptible to hydrolytic attack. Subsequently, the structure of a related enzyme, R.Ecl18kI, which recognizes CCNGG showed that it flips both the bases in its central base pair (7), and strengthening the argument that R.PspGI does the same. More recently, a preliminary report based on 2-aminopurine (2AP) fluorescence enhancement also suggested that R.PspGI flips its central bases out of the duplex (8).
It is generally believed that base flipping is the mechanism by which a DNA methyltransferase is able to gain access to bases to perform chemistry. M.HhaI, a C5-cytosine MTase, was the first MTase crystallized with its substrate DNA and the co-crystal showed that the target base was flipped out of the duplex into the active site of the enzyme (9). Subsequently, two other DNA methyltransferases, M.HaeIII, also a 5-cytosine MTase, and M.TaqI, an N6-adenine MTase, were also crystallized in the presence of DNA and were found to flip the target base out of the double helix (10,11).
In addition to crystallography, fluorescence enhancement of 2AP has been used to study base flipping by a number of MTases. This method takes advantage of the fact that 2AP fluorescence quantum yield increases when its environment becomes more polar (12). MTases that enhance 2AP's fluorescence when the fluorophore replaces the base that is normally methylated include M.EcoRI (13), M.TaqI (14), M.HhaI (14), M.EcoP151 (15), M.EcoKI (16), M.RsrI (17), T4 Dam (18), M.KpnI (19) and E. coli Dam (20). However, some exceptions to this general rule also exist. This includes M.EcoRV, which did not enhance 2AP fluorescence when it substituted the target base for methylation (21). Additionally, MTases M.EcoKI (16), M.EcoRV (21) and M.EcoP151 (15) enhanced 2APs fluorescence when it was placed at positions other than the base to which the methyl group is transferred.
So far, all the MTases for which direct evidence has been presented for base flipping have been either C5-cytosine or N6-adenine methyltransferases. Only one N4-cytosine MTase has been crystallized to date, M.PvuII (22), but crystal structure of M.PvuII was obtained without a DNA substrate. To obtain more direct evidence for base flipping by the N4-cytosine MTases, we studied the ability of M.PspGI to flip 2AP out of DNA and compared it to the ability of R.PspGI to do the same. The results show that while both the enzymes in this R–M system flip DNA bases, there are significant differences between their targets and flipping mechanism.
MATERIALS AND METHODS
Materials
2-Aminopurine-2′-deoxyribose-5′-triphosphate was purchased from TriLink BioTechnologies (San Diego, CA, USA), while 2AP free base was obtained from Sigma-Aldrich (St Louis, MO, USA). S-adenosyl-l-methionine (SAM) was obtained from New England Biolabs (Ipswich, MA, USA) and S-adenosyl-l-homocysteine (SAH) was obtained from Sigma-Aldrich (St Louis, MO, USA). 2AP-containing DNA oligomers were synthesized by the W.M. Keck Oligonucleotide Synthesis Facility at Yale University (New Haven, CT, USA), while the complementary oligomer was obtained from Sigma-Genosys (The Woodlands, TX, USA). Some of these oligomers contained biotin at the 3-end for reasons unrelated to this study. All the oligomers were purified from sequencing gels and were annealed to their complement by heating to 95°C followed by cooling to room temperature. The sequences of the oligomers used in this study and their melting temperature are listed in Table 1.
Table 1.
DNA oligomers
| Name | Sequencea | Melting temperature of duplexb (°C) |
|
|---|---|---|---|
| REase bufferc | MTase bufferd | ||
| 2AP18 | TGCTACC2GGC GAAGATA-biotin | 67 | – |
| O15N | CTTCGCCTGGTAGCA | – | – |
| 2AP21 | CGACGCAAGCCGACGCCA GC2CCACCAGGACGCCGCATA | 87 | 75 |
| 2AP26 | CGACGCAAGCCGACGCCAGCAC CAC2AGGACGCCGCATA | 85 | 71 |
| 2AP27 | CGACGCAAGCCGACGCC AGCACCACC2GGACGCCGCATA | 87 | 75 |
| O2APT | GCGGCGTCCTGGTGGTGCTG GCGTCGGCTTGCGTCG | – | – |
aPosition of 2AP in the sequence is indicated by 2.
bMelting temperatures of duplexes containing a 2AP-containing oligomer hybridized to their complement (i.e. 2AP18 with O15N, and 2AP21, 2AP26 or 2AP27 with O2APT). The melting temperature is the midpoint of 2AP fluorescence enhancement when the temperatures of duplexes were increased from 25°C to 95°C in the absence of any protein.
cREase buffer—10 mM Tris pH 7.9, 50 mM NaCl, 10 mM MgCl2.
dMTase buffer—10 mM Tris, pH 8.0, 1 mM EDTA.
Expression and purification of proteins
Purification of wild-type (WT) PspGI and its D138A mutant has been described previously (1,3). The expression constructs for these proteins, pET21a-PspGI-WT, pET21a-PspGI-D138A and pACYC-M.PspGI, were a generous gift from Vera Pingoud (Justus-Liebig-Universität, Giessen, Germany). These plasmids were introduced in the strain ER2744 (fhuA2 glnV44 e14− rfbD1? relA1? spoT1? endA1 thi-1 Δ(mcrC-mrr)114::IS10 lacZ::T7gene1) and expression of both endonuclease and methyltransferase was induced with 300 μM IPTG and followed by incubation for 16 h at 16°C. Cells were collected by centrifugation at 3000 g, resuspended in 50 ml lysis buffer (50 mM NaCl, 20 mM Tris–HCl pH 8.5) containing Complete EDTA-free Protease Inhibitor Cocktail from Roche Diagnostics (Indianapolis, IN, USA). The cells were broken by sonication and the resulting lysate was centrifuged at 20 000 g for 30 min to remove cell debris. The supernatant was then heated to 70°C for 30 min and then centrifuged for 30 min at 20 000 g to remove the precipitated material. The cleared lysate was then loaded onto a P11 cellulose phosphate column (Whatman, Maidstone, England) equilibrated with the lysis buffer. The column was washed with 3.5 column volumes lysis buffer, followed by 3.5 column volumes of R.PspGI elution buffer (300 mM NaCl, 20 mM Tris pH 8.5). Subsequently, the M.PspGI was eluted from the same column with 1 M NaCl in Tris pH 8.5. Proteins were concentrated in Millipore spin concentrators with MW cutoffs of 10 kDa and dialyzed against storage buffer (10 mM Tris pH 7.9, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT and 50% glycerol). Protein concentrations were determined by using the Protein Assay kit from Bio-Rad (Hercules, CA, USA).
Methylation of DNA
In some initial methylation studies, a molar excess of MTase over methylation sites in the plasmid DNA was used, but in subsequent studies M.PspGI was used at a methylation site to enzyme ratio of 20. The reaction buffer contained 20 mM Tris (pH 7.9), 75 mM NaCl and 160 μM SAM. Divalent metal salts or EDTA was added to the buffers as indicated in figure legends. The reactions were incubated at 70°C for 2 h after which SDS was added to 0.2% to stop the reaction followed by deproteinization using phenol–chloroform extraction. Following ethanol precipitation, the DNA was resuspended in 20 μl of 1 × MvaI buffer containing 5 units of MvaI (Takara Bio USA, Madison, WI, USA). Digestions were continued overnight at 37°C and the products were separated on agarose gels to ascertain the extent of protection.
Fluorescence studies
All fluorescence measurements were done using a Varian Cary Eclipse spectrofluorometer, equipped with a Peltier Multicell Holder for temperature control (Varian Inc., Palo Alto, CA, USA). For the initial scans, slit widths were set at 5 nm in both the excitation and emission windows. During data collection, slit widths were 5 nm in the excitation window and 10 nm in the emission window. Measurements with M.PspGI were done in TE (10 mM Tris pH 8.0, 1 mM EDTA), while those with R.PspGI (WT and D138A mutant) were performed in 10 mM Tris pH 7.9 and 50 mM NaCl. Divalent metals, SAM or SAH were added as indicated in figure legends. Although the optimal excitation wavelength for free 2AP is 300 nm, we chose a longer wavelength (310 nm) to excite the fluorophore in the duplex. This was done because both R.PspGI and M.PspGI contain several tryptophans that contribute significantly to the overall fluorescence emission when 300 nm is used for excitation. We confirmed the effectiveness of this strategy by exciting 2AP21 duplex, which contains 2AP outside the PspGI recognition site (Table 1) at 310 nm and measuring the fluorescence with increasing amounts of R.PspGI D138A mutant. Little increase in fluorescence at 373 nm was observed with increasing amounts of the enzyme (Supplementary Figure S1). Subsequently, all fluorescence measurements were performed with a PMT voltage set at 800 V (unless indicated otherwise in figure legends), the excitation wavelength was set at 310 nm and the emission wavelength was set at 373 nm.
RESULTS
DNA methylation by M.PspGI
M.PspGI was expressed in E. coli along with the WT R.PspGI or its mutant D138A and the endonuclease was separated from the methyltransferase by chromatography. The proteins purified in this manner appeared homogeneous on SDS–PAGE (Supplementary Figure S2). The purification of WT R.PspGI and its D138A mutant has been reported before (1,3), but this is the first report of the purification of M.PspGI. The molecular weight of M.PspGI from SDS–PAGE is ∼50 kDa, which is consistent with the predicted molecular weight of 50 355 Da from its gene sequence (1).
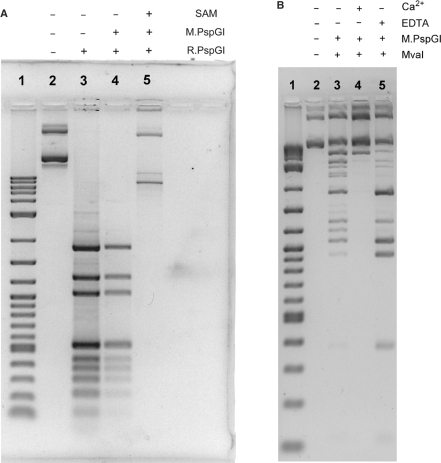
M.PspGI protected plasmid DNA against cleavage by R.PspGI when SAM was included in the methylation reaction (Figure 1A, lane 5), but not without SAM (Figure 1A, lane 4). If DNA methylated with M.PspGI is challenged with the endonuclease ScrFI, which recognizes CCNGG sequences, the resulting pattern of DNA fragments is identical to that created by NciI (recognition sequence CCSGG, S is G or C; Supplementary Figure S3). Based on these data, we conclude that M.PspGI methylates CCWGG sites, but not the related CCSGG sites and hence R.PspGI and M.PspGI have identical sequence specificity.
Figure 1.
Protection of DNA by M.PspGI against endonucleases. (A) Agarose gel of pAB7 DNA digested with R.PspGI is shown. Plasmid pAB7 was first methylated (or not) using molar excess of the MTase followed by REase digestion as indicated above the gel. Lane 1 contains 2-log Ladder (New England Biolabs). (B) The reaction conditions were similar to those in part A except MvaI was used to diegest DNA instead of R.PspGI. Also, SAM was included in all the methylation reactions.
We also challenged M.PspGI methylated plasmid DNA with R.MvaI. R.MvaI cuts unmethylated DNA and C5-methylcytosine containing DNA at CCWGG sites, but does not cut when the DNA contains N4-methylcytosine at the first or the second position of cognate sequence (23,24). We found that the plasmid DNA was protected against R.MvaI digestion by methylation by M.PspGI (Figure 1B and data not shown). These results are consistent with M.PspGI methylating N4-cytosine position of the second cytosine in the cognate sequence, as has been predicted previously (1).
Stimulation of methyltransfer reaction by divalent metal ions
Our early experiments with M.PspGI were plagued with problems. Initially, it was possible to achieve complete protection of DNA against R.PspGI cleavage only by using a molar excess of M.PspGI over CCWGG sites in substrate DNA. However, when we did this, most of the DNA was lost during the manipulations (data not shown). Reducing M.PspGI used in the reactions to catalytic amounts helped with the problem of DNA loss, but led to very poor protection (Figure 1B, lane 3). Unexpectedly, including CaCl2 in the standard MTase buffer increased methylation by M.PspGI to the point where nearly all the CCWGG sites became resistant to MvaI (Figure 1B, lane 4). If EDTA was included in the MTase buffer instead of CaCl2, M.PspGI was unable to protect the DNA against the endonuclease (Figure 1B, lane 5).
We wondered whether this stimulatory effect was specific for Ca2+ or could also be seen with other divalent metal ions such as Mg2+. However, as the first few batches of M.PspGI were purified from cells which also contained WT R.PspGI, and we were concerned that any endonuclease contamination of the MTase preparations would result in DNA cleavage when Mg2+ was included in the MTase reaction buffer. To prevent this problem, we used MTase obtained from cells expressing the D138A mutant of PspGI and tested it for activity in the presence of different divalent metal ions. Addition of Ca2+ or Mg2+, but not Mn2+, Co2+ or Cd2+ to the reaction buffer stimulated the activity of the MTase (Supplementary Figure S4). Thus, only some divalent metals stimulate M.PspGI activity.
Enhancement of 2AP fluorescence by M.PspGI
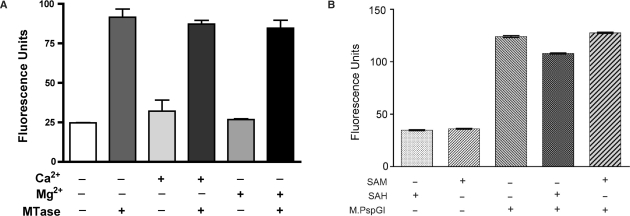
We wondered whether, like the MTases that methylate C5 in cytosine or N6 in adenine (25), M.PspGI also flips its target base out of DNA and whether this could be monitored using environment-specific probe 2AP. When a molar excess of M.PspGI was incubated with duplex DNA containing the oligonucleotide 2AP26 (Table 1), the fluorescence intensity increased 3- to 4-fold suggesting a substantial change in the environment for 2AP. However, unlike the complete methyl transfer reaction, this enhancement did not depend on the presence of divalent metals (Figure 2A). Consequently, we omitted divalent metal ions from buffers used for subsequent fluorescence experiments involving M.PspGI. The fluorescence intensity profile of 2AP26 as a function of concentration of M.PspGI was hyperbolic and was fit to a single-site binding equation yielding a Kd value of 0.5 ± 0.1 μM (Supplementary Figure S5). We interpret these results to mean that M.PspGI binds the 2AP26 duplex and flips the 2AP at the second position within its recognition sequence enhancing its fluorescence signal.
Figure 2.
Base flipping by PspGI enzymes. (A) Fluorescence of 2AP26 duplex (500 nM) under different conditions. Measurements were done in the MTase buffer without SAM or its analogs. MTase refers to M.PspGI (1.5 μM) and the divalent metal ions were added as chloride salts at 10 mM. The results shown are mean and SD from three parallel measurements made at 65°C. (B) Lack of cofactor requirement for base flipping by M.PspGI. Incubation conditions were similar to those in part A except no divalent metals were added. Instead SAM or SAH were added as indicated. The results shown are mean and SD from three measurements made at 65°C.
It should be noted that the ability of M.PspGI to enhance 2AP fluorescence does not depend on SAM or any other exogenous cofactor. M.PspGI increased the fluorescence intensity in the presence of SAM, SAH or in the absence of any coenzyme (Figure 2B). However, the increase was largely eliminated when SDS was included in the reaction (data not shown). These results show that the enhancement in 2AP fluorescence required active enzyme, but it did not require exogenously added divalent metal ions or a methyl donor.
M.PspGI and R.PspGI flip different bases within CCAGG
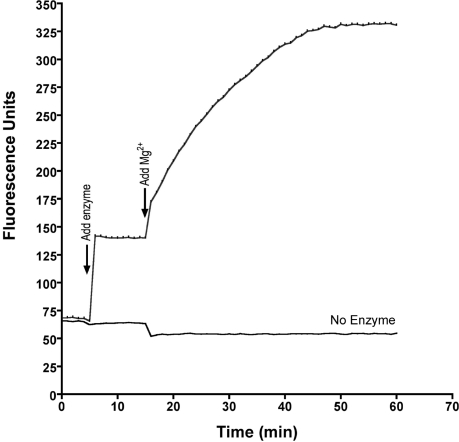
We have argued previously that R.PspGI flips the two bases in the central base pair within CCWGG and CCSGG sequences (6). We confirmed this prediction for CCWGG by using an oligonucleotide in which the adenine in the CCAGG sequence was replaced with 2AP and paired with a thymine (duplex containing oligonucleotide 2AP18; Table 1). When WT R.PspGI was added to such a duplex in the presence of Ca2+ in the buffer, an immediate increase in 2AP fluorescence was observed (Figure 3). This increase was stable over time until MgCl2 was added to the cuvette. This resulted in a much larger time-dependent increase in fluorescence intensity (Figure 3). This increase occurs presumably because when Mg2+ ions replace Ca2+ in the active site of the enzyme, the duplex is cleaved and 2AP becomes part of the single-stranded tail of product DNA. In other experiments, we noticed that contamination of the reaction buffer with even trace amounts of Mg2+ led to DNA cleavage by R.PspGI and a consequent larger fluorescence increase. To avoid confusing base flipping by R.PspGI with endonucleolytic cleavage by the enzyme, we switched to using the catalytically inactive D138A mutant of the enzyme in subsequent experiments. We also switched to using a longer duplex containing 2AP, 2AP27, because it is identical to 2AP26 in sequence except for the placement of 2AP (Table 1).
Figure 3.
2AP fluorescence enhancement by R.PspGI. The fluorescence of 2AP18 duplex (2.25 μM in REase buffer with 10 mM CaCl2) was monitored at 55°C with time in two cuvettes in parallel. At 5 min, WT R.PspGI was added to one cuvette to final concentration 5 μM (upper curve) and fluorescence measurements were continued. At 15 min, MgCl2 was added to both the cuvettes to final concentration of 10 mM and measurements were continued for additional 45 min.
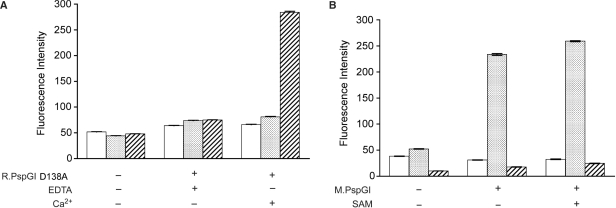
When these duplexes were incubated with D138A mutant of R.PspGI in the presence of Ca2+ ions, only the 2AP27 duplex showed a large fluorescence enhancement (Figure 4A). The observed enhancement in fluorescence of 2AP27 duplex was dependent on the presence of Ca2+ (Figure 4A) or Mg2+ (data not shown) ions in the buffer. A much smaller increase was seen when 2AP26 duplex was incubated with the D138A mutant and little increase in fluorescence was observed when the 2AP21 duplex containing 2AP outside the PspGI recognition sequence (Table 1). These data are qualitatively similar to the observations by Tamulaitis et al. (8) and show that R.PspGI flips 2AP when present in the central, but not the second position of the recognition sequence.
Figure 4.
Base flipping by R.PspGI and M.PspGI. (A) 2AP21 (open box), 2AP26 (stippled) or 2AP27 (hatched) were mixed with R.PspGI D138A mutant in REase buffer containing 10 mM CaCl2. The enzyme was present at 4-fold molar excess over DNA (500 nM). Mean 2AP fluorescence and SD under different buffer conditions at 65°C are shown. (B) 2AP21 (open box), 2AP26 (stippled) or 2AP27 (hatched) were mixed with M.PspGI in MTase buffer. The enzyme was present at one-to-one molar ratio to DNA (500 nM). Mean 2AP fluorescence and SDs under different reaction conditions at 65°C are shown.
In contrast to R.PspGI, M.PspGI caused large enhancement of fluorescence of 2AP only when it was present at the second position in the recognition sequence. It did not enhance fluorescence in 2AP21 duplex where the base analog lies outside the PspGI recognition sequence (Figure 4B) and caused only a small increase in fluorescence of 2AP27 duplex (Figure 4B). These data are consistent with M.PspGI flipping the second C in CCWGG, while R.PspGI flips the purine in the central base pair out of the duplex.
Temperature profiles of base flipping
In some initial experiments, we had measured enhancement of 2AP fluorescence at different temperatures and had found that it generally increased with temperature (Supplementary Table S1 and data not shown). Because the archaeon from which the PspGI enzymes are obtained is thought to grow around 85°C (1,26) and semi-quantitative measurements of endonuclease activity of R.PspGI found it to be optimal between 75°C and 85°C (1), we wondered whether the base-flipping activity of PspGI also peaked around 85°C.
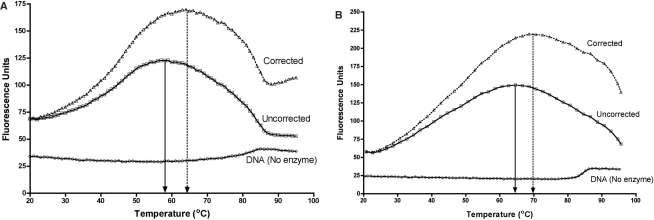
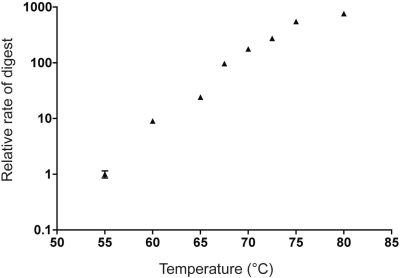
To evaluate the temperature dependence of base flipping by these enzymes, duplexes 2AP26 and 2AP27 were respectively incubated with M.PspGI and R.PspGI (D138A mutant), and the mixtures were heated from 20°C to 95°C with continuous monitoring of fluorescence. The temperature profiles of fluorescence intensity by both the enzymes had a bell-shaped appearance with peaks of fluorescence appearing at temperatures substantially below 85°C (Figure 5A and B). Based on the first derivatives of the intensity curves, the peaks of fluorescence occur respectively at ∼58°C and ∼64°C for the MTase and the REase. These profiles were reproducible and did not show significant hysteresis, i.e. decreasing temperature from 95°C to room temperature and then increasing it again to 95°C created a similar profile (data not shown). Because these results were unexpected, we decided to reconfirm the temperature dependence of cleavage activity of R.PspGI. 2AP27 was incubated with R.PspGI in the presence of Mg2+ ions and the initial velocity of DNA cutting was measured from 55°C to 80°C. The highest activity of the enzyme was found to be at highest temperature used, 80°C, suggesting that the optimal temperature for DNA cleavage by R.PspGI is at 80°C or above (Figure 6).
Figure 5.
Temperature profiles of base flipping. (A) Profile of 2AP26 fluorescence in the absence and presence of M.PspGI. The DNA was in MTase buffer at 500 nM and the enzyme was at 3-fold molar excess. The fluorescence intensities are shown with empty squares. The intensity values were corrected for relative decrease in fluorescence of 2APTP (Supplementary Figure S6). The corrected data are plotted as open triangles. Positions of peaks in each plot are indicated by a vertical arrow. The fluorescence profile of 2AP26 duplex without protein as a function of temperature is shown as circles. (B) Profile of 2AP27 fluorescence in the absence and presence of R.PspGI D138A mutant. The DNA was in REase buffer supplemented with CaCl2 (10 mM) at 500 nM and the enzyme was at 3-fold molar excess. The fluorescence intensities are shown with empty squares. The intensity values were corrected for relative decrease in fluorescence of 2APTP (Supplementary Figure S6). The corrected data are plotted as open triangles. Positions of peaks in each plot are indicated by a vertical arrow. The fluorescence profile of 2AP27 duplex without protein as a function of temperature is shown as circles.
Figure 6.
Dependance of R.PspGI catalytic activity on temperature. The fluorescence of 2AP27 duplex (500 nM in REase buffer with 10 mM MgCl2) was monitored at various temperatures. The reactions were started with the addition of WT R.PspGI to concentration of 80 nM and fluorescence was monitored for a few minutes to a few hours depending on the temperature. The initial velocity of the reactions were calculated and normalized with respect to the velocity at 55°C.
We were concerned that a decrease in 2AP fluorescence intensity with increasing temperature due to nonradiative energy transfers (12) may shift the peak base-flipping activities of PspGI enzymes to a temperature <85°C. To correct for this effect, the temperature profiles of fluorescence of 2AP nucleoside and its triphosphate were determined. Both showed a roughly linear response (Supplementary Figure S6) and the profile of 2APTP was used to correct the 2AP26 and 2AP27 profiles in the presence of PspGI enzymes. The corrections did shift the fluorescence peaks higher by ∼6°C, but they still occurred at temperatures substantially <85°C (Figure 5A and B). Thus the optimal base-flipping activities of PspGI enzymes may not coincide with the growth temperature of the source archaeon and the temperature optimum of catalytic activity of R.PspGI.
DISCUSSION
We purified the modification methyltransferase in the PspGI R–M system to apparent homogeneity and characterized its ability to protect DNA against R.PspGI and to flip DNA bases. The results presented here show that M.PspGI protects DNA against R.PspGI and suggest that it methylates the second cytosine in its recognition sequence CCWGG on the exocyclic nitrogen. They also show that while Ca2+ and Mg2+ ions stimulate the methyltransferase activity of the MTase, they are not required for its ability to flip the target base out of the duplex. The methyl donor, SAM, is also not required for base flipping. In contrast, base flipping or cleavage by the endonuclease does not occur in the absence of divalent metal ions. The former reaction occurs in the presence of either Ca2+ or Mg2+ in the buffer, but the latter reaction strictly requires Mg2+. Furthermore, results from 2AP fluorescence enhancement experiments show that while M.PspGI flips the second cytosine in the cognate sequence, R.PspGI flips the purine in the central base pair. Presumably, the pyrimidine paired with this base is also flipped out, but the experiments did not directly address this possibility. We also found that although the organism in which PspGI R–M system was discovered grows optimally around 85°C, the base-flipping reaction—as monitored through fluorescence enhancement of 2AP—occurs optimally between 60°C and 70°C for both the enzymes.
M.PspGI is the first N4-cytosine methyltransferase (N4C MTase) for which direct biochemical evidence for base flipping has been presented. Previously, the structure of the N4C MTase M.PvuII without DNA was used to predict that the enzyme flips cytosine out of the duplex (22), but this has not yet been confirmed with a co-crystal structure or through biochemical studies. M.PspGI is unusual among base-flipping enzymes in that it is associated with another base-flipping enzyme, R.PspGI, that recognizes the same DNA sequence but flips different DNA bases out of DNA. Additionally, the two enzymes are unrelated at the primary sequence level and have different metal ion requirements for base flipping (Figures 2A and 4A). Thus, the PspGI R–M system represents two different mechanisms for recognizing the same DNA substrate and flipping DNA bases.
Our study of M.PspGI led to at least two unexpected observations. First, we found that the methyl transfer activity of M.PspGI is stimulated by Ca2+ or Mg2+ ions. Consistent with this observation, the MTase sequence does contain a putative magnesium ion binding motif PD(X)n(D/E)XK [PD212…DSK271 or DYK292 or DEK332, where the superscripts are residue numbers in the protein sequence; (27)]. MTase activities that are part of Type I and III R–M systems are known to require Mg2+ for their activity. Additionally, MTases within several members of IIB, IIG and IIH R–M subtypes are also stimulated by Ca2+ or Mg2+ (28). However, unlike M.PspGI, some of these enzymes act as multisubunit complexes with endonuclease and/or recognition subunits and share many similarities with type I systems. For example, M.AhdI is a type IIH enzyme which is tetrameric, contains a recognition subunit and requires Mg2+ for its activity (29). To our knowledge, M.PspGI is the only MTase that is part of a type IIP R–M system (like EcoRI and BamHI) and shares little similarity to type I or type III systems, that is strongly stimulated by divalent metals.
Two different mechanisms have been offered for how divalent metal ions may stimulate MTase activity. One mechanism is exemplified by M.EcoP15I, which is part of a type III R–M system and requires Mg2+ for both base flipping and methyl transfer. Mutational analysis of this enzyme suggests that the metal ion stabilizes a flipped out adenine base and hence is required for specific binding to DNA (30). In contrast, orphan MTase M.SssI has high affinity for DNA in the absence of Mg2+ and this is thought to prevent it from leaving the product. Consequently, it is catalytically active in the absence of Mg2+ but acts in cis, i.e. it methylates multiple sites within a single DNA molecule, but not in different molecules (31).
It is likely that M.PspGI behaves more like M.SssI than M.EcoP15I. One indication of this is the observation that M.PspGI does not require divalent metal ions for base flipping (Figure 2A). It probably has very high affinity to nonspecific DNA in the absence of Mg2+ or Ca2+ and this is the likely reason why we found severe DNA loss following MTase reactions performed without divalent metal ions. One of the manipulations following methyl-transfer step was a phenol–chloroform extraction and we believe that most of the DNA was lost at the polar-hydrophobic interface. Another piece of evidence that this enzyme has high affinity toward nonspecific DNA comes from the fact that it binds tightly to a column that mimics DNA, phosphocellulose. M.PspGI elutes from this column at a very high-salt concentration (1 M NaCl). Consequently, M.PspGI may also act in cis in the absence of divalent metal ions in the reaction buffer reducing its activity when the amount of DNA is in molar excess over the enzyme (referred above as ‘catalytic’ amounts of enzyme). Thus, Mg2+ or Ca2+ appears to reduce the affinity of M.PspGI to nonspecific DNA. M.SssI was reported to have topoisomerase activity that relaxes DNA in the presence of Mg2+ (31). We have not tested M.PspGI for such an activity.
The second unexpected observation from our work was that the temperature profiles of 2AP fluorescence enhancement with R.PspGI and M.PspGI were inconsistent with the growth temperature of the source organism. The peaks of 2AP fluorescence occurred 15–20°C below the growth temperature and the peak of endonuclease activity (Figure 5A and B). This raises some general questions about what the 2AP fluorescence measures in the assay and specifically whether base-flipping activities of the two PspGI enzymes are optimal at the growth temperature of the organism.
It is reasonable to assume that specific binding of PspGI enzymes to their cognate sequence (or its 2AP-containing version) results in base flipping. If this is correct then the temperature profiles of 2AP fluorescence intensities (Figure 5A and B) reflect the temperature profiles of Ka for specific DNA binding by the enzymes. This would mean that PspGI enzymes bind their cognate sequence optimally at a temperature much below the growth temperature of the source organism. One way to explain this discrepancy is to postulate that the enzymes originated in an ancestor that grew at a much lower temperature and that the enzymes have not fully adapted to the new growth environment. It is useful to note that a BLAST search of protein databases identifies several dozen sequence homologs of PspGI enzymes, but none are from a thermophilic organism (data not shown). Alternately, the enzyme pockets that accommodate the flipped base in the two enzymes change structure with temperature and the environment of the flipped 2AP is much more polar in the 60–70°C range than at 85°C. Additional biochemical studies of these enzymes are needed to better understand this phenomenon.
Tamulaitis et al. (8) reported previously that WT R.PspGI enhances 2AP fluorescence when the base analog is present at the central position of PspGI recognition site (equivalent to 2AP18 or 2AP27 duplexes). Our results with R.PspGI are consistent with this report except for the magnitude of the fluorescence enhancement. Tamulaitis et al. (8) found that R.PspGI caused a 64-fold enhancement of 2AP fluorescence using a 5-fold molar excess of protein over DNA at 25°C, while we found only a 2- to 10-fold enhancement with excess enzyme (Figure 3 and data not shown). We find that the D138A mutant causes a much larger fluorescence enhancement than WT enzyme (data not shown), but even this mutant causes only a 5-fold or 22-fold enhancement at 25°C with 9-fold molar excess of protein over DNA (Supplementary Table S1). The extremely large enhancement reported by Tamulaitis et al. (8) is somewhat surprising because the temperature they used for measurement is at least 40°C below the optimal temperature for fluorescence enhancement by the D138A mutant (Figure 5B). It should be noted that sequences flanking the CCWGG site are different in the duplex used by Tamulaitis et al. and in 2AP18 or 2AP27 and it is possible that the efficiency of base flipping by R.PspGI strongly depends on the flanking sequence. Alternately, it is possible that in the experiments described by Tamulaitis et al. the duplex was cleaved by R.PspGI. This enzyme has an unusually low requirement for Mg2+ ions for activity [<1 mM; (3)] and hence even trace contamination of buffers or spectrophotometer cuvettes with Mg2+ can lead to strand cleavage and a much larger enhancement of fluorescence (Figure 3).
When we first proposed in a manuscript (June 2005) that R.PspGI flips the bases in the center of its recognition sequence based entirely on the ability of this enzyme to enhance deamination of cytosine at the central position in CCSGG sequence, it was not accepted. Subsequent publication of the crystal structure for Ecl18kI, which showed that this enzyme flips its central base pair out of the duplex (7) made our hypothesis acceptable (6). More recent demonstration of fluorescence enhancement of 2AP placed in the central position [(8) and this article], enhanced sensitivity to chloroacetaldehyde of cytosine at the central position in CCSGG sequence when bound by R.PspGI (32) and crystal structure of R.PspGI with DNA (Bochtler,M. et al. unpublished results) provides convincing validation for that hypothesis.
In summary, we have shown here that both members of the PspGI R–M system flip at least one base within their common recognition sequence. However, they have strikingly different requirements for divalent metal ions for this reaction suggesting very different mechanisms of sequence recognition and base flipping. These enzymes are highly thermostable making their purification from E. coli-based clones facile. Together, these features make the PspGI system an attractive system for studying the mechanism of DNA base flipping.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health (GM 57200 and CA 97899). Funding for open access charge: GM 57200.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank New England Biolabs (Ipswitch, MA, USA), and Alfred and Vera Pingoud (Justus-Liebig-University, Giessen, Germany) for providing the clones for PspGI genes.
REFERENCES
- 1.Morgan R, Xiao J, Xu S. Characterization of an extremely thermostable restriction enzyme, PspGI, from a pyrococcus strain and cloning of the PspGI restriction-modification system in Escherichia coli. Appl. Environ. Microbiol. 1998;64:3669–3673. doi: 10.1128/aem.64.10.3669-3673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 3.Pingoud V, Conzelmann C, Kinzebach S, Sudina A, Metelev V, Kubareva E, Bujnicki JM, Lurz R, Luder G, Xu S.-Y, et al. PspGI, a Type II restriction endonuclease from the extreme thermophile pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 4.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985;13:1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007;35:4792–4799. doi: 10.1093/nar/gkm513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 10.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase MTaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 11.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of Haelll methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 12.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. formycin, 2-Aminopurine Riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 13.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 14.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy YV, Rao DN. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 16.Su TJ, Connolly BA, Darlington C, Mallin R, Dryden DT. Unusual 2-aminopurine fluorescence from a complex of DNA and the EcoKI methyltransferase. Nucleic Acids Res. 2004;32:2223–2230. doi: 10.1093/nar/gkh531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szegedi SS, Reich NO, Gumport RI. Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res. 2000;28:3962–3971. doi: 10.1093/nar/28.20.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malygin EG, Evdokimov AA, Zinoviev VV, Ovechkina LG, Lindstrom WM, Reich NO, Schlagman SL, Hattman S. A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 2001;29:2361–2369. doi: 10.1093/nar/29.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bheemanaik S, Bujnicki JM, Nagaraja V, Rao DN. Functional analysis of amino acid residues at the dimerisation interface of KpnI DNA methyltransferase. Biol. Chem. 2006;387:515–523. doi: 10.1515/BC.2006.067. [DOI] [PubMed] [Google Scholar]
- 20.Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowher H, Jeltsch A. Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 22.Gong W, O’Gara M, Blumenthal RM, Cheng X. Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butkus V, Klimasauskas S, Kersulyte D, Vaitkevicius D, Lebionka A, Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985;13:5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gromova ES, Kubareva EA, Vinogradova MN, Oretskaya TS, Shabarova ZA. Peculiarities of recognition of CCA/TGG sequences in DNA by restriction endonucleases MvaI and EcoRII. J. Mol. Recognit. 1991;4:133–141. doi: 10.1002/jmr.300040405. [DOI] [PubMed] [Google Scholar]
- 25.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southworth MW, Kong H, Kucera RB, Ware J, Jannasch HW, Perler FB. Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9 degrees N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl Acad. Sci. USA. 1996;93:5281–5285. doi: 10.1073/pnas.93.11.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sistla S, Rao DN. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 29.Marks P, McGeehan J, Wilson G, Errington N, Kneale G. Purification and characterisation of a novel DNA methyltransferase, M.AhdI. Nucleic Acids Res. 2003;31:2803–2810. doi: 10.1093/nar/gkg399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bist P, Rao DN. Identification and mutational analysis of Mg2+ binding site in EcoP15I DNA methyltransferase: involvement in target base eversion. J. Biol. Chem. 2003;278:41837–41848. doi: 10.1074/jbc.M307053200. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo K, Silke J, Gramatikoff K, Schaffner W. The CpG-specific methylase SssI has topoisomerase activity in the presence of Mg2+ Nucleic Acids Res. 1994;22:5354–5359. doi: 10.1093/nar/22.24.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daujotyte D, Liutkeviciute Z, Tamulaitis G, Klimasauskas S. Chemical mapping of cytosines enzymatically flipped out of the DNA helix. Nucleic Acids Res. 2008;36:e57. doi: 10.1093/nar/gkn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.